Abstract
Previous analysis of the Tra1 region of the conjugative element pRS01 from Lactococcus lactis subsp. lactis ML3 suggested that an origin of transfer (oriT) was present. Deletion derivatives of this cloned Tra1 region were assayed for mobilization in the presence of the wild-type pRS01 element in trans. The pRS01 oriT was localized to a 446-nucleotide segment in the intergenic region between open reading frames ltrD and ltrE. Sequence analysis of this region revealed a cluster of direct and inverted repeat structures characteristic of oriT regions associated with other conjugative systems.
Bacterial conjugation is a common mechanism for genetic exchange in nature. Studies on a variety of enterobacterial conjugative elements (11, 29) have resulted in a model in which bacterial conjugation is considered a two-part process; the first component involves bringing donor and recipient cells together to form an effective mating pair, and the second step involves enzymatic transfer of a single strand of the conjugative plasmid into the recipient cell. Initiation of single-stranded transfer involves the action of a specialized nucleoprotein complex called the relaxosome to produce single-stranded cleavage at a specific site (nic) within the origin of transfer (oriT) of the conjugative element (11, 29).
While our understanding of conjugative elements from gram-positive bacteria has increased, only recently have studies begun to define oriT and other relaxosome components. A specific nick site has recently been identified on the broad-host-range conjugative plasmid pIP501 from Streptococcus agalactiae and on the conjugative plasmid pGO1 from Staphylococcus aureus (5, 27). Surprisingly, the sequences of the nic regions of both pGO1 and pIP501 were shown to be quite similar to nic region sequences of a family of IncQ type conjugative elements from gram-negative bacteria (5, 11, 27). Transfer origins have also been identified for several other conjugative systems from gram-positive bacteria, including pheromone-inducible conjugative plasmid pAD1 (1, 4), non-self-transmissible staphylococcal plasmid pC221 mobilized by the conjugative vector pGO1 (17), and non-self-transmissible plasmids pUB110 and pBC16 from Bacillus subtilis, both of which are mobilized by the conjugative plasmid pLS20 (22). In addition, oriT sequences have recently been identified on the broad-host-range conjugative transposon Tn916 (10) and the streptococcal transposon Tn5252 (24).
The conjugative element pRS01 has been found in several Lactococcus lactis subsp. lactis genomes (7). pRS01 has been shown to mediate high-frequency transfer of genes encoding lactose utilization among lactococci and possesses the gene(s) responsible for a cell aggregation phenotype. Mapping of pRS01 identified four distinct regions (Tra1, Tra2, Tra3, and Tra4) involved in conjugative transfer (13). Sequence analysis of the Tra1 region revealed a gene, ltrB, with extensive homology to the genes encoding other plasmid and conjugative relaxases. In this work the oriT of pRS01 was localized and was shown to reside within the Tra1 region upstream of the ltrB gene.
Bacterial strains, media, and matings.
Escherichia coli DH5α, which was used as a cloning host, was grown in Luria-Bertani medium (20). The selective media used for E. coli strains contained 30 μg of chloramphenicol per ml. L. lactis subsp. lactis strains were grown in GM17 (M17 medium [25] containing 0.5% glucose) at 30°C without agitation. The selective media used for L. lactis strains contained antibiotics at the following concentrations: erythromycin, 10 μg/ml; chloramphenicol, 5 μg/ml; rifampin, 50 μg/ml; and spectinomycin, 300 μg/ml. All plating media contained 1.5% Bacto Agar (Difco Laboratories).
To determine transfer frequencies, plate matings were performed on GM17 as described previously (13). The donor strains (L. lactis subsp. lactis DM2036 [13] containing various pLE12 deletion derivatives) and the recipient strain (L. lactis subsp. lactis LM2345 [2]) were generated by using a 2% inoculum and GM17, followed by growth at the appropriate temperature without shaking until exponential growth was achieved. The donor and recipient cells were then centrifuged, and the pellet was resuspended in a 0.02 to 0.01 volume of GM17. Typically, 50 μl of donor cells and 50 μl of recipient cells were mixed, spread onto a GM17 plate, and incubated for 12 h at 30°C. The mating mixture was then washed off the plate with 1 ml of sterile phosphate-buffered saline, diluted appropriately, and plated onto selective medium. To determine pLE12 series plasmid transfer, transconjugants were selected on medium containing 5 μg of chloramphenicol per ml. Transfer frequencies were calculated by determining the number of transconjugants recovered per input donor in at least three independent trials.
DNA manipulation and analysis.
General molecular biology techniques were performed as described previously (20). Plasmid isolation and electroporation were performed as described previously (16). A sequence analysis was performed with the Genetics Computer Group (Madison, Wis.) sequence analysis software.
Characterization of the pRS01 oriT region.
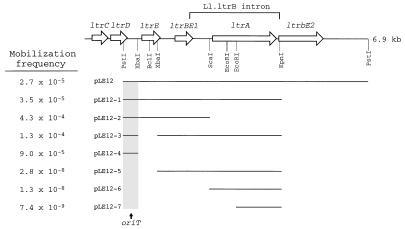
Previous complementation analysis of pRS01 Tra1 region insertions with Tra1 DNA resulted in mobilization of the complementing vector, pLE12, into lactococcal recipients (14, 16). The results suggested that the pRS01 oriT was on the 7.5-kb PstI fragment cloned into pLE12. In an effort to localize this oriT, we analyzed a series of pLE12 deletion derivatives for mobilization in the presence of the Tra+ cointegrate plasmid pM2036 (13). pM2036 is a cointegrate plasmid composed of conjugal element pRS01 and the L. lactis-E. coli shuttle vector pTRK28 (19). All derivatives possessing a 446-nucleotide PstI-XbaI fragment, which contained the intergenic region between ltrD and ltrE, were mobilized by pM2036 at a frequency that was 3 to 4 orders of magnitude higher than the frequencies obtained for subclones lacking the PstI-XbaI fragment (Fig. 1). In addition, derivatives containing the 446-nucleotide PstI-XbaI fragment were mobilized at frequencies ranging from 10−4 to 10−5 transconjugant per donor, values which were similar to the mobilization frequencies previously observed for pLE12 when it was used to complement various Tra− Tra1 region insertions (16).
FIG. 1.
Genetic map of the Tra1 region of pRS01 and results of a pLE12 derivative mobilization analysis. Plasmid pLE12 contains the complete Tra1 region. Plasmids pLE12-1, pLE12-2, and pLE12-4 are deletion derivatives of pLE12. Plasmids pLE12-3, pLE12-5, pLE12-6, and pLE12-7 are deletion derivatives of pLE12-1. The mobilization frequency was calculated by determining the number of transconjugants per input donor.
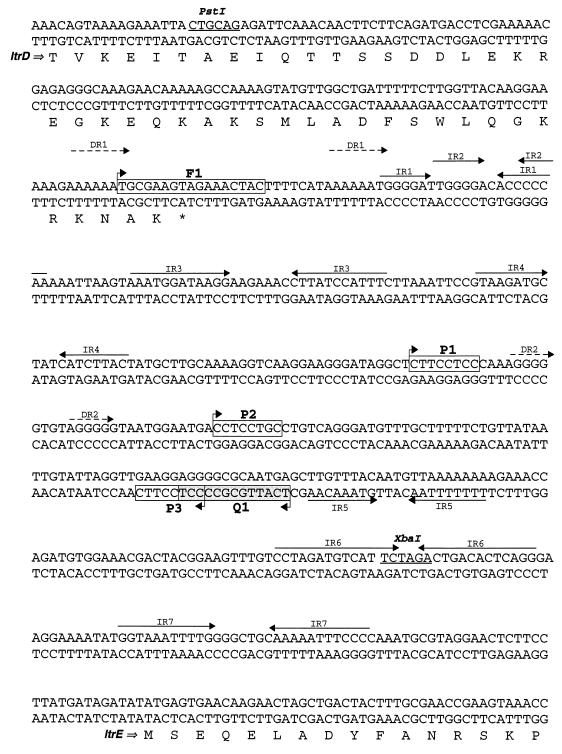
Sequence analysis of the intergenic region between ltrD and ltrE revealed a series of inverted repeat and direct repeat structures (IR1 through IR7, DR1, and DR2) (Fig. 2). Clusters of inverted and direct repeats and a general AT-rich nucleotide composition are characteristics of other bacterial oriT regions (11, 29). Surprisingly, the AT ratio of the pRS01 oriT region (60%) was lower than the AT ratio of the flanking Tra1 region sequence (65%) or the general AT ratio for lactococci (64%) (21).
FIG. 2.
Nucleotide sequence of the pRS01 ltrD-ltrE intergenic region. Inverted repeats and direct repeats are indicated by solid arrows and dashed arrows, respectively. Regions homologous to the IncQ (Q1), IncP (P1 through P3), and F (F1) family nic regions are enclosed in boxes.
In recent years biochemical and sequence data from a variety of conjugal elements have revealed that most nic region sequences fall into one of three consensus sequence classes named for prototype nic regions of each group, namely, IncQ, IncP, and F-like nic sites (11). In addition, all three classes of nic regions (IncQ, IncP, and F) are known to possess an upstream inverted repeat involved in termination of DNA transfer into the recipient cell (11). A search of the ltrD-ltrE intergenic region for these consensus sequences revealed three potential IncP family nic sites (P1 through P3) (Fig. 2), one F family site (F1), and one IncQ family site (Q1) (Fig. 2). One of the three IncP class sequences, P2 exhibited the highest similarity to the consensus IncP nic region sequence, matching it at seven of eight nucleotide positions (88% identity). While the sequences of sites P1 and P3 were less homologous to the consensus IncP nic region sequence (level of identity for each site, 75%), P3 did possess an upstream inverted repeat (IR5) (Fig. 2). The sequence of the single IncQ family homologous site, Q1, matched the consensus sequence at 10 of 12 nucleotide positions (83% identity). In addition, the Q1 site contained an upstream inverted repeat similar in structure to the inverted repeats found in the nic regions of the staphylococcal conjugal plasmid pGO1, the streptococcal plasmid pIP501, and other IncQ family elements (5, 11). Finally, the F family site, F1, exhibited minimal homology to the F family consensus nic region (13 of 17 nucleotides) and overlapped the upstream C-terminal end of the ltrD reading frame.
Discussion.
The conjugal element pRS01 is present in certain lactococci (7). A previous analysis of this element identified four distinct transfer regions (13). Complementation of the Tra1 region resulted in mobilization of the complementing plasmid, suggesting that a cis-acting oriT was present (14, 16). A sequence analysis of the Tra1 region revealed six potential open reading frames, including a putative conjugative relaxase ltrB open reading frame interrupted by the group II intron (15, 16). In this work we localized the pRS01 oriT to a 446-nucleotide segment of the Tra1 region between ltrD and ltrE. Like other bacterial oriT regions, this segment possessed multiple inverted and direct repeats. Recently, a specific nic site within an oriT region was identified for the streptococcal conjugative plasmid pIP501 (27) and the staphylococcal plasmid pGO1 (5). These nic site regions exhibited strong sequence similarity to the IncQ family of nic regions (11), suggesting that there is a possible link between gram-positive and gram-negative bacterial conjugal systems. Analysis of the pRS01 oriT region identified a sequence segment possessing 10 of 12 nucleotides present in a consensus IncQ nic region (11). In addition, three putative IncP family nic region sites and an F class nic region were also identified. The precise location of the pRS01 nick site remains to be determined.
Few transfer origins from conjugative elements in lactococci have been identified. Lucey et al. (12) characterized a region of plasmid pCI528 involved in mobilization by the ML3-712 class of lactococcal strains, which possess the homologous pRS01 or 712 sex factor elements. In that work, a minimal mobilization region was shown to contain a series of inverted repeat structures upstream of a putative mobilization protein (ORF1) (12). Surprisingly, the transfer origin from pRS01 identified in this work is not related to the mobilization region identified in pCI528 even though both regions, when placed in trans, are mobilized by the same ML3-712 class sex factor elements. The mobilization protein encoded on pCI528 and the predicted LtrB relaxase from pRS01 do exhibit some homology (22%), however (16). The difference in putative transfer origins which are mobilized by the same conjugal elements suggests that either (i) the pRS01 and 712 sex factor elements can mobilize different transfer origins or (ii) the pCI528 mobilization region is a site for recombination with conjugative elements, resulting in mobilization by a cointegrate mechanism (18).
In the past decade electroporation has become the method of choice for transforming various lactic acid bacteria, and consequently numerous electroporation protocols have been developed (3, 6, 8, 9, 26, 28). Unfortunately, many lactic acid bacteria are still difficult to transform by electroporation, which results in tedious and sometimes unsuccessful optimization of electroporation conditions. In addition, larger plasmid constructs are generally less effectively transformed by electroporation (23). In contrast to electroporation, conjugative mobilization of pRS01 oriT-containing derivatives may offer an alternative for genetic delivery into lactococci and, potentially, other lactic acid bacteria. Since the transfer is based on conjugal delivery, the size of the mobilizable element probably does not influence the transfer frequency. Moreover, the technical ease with which oriT-containing derivatives can be mobilized, via simple cross-streak matings, makes pRS01-dependent mobilization an attractive alternative to electroporation. Given that pRS01 is an indigenous conjugal element in lactococci, the discovery of the pRS01 oriT should also allow development of food grade mobilizable delivery systems.
Acknowledgments
This work was supported by grants from the Minnesota-South Dakota Dairy Foods Research Center (to G.M.D.) and the Kraft General Foods Chair (to L.L.M.). D.A.M. was supported by a NIGMS Predoctoral Biotechnology Training Grant.
Footnotes
Paper no. 97-1-18-0024 of the contribution series of the Minnesota Agricultural Experiment Station based on research conducted under Project 18-62.
REFERENCES
- 1.An F Y, Clewell D B. The origin of transfer (oriT) of the enterococcal, pheromone-responding, cytolysin plasmid pAD1 is located within the repA determinant. Plasmid. 1997;37:87–94. doi: 10.1006/plas.1996.1270. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D G, McKay L L. Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis ML3. J Bacteriol. 1984;158:954–962. doi: 10.1128/jb.158.3.954-962.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aukrust T W, Brurberg M B, Nes I F. Transformation of Lactobacillus by electroporation. Methods Mol Biol. 1995;47:201–208. doi: 10.1385/0-89603-310-4:201. [DOI] [PubMed] [Google Scholar]
- 4.Clewell D B. Sex pheromones and the plasmid-encoded mating response in Enterococcus faecalis. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 349–364. [Google Scholar]
- 5.Climo M W, Sharma V K, Archer G L. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease of the staphylococcal plasmid pGO1. J Bacteriol. 1996;178:4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dicks L M T. Transformation of Leuconostoc oenos by electroporation. Biotechnol Techniques. 1994;8:901–904. [Google Scholar]
- 7.Gasson M J, Swindell S, Maeda S, Dodd H M. Molecular rearrangement of lactose plasmid DNA associated with high-frequency transfer and cell aggregation in Lactococcus lactis 712. Mol Microbiol. 1992;6:3213–3223. doi: 10.1111/j.1365-2958.1992.tb01776.x. [DOI] [PubMed] [Google Scholar]
- 8.Gautier M, Roualt A, Lemee R. Electrotransformation of Propionibacterium freudenreichii TL110. Lett Appl Microbiol. 1995;20:125–129. [Google Scholar]
- 9.Holo H, Nes I F. Transformation of Lactococcus by electroporation. Methods Mol Biol. 1995;47:195–199. doi: 10.1385/0-89603-310-4:195. [DOI] [PubMed] [Google Scholar]
- 10.Jaworski D D, Clewell D B. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J Bacteriol. 1995;177:6644–6651. doi: 10.1128/jb.177.22.6644-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 12.Lucey M, Daly C, Fitzgerald G. Analysis of a region from the bacteriophage resistance plasmid pCI528 involved in its conjugative mobilization between Lactococcus strains. J Bacteriol. 1993;175:6002–6009. doi: 10.1128/jb.175.18.6002-6009.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills D A, Choi C K, Dunny G M, McKay L L. Genetic analysis of regions of the Lactococcus lactis subsp. lactis plasmid pRS01 involved in conjugative transfer. Appl Environ Microbiol. 1994;60:4413–4420. doi: 10.1128/aem.60.12.4413-4420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills D A, Choi C K, Dunny G M, McKay L L. Characterization of the conjugation system associated with the Lactococcus lactis ssp. lactis plasmid pRS01. Dev Biol Stand. 1995;85:543–548. [PubMed] [Google Scholar]
- 15.Mills D A, Manias D A, McKay L L, Dunny G M. Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J Bacteriol. 1997;179:6107–6111. doi: 10.1128/jb.179.19.6107-6111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills D A, McKay L L, Dunny G M. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Projan S J, Archer G L. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol. 1989;171:1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reimmann C, Haas D. Mobilization of chromosomes and nonconjugative plasmids by cointegrative mechanisms. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 137–188. [Google Scholar]
- 19.Romero D A, Klaenhammer T R. Characterization of insertion sequence IS946, an iso-ISS1 element, isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990;172:4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Sandine W E, Radich P C, Elliker P R. Ecology of the lactic streptococci. A review. J Milk Food Technol. 1972;35:176–185. [Google Scholar]
- 22.Selinger L B, McGregor N F, Khachatourians G G, Hynes M F. Mobilization of closely related plasmids pUB110 and pBC16 by Bacillus plasmid pXO503 requires trans-acting open reading frame beta. J Bacteriol. 1990;172:3290–3297. doi: 10.1128/jb.172.6.3290-3297.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siguret V, Ribba A S, Cherel G, Meyer D, Peitu G. Effect of plasmid size on transformation efficiency by electroporation of Escherichia coli DH5α. BioTechniques. 1994;16:422. [PubMed] [Google Scholar]
- 24.Srinivas P, Kilic A O, Vijayakumar M N. Site-specific nicking in vitro at oriT by the DNA relaxase of Tn5252. Plasmid. 1997;37:42–50. doi: 10.1006/plas.1996.1268. [DOI] [PubMed] [Google Scholar]
- 25.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker D C, Aoyama K, Klaenhammer T R. Electrotransformation of Lactobacillus acidophilus group A1. FEMS Microbiol Lett. 1996;138:233–237. doi: 10.1111/j.1574-6968.1996.tb08163.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang A, Macrina F L. Streptococcal plasmid pIP501 has a functional oriT site. J Bacteriol. 1995;177:4199–4206. doi: 10.1128/jb.177.15.4199-4206.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei M Q, Rush C M, Norman J M, Hafner L M, Epping R J, Timms P. An improved method for the transformation of Lactobacillus strains using electroporation. J Microbiol Methods. 1995;21:97–109. [Google Scholar]
- 29.Wilkins B, Lanka E. DNA processing and replication during plasmid transfer between Gram-negative bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 105–129. [Google Scholar]