Abstract
The ability of Acinetobacter sp. strain BD413(pFG4ΔnptII) to take up and integrate transgenic plant DNA based on homologous recombination was studied under optimized laboratory conditions. Restoration of nptII, resulting in kanamycin-resistant transformants, was observed with plasmid DNA, plant DNA, and homogenates carrying the gene nptII. Molecular analysis showed that some transformants not only restored the 317-bp deletion but also obtained additional DNA.
Bacterial antibiotic resistance genes are still frequently used as markers in transgenic plants. Due to the problems caused by antibiotic-resistant pathogens, the use of antibiotic resistance genes in transgenic plants is subject to debate. It is hypothesized that the introduction of bacterial genes into the plant genome leads to a higher probability of gene transfer from plants to bacteria due to the presence of homologous sequences. However, until now, there has been a lack of clear experimental evidence that successful gene transfer from plants to bacteria can occur at all. Natural transformation, the ability of bacteria to actively take up free DNA, is a way plant DNA can be transferred to bacteria. At present, around 40 species, some of which are soil- or water-borne bacteria, are known to develop the ability (called competence) for natural transformation (13). Prerequisites for natural transformation under soil conditions are the availability of free DNA, the development of competence, and the stable integration of the captured DNA into the bacterial genome. In microcosm experiments, bacterial DNA adsorbed to soil particles was able to transform competent bacteria and to persist in soil (7, 12, 13, 17). DNA adsorbed to soil particles is, to some degree, protected against attacks of nucleases (12, 13, 19, 20). In agricultural soils, the persistence of transgenic plant DNA released into the environment, e.g., from senescent or rotting plant material, has been observed by several researchers (2, 19, 24, 26). Several groups have tried to transform bacteria with transgenic plant DNA from different plants and naturally competent bacteria (2, 3, 16, 23). However, transformation of bacteria with transgenic plant DNA has not yet been demonstrated. In this study, experiments were done under optimized laboratory conditions to examine the ability of Acinetobacter sp. strain BD413 to capture and integrate transgenic sugar beet DNA based on homologous recombination. All transformation experiments were done with a rifampin-resistant mutant of Acinetobacter sp. strain BD413 (obtained from J. D. van Elsas, Institute for Plant Protection [IPO-DLO], Wageningen, The Netherlands) which is known to be naturally transformable (11). Plasmid pGSFR160, which was provided by Plant Genetic Systems, Ghent, Belgium, harbored a gene construct consisting of the phosphinothricin acetyltransferase gene (bar), the neomycin phosphotransferase gene (npt) (both under control of the TR1/TR2 promoter from Agrobacterium tumefaciens [31]), the 3′ocs terminator, and nonspecific parts (Fig. 1). The same construct was present in the transgenic sugar beet plants obtained from Planta GmbH, Einbeck, Germany. Due to low-level expression of nptII controlled by TR1, the Acinetobacter sp. remained kanamycin sensitive even after the introduction of pGSFR160.
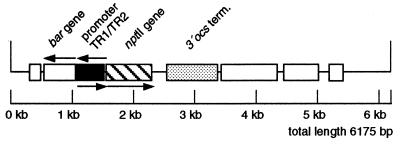
FIG. 1.
Map of the gene construct. The gene construct is part of pGSFR160 and is chromosomally integrated into the genome of the sugar beet. It consisted of the gene bar, the bidirectional promoter TR1/TR2, nptII, the 3′ocs terminator sequence, and nonspecified parts.
Construction of pFG4ΔnptII.
To facilitate the stable integration of transgenic plant DNA in Acinetobacter sp. strain BD413 and the selection of transformants, plasmid pFG4ΔnptII, carrying a part of Tn5 with an incomplete nptII (a deletion of 317 bp), was constructed and introduced in Acinetobacter sp. strain BD413. A 1,693-bp fragment of transposon Tn5 (nucleotide 1236 to nucleotide 2929) consisting of the nptII promoter, the nptII gene (kanamycin resistance), and the bleomycin resistance gene was amplified by PCR with P1 (5′ TGC TAA AGG AAG CGG AAC 3′) and P2 (5′ AGG TCA ACA GGC GGT AAC 3′) as forward and reverse primers, respectively. Primers were designed on the basis of the published Tn5 sequence (accession no. L19385 and U00004) with the Oligo 4.0 program (21). PCR mixtures contained 0.1 μmol of both the forward and reverse primer, 3.75 mM MgCl2, 0.2 mM deoxyribonucleoside triphosphates, 1× Stoffel buffer, and 2.5 U of Taq polymerase Stoffel fragment (Applied Biosystems). Amplification involved a 7-min step at 92°C; subsequently, 35 cycles consisting of 1 min at 92°C (denaturation), 1 min at 58°C (primer annealing), and 1.5 min at 72°C (primer extension) were performed, followed by a 10-min final extension step at 72°C. The 1,693-bp PCR product was cloned into PCR vector pT7-blue (Novagen, Madison, Wis.). Digestion with NcoI and Tth111I introduced a 317-bp deletion into the central part of nptII. After the ends were blunted by integrating deoxyribonucleoside triphosphates with the Klenow fragment, the plasmid was recircularized. A HindIII-BamHI fragment of the resulting plasmid containing the insert was introduced into the multiple cloning site of pMMB190 (15), resulting in pFG4ΔnptII (Fig. 2). The IncQ plasmid pMMB190 was provided by M. Bagdasarian, Michigan State University, East Lansing, Mich. All digestions were carried out with restriction enzymes from Boehringer, Mannheim, Germany, or New England Biolabs, Beverly, Mass., according to the manufacturers’ instructions. All ligations were carried out with Fast-Link ligase (Epicentre Technologies, Madison, Wis.). For blunting of digested fragments, Klenow fragment (Boehringer) was used. Buffer and media were prepared according to Sambrook et al. (22) unless indicated otherwise. Both pMMB190 and pFG4ΔnptII contained an ampicillin resistance gene which could be used for selection. Transformation of Escherichia coli XL-blue cells was carried out by electroporation according to the protocol of Bio-Rad (Hercules, Calif.). Plasmid pFG4ΔnptII was introduced into Acinetobacter sp. strain BD413 by natural transformation as described previously (17).
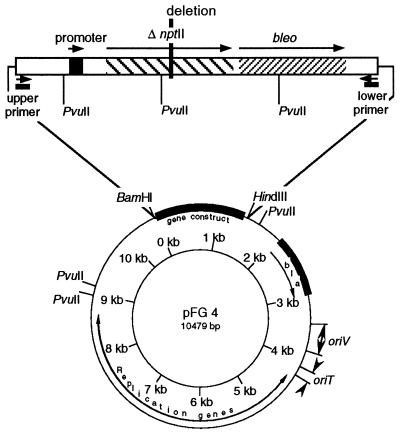
FIG. 2.
Map of pFG4ΔnptII. The insert of the plasmid consisted of the promoter region, the deleted gene nptII (ΔnptII [deletion of 317 bp]), and the bleomycin resistance gene (bleo). Primer binding sites and PvuII restriction sites are indicated.
Transformation experiments.
The resulting Acinetobacter sp. strain BD413(pFG4ΔnptII), which was rifampin and ampicillin resistant but kanamycin sensitive due to the 317-bp deletion in nptII, was used as a recipient in transformation experiments. Transformation experiments were done with either DNA of plasmid pGSFR160, linearized pGSFR160 without an origin of replication (oriV), or DNA extracted from transgenic sugar beet plants harboring the gene construct. In addition, transformations were attempted with leaf homogenates of transgenic sugar beet carrying the gene construct. DNA of plasmid pGSFR160 was isolated with the plasmid extraction kit from Qiagen (Hilden, Germany). To obtain linearized plasmid DNA without oriV, pGSFR160 was treated with SacI and HindIII. DNA from transgenic and nontransgenic sugar beets was extracted by the hexadecyltrimethyl ammonium bromide method (30). Extracted DNA was dissolved in sterile deionized water. To produce plant homogenate, leaves of sugar beet were frozen at −70°C and homogenized afterwards. The liquid phase obtained was suitable for transformation without being treated further. Transformation experiments were carried out basically as described by Nielsen et al. (17) on Mueller-Hinton agar (MHA; Merck, Darmstadt, Germany) containing 50 μg of rifampin per ml and 100 μg of ampicillin per ml. Natural competence of Acinetobacter sp. strain BD413 was developed by growing it in 10 ml of Luria-Bertani medium at 28°C until early stationary phase. Cells were washed once in 10 ml of saline and stored at −70°C in 10 ml of saline containing 15% glycerol until use. After the addition of DNA, competent cells (100 μl = approximately 5 × 107 CFU) were placed on a nylon membrane filter (Millipore, Bedford, Mass.) on top of the agar and grown at 28°C for 24 h. The bacterial layer was resuspended in 4 ml of saline, concentrated by centrifugation for 2 min at 5,500 × g or diluted 10-fold, and plated on MHA supplemented with 50 μg of kanamycin per ml, 50 μg of rifampin per ml, and 100 μg of ampicillin per ml to detect transformants. The number of CFU of recipient Acinetobacter sp. strain BD413(pFG4ΔnptII) was determined by plating on MHA containing 50 μg of rifampin per ml and 100 μg of ampicillin per ml. The colonies were counted after incubation at 28°C for 2 days. The transformation frequency was determined as the ratio of the number of transformants to the number of Acinetobacter sp. strain BD413(pFG4ΔnptII) CFU. All experiments were conducted twice in two (plasmid transformation) or three (plant DNA or homogenate) replicates. Controls were made with water and nontransgenic plant DNA. Transformations with all sources of DNA listed above resulted in kanamycin-resistant Acinetobacter sp. strain BD413(pFG4ΔnptII), indicating the restoration and expression of nptII located on pFG4ΔnptII. Transformation frequencies are shown in Table 1. Compared to transformation with plasmid DNA, transformation frequency with plant DNA was drastically reduced. With plant homogenate of sugar beet leaves, the transformation of Acinetobacter sp. strain BD413(pFG4ΔnptII) occurred at a frequency of 1.5 × 10−10. Controls made with water and nontransgenic plant DNA were negative, indicating that no kanamycin-resistant spontaneous mutants appeared. In contrast, Nielsen et al. (16) failed to demonstrate the transformation of Acinetobacter sp. strain BD413 by transgenic sugar beet DNA due to the absence of homologous sequences.
TABLE 1.
Transformation frequencies of Acinetobacter sp. strain BD413(pFG4ΔnptII) transformed with transgenic DNA and plant homogenate
| Transformed transgenic material | Replicates considered (no. positive/total) | Amt of DNA used | No. of transformants (CFU/plate)a | Avg transformation frequencyb |
|---|---|---|---|---|
| pGSFR160 | ||||
| Circular | 2/2 | 200 ng | 1.1 ± 0.1 × 106 | 9.85 × 10−5 |
| Without oriV | 2/2 | 200 ng | 3.5 ± 0.1 × 105 | 3.15 × 10−5 |
| Sugar beet DNA | ||||
| Transgenic | 6/6 | ∼2 μg | 3.3 ± 0.1 × 101 | 5.36 × 10−9 |
| Nontransgenic | 6/6 | ∼2 μg | 0 | 0 |
| Plant homogenate of transgenic sugar beet leaves | 4/6 | 100 μl of plant homogenate | 1.5 ± 1.9 × 100 | 1.5 × 10−10 |
Values are presented as means ± standard deviations, both with the same order of magnitude.
Transformation frequency is the ratio of the number of kanamycin-resistant transformants to the number of Acinetobacter sp. strain BD413(pFG4ΔnptII) CFU.
Characterization of transformants.
Representative numbers of kanamycin-resistant colonies (a total of 20 transformants) obtained from transformations with the purified DNA samples were screened by PCR (P1/P2 system) for the presence of the restored gene nptII. The appearance of specific PCR products of 1.7 kb for all transformants tested demonstrated the restoration of the deleted nptII (data not shown). The size of the control PCR product obtained with Acinetobacter sp. strain BD413(pFG4ΔnptII) was approximately 1.4 kb. The restoration of the gene nptII was confirmed by Southern blot hybridization of the PCR products with a probe specific for the deletion (ΔnptII). The 1,693-bp PCR product obtained with Tn5 template was digested with Tth111I-NcoI, generating a 317-bp fragment. After separation on a 0.7% agarose gel with Tris-acetate–EDTA buffer, the 317-bp fragment was recovered by the QiaQuick gel extraction kit (Qiagen) and labelled with digoxigenin-11dUTP (Boehringer).
Six transformants obtained after transformation with plant homogenate (pNT1 to pNT6) were characterized in detail because hybridization-positive PCR products which were approximately 650 bp larger than expected were obtained for three of them (pNT2, pNT3, and pNT6).
Plasmid DNA was extracted from all six transformants (pNT1 to pNT6) and pMMB190 according to Holmes and Quigley (9). The plasmid extracts were treated with RNase A (20 μg/ml for 30 min at 37°C) and purified with the Wizard DNA purification kit (Promega, Madison, Wis.). Plasmid DNA was digested with PvuII and analyzed by Southern blot hybridization with the probe for ΔnptII (Fig. 3).
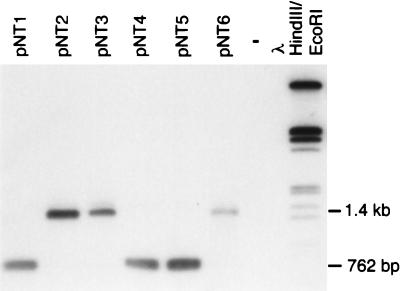
FIG. 3.
Southern blot hybridization of PvuII-digested plasmid DNA extracted from kanamycin-resistant Acinetobacter sp. obtained after transformation with plant homogenates. Hybridization was carried out with the ΔnptII probe.
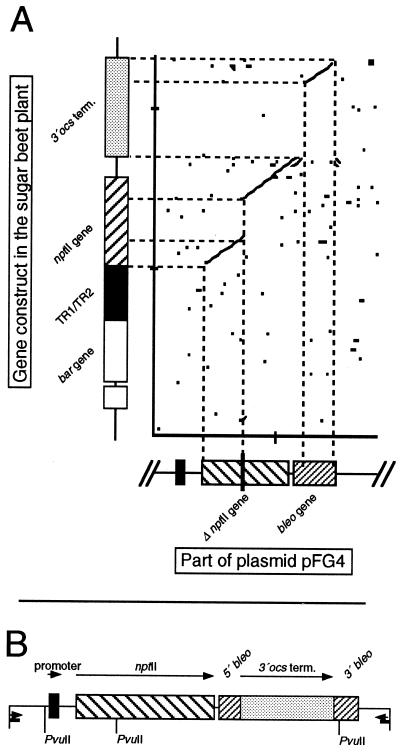
For pNT1, pNT4, and pNT5, an ethidium bromide-stained gel showed a 762-bp fragment as expected for nptII instead of the 445-bp fragment expected from the original pFG4ΔnptII, demonstrating restoration of the gene nptII. However, pNT2, pNT3, and pNT6 PvuII restriction digests generated a 1.4-kb fragment, indicating the unexpected additional transfer of an approximately 650-bp DNA fragment. Sequences of the gene construct in the transgenic sugar beet and pFG4ΔnptII were compared for homology with MacDNASIS Pro 3.6 (Hitachi Software Engineering Ltd., Yokohama, Japan). The comparison was carried out with 8-base segments (Fig. 4A). Homologous segments are indicated by dots in the graph. Larger regions of homology appear as lines in the graph. Sequence comparison of the gene construct and the sequence of pFG4ΔnptII showed unexpected homology between the 3′ end of the 3′ocs terminator region inside the gene construct and the 3′ end of the bleomycin resistance gene inside pFG4ΔnptII (Fig. 4A). Most likely, in the gene construct, the 3′ocs terminator had been inserted into the bleomycin resistance gene and thus each end of the 3′ocs terminator was flanked by at least 172 bp of the bleomycin resistance gene. A second homologous recombination event was assumed, resulting in the integration of the 3′ocs terminator sequence into the bleomycin resistance gene of pFG4ΔnptII, which thus produced pNT2, pNT3, and pNT6. To confirm this assumption, the fragment obtained after the PCR amplification of pNT2 with P1/P2 was cloned into PCR vector pT7-blue (Novagen). A 700-bp fragment of the 5′ end of the insert was removed by HindIII and BstBI digestion. The recircularized plasmid was sequenced in forward and reverse directions by means of the universal sequencing primers T7 promoter primer and U-19-mer primer. Sequencing carried out by 4base GmbH, Reutlingen, Germany, confirmed the assumption that the 3′ocs terminator was integrated in pNT2. Hence, this region provided homology, explaining the ready production of transformants with larger inserts. Combined with the results of the restriction analysis, this finding allowed us to map the insert in pNT2 (Fig. 4B).
FIG. 4.
(A) Sequence comparison of pFG4ΔnptII and the gene construct. Sequences were compared in 8-base segments, and homologous segments are indicated by dots. (B) Map of the insert of pNT2 based on restriction analysis and sequencing. As a result of homologous recombination, the insert of pNT2 consisted of the promoter region, the restored nptII, and the 3′ocs terminator integrated into the bleomycin resistance gene (bleo). Primer sites and restriction sites for PvuII are indicated.
In this study, transformation of naturally competent bacteria by transgenic plant DNA, even with plant homogenates, was demonstrated for the first time. Neither the high content of nonspecific DNA (99.9995% of transgenic plant DNA is usually nontransgenic, based on a study by Arumuganathan and Earle [1]) nor the much higher methylation rate of plant DNA compared with that of bacterial DNA (up to 30% of all guanine bases are methylated [6]) prevented transformation. In comparison with purified plant DNA, transformations with plant homogenate showed a reduced transformation frequency. The state of the DNA (association with chromatin proteins and protection by the core membrane), the relatively low concentration of the target DNA in the mix, and a potentially inhibiting activity of other cell compounds may be responsible for the decreased transformation efficiency.
Recently, horizontal gene exchange between distantly related bacteria (14, 29) as well as gene exchange from bacteria to yeast (8), mammalian cells (4), and plant cells (10) has been reported. Furthermore, based on sequence comparisons, it has been assumed that horizontal gene transfer from plants to bacteria occurred during evolution (5, 28, 32). However, transfer of plant DNA to bacteria has not been demonstrated experimentally until now, perhaps because of an absence of homologous sequences in the bacteria (16) or the use of less efficiently transformable bacteria (3, 23). Another reason for previous failures may be attempts to monitor the transfer of complete genes, whereas bacterial transformation frequently involves the recombination of short DNA segments, resulting in gene mosaics (27).
With the introduction of bacterial genes, bacterial promoter and terminator sequences, and bacterial origins of replication into transgenic plants, the degree of sequence homology between the genomes of competent bacteria and transgenic plant DNA increases. Gene constructs inserted into plant genomes often consist of sequences of mixed origin, e.g., the sequence of one gene is inserted into another gene. Homologous recombination between a recombinant sequence in the plant chromosome and the natural sequence in competent bacteria can result in the stable insertion of the captured DNA. In our experiments, this obviously occurred with the 3′ocs terminator sequence, which was flanked by sequences of the bleomycin resistance gene and therefore transferred into pFG4ΔnptII via homologous recombination.
The successful transfer of transgenic plant DNA to naturally competent Acinetobacter sp. is scientifically interesting, as it shows that the main prerequisite for such transfers is the presence of DNA homology in the recipient genome. However, our findings should not affect the evaluation of the use of antibiotic resistance genes such as nptII as markers in transgenic plants. First, promoter sequences such as NOS or TR1/TR2 are not active in most bacteria. Thus, transfer of the gene nptII from transgenic plants would not endow the recipient bacteria with a kanamycin resistance phenotype. Secondly, most of the antibiotic resistance genes used as marker genes are widely disseminated in environmental bacteria. The gene nptII has been shown to be present in approximately 50% of kanamycin-resistant enteric bacteria isolated from municipal sewage (25). The key to limit the establishment and dissemination of antibiotic resistance in bacteria is restricted use of antibiotics in human and animal therapy and avoidance of antibiotics in animal nutrition and plant protection.
Recently, Nielsen et al. (18) demonstrated that Acinetobacter sp. strain BD413 can be easily induced by nutrients to undergo natural transformation with chromosomal DNA in soil. Although the transformation experiments in this study were done under optimized laboratory conditions, our results suggest that gene transfer from the plant chromosome to bacteria might occur in soil if homologous sequences are present in competent bacteria. However, the in situ transformation frequencies would likely be much lower than those under laboratory conditions.
Acknowledgments
This work was supported by grant 0310642 of the German Ministry of Education, Science, Research and Technology.
We thank K. M. Nielsen for introducing us to the filter transformation technique.
REFERENCES
- 1.Arumuganathan K, Earle E D. Nuclear DNA content of some important plant species. Plant Mol Biol. 1991;9:208–218. [Google Scholar]
- 2.Becker J, Siegert H, Logemann J, Schell J. Bundesministerium für Forschung und Technologie (ed.) Bonn, Germany: Biologische Sicherheit. Bundesministerium für Forschung und Technologie; 1994. Begleitende Sicherheitsforschung zur Freisetzung genetisch veränderter Petunien; pp. 563–578. [Google Scholar]
- 3.Broer I, Dröge-Laser W, Gerke M. Examination of the putative horizontal gene transfer from transgenic plants to agrobacteria. In: Schmidt E R, Hankeln T, editors. Transgenic organisms and biosafety. Berlin, Germany: Springer-Verlag; 1996. pp. 67–70. [Google Scholar]
- 4.Courvalin P. Gene transfer from bacteria to mammalian cells. C R Acad Sci Ser III Sci Vie. 1995;318:1207–1212. [PubMed] [Google Scholar]
- 5.Doolittle R F, Feng D F, Anderson K L, Alberro M R. A naturally occurring horizontal gene transfer from a eukaryote to a prokaryote. J Mol Evol. 1990;31:383–388. doi: 10.1007/BF02106053. [DOI] [PubMed] [Google Scholar]
- 6.Finnegan E J, Brettell R I S, Dennis E S. The role of DNA methylation in the regulation of plant gene expression. In: Jost J P, Saluz H P, editors. DNA methylation. Molecular biology and biological significance. Basel, Switzerland: Birkhäuser Verlag; 1993. pp. 218–261. [DOI] [PubMed] [Google Scholar]
- 7.Gallori E, Bazzicalupo M, Dal Canto L, Fani R, Nannipieri P, Vettori C, Stotzky G. Transformation of Bacillus subtilis by DNA bound on clay in nonsterile soil. FEMS Microbiol Ecol. 1994;15:119–126. [Google Scholar]
- 8.Heinemann J A, Sprague G F. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340:205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 9.Holmes D S, Quigley N. Isolation of plasmid DNA by Mini-Quick-Method. Annu Rev Biochem. 1983;114:193–197. [Google Scholar]
- 10.Hooykaas P J J. Transformation of plant cells via Agrobacterium. Plant Mol Biol. 1989;13:327–336. doi: 10.1007/BF00025321. [DOI] [PubMed] [Google Scholar]
- 11.Juni E. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J Bacteriol. 1972;112:917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanna M, Stotzky G. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl Environ Microbiol. 1992;58:1930–1939. doi: 10.1128/aem.58.6.1930-1939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazodier P, Petter R, Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989;171:3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morales V M, Bäckman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen K M, Gebhard F, Smalla K, Bones A M, Van Elsas J D. Evaluation of possible horizontal gene transfer from transgenic plants to the soil bacterium Acinetobacter calcoaceticus BD413. Theor Appl Genet. 1997;95:815–821. [Google Scholar]
- 17.Nielsen K M, van Weerelt D M, Berg T N, Bones A M, Hagler A N, van Elsas J D. Natural transformation and availability of transforming DNA to Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol. 1997;63:1945–1952. doi: 10.1128/aem.63.5.1945-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen K M, Bones A M, van Elsas J D. Induced natural transformation of Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol. 1997;63:3972–3977. doi: 10.1128/aem.63.10.3972-3977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paget E, Simonet P. On the track of natural transformation in soil. FEMS Microbiol Ecol. 1994;15:109–118. [Google Scholar]
- 20.Romanowski G, Lorenz M G, Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991;57:1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rychlik W, Rhoads R E. A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 1989;17:8543–8551. doi: 10.1093/nar/17.21.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schlüter K, Fütterer J, Potrykus I. Horizontal gene transfer from a transgenic potato line to a bacterial pathogen (Erwinia chrysanthemi) occurs—if at all—at an extremely low frequency. Biotechnology. 1995;13:94–98. doi: 10.1038/nbt1095-1094. [DOI] [PubMed] [Google Scholar]
- 24.Smalla K. Proceedings of the Basel Forum on Biosafety. Safety of transgenic crops. Environmental and agricultural considerations. Biosafety research and assessment of technology impacts of the Swiss Priority Programme Biotechnology, Basel, Switzerland. 1995. Horizontal gene transfer from transgenic plants into plant associated microorganisms and soil microorganisms; pp. 29–34. [Google Scholar]
- 25.Smalla K, van Overbeek L S, Pukall R, van Elsas J D. Prevalence of nptII and Tn5 in kanamycin-resistant bacteria from different environments. FEMS Microbiol Ecol. 1993;13:47–58. [Google Scholar]
- 26.Smalla K, Gebhard F, van Elsas J D, Matzk A, Schiemann J. Proceedings of the 3rd International Symposium on The Biosafety Results of Field Tests of Genetically Modified Plants and Microorganisms. Oakland, Calif: The University of California Division of Agriculture and Natural Resources; 1994. Bacterial communities influenced by transgenic plants; pp. 157–168. [Google Scholar]
- 27.Smith J M, Dowson C G, Spratt B G. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 28.Smith M W, Feng D-F, Doolittle R F. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 29.Trieu-Cuot P, Carlier C, Courvalin P. Plasmid transfer by conjuction from Escherichia coli to gram-positive bacteria. FEMS Microbiol Lett. 1987;48:289–294. [Google Scholar]
- 30.Trinker N A, Fortin M G, Mather D E. Random amplified polymorphic DNA and pedigree relationship in spring barley. Theor Appl Genet. 1993;85:976–984. doi: 10.1007/BF00215037. [DOI] [PubMed] [Google Scholar]
- 31.Velten J, Hain L R, Schell J. Isolation of a dual plant promoter fragment from the Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1984;3:2723–2730. doi: 10.1002/j.1460-2075.1984.tb02202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakabayashi S, Matsubara H, Webster D A. Primary sequence of a dimeric bacterial haemoglobin from Vitreoscilla. Nature. 1986;322:481–483. doi: 10.1038/322481a0. [DOI] [PubMed] [Google Scholar]