Abstract
In women, cervical cancer (CC) is the fourth most common cancer around the world with average cases of 604,000 and 342,000 deaths per year. Approximately 50% of high-grade CC are attributed to human papillomavirus (HPV) types 16 and 18. Chances of CC in HPV-positive patients are 6 times more than HPV-negative patients which demands timely and effective treatment. Repurposing of drugs is considered a viable approach to drug discovery which makes use of existing drugs, thus potentially reducing the time and costs associated with de-novo drug discovery. In this study, we present an integrative drug repurposing framework based on a systems biology-enabled network medicine platform. First, we built an HPV-induced CC protein interaction network named HPV2C following the CC signatures defined by the omics dataset, obtained from GEO database. Second, the drug target interaction (DTI) data obtained from DrugBank, and related databases was used to model the DTI network followed by drug target network proximity analysis of HPV-host associated key targets and DTIs in the human protein interactome. This analysis identified 142 potential anti-HPV repurposable drugs to target HPV induced CC pathways. Third, as per the literature survey 51 of the predicted drugs are already used for CC and 33 of the remaining drugs have anti-viral activity. Gene set enrichment analysis of potential drugs in drug-gene signatures and in HPV-induced CC-specific transcriptomic data in human cell lines additionally validated the predictions. Finally, 13 drug combinations were found using a network based on overlapping exposure. To summarize, the study provides effective network-based technique to quickly identify suitable repurposable drugs and drug combinations that target HPV-associated CC.
Keywords: Cervical cancer, Drug repurposing, Human papillomavirus, Protein interactions, Systems biology
Graphical Abstract
Highlights
-
•
Cervical cancer (CC) is the fourth most common cancer in women, with an average of 604,000 cases and 342,000 deaths a year.
-
•
Human papillomavirus (HPV) types 16 and 18 are responsible for approximately 50% of high-grade cervical cancer cases.
-
•
The chances of developing CC in HPV-positive cases are six times higher than otherwise, showing need for timely treatment.
-
•
An integrative drug repurposing identified 142 potential anti-HPV repurposable drugs to target HPV-induced CC pathways.
-
•
51 drugs used for CC, 33 have antiviral activity, and 13 combinations were identified using overlapping exposure analysis.
1. Introduction
Cervical cancer (CC) remains a significant public health concern and is the fourth most common cancer affecting women globally. According to recent estimates, approximately 604,000 new cases of CC and 342,000 related deaths occur each year. These alarming statistics highlight the need for increased efforts towards CC prevention and management, particularly in low- and middle-income countries, where over 90% of new cases and deaths occur. Human papillomavirus (HPV) is a significant risk factor for CC, with two HPV types (16 and 18) being responsible for almost 50% of high-grade cervical pre-cancers [1]. The virus is predominantly transmitted through sexual contact, and the majority of people become infected with HPV shortly after becoming sexually active [2]. However, it's important to note that over 90% of HPV infections will eventually clear on their own without causing any harm [3], [4]. Women living with HPV face a significantly increased risk of developing CC, with a six-fold higher likelihood compared to women without HPV [5]. Therefore, it's crucial that healthcare providers prioritize CC screening and prevention measures for women living with HPV to reduce their risk of developing this type of cancer. Fortunately, CC can be prevented, and early diagnosis and treatment can lead to a high cure rate. A combination of primary and secondary prevention strategies such as HPV vaccination, screening, and treatment of pre-cancerous lesions have been shown to be effective in preventing CC at initial stages of disease [6]. However, the high death ratio to infected personnels demands the introduction of new and more effective therapies to be developed. In addition, current treatments for advanced CC are often ineffective and can cause serious side effects, underscoring the urgent need for more effective therapies. Given the high burden of disease and limited treatment options, there is a critical need for continued research to develop new, effective treatments for HPV-associated CC [7], [8].
In this connection, drug discovery comes into picture where a new drug molecule is brought from idea to a market ready product. The process of drug discovery is a long and expensive journey that can take up to 12–17 years with an investment of $2-$3 billion [9], [10], [11]. In addition, the success rate of drug discovery is relatively low, standing at less than 10%. To overcome issues with drug discovery, drug repurposing (DR) is increasingly being recognized as a promising alternative to traditional drug discovery. The approach involves identifying new therapeutic uses for existing drugs that have already been approved by regulatory authorities (FDA). This means that much of the time-consuming and costly preclinical work has already been done, allowing researchers to focus on repurposing existing drugs for new therapeutic purposes. It takes only 2–5 years and requires an investment of $200-$300 million, which is significantly lower than that of drug discovery. Moreover, DR has a higher success rate, standing at greater than 30% [12], [13]. As a result, DR is becoming an attractive option for pharmaceutical companies and researchers looking to bring new treatments to patients more quickly and efficiently. Moreover, DR can also help to address the growing problem of drug resistance, as it can uncover new uses for drugs that are already available and have a known safety profile. These statistics highlight the significant advantages of DR over drug discovery in terms of cost, time, and success rates. Many studies have found multiple potential drugs repurposable for CC including the anti-inflammatory drugs, immunomodulators, and DNA-damaging agents. Moreover, current treatment options for cervical cancer, including radiotherapy and chemotherapy, have limited efficacy and can cause significant side effects [14]. Platinum-based chemotherapy is the standard of care for advanced cervical cancer, but its efficacy is limited, and it can cause significant toxicity. There is a need for novel agents and treatment strategies to improve the therapeutic effect of current treatments and overcome the persistence of HPV [15].
Thus, several recent studies have explored the potential of drug repurposing for the treatment of various cancers using different approaches, including machine learning, virtual screening, and network-based methods. Moreover, there are ongoing studies and clinical trials on the repurposing of approved drugs for cancer therapy, including cervical cancer [16], [17], [18]. Some of the drugs that have shown potential for repurposing in cervical cancer therapy include metformin [19], thalidomide [20], and sonidegib [21]. Additionally, the US FDA has approved three PD-L1 inhibitors, namely Atezolimumab, Durvalumab, and Avelumab, that have been used in some solid tumors, including cervical cancer [22]. The use of immune checkpoint inhibitors, such as pembrolizumab and dostarlimab, in combination with standard chemotherapy has also shown promising results in recent clinical trials [23]. However, more research is needed to determine the effectiveness of these drugs for cervical cancer therapy. A study used a machine learning algorithm to identify drug candidates for the treatment of glioblastoma, a type of brain cancer [24]. The algorithm identified the anti-inflammatory drug celecoxib and the anti-psychotic drug fluphenazine as potential candidates. Similarly, a study used a virtual screening method to identify potential drug candidates for the treatment of triple-negative breast cancer [25]. The screening process identified several FDA-approved drugs, including the anti-malarial drug chloroquine, as potential candidates. While these approaches have shown promise, there are some drawbacks to machine learning and virtual screening, including the potential for false positives and a limited ability to identify novel targets [26], [27], [28]. In contrast, network-based approaches leverage existing knowledge of biological pathways and interactions to identify new drug targets, repurposable drugs, and potential drug combinations. In one study, a network-based approach was used to identify a combination of repurposed drugs that showed promising anti-cancer activity in preclinical models of triple-negative breast cancer [29]. Overall, while machine learning and virtual screening approaches have shown some success in identifying potential drug candidates, network-based approaches appear to be a more promising and robust approach for drug repurposing. Network-based drug repurposing studies have been utilized in the identification of potential antiviral drugs. For example, some recent study used network-based approaches to identify potential drugs for the treatment of COVID-19, including the repurposing of the antiviral drugs remdesivir and favipiravir [30], [31], [32]. Additionally, our recent review paper highlighted the use of these approaches for drug repurposing of viral cancers [33], [34].
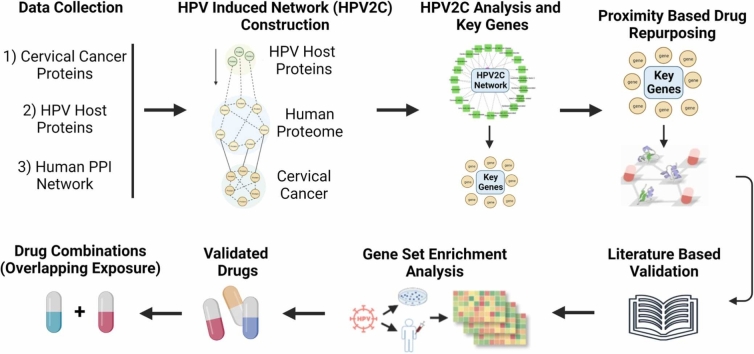
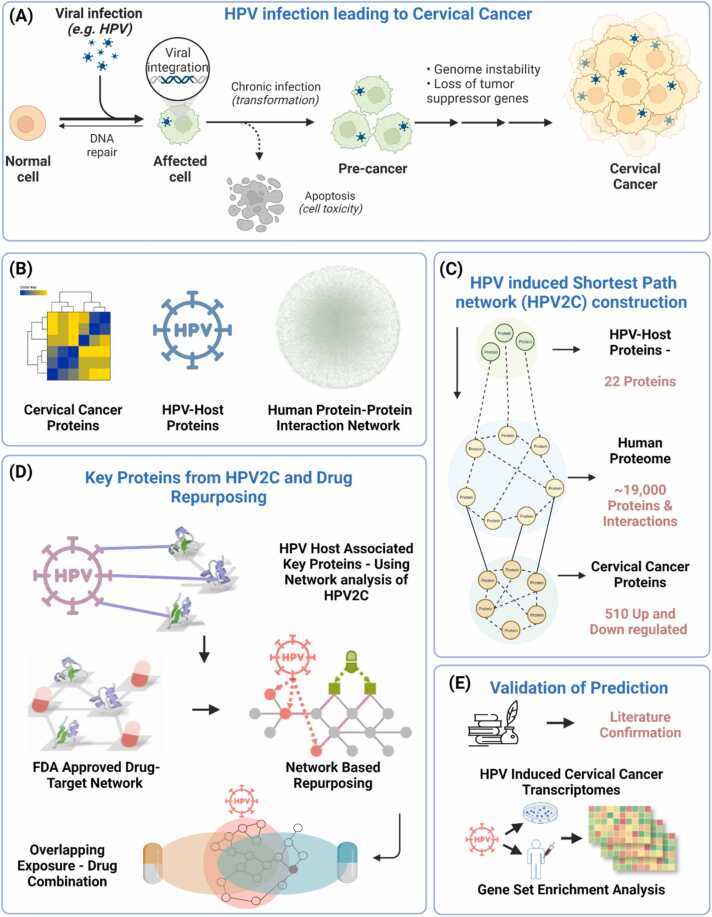
Here this research introduces and applies an integrative approach to drug repurposing, utilizing a network medicine platform enabled by systems biology. The study proposed the drug repurposing for HPV-associated CC (Fig. 1A). We established a protein interaction network of CC induced by HPV following the CC disease signatures determined by multi-omics data (Figure 1BC). The analysis of drug target network proximity based on HPV-host and drug target interactions in the human protein interactome identified 142 possible repurposable drugs for treating HPV-induced CC pathways (Fig. 1D). Among the predicted drugs, 51 have already been used for CC treatment and 33 possess anti-viral activity. The validity of the predictions was confirmed by literature survey and gene set enrichment analysis of potential drugs in drug-gene signatures and HPV-induced CC-specific transcriptomic data in human cell lines (Fig. 1E). Furthermore, 13 potential drug combinations were discovered using network-based overlapping exposure (Fig. 1D). Finally, this study provides a powerful network-based methodology to rapidly identify suitable repurposable drugs and drug combinations targeting HPV-associated CC.
Fig. 1.
Introductory figure showing network-based drug repurposing. A) Mechanism of HPV infection leading to cervical cancer. B) Data collection for HPV-hosts, cervical cancer, and Human protein-protein interactions. C) Construction of shortest path source (HPV-host) to target (Cervical cancer) network (HPV2C). D) Obtaining key proteins and performing drug repurposing using network proximity analysis. E) Validation of the prediction.
2. Materials and methods
2.1. HPV host proteins & cervical cancer DEGs
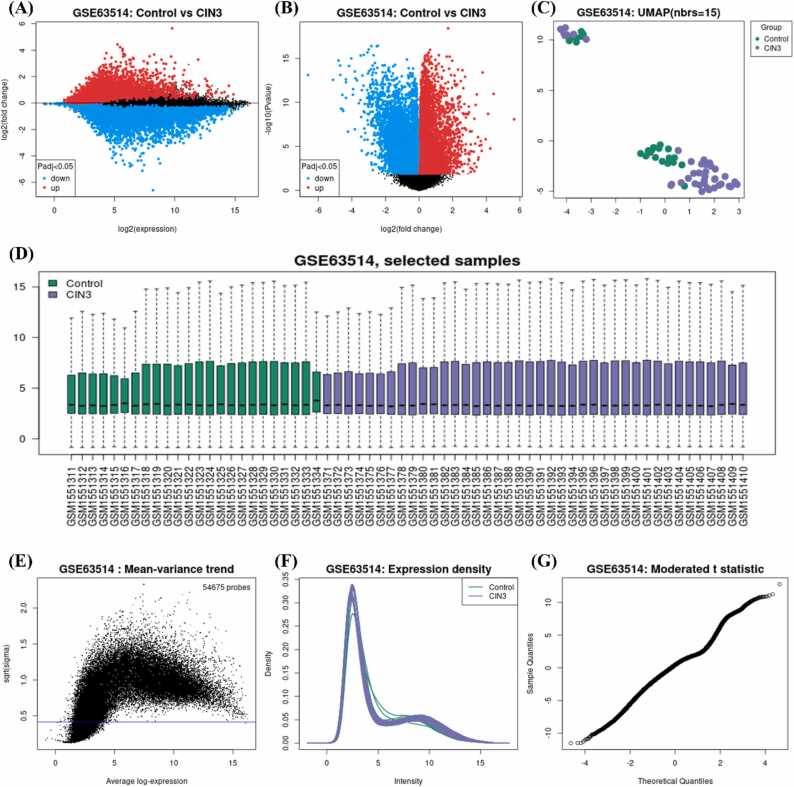
The data for HPV-host proteins and their interaction based on sizeable efforts was obtained from literature. The host proteins of HPV type 16 and 18 were mapped to Ensemble [35] and Uniprot IDs [36]. These proteins were either the direct targets of HPV proteins or were involved in critical pathways of HPV infection identified by multiple experimental sources [37], [38], including high-throughput yeast-two-hybrid (Y2H) systems, viral protein pull-down assay, in vitro co-immunoprecipitation and RNA knock down experiment [39]. Finally, a total of 22 HPV-associated host proteins were identified (Table S1). Additionally, a transcriptomic dataset (GSE63514) of HPV associated CC was downloaded from Gene Expression Omnibus (GEO) database [40]. The dataset contains total of 128 samples, of which 24 are normal (non-cancerous) samples and remaining 104 are disease (cancerous) samples. Differential gene expression analysis resulted in 536 up and downregulated genes (Table S2) (Fig. 2).
Fig. 2.
Cervical cancer expression analysis results using GEO2R web-based tool. A-B) Expression results in log2(expression) and fold change respectively where red are upregulated, and blue are downregulated. C) UMAP grouping between controls and CIN3 samples followed by D) total control and CIN3 samples. E-G) shows the statistical analysis results.
2.2. Functional enrichment analysis
Followed by data collection, we conducted the enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) [41] and Gene Ontology (GO) [42] for the HPV-host proteins and DEGs using ShineyGo 0.77 [43], DAVID [44], and gProfiler [45] tools respectively. Also, disease-enrichment was performed using DAVID functional annotation tool.
2.3. Human PPI network construction
For developing an exhaustive yet comprehensive set of PPIs, data from multiple bioinformatics and ontological databases was assembled such as STRING [46], SNAP [47], DisGeNET [48] etc. The constructed PPIs were screened to have three different types of supporting evidence. (i) high-throughput yeast-two-hybrid (Y2H) systems tested binary PPIs, (ii) analysis of binary physical PPIs obtained from protein 3D structures, and (iii) low-throughput experiments reported in the literature to construct a signaling network, inspired from a recent study mentioned in [30]. All data obtained from gene expression, metabolic associations, and evolutionary analysis were disregarded. The genes were matched to their official gene symbols and Uniprot and Ensemble ID utilizing GeneCards. The human PPI network constructed [49] had a total of 17516 proteins (nodes) and 893,876 edges linking the nodes (Fig. S1). The construction and procedure was also inspired from study done in [31] where 5 source types were used to finalize the high confidence PPIs to create human protein interactome.
2.4. HPV-induced cervical cancer (HPV2C) subnetwork construction
The constructed HPV2C network consisted of all the shortest paths between the HPV-host proteins and CC differentially expressed proteins (DEPs) in the human PPI network developed [31] using the STRING, SNAP, and DisGeNET protein interactions data. The key idea behind developing HPV2C network construction in this study was to come up with HPV related CV–associated proteins. The STRING database along with other databases was selected for a reason that selecting the STRING database as the PPI database is supported by prior research, which has shown that it offers a more extensive range of information on disease-associated protein sets than other databases. We only considered interactions with a confidence score greater than 0.4 [50], which is the default setting and represents the medium level of confidence for PPI searches in the STRING database. The Dijkstra algorithm [51] was employed to identify the shortest paths connecting every pair of proteins in the human PPI network between HPV-host and CC related DEPs. We utilized the NetworkX Python package [52] for carrying out the shortest path search. Gephi 0.9.2 [53] was used to visualize networks. Here network proximity [54] and RWR network algorithm [55] was used to find the HPV-host associated key proteins from the HPV2C network. To identify significant proteins for each of the network RWR algorithms, permutation tests were carried out 10,000 times. In each of the 10,000 tests [56], a random network was generated while preserving the degree distribution of the original HPV2C network [30], [31]. The random network was created by reconnecting edges in the main network and swapping nodes. The network algorithm was then applied in each permutation test to obtain the results. These results were used to calculate the empirical P value and Z-value of the network algorithm. Finally, the RWR permutation test results were analyzed to determine the final set of key proteins with an empirical P value of ≤ 0.05 [57].
Finally, to summarize for clarity, the key technical aspects of the HPV2C are Data Sources: Specify the version and sources of the databases used, such as STRING, SNAP, and DisGeNET, to ensure transparency and reproducibility. Algorithm Parameters: Provide details of specific parameters used in the Dijkstra algorithm, NetworkX Python package, and Gephi for network construction and visualization, such as distance metrics, scoring thresholds, and layout algorithms. Network Proximity and RWR: Explain the rationale for selecting network proximity and the Random Walk with Restart (RWR) algorithm, and describe any parameters or settings used in these algorithms, including restart probabilities or convergence criteria. Permutation Tests: Elaborate on the methodology for the permutation tests, including how random networks were generated while preserving the degree distribution, and any specific tools or libraries used to perform these tests. Statistical Significance: Clarify the criteria for identifying statistically significant key proteins, such as the choice of an empirical p-value threshold (≤0.05) and the interpretation of the results.
2.5. Drug-target network construction
To build drug-target interaction (DTI) network information of DTI was collected from databases including DrugBank [58], ChEMBL [59], PharmGKB [60], and Therapeutic Target Database (TTD) [61]. The DTI information was obtained from DrugBank. Interactions with binding affinities ≤ 10 μM, drug targets having status reviewed from Uniprot database, and having unique accession Uniprot IDs were kept as valid and used for developing DTI network.
2.6. Network proximity analysis
A DTI network using networkx library was constructed and visualized as shown in Fig. S2. To analyze key proteins from the HPV2C subnetwork, a network-based proximity analysis was conducted using networkx. This involved determining the network proximity inspired from [31] (using Eq. i) between a set of key proteins (K) and drug targets (T) for each approved drug. The shortest path length (d(k,t)) between nodes k ∈ K and t ∈ T in the human DTI network was used to calculate the distance between the drug targets and our key proteins in the HPV2C network.
| (i) |
To evaluate the importance of the gap between a crucial protein in the HPV2C network and a drug dc (K, T), the z score was obtained from the distance through permutation tests utilizing Eq. ii. The corresponding P value was calculated based on the permutation test results using python function.
| (ii) |
2.7. Gene set enrichment analysis
For validation, we carried out gene set enrichment analysis (GSEA) [62]. Specifically, we obtained two distinct data sets showing differential gene expression in individuals with HPV infection, which were retrieved from the GEO database. To conduct an analysis of differential expression, genes were considered differentially expressed if their adjusted P-value < 0.05. Data on the differential expression of genes in cells that were treated with different drugs were obtained from the Connectivity Map (CMAP) database [63]. The gene profiles of the drugs were utilized to calculate an enrichment score (ES) using CMAP Clue for each drug that was present in both the CMAP dataset and our drug-target network [9], [64].
2.8. Literature validation
To further confirm the indications, literature-based validation of the repurposable drugs is carried out. For each drug, original indication and its use for CC is searched in PubMed, patents, and databases [18], [65], [66]. As a result, a drug whose original indication is either CC or it is preclinically or clinically tried or under clinical trials for CC is labeled as validated. For the remaining drugs anti-viral activity is checked and those which possess antiviral activity are prioritized for preclinical validation.
2.9. Network based drug combination prediction
To ensure the success of this method of combining drugs based on their network, it is necessary to confirm whether the network relationship between two drug-target modules indicates their biological and pharmacological relevance and to measure the network relationship between drug targets and host proteins associated with HPV. To identify possible drug combinations [67], we integrated the top ranked drug lists. Our primary approach is based on the idea that a drug combination can be considered therapeutically effective only if it adheres to a particular relationship with the disease module. This relationship is identified by the Complementary Exposure or overlapping patterns observed in the target modules of both drugs (Eq. iii).
| (iii) |
Finally, 14 drug pairs are suggested to be pre-clinically and clinically validated for potential application in HPV associated CC.
3. Results
3.1. HPV2C network construction
Here, the HPV2C network was generated by connecting the shortest paths between HPV-hosts and CC-associated DEGs [68] in a human PPI network that was constructed from various sources, as described in the methods section. The primary objective of constructing the HPV2C network was to discover proteins that are linked to CC disease [16]. The STRING database was preferred for PPIs due to prior evidence suggesting that it offers more comprehensive information on diverse sets of disease-associated proteins. We used 22 HPV-human host proteins as source proteins and 536 CC related proteins (Expressed) from GEO Dataset (GSE63514) (Fig. 2) as target nodes. Degree, betweenness and closeness centralities values were calculated and were assigned to nodes as attributes followed by application of shortest path from source to target nodes traversing through human interactome which resulted in HPV2C network. The resulting network contained 1153 nodes including source, target, and intermediate or nodes from hidden network. Further analysis showed that, roughly 99% of the routes from HPV-associated proteins to CC-associated expressed proteins in HPV2C network are through multiple proteins. Moreover, the overlap between HPV-associated proteins and CC-associated proteins is merely 2%. These findings imply that the concealed stratum in our network holds significance in comprehending the routes that join HPV and CC. Further, differential expression analysis of GEO dataset (GSE63514) was performed using GEO2R web-based tool [9] with Padj< 0.05. The upregulated genes expression values are shown in Fig. 2A followed by fold change and UMAP values in Fig. 2 BC. The dataset contained samples for CIN1 (a grade 1 pre-cancerous condition of the cervix), CIN2 (a grade 2pre-cancerous condition of the cervix), and CIN3 a high-grade lesion and has a much higher risk of progressing to CC than CIN1 or CIN2 [69]. The sample count CIN3 are analyzed and used in this study (Fig. 2D) followed by mean variance trend, density of expression, and t-statistics are shown in Fig. 2E-G respectively [70]. The analysis results for the CIN1 and CIN2 pre-cancerous conditions are given in Fig. S3 and Fig. S4 respectively.
3.2. Interrogation of HPV2C network to find key proteins and pathways
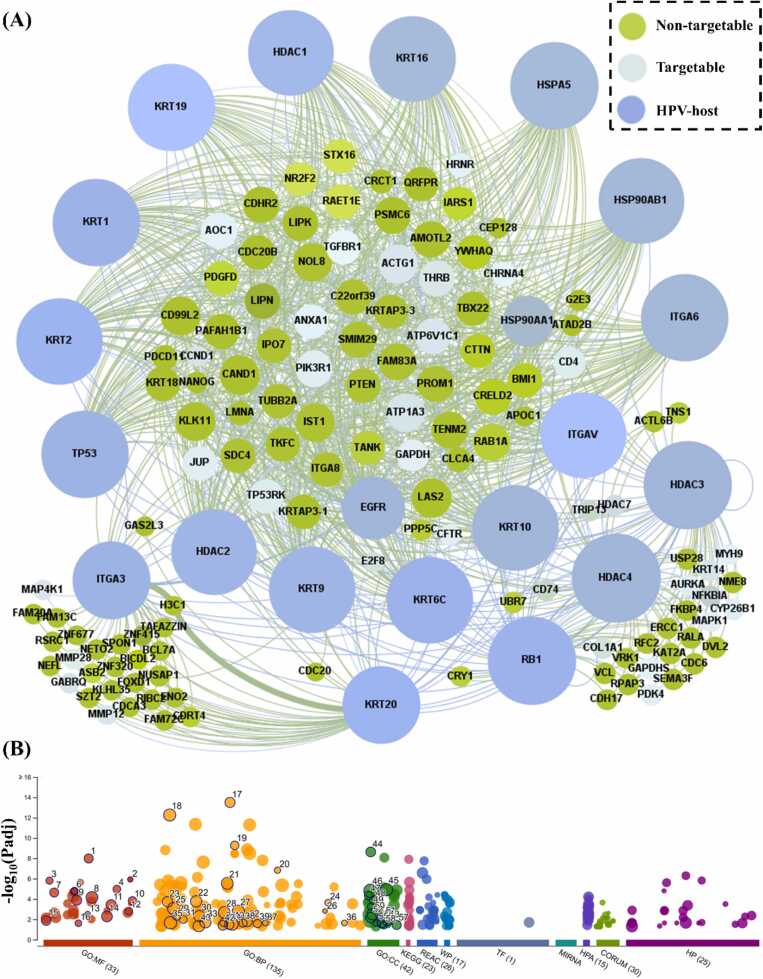
HPV2C networks contain proteins that are crucial to a pathway and can be targeted by drugs since they can significantly affect the pathway's function. To pinpoint the key proteins and disease pathways in the HPV2C network (Fig. 1C), various network algorithms like degree centrality, betweenness centrality, eigenvector centrality, and RWR were employed. To detect proteins that have statistical significance, we conducted 10,000 permutation tests for every network algorithm and picked proteins with empirical P values that were below 0.05. We combined the proteins selected by each network algorithm and classified them as key proteins (Mentioned in materials and methods). As a result, we identified 170 key common proteins against 22 HPV (Table S1 and Fig. S9) as shown in Fig. 3A. Identified key proteins are roughly 14.7% of the total proteins in the HPV2C network. HPV-host proteins are shown interacting with key proteins found from network analysis. The targetable and non-targetable proteins are highlighted using light blue and green color. The criteria was, if the protein is known target of drug, then it is considered as targetable and vice versa. Biological pathway enrichment of the HPV-host proteins resulted in viral carcinogenesis (Fig. S5), other cancers (Fig. S6) and HPV infection leading to cancer (Fig. S7) [71]. To confirm the biological relevance of these proteins to the symptoms of CC, an analysis was conducted. The analysis involved a GO enrichment study of the HPV-proteins, which revealed that the top enriched molecular functions, biological processes, and pathways involved that were potentially associated with the HPV induced viral carcinogenesis and related cancers (Fig. 3B). Detailed description of the GO findings are provided in Fig. S8 (Padj<0.05) and information related HPV-host protein related pathways are given in Fig. 4C and in detail are provided in Table S4.
Fig. 3.
A) HPV-host to key protein interactions where HPV-host are shown using the blue colored nodes, targetable proteins are presented using light blue color, and non-targetable are shown using green color. B) Geno ontology (GO) enrichment analysis of the HPV-host proteins using gProfiler web-based tool.
Fig. 4.
Repurposed drug information. A) Drug-target network of potentially repurposable drugs plotted based on node degree. B) ShinyGO 0.77 based pathway enrichment results of key proteins from HPV2C network, C) shows the pathway enrichment of HPV-host associated key targetable proteins using ShinyGO 0.77.
3.3. In-silico network proximity analysis of drug-target network identifies drug candidates
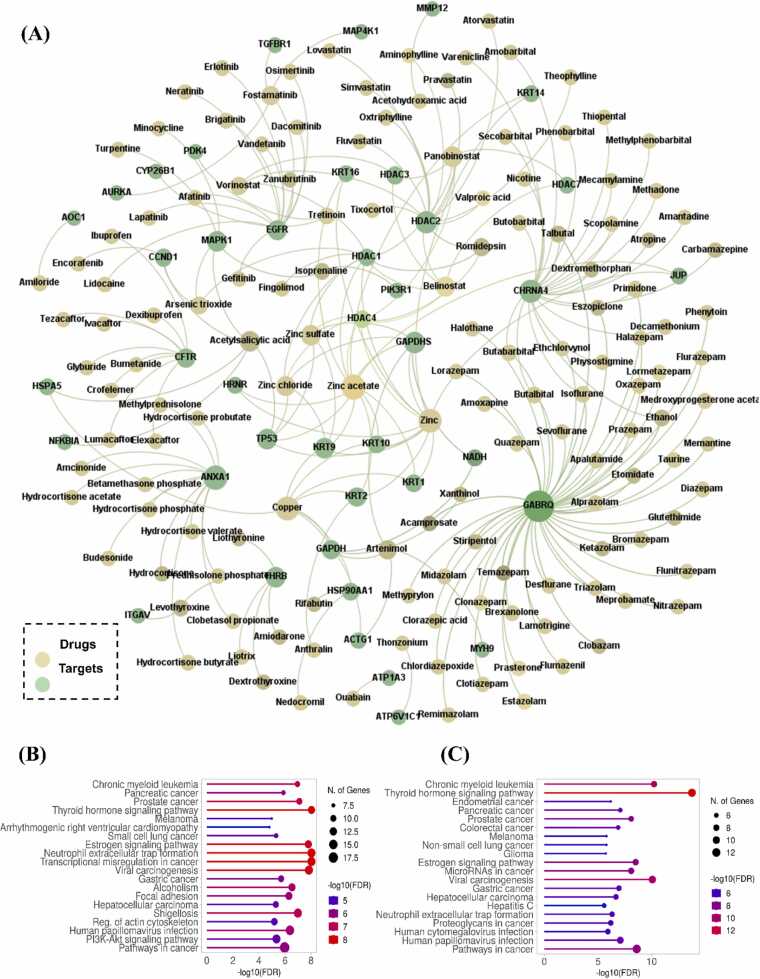
After the identification and analysis of key HPV2C proteins, we were inspired to seek approved drugs that could attach to many of these host proteins. This could enhance the efficacy of the drugs in preventing HPV-induced CC. We utilized an in-silico network-based proximity measure analysis to evaluate the key proteins in the HPV2C network. We gathered 1804 approved drugs from public databases, such as ChEMBL and DrugBank. Next drug-target interaction network was constructed followed by proximity analysis. The virtual screening process enabled us to predict 142 drugs (as shown in Table S3) that are likely to target the key proteins of the HPV2C network (P < 0.05). These network proximity analyses offer putative repurposable candidates for potential prevention and treatment of HPV associated CC. Moreover, the predicted drugs are roughly 7.8% of the total drugs in network. Anatomical Therapeutic Chemical (ATC) codes were then checked to find out the therapeutic areas for which these drugs have been approved. The top therapeutic areas included cancer, anti-viral or infectious disease, and immune system etc. The predicted drugs are plotted in the form of drug-target interaction network as shown in Fig. 4A. The green color nodes represent the drugs whereas other nodes are targets. The interaction network is generated using Gephi 0.9.2 and is shaped based on degree connectivity. This helps us find the drugs which are connected to multiple targets and targets which are inhibited by multiple drugs. Such as “GABRQ” is obviously the protein target with high number of drugs followed by “Zinc acetate” and “copper” with high number of targets in the number. Additionally, pathway enrichment analysis is performed for the druggable targets using ShinyGo web-based tools which show their involvement in cancers and HPV-infection (Fig. 4B). The detailed analysis for GO enrichment of HPV2C key proteins is provided in Fig. S10. Different GO processes such as molecular function (MF), biological processes (BP), and KEGG pathways are mentioned [72]. Additionally, pathway enrichment analysis of key proteins shows the significant enrichment in various cancer signaling pathways (Fig. S11) playing crucial role in cancer progression. Similarly, pathway enrichment analysis of HPV-host proteins is also given in Fig. 4C, and details are provided in Fig. S8. The pathway enrichment results for key proteins and HPV-host proteins coincide and thus it shows that key proteins and HPV-host proteins lies in close vicinity, and it further strengthen the hypothesis that similar targets can by targeted by similar drugs.
3.4. Validation of repurposable drugs
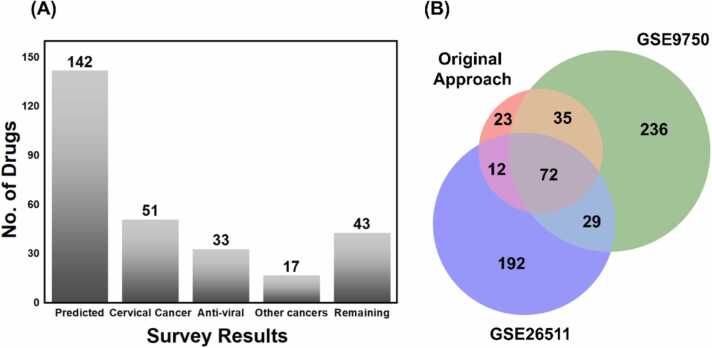
Here we used literature based and GSEA based validation of the repurposed drugs. Initially, a literature survey was performed to confirmed if the predicted drugs have been used already for CC or not. Surprisingly, 51 of 142 (35.9%) were found to have been already used in the CC (Table 1). Additionally, from the remaining 91 drugs, 33 of were found to possess the anti-viral activity (Table 2) but have not been used in CC and could be possible repurposable drug candidates for HPV-associated CC. Antivirals account for 26.05% of the total predicted drugs and 40.65% of remaining 91 drugs. Moreover, among the remaining 58 drugs, 17 (29.3% of 58% and 11.9% of 142 drugs) were anti-cancer drugs used for variety of cancer followed by remaining 43 drugs which were neither anti-cancer nor anti-viral as shown in Fig. 5A. Additionally, our integrated computational approaches were found to be effective in prioritizing compounds, as demonstrated by a hypergeometric test indicating a probability of 35.9% for our 142 drugs being in preclinical or clinical trials, with a resulting P value of 2.3 × 10 − 3 [31].
Table 1.
Literature based validated drugs, shown to have been tested for endometrial cancer.
| DrugBank ID | Name | Uniprot Name | Uniprot ID | |
|---|---|---|---|---|
| 1 | DB00157 | NADH | Glyceraldehyde-3-phosphate dehydrogenase | P04406 |
| 2 | DB00175 | Pravastatin | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | O14556 |
| 3 | DB00184 | Nicotine | Histone deacetylase 2 | Q92769 |
| 4 | DB00227 | Lovastatin | Neuronal acetylcholine receptor subunit alpha 4 | P43681 |
| 5 | DB00281 | Lidocaine | Histone deacetylase 2 | Q92769 |
| 6 | DB00313 | Valproic acid | Epidermal growth factor receptor | P00533 |
| 7 | DB00317 | Gefitinib | Histone deacetylase 2 | Q92769 |
| 8 | DB00333 | Methadone | Epidermal growth factor receptor | P00533 |
| 9 | DB00451 | Levothyroxine | Neuronal acetylcholine receptor subunit alpha 4 | P43681 |
| 10 | DB00475 | Chlordiazepoxide | Thyroid hormone receptor beta | P10828 |
| 11 | DB00530 | Erlotinib | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 12 | DB00543 | Amoxapine | Epidermal growth factor receptor | P00533 |
| 13 | DB00551 | Acetohydroxamic acid | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 14 | DB00594 | Amiloride | Macrophage metalloelastase | P39900 |
| 15 | DB00599 | Thiopental | Amiloride-sensitive amine oxidase [copper-containing] | P19801 |
| 16 | DB00603 | Medroxyprogesterone acetate | Neuronal acetylcholine receptor subunit alpha 4 | P43681 |
| 17 | DB00641 | Simvastatin | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 18 | DB00690 | Flurazepam | Histone deacetylase 2 | Q92769 |
| 19 | DB00741 | Hydrocortisone | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 20 | DB00753 | Isoflurane | Annexin A1 | P04083 |
| 21 | DB00755 | Tretinoin | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 22 | DB00898 | Ethanol | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, mitochondrial | Q16654 |
| 23 | DB00915 | Amantadine | Neuronal acetylcholine receptor subunit alpha 4 | P43681 |
| 24 | DB00959 | Methylprednisolone | Neuronal acetylcholine receptor subunit alpha 4 | P43681 |
| 25 | DB01013 | Clobetasol propionate | Annexin A1 | P04083 |
| 26 | DB01016 | Glyburide | Annexin A1 | P04083 |
| 27 | DB01050 | Ibuprofen | Cystic fibrosis transmembrane conductance regulator | P13569 |
| 28 | DB01076 | Atorvastatin | Cystic fibrosis transmembrane conductance regulator | P13569 |
| 29 | DB01092 | Ouabain | Histone deacetylase 2 | Q92769 |
| 30 | DB01095 | Fluvastatin | Sodium/potassium-transporting ATPase subunit alpha-3 | P13637 |
| 31 | DB01118 | Amiodarone | Histone deacetylase 2 | Q92769 |
| 32 | DB01169 | Arsenic trioxide | Thyroid hormone receptor beta | P10828 |
| 33 | DB01189 | Desflurane | G1/S-specific cyclin-D1 | P24385 |
| 34 | DB01236 | Sevoflurane | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 35 | DB01259 | Lapatinib | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 36 | DB01303 | Oxtriphylline | Epidermal growth factor receptor | P00533 |
| 37 | DB01593 | Zinc | Histone deacetylase 2 | Q92769 |
| 38 | DB01708 | Prasterone | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | O14556 |
| 39 | DB01956 | Taurine | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 40 | DB02546 | Vorinostat | Gamma-aminobutyric acid receptor subunit theta | Q9UN88 |
| 41 | DB05015 | Belinostat | Histone deacetylase 1 | Q13547 |
| 42 | DB06176 | Romidepsin | Histone deacetylase 1 | Q13547 |
| 43 | DB06603 | Panobinostat | Histone deacetylase 1 | Q13547 |
| 44 | DB08916 | Afatinib | Histone deacetylase 1 | Q13547 |
| 45 | DB09130 | Copper | Epidermal growth factor receptor | P00533 |
| 46 | DB09213 | Dexibuprofen | Glyceraldehyde-3-phosphate dehydrogenase | P04406 |
| 47 | DB09330 | Osimertinib | Cystic fibrosis transmembrane conductance regulator | P13569 |
| 48 | DB11638 | Artenimol | Epidermal growth factor receptor | P00533 |
| 49 | DB11828 | Neratinib | Actin, cytoplasmic 2 | P63261 |
| 50 | DB14539 | Hydrocortisone acetate | Epidermal growth factor receptor | P00533 |
Table 2.
Drugs having anti-viral activity.
| DrugBank ID | Name | Uniprot Name | Structures | Uniprot ID | |
|---|---|---|---|---|---|
| 1 | DB00231 | Temazepam | Gamma-aminobutyric acid receptor subunit theta | 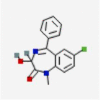 |
Q9UN88 |
| 2 | DB00277 | Theophylline | Histone deacetylase 2 | 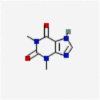 |
Q92769 |
| 3 | DB00279 | Liothyronine | Thyroid hormone receptor beta |  |
P10828 |
| 4 | DB00349 | Clobazam | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 5 | DB00404 | Alprazolam | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 6 | DB00514 | Dextromethorphan | Neuronal acetylcholine receptor subunit alpha 4 |  |
P43681 |
| 7 | DB00572 | Atropine | Neuronal acetylcholine receptor subunit alpha 4 |  |
P43681 |
| 8 | DB00615 | Rifabutin | Heat shock protein HSP 90-alpha |  |
P07900 |
| 9 | DB00659 | Acamprosate | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 10 | DB00747 | Scopolamine | Neuronal acetylcholine receptor subunit alpha 4 |  |
P43681 |
| 11 | DB00842 | Oxazepam | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 12 | DB00887 | Bumetanide | Cystic fibrosis transmembrane conductance regulator |  |
P13569 |
| 13 | DB00981 | Physostigmine | Neuronal acetylcholine receptor subunit alpha 4 |  |
P43681 |
| 14 | DB01043 | Memantine | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 15 | DB01159 | Halothane | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 16 | DB01174 | Phenobarbital | Neuronal acetylcholine receptor subunit alpha 4 |  |
P43681 |
| 17 | DB01223 | Aminophylline | Histone deacetylase 2 |  |
Q92769 |
| 18 | DB01273 | Varenicline | Neuronal acetylcholine receptor subunit alpha 4 |  |
P43681 |
| 19 | DB01544 | Flunitrazepam | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 20 | DB01587 | Ketazolam | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 21 | DB01588 | Prazepam | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 22 | DB01589 | Quazepam | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 23 | DB08868 | Fingolimod | Histone deacetylase 1 |  |
Q13547 |
| 24 | DB09280 | Lumacaftor | Cystic fibrosis transmembrane conductance regulator |  |
P13569 |
| 25 | DB09552 | Thonzonium | V-type proton ATPase subunit C 1 |  |
P21283 |
| 26 | DB11157 | Anthralin | Keratin, type II cytoskeletal 2 epidermal |  |
P35908 |
| 27 | DB11901 | Apalutamide | Gamma-aminobutyric acid receptor subunit theta |  |
Q9UN88 |
| 28 | DB12010 | Fostamatinib | TGF-beta receptor type-1 |  |
P36897 |
| 29 | DB14487 | Zinc acetate | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific |  |
O14556 |
| 30 | DB14533 | Zinc chloride | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific |  |
O14556 |
| 31 | DB14548 | Zinc sulfate, unspecified form | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific |  |
O14556 |
| 32 | DB14669 | Betamethasone phosphate | Annexin A1 |  |
P04083 |
| 33 | DB15035 | Zanubrutinib | Epidermal growth factor receptor |  |
P00533 |
Fig. 5.
Validation results. A) shows literature-based validation results of the predicted drugs and B) shows GSEA based validation results of the repurposable drugs.
Moreover, in order to validate 142 drugs that can be repurposed against HPV-induced cervical cancer (CC) [73], we conducted GSEA on the transcriptome data of host cells infected with HPV-associated CC. We obtained two transcriptomics datasets, identified as GSE9750 and GSE26511, from GEO. These transcriptome data served as gene signatures for HPV-induced CC. Among the selected datasets, GSE9750 contained a total of 66 samples. These include 33 primary tumors, 9 cell lines, and 24 normal cervical epitheliums. Similarly, GSE26511 consisted of 20 negative samples and 19 positive CC samples making a total of 39 samples in dataset. Both datasets were analyzed using GEO2R and DEGs were calculated manually. Next, GSEA was performed for GSE9750 (Fig. S12) which provided useful information of the enriched pathways and biological process. Additionally, we downloaded the gene expression data of drug-treated human cell lines from the CMAP database to obtain drug–gene signatures followed by using clue database to perform drug repurposing against the mentioned datasets [74]. The results obtained from CLUE for both datasets were compared with obtained from our approach as shown in Fig. 5B. Surprisingly, 72 drugs were shared between all three datasets which testifies the significance of this study.
Although our work used methodologies such as GSEA and literature-based validation, there are a number of limitations that must be recognized. Incomplete or biased results could result from literature-based validation since it depends on data availability, publication bias, and data quality. We conducted a thorough literature search and used several sources (PubMed, patents, and databases) to find medications used for CC to overcome these constraints. Despite these attempts, it is important to recognize that certain crucial data may still be missed or distorted because of publication trends. Subjectivity and inter-observer variation may be introduced during the manual estimation of differentially expressed genes in GSEA. The findings' generalizability can be restricted because they are unique to the datasets and methodologies used. The effects of integration might be impacted by complexity. The choice of the dataset also creates potential bias. These restrictions point out areas that need thought and additional investigation in the domain of medication repurposing for HPV-induced cervical cancer.
3.5. Network-based location of potential drug combinations for HPV-led CC
To enhance therapeutic effectiveness and minimize toxicity, drug combinations are frequently employed in the treatment of different viral infections. Nevertheless, identifying and verifying potent drug combinations is constrained by the vast number of possible drug pairs and dosage combinations, leading to a combinatorial explosion. Considering this challenge, we drew inspiration from a study [67] that suggested a novel network-based approach for identifying clinically effective drug combinations. To achieve this, we employed approved drug combinations. We used network based “overlapping exposure” pattern approach to find the drug combinations that may create synergistic effect. After obtaining the possible drug combinations, additional criteria of having minimum of 3 shared targets. As a result, 13 potential drug combinations are obtained as shown in Table 3. Among the mentioned combinations, Belinostat and Vorinostat are effectively tested as pan-HDAC inhibitors against HPV-18 and thus their combination is also found effective in various viral cancers such as CC [75].
Table 3.
Predicted potential drug combinations.
| Drug 1 | Drug 2 | Overlapped proteins | |
|---|---|---|---|
| 1 | Belinostat | Vorinostat | HDAC1, HDAC2, HDAC3 |
| 2 | Belinostat | Panobinostat | HDAC1, HDAC2, HDAC4, HDAC7, HDAC3 |
| 3 | Belinostat | Romidepsin | HDAC1, HDAC2, HDAC4 |
| 4 | Vorinostat | Panobinostat | HDAC1, HDAC2, HDAC3 |
| 5 | Zinc | Zinc chloride | HDAC1, TP53, HDAC4, GAPDHS, KRT9 |
| 6 | Zinc | Copper | KRT10, KRT2, KRT1, KRT9 |
| 7 | Zinc | Zinc sulfate | HDAC1, TP53, HDAC4, GAPDHS |
| 8 | Zinc | Zinc acetate | HDAC1, KRT1, TP53, KRT2, KRT16, HDAC4, KRT14, HRNR, JUP, GAPDHS, KRT10, KRT9 |
| 9 | Zinc chloride | Zinc acetate | HDAC1, TP53, HDAC4, GAPDHS |
| 10 | Zinc chloride | Zinc sulfate | HDAC1, TP53, HDAC4, GAPDHS |
| 11 | Copper | Zinc acetate | KRT10, KRT2, KRT1, KRT9 |
| 12 | Panobinostat | Romidepsin | HDAC1, HDAC2, HDAC4 |
| 13 | Zinc sulfate | Zinc acetate | HDAC1, TP53,HDAC4, GAPDHS |
4. Discussion
Here, various computational techniques, including bespoke methods for integrating data, analyzing networks, and simulating computer models were utilized (Fig. 1B and D). The goal was to discover pathways caused by HPV that contribute to CC. These pathways could potentially be treated with currently available drugs by repurposing them for therapeutic purposes. While network analysis has become a popular method for analyzing genetic datasets to identify disease indicators, our approach, which involved creating the HPV2C network (Fig. 1C) and applications of network algorithms without any preconceived notions, was crucial in uncovering these novel targets. HPV2C was created by three types of protein nodes, first HPV-host proteins (obtained from literature), second CC associated expressed proteins (Fig. 2), and middle or human interactome proteins capturing key pathways and processes involved in HPV infection leading to CC. HPV2C analysis (Materials and Methods) provided 170 key proteins against 22 HPV-host proteins (Fig. 3A). David, ShinyGo, gProfiler based enrichment analysis of HPV-host proteins (Fig. 3B) and key proteins (Fig. 4B and Fig. S10) provided key information of the MP, BP, and enriched pathways. This was followed by DTI network construction of approved drugs from multiple databases and proximity analysis between created DTI network and key proteins from HPV2C led to perform drug repurposing for HPV induced CC (Fig. 1D).
After conducting our analysis, we have identified 142 drugs that have been approved for other uses but may also be effective in treating CC caused by HPV (refer to Table S3). Based on our findings, we believe that these drugs could be repurposed for CC treatment since 51 of 142 (35.9%) were found to have been already used in the CC (Table 1). Additionally, from the remaining 91 drugs, 33 of were found to possess the anti-viral activity (Table 2) (Fig. 5A) but have not been used in CC and could be possible repurposable drug candidates for HPV-associated CC. Moreover, one important aspect of our analysis involves utilizing drugs that have already been approved for other uses. This approach enables us to quickly advance the remaining 91 drugs containing 33 anti-viral that have not entered in clinical trials for CC as the most effective treatments for CC caused by HPV.
Moreover, Avastin with brand name of bevacizumab has been used to treat advanced CC by inhibiting the Vascular endothelial growth factor (VEGF) [76]. Similarity our approach predicted the statins like Pravastatin and Lovastatin for CC supported by literature confirmation [77] (Fig. 4A). Pravastatin and Lovastatin are shown targeting HDAC-2 (Fig. 4A) whereas it is suggested that HDAC inhibitors are promising therapeutic agents to treat benign HPV infections, abrogate progeny virus production [75]. This emphasizes to test the other predicted drugs and raises the exciting possibility of their potential use in CC. Other predicted drugs that were confirmed to have entered clinical trials for CC are Lidocaine [78], Methadone [79], and Levothyroxine [80] etc. Additionally, we performed GSEA using two datasets of transcriptomic signatures in CC and queried the Clue database to find the drugs that could possibly reverse the disease signatures. Upon comparison, we found that 72 drugs are common among these two datasets and our approach as shown in Fig. 5B. In addition, we utilized “overlapping exposure” pattern between the drug-targets and identified potential drug combinations which were latter further refined by setting criteria of significance (targets >= 3) (Table 3).
A literature based complementary study queried the PubMed for the drugs available in ReDO_DB [81] and CDcervix_DB [21] and found that 174/534 (33%) were having greater than or equal to one related abstract or registered trial history in CC. 94/534 (18%) drugs were having human data available, and 52/534 (10%) drugs had been assessed in registered trials [21]. The list of drugs significantly overlaps with our list with having 33 of 142 (23.2%) approved drugs being present in this list. The main reason is that 100% of our drugs are approved whereas the drug list in study [21] contains unapproved drugs as well. The statement points out a significant contrast between the two studies. Our research conducted in-silico has detected potential antiviral medicines that have already been approved and are therefore in an advanced stage of repurposing. On the other hand, the research cited in reference [21] has identified substances found in databases such as ReDO_DB and CDcervix_DB, which include both approved and unapproved compounds. Moreover, our study is based on computational methods that employ cutting-edge network-based techniques and algorithms to identify crucial proteins through network proximity analysis, which leads to drug repurposing in contrast to manual literature search.
Several computational studies have been conducted to find potential cancer drugs through multi-stage analyses, including network proximity measure analysis that focuses on a specific target and its interactomes. In contrast, we have taken a more holistic approach, constructing the entire protein pathway that is significantly affected during HPV infection. This was achieved by uncovering the hidden layer between HPV-host and CC-associated regulated genes, which were identified from literature and proteomic data obtained from gene expression analysis of laser-captured epithelium (GSE63514) from 128 cervical tissue. To identify the key proteins systematically, we used different network algorithms (as described in Materials and Methods). Our approach not only detected 142 drugs but also 13 drug combinations. However, like other studies, our study also has limitation of directionality in PPI network that brings additional information about the type of interaction that takes place such as activation or inhibition. Moreover, lack of experimental validation, limited coverage to chemical space, and lack of efficient integration of different network algorithms are also the major limitations to be addressed in future studies. To overcome these challenges, Computational predictions need to be validated through robust in vitro and in vivo assays at each step of the pipeline. Additionally, expanding the screening of on-demand libraries by several orders of magnitude to billions and more of previously unexplored drug-like compounds, either physical or virtual, is expected to change the drug discovery model in several ways and finally, efficient algorithms need to be integrated properly to achieve the comprehensive study of biological processes at multiple scales.
5. Conclusion
Cervical cancer (CC) remains a significant public health concern and is the fourth most common cancer affecting women globally. According to recent estimates, approximately 604,000 new cases of CC and 342,000 related deaths occur each year, which necessitates the development of more efficient treatments at a faster pace. Due to high cost and risk associated with development of de-novo drug discovery, drug repurposing emerges as an effective alternative and helps in the development of potential therapies for Cervical cancer (CC). The proposed network-based drug repurposing strategy presented in this study has identified 142 FDA-approved drugs as potentially repurposable for CC. The analysis utilized a set of HPV-host proteins and CC associated differentially expressed proteins to create a subnetwork of HPV induced CC (HPV2C) followed by the identification of key targets in HPV2C network. Next, with the help of proximity analysis of drug-target network and key proteins, drug repurposing of approved drugs was performed followed by drug combination prediction (13 potential drug combinations predicted). Additionally, literature-based validation confirmed that 51 of 142 drugs have already been used for CC and have shown inhibition potential. 33 of remaining 99 drugs have been confirmed to have anti-viral activity. Further validation was performed using GSEA where 72 drugs were found overlapped among all datasets. Since the drugs are validated by computational methods, these drugs should be first tried preclinically prior to use them in human or in clinical trials. This analysis creates new opportunities for the quick repurposing of FDA-approved drugs. Moreover, the limitations of current study are highlighted and acknowledged in discussion. Finally, by employing a data-driven unsupervised approach and biological validation, our research has provided useful insights into the mechanisms of HPV-associated CC disease and has identified potential drug repurposing opportunities for the treatment of HPV infection and CC.
CRediT authorship contribution statement
Faheem Ahmed: Conceptualization, Methodology, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, and images. Young Jin Yang: Conceptualization, Methodology, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, and images, Anupama Samantasinghar: Methodology, Data curation, Writing - review & editing. Young Woo Kim: Methodology, Data curation, Writing - review & editing, Jeong Beom Ko: Writing - review & editing. Kyung Hyun Choi: Conceptualization, Validation, Formal analysis, Writing - review & editing, Resources, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflict of interest at this point.
Acknowledgments
This study has been conducted with the support of the Korea Institute of Industrial Technology as “Development of core technology for smart wellness care based on cleaner production process technology (Kitech-PEH23030).
Code
Python codes used for this research will be provided upon request.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.10.038.
Contributor Information
Faheem Ahmed, Email: faheeemahmedlangah@gmail.com.
Young Jin Yang, Email: yangyj23@kitech.re.kr.
Kyung Hyun Choi, Email: amm@jejunu.ac.kr.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Sung H., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;vol. 71(3):209–249. doi: 10.3322/CAAC.21660. [DOI] [PubMed] [Google Scholar]
- 2.Panatto D., et al. Sexual behaviour and risk factors for the acquisition of human papillomavirus infections in young people in Italy: suggestions for future vaccination policies. BMC Public Health. 2012;vol. 12:623. doi: 10.1186/1471-2458-12-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best S.R., Niparko K.J., Pai S.I. Biology of human papillomavirus infection and immune therapy for HPV-related head and neck cancers. Otolaryngol Clin North Am. 2012;vol. 45(4):807–822. doi: 10.1016/j.otc.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shanmugasundaram S., You J. Targeting persistent human papillomavirus infection. Viruses. 2017;vol. 9(8) doi: 10.3390/v9080229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G., Sharma M., Tan N., Barnabas R.V. HIV-positive women have higher risk of human papilloma virus infection, precancerous lesions, and cervical cancer. AIDS. 2018;vol. 32(6):795–808. doi: 10.1097/QAD.0000000000001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGraw S.L., Ferrante J.M. Update on prevention and screening of cervical cancer. World J Clin Oncol. 2014;vol. 5(4):744–752. doi: 10.5306/wjco.v5.i4.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z., Ma D. The precision prevention and therapy of HPV-related cervical cancer: new concepts and clinical implications. Cancer Med. 2018;vol. 7(10):5217–5236. doi: 10.1002/cam4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang R., et al. Human papillomavirus vaccine against cervical cancer: opportunity and challenge. Cancer Lett. 2020;vol. 471:88–102. doi: 10.1016/j.canlet.2019.11.039. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed F., et al. Drug repurposing in psoriasis, performed by reversal of disease-associated gene expression profiles. Comput Struct Biotechnol J. 2022;vol. 20:6097–6107. doi: 10.1016/J.CSBJ.2022.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues R., Duarte D., Vale N. Drug repurposing in cancer therapy: influence of patient’s genetic background in breast cancer treatment. Int J Mol Sci. 2022;vol. 23(8) doi: 10.3390/IJMS23084280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui C., et al. Drug repurposing against breast cancer by integrating drug-exposure expression profiles and drug–drug links based on graph neural network. Bioinformatics. 2021;vol. 37(18):2930–2937. doi: 10.1093/BIOINFORMATICS/BTAB191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnamurthy N., Grimshaw A.A., Axson S.A., Choe S.H., Miller J.E. Drug repurposing: a systematic review on root causes, barriers and facilitators. BMC Health Serv Res. 2022;vol. 22(1) doi: 10.1186/s12913-022-08272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed F., et al. SperoPredictor: an integrated machine learning and molecular docking-based drug repurposing framework with use case of COVID-19. Front Public Heal. 2022;vol. 0:1484. doi: 10.3389/FPUBH.2022.902123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burmeister C.A., et al. Cervical cancer therapies: current challenges and future perspectives. Tumour Virus Res. 2022;vol. 13 doi: 10.1016/j.tvr.2022.200238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L., Wang M., Li X., Yin S., Wang B. An overview of novel agents for cervical cancer treatment by inducing apoptosis: emerging drugs ongoing clinical trials and preclinical studies. Front Med. 2021;vol. 8(July):1–11. doi: 10.3389/fmed.2021.682366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samantasinghar A., Ahmed F., Rahim C.S.A., Kim K.H., Kim S., Choi K.H. Artificial intelligence-assisted repurposing of lubiprostone alleviates tubulointerstitial fibrosis. Transl Res. 2023 doi: 10.1016/j.trsl.2023.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed F., Samantasinghar A., Soomro A.M., Kim S., Choi K.H. A systematic review of computational approaches to understand cancer biology for informed drug repurposing. J Biomed Inform. 2023;vol. 142 doi: 10.1016/J.JBI.2023.104373. [DOI] [PubMed] [Google Scholar]
- 18.Samantasinghar A., et al. A comprehensive review of key factors affecting the efficacy of antibody drug conjugate. Biomed Pharmacother. 2023;vol. 161 doi: 10.1016/j.biopha.2023.114408. [DOI] [PubMed] [Google Scholar]
- 19.Kim H.M., Kang M.J., Song S.O. Metformin and cervical cancer risk in patients with newly diagnosed type 2 diabetes: a population-based study in Korea. Endocrinol Metab. 2022;vol. 37(6):929–937. doi: 10.3803/enm.2022.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C., Feng H., Song L., Li S., Wu Y., Yang L. Synergistic effects of thalidomide and cisplatin are mediated via the PI3K/AKT and JAK1/STAT3 signaling pathways in cervical cancer. Oncol Rep. 2022;vol. 48(4) doi: 10.3892/or.2022.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capistrano I R., et al. Drug repurposing as a potential source of innovative therapies in cervical cancer. Int J Gynecol Cancer. 2022;vol. 32(11):1377–1386. doi: 10.1136/ijgc-2022-003585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiravand Y., et al. Immune checkpoint inhibitors in cancer therapy. Curr Oncol. 2022;vol. 29(5):3044–3060. doi: 10.3390/curroncol29050247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colombo I., Karakasis K., Suku S., Oza A.M. Chasing immune checkpoint inhibitors in ovarian cancer: novel combinations and biomarker discovery. Cancers (Basel) 2023;vol. 15(12) doi: 10.3390/cancers15123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y., Xu R. Drug repurposing for glioblastoma based on molecular subtypes. J Biomed Inform. 2016;vol. 64:131–138. doi: 10.1016/j.jbi.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sultana R. Molecular docking based virtual screening of the breast cancer target NUDT5. Bioinformation. 2019;vol. 15(11):784–789. doi: 10.6026/97320630015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh D.-M., et al. Artificial intelligence and machine learning in cancer imaging. Commun Med. 2022;vol. 2(1):133. doi: 10.1038/s43856-022-00199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter K.A., Huang X. Machine learning-based virtual screening and its applications to Alzheimer’s drug discovery: a review. Curr Pharm Des. 2018;vol. 24(28):3347–3358. doi: 10.2174/1381612824666180607124038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adeshina Y.O., Deeds E.J., Karanicolas J. Machine learning classification can reduce false positives in structure-based virtual screening. Proc Natl Acad Sci. 2020;vol. 117(31):18477–18488. doi: 10.1073/pnas.2000585117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L., Fan S., Vera J., Lai X. A network medicine approach for identifying diagnostic and prognostic biomarkers and exploring drug repurposing in human cancer. Comput Struct Biotechnol J. 2023;vol. 21:34–45. doi: 10.1016/j.csbj.2022.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;vol. 6(1):1–18. doi: 10.1038/s41421-020-0153-3. 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han N., et al. Identification of SARS-CoV-2–induced pathways reveals drug repurposing strategies. Sci Adv. 2021;vol. 7(27) doi: 10.1126/SCIADV.ABH3032/SUPPL_FILE/ABH3032_TABLE_S9.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed F., et al. A comprehensive review of artificial intelligence and network based approaches to drug repurposing in Covid-19. Biomed Pharmacother. 2022;vol. 153 doi: 10.1016/J.BIOPHA.2022.113350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddiqui S., Deshmukh A.J., Mudaliar P., Nalawade A.J., Iyer D., Aich J. Drug repurposing: re-inventing therapies for cancer without re-entering the development pipeline—a review J Egypt Natl Cancer Inst. 2022;341:1–12. doi: 10.1186/S43046-022-00137-0. , vol. 34, no. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed F., et al. Drug Repurposing for viral cancers: a paradigm of machine learning, deep learning, and Virtual screening-based approaches. J Med Virol. 2023;vol. n/a(n/a) doi: 10.1002/jmv.28693. [DOI] [PubMed] [Google Scholar]
- 35.Hubbard T., et al. The Ensembl genome database project. Nucleic Acids Res. 2002;vol. 30(1):38. doi: 10.1093/NAR/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bateman A., et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;vol. 49(D1):D480–D489. doi: 10.1093/NAR/GKAA1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao G., et al. Multi-infection patterns and co-infection preference of 27 human papillomavirus types among 137,943 gynecological outpatients across China. Front Oncol. 2020;vol. 10 doi: 10.3389/fonc.2020.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asif A., et al. Microphysiological system with continuous analysis of albumin for hepatotoxicity modeling and drug screening. J Ind Eng Chem. 2021;vol. 98:318–326. doi: 10.1016/j.jiec.2021.03.035. [DOI] [Google Scholar]
- 39.Brückner A., Polge C., Lentze N., Auerbach D., Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;vol. 10(6):2763–2788. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;vol. 30(1):207–210. doi: 10.1093/NAR/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.“KEGG: Kyoto Encyclopedia of Genes and Genomes.” 〈https://www.genome.jp/kegg/〉 (accessed May 23, 2022).
- 42.Ashburner M., et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;(1):25–29. doi: 10.1038/75556. 251, vol. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;vol. 36(8):2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang D.W., et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;vol. 8(9):1–16. doi: 10.1186/GB-2007-8-9-R183/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raudvere U., et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;vol. 47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szklarczyk D., et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;vol. 51(D1):D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrows C., Saunders W., Austin R., Putnam G., Mansbach H. The sumatriptan/naratriptan aggregated patient (SNAP) database: aggregation, validation and application. Cephalalgia. 2004;vol. 24(7):586–595. doi: 10.1111/j.1468-2982.2003.00722.x. [DOI] [PubMed] [Google Scholar]
- 48.Piñero J., et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;vol. 45(Database issue):D833. doi: 10.1093/NAR/GKW943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S.-J., Liao D.-L., Chen C.-H., Wang T.-Y., Chen K.-C. Construction and analysis of protein-protein interaction network of heroin use disorder. Sci Rep. 2019;vol. 9(1):4980. doi: 10.1038/s41598-019-41552-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thieme S., Walther D. Biclique extension as an effective approach to identify missing links in metabolic compound–protein interaction networks. Bioinforma Adv. 2022;vol. 2(1):vbac001. doi: 10.1093/bioadv/vbac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adnan S., Abood E.W., Abdulmuhsin W. The multi-point delivery problem: shortest path algorithm for real roads network using Dijkstra. J Phys Conf Ser. 2020;vol. 1530(1) doi: 10.1088/1742-6596/1530/1/012040. [DOI] [Google Scholar]
- 52.“Exploring network structure, dynamics, and function using networkx (Conference) | OSTI.GOV.” 〈https://www.osti.gov/biblio/960616〉 (accessed Mar. 15, 2023).
- 53.M. Bastian, S. Heymann, and M. Jacomy, Gephi: An open source software for exploring and manipulating networks. BT - International AAAI Conference on Weblogs and Social,” Int. AAAI Conf. Weblogs Soc. Media, pp. 361–362, 2009.
- 54.Stolfi P., Manni L., Soligo M., Vergni D., Tieri P. Designing a network proximity-based drug repurposing strategy for COVID-19. Front Cell Dev Biol. 2020;vol. 8:1021. doi: 10.3389/FCELL.2020.545089/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W., Sun X., Peng L., Zhou L., Lin H., Jiang Y. RWRNET: a gene regulatory network inference algorithm using random walk with restart. Front Genet. 2020;vol. 11 doi: 10.3389/fgene.2020.591461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fredrickson M.M., Chen Y. Permutation and randomization tests for network analysis. Soc Netw. 2019;vol. 59:171–183. doi: 10.1016/j.socnet.2019.08.001. [DOI] [Google Scholar]
- 57.Wang T., Gu J., Li Y. Inferring the perturbed microRNA regulatory networks from gene expression data using a network propagation based method. BMC Bioinforma. 2014;vol. 15(1):255. doi: 10.1186/1471-2105-15-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart D.S., et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;vol. 34(Database issue):D668. doi: 10.1093/NAR/GKJ067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaulton A., et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;vol. 40(Database issue):D1100. doi: 10.1093/NAR/GKR777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L., Liu H., Chute C.G., Zhu Q. Cancer based pharmacogenomics network supported with scientific evidences: from the view of drug repurposing. BioData Min. 2015;vol. 8(1) doi: 10.1186/S13040-015-0042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y., et al. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022;vol. 50(D1):D1398–D1407. doi: 10.1093/nar/gkab953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;vol. 102(43):15545–15550. doi: 10.1073/PNAS.0506580102/SUPPL_FILE/06580FIG7.JPG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He J., et al. Statistically controlled identification of differentially expressed genes in one-to-one cell line comparisons of the CMAP database for drug repositioning. J Transl Med. 2017;vol. 15(1):198. doi: 10.1186/s12967-017-1302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corsello S.M., et al. The drug repurposing hub: a next-generation drug library and information resource. Nat Med. 2017;vol. 23(4):405. doi: 10.1038/NM.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmed F., et al. Decade of bio-inspired soft robots: a review. Smart Mater Struct. 2022;vol. 31(7) doi: 10.1088/1361-665X/AC6E15. [DOI] [Google Scholar]
- 66.Ahmed F., et al. Multi-material bio-inspired soft octopus robot for underwater synchronous swimming. J Bionic Eng. 2022;vol. 19(5):1229–1241. doi: 10.1007/S42235-022-00208-X. 2022 195. [DOI] [Google Scholar]
- 67.Cheng F., István I., Kovácskovács A., László A.-L., Lászlóbarabási L. Network-based prediction of drug combinations. Nat Commun. 2019;vol. 10(1):1–11. doi: 10.1038/s41467-019-09186-x. 2019 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei J., Wang Y., Shi K., Wang Y. Identification of core prognosis-related candidate genes in cervical cancer via integrated bioinformatical analysis. Biomed Res Int. 2020;vol. 2020 doi: 10.1155/2020/8959210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allahqoli L., et al. Diagnosis of cervical cancer and pre-cancerous lesions by artificial intelligence: a systematic review. Diagn (Basel, Switz) 2022;vol. 12(11) doi: 10.3390/diagnostics12112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y., et al. GRAND: a large-scale dataset and benchmark for cervical intraepithelial Neoplasia grading with fine-grained lesion description. Med Image Anal. 2021;vol. 70 doi: 10.1016/j.media.2021.102006. [DOI] [PubMed] [Google Scholar]
- 71.Bansal A., Singh M.P., Rai B. Human papillomavirus-associated cancers: a growing global problem. Int J Appl Basic Med Res. 2016;vol. 6(2):84. doi: 10.4103/2229-516X.179027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dai Z., Tang H., Pan Y., Chen J., Li Y., Zhu J. Gene expression profiles and pathway enrichment analysis of human osteosarcoma cells exposed to sorafenib. FEBS Open Bio. 2018;vol. 8(5):860–867. doi: 10.1002/2211-5463.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schiffman M., Castle P.E., Jeronimo J., Rodriguez A.C., Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;vol. 370(9590):890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 74.Jiang P., et al. SNX10 and PTGDS are associated with the progression and prognosis of cervical squamous cell carcinoma. BMC Cancer. 2021;vol. 21(1):694. doi: 10.1186/s12885-021-08212-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Banerjee N.S., Moore D.W., Broker T.R., Chow L.T. Vorinostat, a pan-HDAC inhibitor, abrogates productive HPV-18 DNA amplification. Proc Natl Acad Sci. 2018;vol. 115(47):E11138–E11147. doi: 10.1073/pnas.1801156115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tewari K.S., et al. Improved survival with bevacizumab in advanced cervical cancer. N Engl J Med. 2014;vol. 370(8):734–743. doi: 10.1056/NEJMoa1309748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y., Qin A., Li T., Qin X., Li S. Effect of statin on risk of gynecologic cancers: a meta-analysis of observational studies and randomized controlled trials. Gynecol Oncol. 2014;vol. 133(3):647–655. doi: 10.1016/j.ygyno.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 78.Li D., Zhang J., Yin L., Jin Z., Chen X., Meng X. Etomidate inhibits cell proliferation and induces apoptosis in A549 non-small cell lung cancer cells via downregulating WWP2. Exp Ther Med. 2021;vol. 22(5):1254. doi: 10.3892/etm.2021.10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palat G., Chary S. Practical guide for using methadone in pain and palliative care practice. Indian J Palliat Care. 2018;vol. 24(Suppl 1):S21–S29. doi: 10.4103/IJPC.IJPC_186_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu C.-C., et al. Risk of cancer in long-term levothyroxine users: retrospective population-based study. Cancer Sci. 2021;vol. 112(6):2533–2541. doi: 10.1111/cas.14908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pantziarka P., et al. ReDO_DB: the repurposing drugs in oncology database. Ecancermedicalscience. 2018;vol. 12:886. doi: 10.3332/ecancer.2018.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material