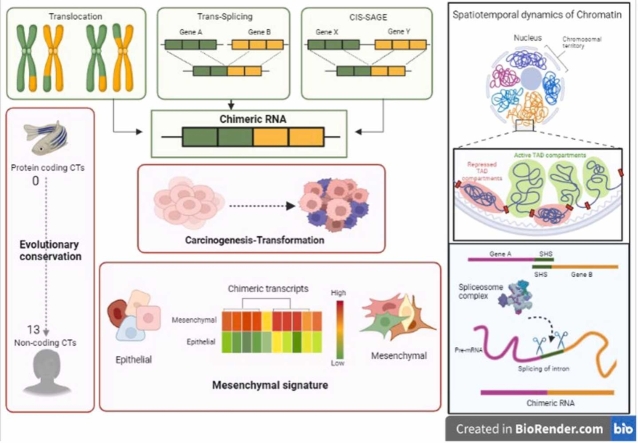
Abstract
The molecular and functional diversity generated by chimeric transcripts (CTs) that are derived from two genes is indicated to contribute to tumor cell survival. Several gaps yet exist. The present research is a systematic study of the spectrum of CTs identified in RNA sequencing datasets of 160 ovarian cancer samples in the The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov). Structural annotation revealed complexities emerging from chromosomal localization of partner genes, differential splicing and inclusion of regulatory, untranslated regions. Identification of phenotype-specific associations further resolved a dynamically modulated mesenchymal signature during transformation. On an evolutionary background, protein-coding CTs were indicated to be highly conserved, while non-coding CTs may have evolved more recently. We also realized that the current premise postulating structural alterations or neighbouring gene readthrough generating CTs is not valid in instances wherein the parental genes are genomically distanced. In addressing this lacuna, we identified the essentiality of specific spatiotemporal arrangements mediated gene proximities in 3D space for the generation of CTs. All these features together suggest non-random mechanisms towards increasing the molecular diversity in a cell through chimera formation either in parallel or with cross-talks with the indigenous regulatory network.
Keywords: Chimeric transcripts, Inter- and intra-chromosomal, Structural annotation, Evolution, Mesenchymal signature, Transformation, Chromatin architecture, Chimera formation
Graphical Abstract
Highlights
-
•
Chimeric Transcripts (CTs) are a non-random mechanism of generation of molecular diversity in a cell.
-
•
A CT-based expression signature was associated with the mesenchymal subgroup of ovarian tumours.
-
•
Protein-coding chimaeras are highly conserved while pseudogenes have evolved recently.
-
•
Correlation of CTs with patient survival indicate a role in transformation and cancer progression.
-
•
In 3D chromatin space, proximal genes with short homologous sequences may give rise to chimaeras.
1. Introduction
Spontaneous and inheritable genetic changes have a steadfast association with cancer. It is realized that these culminate in a plasticity of regulatory mechanisms of transcription and translation to create diverse landscapes of expression from a limited number of genes. Within this scope, perturbed cellular functions through epigenetically altered chromatin structure affects transcription - translation kinetics and consequently generates non-canonical and non-coding RNA through alternative / back splicing, RNA recombination, read-through mechanisms, besides altering transcript and protein stability and protein modifications [1], [2]. Oncogenic effects of splice variants and isoforms are reported [3], [4], while molecular cross-talks between these mechanisms are currently being defined [5] Taken together, de novo expression may well challenge the dogma of genomic alterations being the sole, all-encompassing feature of cancer.
Chimeric transcripts (CTs) have received considerable attention in several normal tissues and in cancer wherein such they contribute to specific biological functions including migration, invasion and transformation [6], [7], [8], [9]. Canonically known to be generated through gross chromosomal structural alterations (chromothripsis, translocations, insertions/deletions etc.), non-canonical CTs generated by purported cis-splicing of adjacent genes or readthrough (cis-SAGe or RT) produces a longer transcript of the 5′ gene with additional C-terminal sequences derived from its downstream 3′ gene, or through cross-strand / trans-splicing mechanisms are also reported [10], [11], [12], [13], [14], [15].
Epithelial to mesenchymal plasticity in ovarian cancer results in tumour heterogeneity leading to difficulty in disease treatment. These processes are carried out by several metabolic drivers. Earlier studies conducted by Gao Q. et. al. 2018 [16] explained how chimeric RNAs contribute to drug resistance in many tumours. Due to resistance to the drugs caused by CTs, chimeric proteins can cause changes in cellular mechanisms and functioning. These numerous alterations to the cells result in phenotypic plasticity, which further improves the cells' survival rates. [17].
In the present study, we assembled chimeric sequences from the TCGA RNA-seq data of 160 high-grade serous ovarian cancer (HGSC) samples towards generation of a comprehensive profiling of de novo and non-canonical transcripts in the disease. Closer examination revealed a structural complexity arising either from differential splicing of partner genes, inclusion of noncoding partners and/or untranslated sequences in the transcript. Further annotation suggested tractability of gene boundaries mediated by transcriptional plasticity to enhance transcript diversity within a cell. Alternatively, the altered spatiotemporal chromatin structure may facilitate the transcription of distant genes brought into proximity and which harbour short homologous sequences. Constancy of structural features across tumor samples suggests non-randomness in generation of CTs. Together, all these mechanisms potentiate a wide molecular diversity within tumors, which almost is likely to represent a parallel expression to the canonical gene - transcript variants - protein isoforms cascade. In some of these may provide growth and survival advantages as a path of least resistance in tumour evolution, while some others displayed a specific association with the mesenchymal phenotype. The possibility of harnessing such mechanisms and their directed targeting to yield clinical benefit could be a new and attractive strategy in the future.
2. Materials and methods
2.1. Development of a computational pipeline on the seven bridges genomics cloud
We developed and executed a computational workflow on the Seven Bridges Cancer Genomics Cloud (https://www.sevenbridges.com/; [18]), for the iterative processing of 160 OvCa RNA-Seq datasets from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/; Supplementary Table 1). After evaluating several algorithims for detection and identification of fusion transcripts based on their outputs and reproducibility in stand-alone runs vs. those in the cloud-based pipeline (FusionHunter, Defuse, STAR, TopHat-fusion), ChimeraScan was incorporated in this pipeline for processing RNA sequencing data via alignment, indexing, extraction of breakpoint sequences and filtration based on reference genome build (GRCh37/hg19), transcriptome reference (hg19 reference assembly) and chimeras in normal tissues (Genotype Expression Project, GTExportal; Supplementary Figs. 1a,1b; [19]). Trimmed alignments were scanned for discordant read pairs and an output list of putative 5′–3′ transcript chimera pairs was identified along with details of each chimera in each sample. We considered 3 nucleotides spanning the chimeric junctions on each side of the fusionpoint as a cutoff. All the CT annotations were manually inspected several times to remove possible pseudotranscripts and non-functional transcripts.
2.2. Chromosome-wise gene associations with cancer
Chromosome-wise association of genes with ovarian and other cancers were examined on Cancer GeneticsWeb (http://www.cancerindex.org/geneweb/); Ensembl East database (http://useast.ensembl.org/Homo sapiens/Location/Genome) provided the gene abundance data. Coding and non-coding gene densities were calculated considering genes along the entire chromosome length as follows,
2.3. Analysis of CT-associated data from ChimeraScan outputs
Spanning reads and isoform fractions were derived from the ChimeraScan outputs. We consolidated all 75 bp spanning reads of each CT to derive longest read sequences (LR; ranging between 71 and 148 bp), which were used in further analyses. Isoform fractions of 5′ and 3′ parental genes (ranging from 0 to 1) were used to generate heatmaps using MeV (Multiple Experiment Viewer v4.9) tool-multiple array software. Cis- and cross- strand CTs for intra- and inter- chromosomal chimeras were also computed and representative data plots/graphs generated using Microsoft Excel 365 and GraphPad prism.
2.4. Cell culture, RNA isolation, cDNA synthesis and RNA sequencing of HGSC cells
A2, A4EP, A4LP and G1M2 cell lines were earlier established in the lab from patient ascites [20] and maintained in Minimal Essential Medium/MEM (Gibco #11095080) supplemented with 5% fetal bovine serum (MP Biomedicals #092910154) and 1% nonessential amino acids (Gibco #11140050). OVCAR3, OV90, OVCA432, CAOV3, OVCA420, PEO14, OVCA420, CAOV3, OVMZ6, IOSE364, CP70, OVCAR4, and A2780 cell lines were sourced and maintained and RNA sequencing data obtained as described in [21], [22]. All cell lines were authenticated by NCCS Repository. RNA extraction of cell lines was performed at 80% confluency using TRIzol™ Reagent (Invitrogen #15596026), and quantified using DeVOVIX DS11-spectrophotometer. Reverse transcription reaction was set for synthesis of complementary DNA (cDNA) using 2 ug RNA as a starting product with cDNA synthesis kit (Thermo Scientific #AB1453A). Correlation plots, alluvial plots and quadrant scatter plots were generated using https://www.bioinformatics.com.cn/en, a free online platform for data analysis and visualization.
2.5. Validation and profiling of chimeric transcripts in OvCa cell lines
Forward and reverse primers were designed for amplification of the sequences around the fusionpoint with reference to specific exon sequences of parental genes involved in chimera formation. Oligos were designed using GeneRunner Version 5.1.01. Beta, or Primer Express (ThermoFisher) and synthesized at 25 nM from Integrate DNA technology (IDT;details can be provided on request). DNA Polymerase (TaKaRa #R050B) was used in PCR reactions; products resolved on a 1.8% agarose gel (Sigma-Aldrich #A9539–100 G). Amplicons bands were cut, DNA extracted using the QIAEX II Gel Extraction Kit (Qiagen #20021, #20051), and sequenced at the National Centre for Microbial Research (NCMR)-facility, Pune. Expression profiling of chimeras was performed on StepOne™ Real-Time PCR System using 1pMol primer mix, 1:10 diluted HGSC and 2x SyBr green pre-mix; HPRT1 was used as the endogenous control. Relative mRNA expression was computed, values normalized and data represented on a log2 scale.
2.6. Identification of CTs pre-annotated on public databases
Longest read of each CT was probed for uniqueness and similarities through comparison with human transcripts in NCBI and Ensembl GRCh38 BLAST (https://blast.ncbi.nlm.nih.gov, http://www.ensembl.org/Homosapiens/Tools/Blast). Sequences with a query cover as well as identity > 90% were considered as pre-annotated and were grouped based on their biotype. Ovarian histopathology data available for 24 pre-annotated CTs in the Human Protein Atlas (www.proteinatlas.org), was accessed and represented using R, while the survival statistics available for 31 CTs was used to plot survival graphs. GenTree (https://gentree.org/) was used to screen vertebrate -specific evolutionary conservation of pre-annotated CTs.
2.7. Analyses of spatiotemporal chromatin dynamics
Genomic distance between intra-chromosomal CTs (n = 3291) was calculated using genomic co-ordinates of their parental genes. Candidates involved with more than 7 partners in CT generation (hub genes) were identified, and 3D Hi-C browser (YEU Lab,http://3dgenome.fsm.northwestern.edu/) was used to visualize Hi-C interaction map of hub genes and their intra-chromosomal partners, as Hi-C contact matrices (heat-maps) coupled with TADs. Briefly, interaction of two genes was initially scanned at 25 kb resolution in ovary tissue and then examined deeper at 5 kb resolution. The 4D- Nucleome data portal (http://vis.nucleome.org/entry/; [23], [24]), was used to generate 3D chromatin maps within which candidate genes were highlighted. Splice site analysis of hub partner genes having homologous sequences in corresponding exon and intron was performed using Spliceator (http://www.lbgi.fr/spliceator /;[25]).
2.8. Data analysis, statistics and graphical representation
Unless otherwise mention, all experiments and data generated were from at least three replicates. One way ANOVA (with repeated measures) test was performed for profiling of CTs in HGSC cells using SigmaStat version 3.0, paired-students ‘t′ test was performed wherever required. Graphs were generated using SigmaPlot version 10.0; heatmap plots were generated in MeV 4 9 0 tool (multiple array software) with median values were given as mid-range for the heatmap. Statistical significance values are p = *< 0.05, * *< 0.01, and * ** <0.001.
3. Results
3.1. Chimeric transcripts represent diverse structural and transcriptional features that occur throughout the genome
We developed a computational workflow on the Seven Bridges Cancer Genomics Cloud platform to enable identification of chimeric transcripts (CTs) through iterative processing of OvCa RNA-Seq datasets from the TCGA using ChimeraScan (Methods; [18]; http://tcga-data.nci.nih.gov; [19]; Supplementary Figs. 1a,1b). 42–414 candidate CTs were identified in each tumour sample, which presented with unique 5′ and 3′ parental sequence alignments. The chromosome-wide distribution of CTs revealed the highest involvement of Chr1 in CT events, while maximum frequency of individual CT spanning reads involving Chr2 and Chr22 (Fig. 1a; Supplementary Table. 2). Higher CT-spanning read frequencies were not necessarily associated with chromosomes harbouring higher gene densities, as exemplified by Chr22 which despite being ‘gene-poor’ had a moderate to high number of CT-spanning reads, while Chr17 and Chr19 display a relatively lower CT frequency despite being ‘gene-rich’. Further exploring chromosome-wise frequencies of pan-cancer and OvCa - associated genes highlighted the involvement of Chr1, Chr2, Chr7 and Chr22 in OvCa as well as CT formation, while Chr18 presented with very few OvCa-associated genes and least number of CTs (www.cancer-genetics.org; Fig. 1b; Supplementary Table 3). Further probing the association of cancer-linked genes, we identified specific tumour suppressors (Chr 2-MSH2, Chr 5-RAD50, Chr 17-BRCA1, Chr19-SMARCA4) and oncogenes (Chr 12-KRAS, Chr17-ERBB2, Chr19-AKT2) as partners in CT formation, suggesting loss/gain of function(s) through these perturbations during transformation.
Fig. 1.
Diversity of molecular rearrangements in chimeric transcripts (CTs). a. Heatmap representing chromosome-wise, genome-wide distribution of gene density, CT frequencies and spanning reads; b. Heatmap representing the distribution of, I. Number of cancer-linked genes, II. Number of ovarian cancer-linked genes, iii. Number of CTs i each chromosome;; c. Heatmap representing isoform fractions (IF) indicating CT enrichment over 5′ and 3′ parental genes for (i) inter-chromosomal and (ii) intra-chromosomal CTs; d. Distribution of cis (++/--) and trans (+-/-+) strands in i. intrachromosomal and ii. interchromosomal CTs; e. Pie-chart representing distribution of the major types of CTs; f. Distribution of spanning reads for 13 CT subtypes viz. Inter-chromosomal (1), Intrachromosomal (2), Intrachromosomal_Complex (3), Intrachromosomal_Converging (4), Intrachromsomal_Diverging (5), Read-through (6), Adjacent_Complex (7), Adjacent_Converging (8), Adjacent_Diverging (9), Overlapping_Complex (10), Overlapping_Converging (11), Overlapping_Diverging (12), Overlapping_Same (13), grey and black circles indicate CT distribution across these subtypes.
Strikingly, isoform fractions of most CTs were higher than both or at least one parental transcript (Clusters 1, 2, 3 respectively; Fig. 1c; Supplementary Fig. 1c). Primary sequence annotation to characterize the involved parental genes indicated their localization either on same or across different chromosomes for each CT (intra- chromosomal or inter-chromosomal respectively; Supplementary Fig. 1d). Further exploring specific DNA strand association in formation of CTs revealed an equal probability of trans (+/- or -/+) or cis (+/+ or -/-) events in formation of inter-chromosomal CTs, whereas a majority of intra-chromosomal CTs events are generated in cis (Fig. 1d; Supplementary Fig. 1e). All these features together suggest CT formation to not be a random cellular event.
CTs were first classified as either interchromosomal or intrachromosomal; the latter presented with very diverse yet discrete features (Fig. 1e, Supplementary Fig. 2) including,
-
(i)
Parental genes often include neighboring genes (currently reported as readthrough, RT), but also include overlapping genes (Ov, generating novel intragenic splice variants), adjacent (Adj, 5′ parent is downstream of 3′ parent on the chromosome), or those separated by long distances as much as being across the centromere on 2 arms of the chromosome (inD-CTs);
-
(ii)
inferring direction of transcription of the parental genes further revealed CTs to be generated either in the same orientation (wherein the strand as well as direction of transcription was same, for example in RT-CTs), complex (wherein the downstream parent on the same strand of the chromosome presents as the 5′ partner), convergent (opposite direction of transcription of the 2 genes that converges at the fusionpoint) or divergent (direction of transcription of the 2 genes diverges from the fusionpoint towards opposite ends.
These structural and transcription-associated features led to the resolution of 12 CT subtypes within the intrachromosomal type that were either intrachromosomal, distant (In_D), overlapping (Ov), readthrough or adjacent; these were further subtyped as being either convergent, divergent, and complex (Supplementary Fig. 2). Examination of their chromosomal abundance and spanning read frequency revealed that while overall inD and RT-CTs were most frequently expressed, enrichment of all subtypes of inD-, RT-, Adj- and Ov-CTs was within in the first quartile (log range 1–10), while those of inD_same and Adj_complex extended into a higher quartile (Fig. 1f).
3.2. Recurrent OvCa-associated CTs display distinct sequence rearrangements of parental genes
Some CTs were expressed in at least 10% tumor samples within the TCGA cohort. These included 3 inter-chromosomal, 3 intragenic variants, 96 RTs and 17 inD-CTs (n = 119; Supplementary Table 4), and were termed as recurrent CTs. To study these at a deeper level, we consolidated all spanning reads of each CT to derive longest read sequences (LR;ranging between 71 and 148 bp). A detailed structural annotation of these LR sequences of each recurrent CT vis-à-vis its comparison with its parental gene transcripts revealed discrete patterns of CT formation–.
1.Coding region sequence (CDS) rearrangements - Cis-splicing involving deletion of the last exon from the 5′ gene (ALE) and first exon from the 3′ gene (AFE) as earlier reported is a dominant structural feature; variations in the theme included deletion of additional exons (mid-CDS) through splicing.
2.Inclusion of Untranslated regions (UTR) - 5′ UTR and in rare cases 3′UTR sequences of either one or both partners were involved in generation of 43 CTs along with CDS-derived sequences. UTRs are associated with regulatory roles and their inclusion in a chimera may reflect on transcript expression and/or stability.
3.Inclusion of non-coding (NC) sequences - One or both the parental transcripts are known as either anti-sense/non-coding RNA, not reported to be translated (no existing protein variants) or identified as nonsense-mediated decay (NMD) variants.
Notably, our study did not reveal intronic fusions since the input data used was of RNA sequencing. However, the above varied sequence arrangements reveal a diversity of genomic attributes of parental genes and transcriptomic complexities associated with CTs in OvCa that could reflect on their emergence through distinct mechanisms.
3.3. CTs exhibit a phenotype-specific association which is likely to be modulated dynamically during transformation
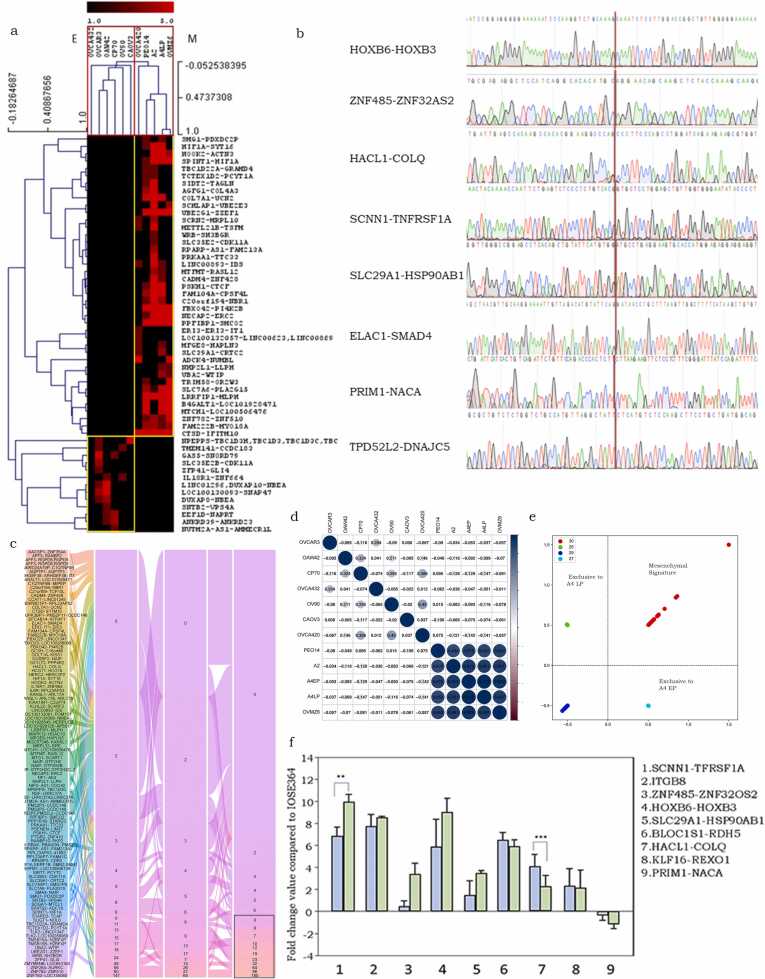
Validation of RNA-sequencing data across a panel of 10 HGSC cell lines revealed expression of 549 CTs (Supplementary Fig. 3a; Supplementary Table.5), 121 of which were shared at least between any two cell lines, while others were individualised and occasionally expressed at a high frequency in a single cell line, one such instance is UBE2Q1-RRNAD1 for which 373 spanning reads were identified in OVCA432 cells (Supplementary Table. 6). Interestingly, a phenotype-specific enrichment of CTs was revealed in cell lines (epithelial vs mesenchymal; [26], [21], [22]; Fig. 2a); a few of these were consequently validated through Sanger sequencing (Fig. 2b). Strikingly, mesenchymal CTs displayed a higher number of spanning reads as compared with epithelial CTs.
Fig. 2.
Identification and expression analysis of CTs in HGSC cell lines. a. A heatmap and two-way hierarchical clustering of CTs identified in cells of epithelial and mesenchymal phenotypes.; b. Electropherograms of the validated Chimeric transcripts in HGSC cell lines through Sanger Sequencing, the vertical line represents the fusion point (fusion point is the junction in the sequence at which two transcript fuse/join); c. Alluvial plot of CTs present in A4 EP, LP and A2 cells. Vertical boxes represent chimeras and the spanning read number while the links represents the expression changes in the cell lines, the black box in the lower right corner highlights the CTs contributing to the mesenchymal signature; d. Correlation plot of CTs expression in 11 HGSC cell lines (scale: blue-Positively correlated, Red: Negatively correlated); e. Quadrant scatter plot for 4 group screened (X and Y axis represents the normalized expression of RNA seq read values); f. Positive and negative fold expression of CTs in A4 Ep and LP cells compared to IOSE364, Scale: Y axis represents the fold change value. X axis represents CTs screened.
To delve towards a deeper understanding of the association of CTs with transformation in the mesenchymal subtype, we examined the CT expression profiles in a progression model established earlier in the lab [20]. Briefly, 19 single cell clones established from a spheroid isolated from HGSC patient tumor ascites underwent spontaneous immortalization of which the A2 clone was identified as the tumor-initiating clone (TIC), while another A4 clone underwent transformation after ∼ 20 passages in culture, providing a matched pair of untransformed and transformed cell lines with distinct molecular profiles (A4-EP and A4-LP respectively;[27]). RNA-sequencing revealed a higher number of common than exclusive CTs between the 2 stages of transformation (Supplementary Table.6). Common CTs displayed a higher number of spanning reads in A4-EP than in A4-LP, suggesting an association with driver events of transformation, while their continuing expression could signify maintenance of the transformed state under a steady, optimal in vitro environment. Strikingly, 24 of these common CTs were also expressed in the A2 TIC, and at comparable number of spanning reads as in the A4EP state. This similarity possibly arises in lieu of A2 and A4EP being single cell clones from the patient sample, while A4LP is an in vitro derivative of A4EP (Fig. 2c). A2 and A4EP represent the mesenchymal subtype and a subset of their common CTs expressed a very strong correlation with each other across other mesenchymal (PEO14, OVMZ6), but not epithelial cell lines (Figs. 2d,2e). These could very well define a ‘mesenchymal signature’ with CTSD-IFITM10 being additionally expressed in OVCA420 which represents the hybrid / mixed phenotype (Table 1).
Table 1.
Mesenchymal signature of chimeric transcripts in HGSC*.
| CT | A4EP | A4LP | A2 | PEO14 | OVMZ6 | OVCA420 | OAW42 | CP70 | OV90 | CAOV3 | OVCA432 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LRRFIP1-MLPH | 147 | 65 | 180 | 70 | 198 | 0 | 0 | 0 | 0 | 0 | 0 |
| PPFIBP1-SMCO2 | 60 | 27 | 96 | 25 | 31 | 0 | 0 | 0 | 0 | 0 | 0 |
| UBE2G1-ZZEF1 | 48 | 17 | 64 | 40 | 59 | 0 | 0 | 0 | 0 | 0 | 0 |
| FBXO42-PI4K2B | 18 | 14 | 32 | 16 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| SLC7A6-PLA2G15 | 18 | 13 | 10 | 3 | 31 | 0 | 0 | 0 | 0 | 0 | 0 |
| SPINT1-HIF1A | 17 | 13 | 7 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| NECAP2-ERC2 | 15 | 9 | 19 | 2 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| CTSD-IFITM10 | 13 | 9 | 12 | 19 | 21 | 8 | 0 | 0 | 0 | 0 | 0 |
| KANSL1-ARL17A | 6 | 5 | 8 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| PSKH1-CTCF | 15 | 9 | 19 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HIF1A-SYT16 | 13 | 8 | 6 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FAM104A-CPSF4L | 11 | 8 | 23 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZNF782-ZNF510 | 0 | 2 | 3 | 2 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| COL7A1-UCN2 | 6 | 5 | 8 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MTCH1-LOC100506476 | 56 | 26 | 0 | 23 | 31 | 0 | 0 | 0 | 0 | 0 | 0 |
| B4GALT1-LOC101928471 | 24 | 17 | 0 | 23 | 24 | 0 | 0 | 0 | 0 | 0 | 0 |
Numbers in columns represent number of spanning reads of CTs identified in HGSC cell lines; the signature considers at least 2 spanning reads per sample in at least 3 cell lines
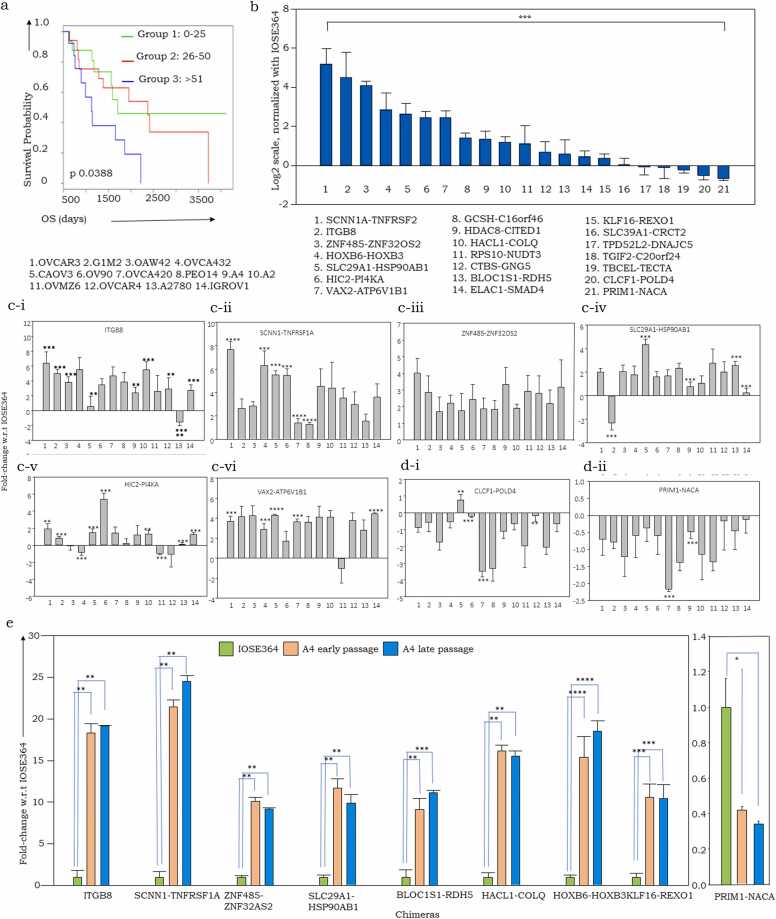
3.4. Expression of CTs contributes to the process of transformation in OvCa and may be associated with patient prognosis
To assess the non-randomness of suggested involvement of CTs in transformation, we further examined the association of recurrent CTs (Supplementary Table 4) with overall survival (OS). Thus, three groups of patients were identified within the TCGA cohort based on the number of tumor-associated CTs, viz. Group1 (0−25), Group2 (26−50), Group3(>51), of which Group1 was associated with a significantly higher OS than Group3 (Fig. 3a). As a corollary, we considered two patient groups in the TCGA cohort (n = 160) with Cohort1 (worst prognosis, OS<6 months) and Cohort2 (best prognosis, OS>60 months) and noted a significant differential means of CT frequency between these cohorts indicating that a higher number of CTs may correlate with lowered survival in HGSC (Supplementary Figs. 3b-i, 3b-ii). Further, since the functional contribution of individual CTs will actually be a determinant of survival, we assessed the association of each recurrent CT on prognosis. This effectively identified a significant association of 12 CTs with OS (9 correlate negatively while 3 exhibit a positive correlation; p < 0.05; Supplementary Fig. 3c).
Fig. 3.
Association of CTs with transformation. a. Kaplan Meier plot for designated patient group: group 1(0−25) group 2 (25−50) group 3 (>51) in the TCGA cohort; b. Relative mRNA expression comparison of 21 CTs in HGSC (average expression across a panel of cell lines) compared with IOSE364 (normal ovarian surface epithelial cell line); c. Fold-change of CT expression in HGSC cell lines normalized with IOSE364 (Y axis: Log2 normalized value), i. HIC2-PI4KA, ii. VAX2-ATP6V1B1, iii. HDAC8-CITED1; iv: CLCF1-POLD2, v- PRIM1-NACA; vi. CTBS-GNG5, vii SLC39A1-CRTC2; d. Comparison of a progression model (HGSC cell line A4) comprising of early, untransformed and late transformed cells, Statistics: Students T test (panel a), One way ANOVA with repeated measures (Holm-Sidak test: panel b and c: Significance values for individual comparison between the cell lines) p < 0.0001 * ** *, p < 0.001 * ** , p < 0.01 * *, p < 0.05 * (Y axis: Normalized values of relative mRNA expression; X axis: chimeras.
Further profiling the expression of 21 recurrent OvCa-CTs in a normal immortalized ovarian surface epithelium cell line (IOSE364) revealed the positive association of HIC2-PI4KA, SLC39A1-CRTC2, RPS10-NUDT3, PRIM1-NACA, CLCF1-POLD4 and ELAC1-SMAD4 along with weak / lack of expression of the remaining (Fig. 3b;Supplementary Fig. 4a). Comparing these profiles across the panel of transformed cell lines identified a ∼2-fold higher expression of SCNN1-TNFRSF1A, ITGB8-ITGB8, ZNF485-ZNF32OS2, HOXB6-HOXB3, SLC29A1-HSP90AB1, HIC2-PI4KA and VAX2-ATP6V1B1, while CLCF1-POLD4 and PRIM1-NACA were downregulated in HGSC over normal cells (Fig. 3c;Supplementary Fig. 4b). These may hence be considered as tumor-associated changes in expression. Moreover, the association of VAX2-ATP6V1B1 and SLC39A1-CRTC2 with poor and good prognosis respectively suggests a future role in prognostication. Besides these associations, expression of comparable levels of some CTs in normal as well as transformed cells could indicate a role in cellular housekeeping functions. All these CTs except PRIM1-NACA were upregulated in the progression model affirming our previous premise that increased CT expression is likely to be an early event in the process of transformation (Fig. 3d).
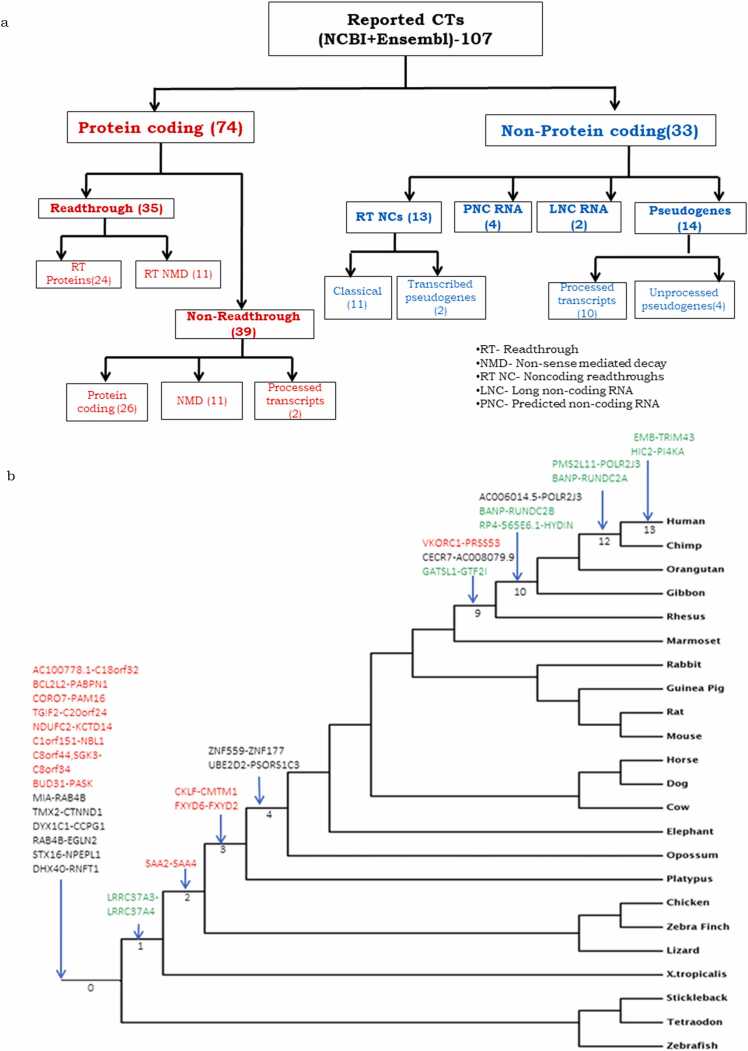
3.5. Biotype-based classification of CTs correlates with their evolutionary conservancy
While most of the CTs identified in our data were novel, we realized that some were reported earlier and/or archived in databases (ChimerDB [28], COSMIC [29], FusionGDB [30], Mitelman database [31], ChiTars 5.0 [32], Tumor fusion gene data portal [33], www.ensembl.org, www.ncbi.nlm.nih.gov, www.genecards.org). Of the 107 CT sequences thus identified as being pre-annotated on the Ensembl and/or NCBI databases (similarity based on >90% query cover and identity; Supplementary Table 7), most are listed as readthrough (RT), assigned distinct loci with unique transcript variant IDs and predicted biotypes (either as protein-coding, PC-CTs or non-coding, NC-CTs; Fig. 4a). Expression of some PC-CTs is also reported in Human Protein Atlas (HPA, https://www.proteinatlas.org; Supplementary Fig. 5a, 5b). Surprisingly, the pseudogenes identified as CTs in our study, were often represented as single parental derivatives, for example, GTF2IP1 (GTF2I pseudogene) aligned with our LR sequences of GATSL1-GTF2I as it harbours 405 bp from GATSL1.
Fig. 4.
a. Biotype based classification of 107 reported transcripts on Ensembl and NCBI databases; b. Primate specific phylogenetic investigation of reported CTs using GenTree database (Red, protein coding CTs, Green, pseudogenes, black, Readthrough NMD and other non-coding, Branch Support Values depicted at each divergent point).
To predict functional relevance of 107 pre-annotated protein coding and non-coding CTs, conservation study of sequences across 23 vertebrate species was performed (GenTree, https://www.gentree.ioz.ac.cn) [7], [34]. Analyses of the CTs and the participating parents was done separately. The majority of "readthrough protein coding" CTs were reported in the database (n = 14) (Fig. 4b), while many "other protein coding" CTs were not. However, many species had their parental genes reported (low BSV, n = 71). Indicating that in vertebrates, both the parental gene and the readthrough protein coding chimaera are conserved. Similar analyses were done for non-coding CTs as well. A small number of readthrough non-coding CTs revealed 0 BSV (n = 4). However, GenTree did not find the remaining categories, such as predicted non-coding and long non-coding categories. In pseudogene categories, only reported pseudogenes were found on GenTree with a wide range of BSV from 0 to 13 (maximum at 9–13) (Fig. 4b). An interesting correlation was that most PC-CTs were present with low BSV (maximum conservancy) suggesting an involvement in key cellular and/or housekeeping functions, while pseudogenes had a higher BSV indicating recent evolution emerging like paralogues. Functional annotation of reported CTs suggest roles in mRNA surveillance, TGF-β, FoxO, PI3-Akt signaling pathways, or divergent biological processes including nucleic acid binding, ribosome, and carbohydrate binding, etc. thereby assigning cellular relevance for expression of these chimeras (https://www.genecards.org, Supplementary Figs. 5b,5c,5d). The emergence of the species at different timepoint is denoted by branch support value and their timeline of emergence are given in Supplementary Table 8. Taken together, besides revealing the non-random occurance of CTs, these findings indicate one more situation in which cellular processes / molecules driving normal homeostasis are hijacked by cancer cells towards their own survival or disease progression.
3.6. Spatiotemporal dynamics of chromatin organization may drive the formation of CTs
While each known mechanism (chromosomal structural alterations / cis-SAGE-RT / trans-splicing) can account for some of the CTs in our study, the large majority of the inD-CTs presenting in cis with median parental gene distances of 104-107 bp remained a mystery (Supplementary Fig. 6a). Specifically, these cannot be accounted for by RT between neighbouring genes that implies genomic proximity as an essential feature in enabling continual transcription. We hence hypothesized that partnering preferences across and within chromosomes may be mediated by specific spatiotemporal base-pairing of gene pairs through chromatin looping and topology-associated domain (TAD) - facilitated proximities irrespective of genomic distances [35], [15], and their presentation to the transcriptional machinery as continuous entities. In support of this premise, we examined the location and proximity of active transcription gene hubs in 3D contact maps of Hi-C chromatin data for ovarian tissue using Hi-C data browser (northwestern.edu) [37] and visualized these within 3D nuclear chromatin architectures in multimodal datasets on the 4D Nucleome (http://vis.nucleome.org/entry/) [38]. This facilitated visualization of data at 5–40 kb resolution, with ∼4 Mb region of same chromosome being covered at 40 kb resolution, which could support our hypothesis by inspecting interactions between hub genes (that frequently gave rise to CTs).
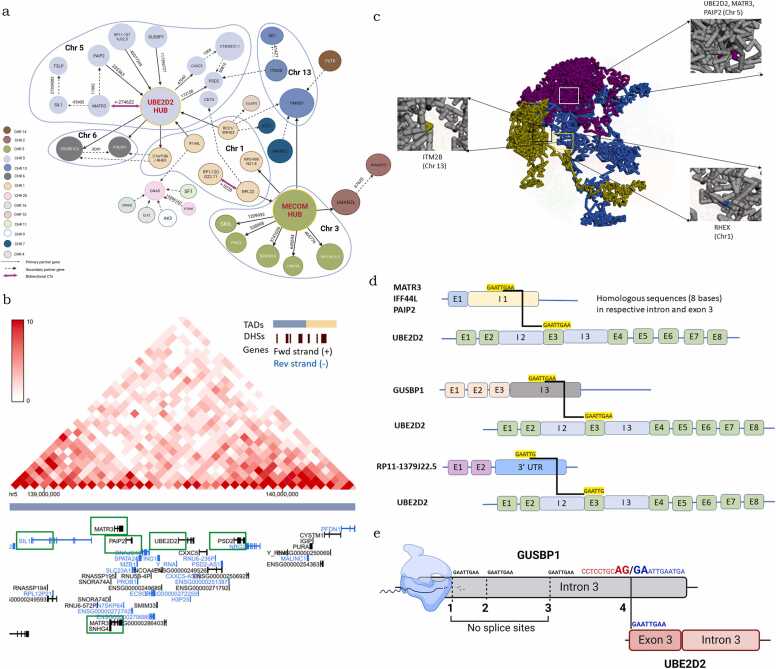
A prominent hub in our study UBE2D2, is involved in 12 CTs including 5 inter-chromosomal, 1 RT and 6 inD-CTs, while several of its partners interact with each other and/or other genes in their proximities to generate satellite CT hubs. We indeed could establish proximities between UBE2D2, MATR3, PAIP2, SIL1, CXXC5, SNGH4 and PSD2 in Hi-C data (genomic distance 2–3 *105 bases; Fig. 5a,5b; Supplementary Fig. 6b; Supplementary Table 9). Multiple UBE2D2 contacts were also revealed on 4D Nucleome browser suggesting proximity with its identified partners across the many available structures, of which MATR3 and PAIP2 are on same chromosome, while ITM2B and RHEX are on other chromosomes (Fig. 5c). Moreover, the UBE2D2-MATR3 and MATR3-UBE2D2 pair of CTs suggests bidirectional transcription between this and few other gene pairs (Supplementary Table 10). In other cancers also, multiple partners of our hub genes are reported. For example, UBE2D2 forms multiple chimeras in Lung Adenocarcinoma (LUAD) [36]. NUDUFS4 (chr 5), EXTL3 (chr 8), SND1 (chr 5), JHDMID (chr 7), ZFR (chr 5), AHCYL2 (chr 7), BRAF (chr 7) are the genes involved in primary and secondary contacts of UBE2D2. Most of these are on chromosome 5 and 7 indicating chromosome 5 and 7 might be in proximity in lung tissue based on our hypothesis. We were able to find contact maps of JHDMID, BRAF (these two genes form chimera with common partner SND1) and SND1, AHCYL2 in A549 (lung cancer cell line) (Supplementary Fig. 6c). We could not show all interactions due to limitation of 4 Mb window in Hi-C data. In the same publication we found another fusion ITM2B-MATR3 (these genes are present in UBE2D2 hub but do not form fusion in our database) in ovarian cancer chimeric transcript data. This strongly supports our hypothesis. The most striking observation of CTs involving UBE2D2 was that, its participation as a 3′ partner invariantly involved Exon3 at the fusionpoint, while that as a 5′ partner was variable (Exon 1 / 6 / 7). On examining sequences of both partners around the fusionpoint, a homologous (GAATTG / GAATTGAA) stretch present at the beginning of Exon3 of UBE2D2 was identified in the last participating intron or the 3′UTR of several of the 5′ partner genes (Fig. 5d). Such sequence homology may facilitate preferential RNA polymerase slippage and transcription across parental genes followed by generation of new splice sites that facilitate alternative splicing. In exploring this possibility, we specifically identified 4 GAATTGAA repeats in Intron 2 of GUSBP1. Only the last repeat predictably can function as an acceptor in the alternative intron/exon boundary and splice junction is generated (Fig. 5e).
Fig. 5.
Higher-order chromatin organization in the nucleus and gene proximity could facilitate RNA polymerase slippage and CT formation. a. UBE2D2 hub with its primary and secondary partner genes; b. Contact maps of UBE2D2 and its partner genes MATR3, PAIP2, CXXC5, SNHG, SIL1 and PSD2 in Hi-C data from ovarian tissue (Hi-C genome browser); c. Global, chromosome and gene view of chromatin arrangement captured from 4D nucleome browser. In which, chr5 (purple): UBE2D2, MATR3, PAIP2, Chr13 (green): ITM2B, Chr 1 (blue) RHEX have been highlighted; d. Schematic representing a homologous sequence (GAATTGAA/GAATTG) at beginning of UBE2D2 Exon3, and in the middle of Intron1 of MATR3/IFF44L/PAIP2 or Intron3 of GUSBP1 or 3′UTR of RP11–1379J22.5; e. Schematic representing new splice site generation and alternative splicing at the 4th homologous repeat (GAATTGAA) in GUSBP1 Intron3 and UBE2D2 Exon 3 of (at start) that may generate GUSBP1- UBE2D2 through RNA polymerase slippage. Red and blue text indicates the intron/exon boundary at the new splice site within GUSBP1 Intron3.
Surprisingly, these sequences are present in the middle of the intron and not at end. RNA polymerase slippage can be facilitated due to such sequence homology and hence leading to transfer of RNA polymerase from 5′ transcript to 3′ transcript. Later splicing mechanism remains unclear. Here, we cannot deny the possibility that the same stretch of SHS can also be present in other introns of the same gene. For example, in case of MATR3, GAATTGAA is also present in downstream introns (intron 7 and 10). But we suspect the spatial proximity is also equally important to drive this phenomenon. This is a hypothesis which might not be applied to all intra-chromosomal distant CTs and needs further experimental validation.
MECOM is another active hub that comprises of 9 unique CTs, wherein although its partner genes do not interact with each other, they generate satellite CT hubs with other genes including a pair of bidirectional chimeras viz. RPL22-RP1–120G22.11 and RP1–120G22.11-RPL22 (Supplementary Fig. 7a; Supplementary Table 11). The satellite hubs of HMGB1 (Chr1, Chr13) and RPL22 (Chr1, Chr5) also display cross-linkages with the UBE2D2 secondary network. Examining ovarian tissue contact maps on the Hi-C browser revealed multiple MECOM contacts suggesting spatio-temporal proximity with its partners (LRRC34, PHC3, SKIL; Supplementary Fig. 7b). Strikingly, the participation of MECOM as a 5′ partner invariantly involved a (GAATTG / GAATTGAA) stretch present at the end of it Exon1, which is the fusionpoint; these sequences were also present in the upstream introns of the 3′ gene partners involved and thereby may trigger RNA polymerase slippage, new splice site generation and alternative splicing (Supplementary Fig. 7c). Conclusively, this assigns a silent role to intronic sequences that possibly complement spatio-temporal orientation of chromosome territories, which could culminate in formation of specific CTs, otherwise deemed impossible.
4. Discussion
Our analyses of the RNA sequencing data for serous ovarian adenocarcinomas in the TCGA revealed an entire landscape of CTs that we sought to characterize. Previously predicted gene fusions in ovarian cancer transcriptomes have contributed to understanding of tumor physiology [39]. Some of these include BCAM-AKT2 (constitutive activation of AKT2 [39], MUC1-TRIM46-KRTCAP2 and SPON1-TRIM29 (chemoresistance / therapy [40], [41]), URB1-C21ORF45 and CTBS-GNG5 (suppression of tumor growth, [42], [43]), DPP9-PPP6R3 and DPP9-PLIN3 (adverse effects on tumor suppressor functioning, [44]), several fusions of ABCB1 and CCNL2 [45], [46], while CDKN2D-WDFY2, SPON1-TRIM29 and ESRRA-C11orf20 have diagnostic potential [47], [48],
The current mechanisms of splicing defects and / or readthrough transcription posits that higher gene densities may influence CT formation. Contrarily, we identified that the frequency of CT synthesis is not dependent on the number of genes present on a specific chromosome, but suggests that chromosomes with cancer-related genes are likely to be involved in chimera generation. The regulation of transcription (direction as well as dynamics), and splicing relies on RNA secondary structures in enhancing sequence complexities. Structural annotation of CTs identified in our study indicated the involvement of coding as well as non-coding transcripts as well as UTRs of genes. The latter are important determinants of transcript stability and gene expression regulation since altered 5′ / 3′ UTRs may either be protective or expose the transcript to degradation, while downstream exons of CTs may co-opt promoters from upstream exons from a different locus. The evolutionary conservation of protein-coding CTs may indicate a proclivity of enrichment for longevity [49].
Survival data of ovarian cancer patients in the Human Protein Atlas correlates CT expression with overall patient survival, which was also found reiterated in our study. Examining the specific CTs in malignant vs normal cell lines revealed altered expression of some of these in transformation, as earlier reported in association with cellular plasticity and altered cellular functionality [17]. The association of a few CTs as a mesenchymal signature could indicate a role for some of these CTs in maintaining a specific phenotype, which further strengthens our hypothesis that CTs play a crucial role in phenotypic plasticity and their related functions. It has been demonstrated that CT-generated proteins may compete with their parental proteins and perturb entire cascades through altered protein-protein interactions [50]. However, this will necessitate testing of the translational potential of the CTs, which is the obvious next phase of our study.
We also explored the theme that short homologous sequences (SHS) within genes separated by large genomic distances could be brought into proximity through higher order spatiotemporal alterations of the chromosome territories, chromatin looping and TAD formation that could lead to aberrant RNA polymerase slippage events. Although Hi-C data analysis posed a few restrictions including coverage within genomic distances of 4 Mb at a single time (at 40 kb resolution) and unavailability of inter-chromosomal Hi-C data for ovarian tissue, we did identify proximities within the UBE2D2 and MECOM hubs to generate CTs that otherwise cannot be accounted for by cis-SAGE or trans-splicing. Moreover, all CTs were detected in ovarian cancer RNA-seq data and most were absent in GTEX, which suggests that despite spatial proximities, a vulnerability to be co-transcribed through RNA polymerase slippage may occur only in cancer cells, and may be additionally influenced by epigenetic processes such as histone modifications, DNA methylation, etc. A recent report emphasizes the distinction of fusion transcripts generated through cis-SAGe or chromosomal rearrangements from those involving trans-splicing, by assigning a role to the poly-A tail at the terminal end of the 3′ gene in facilitating CT generation [51]. It is definite that the involvement of arduous transcription events and chromatin-RNA structures in CT formation would involve more than a single mechanism. These are exciting findings and hold potential for future investigations in exploring the diverse mechanisms of generation of CTs and their contribution to cellular physiology.
Author contributions
AS: developed, tested and validated the cloud-based pipeline; ASM: methodology, analysed and interpreted data and contributed to original draft preparation and its revision; ARJ: methodology, analysed and interpreted data and contributed to original draft preparation and its revision; KP: Data Curation; AM: generated the survival plots; VMD: analysed data represented in Fig. 1f) SAB conceptualized and planned the study design, developed methodology, analysed and interpreted data, supervised the project and edited the manuscript draft to its final form.
Funding
This work was supported by the Tata Innovation Fellowship of the Department of Biotechnology, Government of India [BT/HRD/35/01/04/2017], Fulbright-Nehru grants of USIEF [Award No. 2055/F-NAPE/2015] and NCCS Intramural grants to SAB, KP was granted project research fellowship by Department of Biotechnology, India [BT/HRD/35/01/04/2017]. Research fellowships were availed by ASM and ARJ from Council of Scientific and Industrial Research, New Delhi, India and from NCCS, Pune, India respectively.
Declaration of Competing Interest
None of the authors have any competing financial interests to declare in this work.
Acknowledgements
Data used in this publication were generated by the Tumor Genome Atlas Consortium [NCI/NIH]. We gratefully acknowledge the role of Mr. Anil Srivastava, Open Health Systems Laboratory, Rockville, US in accessing and processing TCGA data. Discussions and guidance received from Dr. Erik Lehnert (Seven Bridges Genomics) enabled crucial trouble shooting during the development and implementation of our computational pipeline on the CGC Cloud. The collaborative grant with Seven Bridges Genomics ($10,000 of compute and storage credits is deeply appreciated. The Seven Bridges Cancer Research Data Commons Cloud Resource has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, Contract No. HHSN261201400008C and ID/IQ Agreement No. 17 ×146 under Contract No. HHSN261201500003I and 75N91019D00024. Technical assistance from Mr. Avinash Mali and Ms. Snehal Nimdeokar is gratefully acknowledged. We also thank Dr. Vidisha Tripathi, NCCS, Pune for generously providing 2X SyBr Green premix, and the sequencing facility of National Centre for Microbial Resource (NCMR), Pune, India for Sanger sequencing of the PCR products. Some figures in this manuscript were created using BioRender.com.
Supplementary information
-
1.
Supplementary Information 1. Supplementary Figures S1-S7
-
2.
Supplementary Information 2. Supplementary Tables 1-11
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.10.028.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Gingeras T.R. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461(7261):206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuehner J.N., Pearson E.L., Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12(5):283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briones-Orta M.A., Avendaño-Vázquez S.E., Aparicio-Bautista D.I., Coombes J.D., Weber G.F., Syn W.K. Osteopontin splice variants and polymorphisms in cancer progression and prognosis. Biochim Et Biophys Acta (BBA)-Rev Cancer. 2017;1868(1):93–108. doi: 10.1016/j.bbcan.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Chen H., Gao F., He M., Ding X.F., Wong A.M., Sze S.C., Wong N. Long‐read RNA sequencing identifies alternative splice variants in hepatocellular carcinoma and tumor‐specific isoforms. Hepatology. 2019;70(3):1011–1025. doi: 10.1002/hep.30500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, H., Brown, R.L., Wei, Y., Zhao, P., Liu, S., Liu, X.,. & Cheng, C. (2019). CD44 splice isoform switching determines breast cancer stem cell state. Genes & development, 33(3–4), 166–179. [DOI] [PMC free article] [PubMed]
- 6.Kim R.N., Kim A., Choi S.H., Kim D.S., Nam S.H., Kim D.W., Park H.S. Novel mechanism of conjoined gene formation in the human genome. Funct Integr Genom. 2012;12:45–61. doi: 10.1007/s10142-011-0260-1. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Xie G.F., He Y., Deng L., Long Y.K., Yang X.H., Shao J.Y. Interfering expression of chimeric transcript SEPT7P2-PSPH promotes cell proliferation in patients with nasopharyngeal carcinoma. J Oncol. 2019;2019 doi: 10.1155/2019/1654724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H., Gao F., He M., Ding X.F., Wong A.M., Sze S.C., Wong N. Long‐read RNA sequencing identifies alternative splice variants in hepatocellular carcinoma and tumor‐specific isoforms. Hepatology. 2019;70(3):1011–1025. doi: 10.1002/hep.30500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pflueger D., Mittmann C., Dehler S., Rubin M.A., Moch H., Schraml P. Functional characterization of BC039389-GATM and KLK4-KRSP1 chimeric read-through transcripts which are up-regulated in renal cell cancer. BMC Genom. 2015;16:1–14. doi: 10.1186/s12864-015-1446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chwalenia K., Facemire L., Li H. Chimeric RNAs in cancer and normal physiology. Wiley Interdiscip Rev: RNA. 2017;8(6) doi: 10.1002/wrna.1427. [DOI] [PubMed] [Google Scholar]
- 11.Jia Y., Xie Z., Li H. Intergenically spliced chimeric RNAs in cancer. Trends Cancer. 2016;2(9):475–484. doi: 10.1016/j.trecan.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherbenou D.W., Hantschel O., Turaga L., Kaupe I., Willis S., Bumm T., Deininger M.W. Characterization of BCR-ABL deletion mutants from patients with chronic myeloid leukemia. Leukemia. 2008;22(6):1184–1190. doi: 10.1038/leu.2008.65. [DOI] [PubMed] [Google Scholar]
- 13.Akiva P., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16(1):30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Zou Q., Li F., Zhao W., Xu H., Zhang W., Yang X. Identification of the cross-strand chimeric RNAs generated by fusions of bi-directional transcripts. Nat Commun. 2021;12(1):4645. doi: 10.1038/s41467-021-24910-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Q., Liang W.W., Foltz S.M., Mutharasu G., Jayasinghe R.G., Cao S., Cope L. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23(1):227–238. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee S., Heng H.H., Frenkel-Morgenstern M. Emerging role of chimeric RNAs in cell plasticity and adaptive evolution of cancer cells. Cancers. 2021;13(17):4328. doi: 10.3390/cancers13174328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau J.W., Lehnert E., Sethi A., Malhotra R., Kaushik G., Onder Z., Davis-Dusenbery B. The Cancer Genomics Cloud: collaborative, reproducible, and democratized—a new paradigm in large-scale computational research. Cancer Res. 2017;77(21):e3–e6. doi: 10.1158/0008-5472.CAN-17-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Moore H.F. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bapat S.A., Mali A.M., Koppikar C.B., Kurrey N.K. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65(8):3025–3029. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 21.Varankar S.S., More M., Abraham A., Pansare K., Kumar B., Narayanan N.J., Bapat S.A. Functional balance between Tcf21–Slug defines cellular plasticity and migratory modalities in high grade serous ovarian cancer cell lines. Carcinogenesis. 2020;41(4):515–526. doi: 10.1093/carcin/bgz119. [DOI] [PubMed] [Google Scholar]
- 22.Kamble S.C., Sen A., Dhake R.D., Joshi A.N., Midha D., Bapat S.A. Clinical stratification of high-grade ovarian serous carcinoma using a panel of six biomarkers. J Clin Med. 2019;8(3):330. doi: 10.3390/jcm8030330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dekker J., Belmont A.S., Guttman M., Leshyk V.O., Lis J.T., Lomvardas S., 4D Nucleome Network The 4D nucleome project. Nature. 2017;549(7671):219–226. doi: 10.1038/nature23884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiff S.B., Schroeder A.J., Kırlı K., Cosolo A., Bakker C., Mercado L., Park P.J. The 4D nucleome data portal as a resource for searching and visualizing curated nucleomics data. Nat Commun. 2022;13(1):2365. doi: 10.1038/s41467-022-29697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scalzitti N., Kress A., Orhand R., Weber T., Moulinier L., Jeannin-Girardon A., Thompson J.D. Spliceator: multi-species splice site prediction using convolutional neural networks. BMC Bioinforma. 2021;22(1):1–26. doi: 10.1186/s12859-021-04471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardi N.L., Deshpande T.U., Kamble S.C., Budhe S.R., Bapat S.A. Discrete molecular classes of ovarian cancer suggestive of unique mechanisms of transformation and metastases. Clin Cancer Res. 2014;20(1):87–99. doi: 10.1158/1078-0432.CCR-13-2063. [DOI] [PubMed] [Google Scholar]
- 27.Kalra R.S., Bapat S.A. Expression proteomics predicts loss of RXR-γ during progression of epithelial ovarian cancer. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0070398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim N., Kim P., Nam S., Shin S., Lee S. ChimerDB—a knowledgebase for fusion sequences. Nucleic Acids Res. 2006;34(suppl_1):D21–D24. doi: 10.1093/nar/gkj019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forbes S., Clements J., Dawson E., Bamford S., Webb T., Dogan A., Stratton M.R. COSMIC 2005. Br J Cancer. 2006;94(2):318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim P., Zhou X. FusionGDB: fusion gene annotation DataBase. Nucleic Acids Res. 2019;47(D1):D994–D1004. doi: 10.1093/nar/gky1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitelman, F. (2005). Mitelman database of chromosome aberration in cancer. http://cgap. ncbi.nih. gov/chromosomes/mitelman.
- 32.Balamurali, D., Gorohovski, A., Detroja, R., Palande, V., Raviv-Shay, D., & Frenkel-Morgenstern, M. (2020). ChiTaRS 5.0: the comprehensive database of chimeric transcripts matched. [DOI] [PMC free article] [PubMed]
- 33.Hu Xin, Wang Qianghu, Tang Ming, Barthel Floris, Amin Samirkumar, Yoshihara Kosuke, Lang Frederick M., et al. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 2018;46(D1):D1144–D1149. doi: 10.1093/nar/gkx1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponting C.P. Biological function in the twilight zone of sequence conservation. BMC Biol. 2017;15(1) doi: 10.1186/s12915-017-0411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tena J.J., Santos-Pereira J.M. Topologically associating domains and regulatory landscapes in development, evolution and disease. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.702787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34(37):4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Yue F. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19(1):1–12. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu X., Zhang Y., Wang Y., Tian D., Belmont A.S., Swedlow J.R., Ma J. Nucleome browser: an integrative and multimodal data navigation platform for 4D nucleome. Nat Methods. 2022;19(8):911–913. doi: 10.1038/s41592-022-01559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzyzanowski P.M., Sircoulomb F., Yousif F., Normand J., La Rose J., E. Francis K., Rottapel R.K. Regional perturbation of gene transcription is associated with intrachromosomal rearrangements and gene fusion transcripts in high grade ovarian cancer. Sci Rep. 2019;9(1):3590. doi: 10.1038/s41598-019-39878-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kannan K., Coarfa C., Chao P.W., Luo L., Wang Y., Brinegar A.E., Yen L. Recurrent BCAM-AKT2 fusion gene leads to a constitutively activated AKT2 fusion kinase in high-grade serous ovarian carcinoma. Proc Natl Acad Sci. 2015;112(11):E1272–E1277. doi: 10.1073/pnas.1501735112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kannan K., Kordestani G.K., Galagoda A., Coarfa C., Yen L. Aberrant MUC1-TRIM46-KRTCAP2 chimeric RNAs in high-grade serous ovarian carcinoma. Cancers. 2015;7(4):2083–2093. doi: 10.3390/cancers7040878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagasawa S., Ikeda K., Shintani D., Yang C., Takeda S., Hasegawa K., Inoue S. Identification of a novel oncogenic fusion gene SPON1-TRIM29 in clinical ovarian cancer that promotes cell and tumor growth and enhances chemoresistance in A2780 Cells. Int J Mol Sci. 2022;23(2):689. doi: 10.3390/ijms23020689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plebani R., Oliver G.R., Trerotola M., Guerra E., Cantanelli P., Apicella L., Alberti S. Long-range transcriptome sequencing reveals cancer cell growth regulatory chimeric mRNA. Neoplasia. 2012;14(11):1087. doi: 10.1593/neo.121342. -49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smebye M.L., Agostini A., Johannessen B., Thorsen J., Davidson B., Tropé C.G., Micci F. Involvement of DPP9 in gene fusions in serous ovarian carcinoma. BMC Cancer. 2017;17(1):1–10. doi: 10.1186/s12885-017-3625-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christie E.L., Pattnaik S., Beach J., Copeland A., Rashoo N., Fereday S., Bowtell D.D. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nat Commun. 2019;10(1):1295. doi: 10.1038/s41467-019-09312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agostini A., Brunetti M., Davidson B., Göran Tropé C., Heim S., Panagopoulos I., Micci F. Identification of novel cyclin gene fusion transcripts in endometrioid ovarian carcinomas. Int J Cancer. 2018;143(6):1379–1387. doi: 10.1002/ijc.31418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kannan K., Coarfa C., Rajapakshe K., Hawkins S.M., Matzuk M.M., Milosavljevic A., Yen L. CDKN2D-WDFY2 is a cancer-specific fusion gene recurrent in high-grade serous ovarian carcinoma. PLoS Genet. 2014;10(3) doi: 10.1371/journal.pgen.1004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salzman J., Marinelli R.J., Wang P.L., Green A.E., Nielsen J.S., Nelson B.H., Brown P.O. ESRRA-C11orf20 is a recurrent gene fusion in serous ovarian carcinoma. PLoS Biol. 2011;9(9) doi: 10.1371/journal.pbio.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oz N., Vayndorf E.M., Tsuchiya M., McLean S., Turcios-Hernandez L., Pitt J.N., Kaya A. Evidence that conserved essential genes are enriched for pro-longevity factors. GeroScience. 2022;44(4):1995–2006. doi: 10.1007/s11357-022-00604-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frenkel-Morgenstern M., Lacroix V., Ezkurdia I., Levin Y., Gabashvili A., Prilusky J., Valencia A. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012;22(7):1231–1242. doi: 10.1101/gr.130062.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedrich S., Sonnhammer E.L. Fusion transcript detection using spatial transcriptomics. BMC Med Genom. 2020;13:1–11. doi: 10.1186/s12920-020-00738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material