Ex vivo screening of primary AML samples for susceptibility to venetoclax-based drug combinations identify potentially effective combinations and patient characteristics associated with response.
Abstract
The BCL2 inhibitor venetoclax combined with the hypomethylating agent azacytidine shows significant clinical benefit in a subset of patients with acute myeloid leukemia (AML); however, resistance limits response and durability. We prospectively profiled the ex vivo activity of 25 venetoclax-inclusive combinations on primary AML patient samples to identify those with improved potency and synergy compared with venetoclax + azacytidine (Ven + azacytidine). Combination sensitivities correlated with tumor cell state to discern three patterns: primitive selectivity resembling Ven + azacytidine, monocytic selectivity, and broad efficacy independent of cell state. Incorporation of immunophenotype, mutation, and cytogenetic features further stratified combination sensitivity for distinct patient subtypes. We dissect the biology underlying the broad, cell state–independent efficacy for the combination of venetoclax plus the JAK1/2 inhibitor ruxolitinib. Together, these findings support opportunities for expanding the impact of venetoclax-based drug combinations in AML by leveraging clinical and molecular biomarkers associated with ex vivo responses.
Significance:
By mapping drug sensitivity data to clinical features and tumor cell state, we identify novel venetoclax combinations targeting patient subtypes who lack sensitivity to Ven + azacytidine. This provides a framework for a taxonomy of AML informed by readily available sets of clinical and genetic features obtained as part of standard care.
See related commentary by Becker, p. 437 .
This article is featured in Selected Articles from This Issue, p. 419
INTRODUCTION
Acute myeloid leukemia (AML), a common adult leukemia, is characterized by considerable genetic heterogeneity that contributes to disease and complicates effective treatments (1). The standard of care for fit patients with minimal comorbidities is chemotherapy with or without hematopoietic stem cell transplantation. Patients who are older or less fit tolerate chemotherapy poorly, and effective treatment options are limited to hypomethylating agent (HMA) therapies (2–4). Older patients treated with azacytidine achieve complete remission (CR) in approximately a third of cases, and until recently, azacytidine was the only agent with a survival advantage for these patients outside the context of stem cell transplantation (5).
To improve on azacytidine therapy, earlier combination studies with azacytidine backbones were limited by additional toxicity (6). However, newer agents have increased response rates with minimal added toxicity when combined with azacytidine. AML cells evade proapoptotic stress signals by upregulating antiapoptotic proteins such as BCL2, BCL2L1 (BCL-xL), or MCL1, a dependency that has prompted the development of drugs that mimic the activity of proapoptotic BH3-only proteins or “BH3 mimetics.” Venetoclax, a specific BCL2 inhibitor, was developed to restore activation of apoptosis in AML and other cancers (7, 8). In a seminal study for older unfit patients with AML randomized to receive either standard azacytidine or venetoclax and azacytidine (Ven + azacytidine), overall survival and rates of remission were significantly higher for Ven + azacytidine patients (9). Consequently, Ven + azacytidine was approved as the new standard for first-line treatment of older or unfit patients with AML. Yet a clinical challenge remains: the ability to identify responsiveness to this therapy a priori.
Molecular patterns associated with clinical responses to Ven + azacytidine in patients with newly diagnosed AML (10) and in patients with relapsed/refractory AML (11) indicate that the presence of mutations in NPM1 or IDH1/2 predict higher response rates, whereas adverse cytogenetics and mutations in TP53, KRAS/NRAS, FLT3, and SF3B1 predict worse overall survival, although patients harboring these mutations in some cases show response prior to relapse. Further elucidation of patterns of resistance will help to optimize HMA/venetoclax “doublets” by adding a third agent.
BCL2 is highly expressed in subpopulations of AML cells enriched for stem/progenitor cell phenotypes, which contributes to venetoclax sensitivity, whereas differentiated tumor cells with a monocytic phenotype rely on other BCL2 family members and are less sensitive to venetoclax (12–15). Due to the hierarchical organization of hematopoiesis, the composition of leukemia reflects a point in hematologic maturation where a differentiation block gives rise to a blast population. The block, which can occur at early or late differentiation stages, is mirrored in the transcriptomic profiles of the constituent cells (16, 17). Ven + azacytidine effectively targets the leukemia stem cell population (18). Initial response rates with this combination are transient and can result in relapse through intrinsic molecular characteristics of the tumor (15). Kinase-activating mutations such as FLT3-ITD, or TP53 alterations can also produce adaptive drug resistance to Ven + azacytidine therapy (10).
Patient responses to Ven + azacytidine coupled with the transcriptomic signatures that underlie hematopoietic differentiation stages have led to the concept that leukemia treatments may be more effective by integrating gene-expression profiles and functional drug responses, as sensitivities for certain drugs have preferential associations with tumor differentiation states (19, 20). These observations provide the context for aligning ex vivo drug combination sensitivities from primary AML patient specimens with clinical features determined at the time of diagnosis. We hypothesize that identifying combination strategies whereby drug sensitivities are associated with discrete clinical features inherent to leukemic cells may enhance therapeutic outcomes. The efficacy of Ven + azacytidine provides impetus to investigate other venetoclax-inclusive combinations for patients who are ineligible for standard-of-care therapy. Using Ven + azacytidine as a reference, we evaluated a set of 25 venetoclax combinations, identified several with enhanced efficacy relative to Ven + azacytidine, and found associations of combination sensitivities with discrete patient characteristics.
RESULTS
Ex Vivo Drug Sensitivity Recapitulates Clinical Experience with Ven + Azacytidine in AML
AML therapy has improved with the use of Ven + azacytidine, and it has become the standard of care for elderly patients. Using an ex vivo assay, we evaluated Ven + azacytidine on primary specimens from AML patient samples (n = 142) and observed greater sensitivity for the combination compared with azacytidine and to a lesser degree relative to venetoclax alone (Fig. 1A). Sensitivity to Ven + azacytidine was measured by determining the area-under-the-dose response curve (AUC) following drug exposure. The Ven + azacytidine combination showed enhanced activity in 47% of the matched samples tested ex vivo (Fig. 1B), as measured by a combination ratio (defined as the combination AUC value divided by that of the most potent single-agent). Consistent with previous clinical observations showing Ven + azacytidine sensitivity on primitive but not mature tumor cells (17), we found that ex vivo sensitivity to Ven + azacytidine associated significantly with surface expression of CD117, a marker of hematopoietic stem cells, whereas resistance to Ven + azacytidine associated with expression of the monocytic marker CD14 (Fig. 1C).
Figure 1.
Ex vivo drug-sensitivity recapitulates clinical experience with Ven + azacytidine in AML. A,Ex vivo sensitivities for venetoclax (Ven), azacytidine and Ven + azacytidine on matched AML primary samples (n = 142). Sensitivity is represented as % of the maximum (max) area-under-the-dose response curve (AUC) derived for a 7-point concentration series ranging from 10 μmol/L to 10 nmol/L. B, Distribution of combination ratio (CR) values for patient samples treated with Ven + azacytidine ex vivo. CR is defined as the AUC of the combination divided by the AUC of the most potent single agent, where AUC CR <1 denotes the enhanced efficacy of the combination. C, Differences in Ven + azacytidine ex vivo sensitivity among patient samples with respect to expression of select immunophenotypic markers of primitive (CD117) and monocytic tumor cells (CD14). Neg, negative; Pos, positive. D, Differences in ex vivo sensitivity of Ven + azacytidine among patient samples based on the presence of prognostically relevant mutations in IDH1, NPM1, and either NRAS or KRAS. WT, wild-type. E, For 10 newly diagnosed patients with AML with both ex vivo screening and subsequent clinical treatment with Ven + azacytidine, ex vivo sensitivity collected at baseline prior to treatment is compared based on subsequent clinical response achieved on treatment. P values shown were determined using Mann–Whitney tests.
Clinical response to Ven + azacytidine correlates with select genetic subtypes such as NPM1 and IDH mutations, but not others such as RAS mutations (9, 11). These associations were recapitulated in data from ex vivo screening as samples harboring IDH1 or NPM1 mutations demonstrated significantly greater sensitivity to Ven + azacytidine, whereas samples with RAS mutations exhibited reduced sensitivity (Fig. 1D). Although it is a formal possibility that inherent chemosensitivity of some patient sample cells could explain their sensitivity ex vivo, screening data obtained at the time of diagnosis for a limited number of patients who received treatment with Ven + azacytidine correlated with subsequent clinical response (Fig. 1E), indicating concordance between the ex vivo and clinical data.
Novel Partners with Venetoclax Augment Combination Therapy Across a Spectrum of AML Clinical Features
Although Ven + azacytidine has improved patient outcomes, its effectiveness is limited by its restricted sensitivity for primitive tumor cell states and by acquired resistance, indicating a need for additional venetoclax-inclusive combinations (15). This limitation prompted us to evaluate a set of 25 new molecularly-targeted venetoclax-inclusive combinations by profiling ex vivo drug sensitivities on primary specimens from 433 patients with AML. The majority of these cases were collected at the time of initial AML diagnosis (n = 318), though analysis also encompassed patients with relapsed/refractory AML (n = 85). Combinations formed using FDA-approved drugs or late-stage development compounds represent a range of molecular targets (Supplementary Table S1). Due to the nature of combinations being added to screening panels over time, not all samples were evaluated with each combination. Among the new drug combinations tested with venetoclax as a backbone, the majority exhibited enhanced efficacy and potency relative to Ven + azacytidine (Fig. 2A; Supplementary Fig. S1). Similar overall patterns of sensitivity were observed for combination partner drugs within the same drug class, such as BET inhibitors (JQ1 and BETi), PI3K inhibitors (idelalisib and taselisib), and p38MAPK inhibitors (doramapimod and PH-797804).
Figure 2.
Novel partners with venetoclax augment the sensitivity of Ven + azacytidine. A, Comparisons of overall ex vivo potency (as measured by median AUC) and enhanced efficacy [median AUC combination ratio (CR)] for venetoclax (Ven) combined with azacytidine and 30 novel drug partners among primary AML patient specimens. Numbers of evaluable samples and fractions exhibiting the highest single-agent (HSA) synergy for each combination are shown as point size and color, respectively. Note that absolute numbers of samples tested with a particular combination vary as they were added to drug panels over time. Ven + azacytidine is highlighted in red for reference comparison purposes. B, Heat map of matched ex vivo sensitivity data [AUC % of maximum (max)] for 95 AML patient samples tested with Ven + azacytidine, a subset of novel Ven combinations, and their respective single agents. C–E, Comparisons of Ven combination sensitivities with respect to AML type (C), ELN 2017 risk (D), and peripheral blood (PB) monocyte percentage from differential blood counts obtained at the time of sample collection (E). For categorical variables (de novo/secondary diagnosis and ELN 2017 risk) are compared by Mann–Whitney test; points indicate the difference of median AUC and bars indicate the 95% confidence interval around this estimate. Negative and positive differences in median values reflect greater sensitivity and resistance, respectively, in patient samples with secondary AML or adverse risk. Percent monocytes are correlated with combination AUC by Spearman rank test, where negative and positive Spearman r values denote correlation with sensitivity and resistance, respectively. Blue and orange coloring represent statistically significant associations with sensitivity and resistance, respectively; gray color indicates the comparison was not statistically significant. Secondary AML denotes instances in which a patient's AML transformed from one of multiple disease states.
Alignments of AUC sensitivity values for a subset of patient samples with matched drug combination data revealed discrete patterns of enhanced efficacy. Potent sensitivity of single-agent venetoclax was evident in approximately 30% of the samples, whereas azacytidine was weakly effective across all samples ex vivo (Fig. 2B). Inhibitors such as ibrutinib, idelalisib, and ruxolitinib demonstrated similar potency profiles to azacytidine. In contrast, inhibitors including BETi, doramapimod, and taselisib showed single-agent sensitivity profiles with greatest potency on samples insensitive to venetoclax (Fig. 2B). Among matched samples, those with sensitivity to the Ven + azacytidine combination largely overlapped those with sensitivity to venetoclax alone. This was also true for Ven + ibrutinib. In contrast, venetoclax combined with idelalisib, ruxolitinib, BETi, doramapimod, or taselisib showed broader sensitivity profiles indicative of complementary or enhanced efficacy relative to Ven + azacytidine (Fig. 2B).
To evaluate potential combination toxicity to nonleukemic compartments, we tested Ven + azacytidine and these 25 combinations on human stromal cell lines HS-5 (Supplementary Fig. S2A) and HS-27 (Supplementary Fig. S2B). Although there were differences in sensitivity for some combinations with respect to the two stromal cell lines, the majority of combinations showed comparable, limited toxicity to either cell line. In addition, we tested freshly isolated mononuclear cells (MNC) from nonleukemic bone marrow and observed modest effects for some combinations; nearly all combinations showed reduced activity against these cells compared with primary AML counterparts (Supplementary Fig. S2C and S2D). For one AML patient specimen (22-00043), matched ex vivo sensitivity data for a limited set of single agents and combinations tested on both the primary AML blasts and stromal cells isolated from this sample indicated stromal cells have reduced sensitivity relative to the leukemic blasts (Supplementary Fig. S2E).
The different sensitivity profiles of the combinations were evaluated with respect to clinical features obtained at the time of diagnosis. The majority of venetoclax combinations did not show a difference in sensitivity between de novo and secondary AML diagnosis [denoting AML transformed from myelodysplastic syndrome (MDS) or myeloproliferative neoplasm (MPN)]. Notably, although Ven + azacytidine trended toward less sensitivity in secondary AML samples but did not achieve significance, venetoclax combinations with ARRY-382, BETi, and ibrutinib were significantly less effective in secondary AML (Fig. 2C). In contrast, Ven + quizartinib was more effective on secondary AML samples. With respect to ELN2017 risk, Ven + quizartinib was more effective on samples from patients with adverse prognostic risk; venetoclax combinations with ARRY-382 and olaparib were preferentially effective on favorable risk samples (Fig. 2D). Consistent with the reported resistance to Ven + azacytidine in patients with monocytic AML subtypes (15), ex vivo Ven + azacytidine sensitivity was decreased in samples with high monocyte percentages (Fig. 2E). Across all venetoclax combinations tested, more than half demonstrated a significant correlation with monocyte levels, either with resistance similar to Ven + azacytidine or with increased sensitivity. In contrast, combinations such as Ven + idelalisib and Ven + ruxolitinib showed little to no difference in sensitivity based on monocyte levels in the peripheral blood (Fig. 2E).
Venetoclax-Combination Sensitivity Correlates with AML Tumor Cell State Signatures
The associations of drug combination sensitivities with respect to immunophenotypic markers and peripheral blood monocyte levels led us to evaluate sensitivities across the range of cell state–specific gene-expression signatures defined for AML tumors (17). Training and test subsets of samples from our full sample cohort were stratified and balanced with respect to clinical and genetic features (Supplementary Table S2). Cell state signature scores were deconvoluted from bulk RNA sequencing (RNA-seq) and computed using an eigengene methodology as described (19, 20). Comparison of the associations of AML cell state signatures with venetoclax-combination sensitivities revealed overall concordance between the training and test sets (Fig. 3A), allowing for the merger of the two datasets for subsequent analyses to increase sample numbers. Clustering of correlations between venetoclax-combination sensitivities and cell state transcription signatures revealed three distinct patterns of sensitivity tracking with the type of partner drug (Fig. 3B, left; Supplementary Table S3). Type 1 partners exhibited combination sensitivity resembling Ven + azacytidine in that their respective combinations were effective on primitive cell states. Type 2 partners showed preferential combination sensitivity on more differentiated monocyte-like and conventional dendritic cell (cDC)-like cell states. Type 3 partners had a broad profile that did not align with particular cell state signatures. These partner-type associations were lessened or not at all apparent with respect to the single-agent sensitivities and cell state signatures (Fig. 3B, right). To examine the combination sensitivities at the patient samples level, AUC values for selected combinations of each type were plotted relative to progenitor-like and monocyte-like expression scores (Fig. 3C). For type 1 partner combinations (e.g., Ven + azacytidine and Ven + ibrutinib), greater ex vivo sensitivity associated with samples featuring high progenitor-like and low monocyte-like expression scores. Sensitivity to type 2 partner combinations (e.g., Ven + dasatinib) was increased in samples exhibiting high monocyte-like and low progenitor-like expression scores. Combinations involving type 3 partners (e.g., Ven + idelalisib and Ven + ruxolitinib) demonstrated diffuse sensitivity distributions that did not associate with either progenitor-like or monocyte-like gene set expression.
Figure 3.
Varying venetoclax partner drugs alters associations of sensitivity with AML tumor cell state. A, Concordance of drug-sensitivity cell state Pearson correlations for training and test sample cohorts. Colors indicate significant P values [after adjustment for family-wise error rate (FWER)] for both cohorts (purple), training set only (green), and test set only (orange). B, Clustered heat map of Pearson correlation r values for venetoclax (Ven) combination sensitivities (left) and respective single-agent sensitivities (not clustered, right) with respect to tumor cell state gene-expression signatures. Negative (blue) and positive (orange) correlation values correspond to sensitivity and resistance, respectively. C, Sensitivity of Ven + azacytidine and selected Ven combinations with respect to individual patient sample cell state. Dots represent individual patient specimens across the cell state spectrum defined by monocyte-like versus progenitor-like expression scores. Ex vivo drug sensitivity (AUC) is denoted in the color scale hue of each dot. Max, maximum.
Clinical and Genetic Patient Features Associate with Differences in Venetoclax-Combination Sensitivities
We further dissected ex vivo drug response with respect to panels of clinical and genetic patient features routinely collected in standard care. Analysis of mutations detected via clinical sequencing across a core panel of 42 genes revealed several significant associations with combination sensitivity and resistance (Fig. 4A; Supplementary Table S4). The sensitivity of Ven + azacytidine was greater among patient samples featuring NPM1 or IDH1/2 mutations and decreased for those with RAS and TP53 mutations. Venetoclax combinations such as with ruxolitinib, dasatinib, BET inhibitors, and palbociclib retained sensitivity on RAS and TP53-mutant samples.
Figure 4.
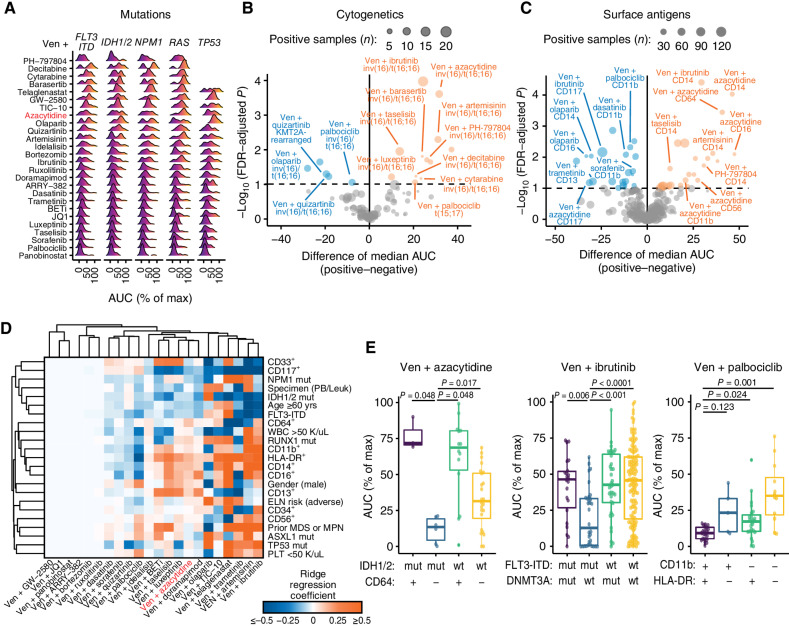
Clinical and genetic features map to associations with combination sensitivity. A, Ridge density plots of venetoclax (Ven) combination sensitivity differences with respect to indicated gene mutations. Color gradients indicate AUC-based ex vivo sensitivity (dark blue/purple) through resistance (orange/yellow). Max, maximum. B and C, Volcano plots showing a comparison of Ven combination sensitivity differences with respect to recurrent cytogenetic rearrangements (B) and immunophenotype surface markers (C). Negative and positive tails of each plot correspond to significantly increased (blue) and decreased sensitivity (orange), respectively, in samples positive for the tested feature. Size of each point represents the number of samples in the comparison that were positive for the tested feature for the given combination. The difference in median AUC was computed by the Hodges–Lehmann; P values were determined by Mann–Whitney tests and corrected for false discovery using the Benjamini–Hochberg method. D, Clustered heat map of multivariate ridge regression coefficient estimates for clinical and genetic feature associations with ex vivo combination sensitivity among newly diagnosed AML patient specimens. Blue and orange shading corresponds to associations of the indicated feature with sensitivity and resistance, respectively, for the corresponding combination. Feature panels and combinations included reflect limitations involving sample numbers and model requirements for minimizing missing data for analysis. E, Bivariate subgroup stratification of ex vivo sensitivity for select Ven combinations for indicated features. Combination AUC was compared across subgroups by the Kruskal–Wallis test; post-hoc pairwise Mann–Whitney tests were performed, with FWER-adjusted P values shown. Abbreviations: mut, mutated; wt, wild-type.
Comparison of combination sensitivity among a panel of prognostically impactful cytogenetic rearrangements in AML found Ven + azacytidine sensitivity decreased in samples harboring the CBFB–MYH11 fusion (inv(16) or t(16;16)); similar findings were observed for this rearrangement for several type 1 partner combinations with venetoclax (decitabine, cytarabine, ibrutinib, barasertib, artemisinin, taselisib, luxeptinib, and PH-797804; Fig. 4B). The presence of the inv(16) or t(16;16) rearrangement tracked with increased sensitivity to the type 2 partner combinations of Ven + palbociclib, Ven + quizartinib, and Ven + olaparib. Ven + quizartinib also demonstrated increased sensitivity in samples harboring the KMT2A rearrangements.
The association of ex vivo sensitivity with respect to common immunophenotypic markers showed results consistent with those of tumor cell state for some but not all combinations. Patient specimens expressing key surface markers of monocytic and/or mature myeloid cells (CD11b, CD13, CD14, CD16, CD56, and CD64) exhibited decreased sensitivity to Ven + azacytidine and other type 1 partner combinations including venetoclax combined with ibrutinib, artemisinin, taselisib, luxeptinib, and PH-797804, while conversely exhibiting increased sensitivity for multiple type 2 combinations such as Ven + dasatinib, Ven + palbociclib, and Ven + olaparib (Fig. 4C). Notably, several combinations of the type 3 pattern, such as Ven + ruxolitinib, Ven + doramapimod, Ven + BETi, and Ven + ARRY-382, showed no significant differences in sensitivity across any of the mutation, cytogenetic, and surface antigen panels (complete details are included in Supplementary Table S4). Together, these findings highlight the partial overlap of genetic aberrations and effects on cell state and their ability to inform more targeted venetoclax-combination efficacy.
Multivariable ridge regressions were fit to evaluate the independent strength of predictors of venetoclax combination ex vivo sensitivity and identify effective combinations for specific newly diagnosed AML patient attributes (Fig. 4D; Supplementary Table S5). Given the limitations of sample numbers per feature for each combination screened, analysis was confined to 22 combinations with complete data for 23 features. Ven + azacytidine had several pronounced feature-based associations among newly diagnosed AML cases, with sensitivity associated with IDH1/2- and NPM1-mutated samples and with CD117 expression while markers of mature myeloid cells (CD11b, CD14, CD16, and CD64), mutations of TP53 and RUNX1, and adverse ELN risk associated with resistance. Notable associations with sensitivity for other combinations included Ven + trametinib and Ven + BETi for RUNX1-mutant patients, Ven + palbociclib for adverse ELN risk, Ven + olaparib for patients with prior MDS or MPN, Ven + telaglenastat and Ven + doramapimod for ASXL1-mutant patients, and venetoclax paired with quizartinib, dasatinib, palbociclib, or luxeptinib for TP53-mutated patients (Fig. 4D). Based on these associations, combination sensitivity was evaluated via pairwise feature comparisons of mutations, cytogenetics, and markers of cell state (Supplementary Table S6). This revealed many instances of further stratification of ex vivo sensitivity, such as increased sensitivity to Ven + azacytidine in patients who both harbor IDH1/2 mutations and lack expression of CD64. Enhanced sensitivity was observed for Ven + ibrutinib in patients with FLT3-ITD but no DNMT3A variant and for Ven + palbociclib in patients positive for expression of both CD11b and HLA-DR (Fig. 4E).
Analysis of a Type 3 Partner Combination: Ven + Ruxolitinib
Although the enhanced efficacy of several of the venetoclax combinations derives from combining partner drugs with complementary cell state selectivity profiles to that of venetoclax, certain type 3 partner combinations enhance venetoclax efficacy across a broad swath of AML patient samples irrespective of cell state and other clinical and genetic variables. For the Ven + ruxolitinib combination, ruxolitinib's efficacy ex vivo is very limited as a single agent yet the ex vivo combination effectiveness was broad and did not correlate with the range of clinical and genetic features surveyed. Given these qualities, the established safety profiles of the two single agents, and that this combination is currently under evaluation in a phase I clinical trial in patients with relapsed/refractory AML (NCT03874052), we performed additional mechanistic studies for Ven + ruxolitinib to understand its combination efficacy and mechanisms of resistance.
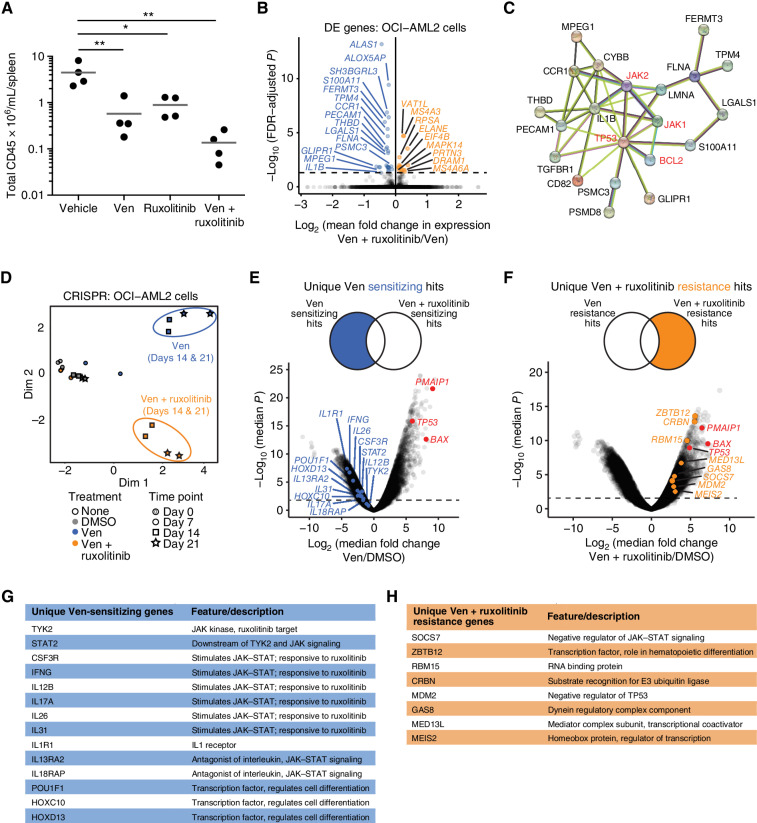
To confirm the effectiveness of Ven + ruxolitinib in vivo, primary AML patient MNCs were injected into NSGS mice, monitored for engraftment, and then treated with vehicle, venetoclax, ruxolitinib, or Ven + ruxolitinib daily (n = 4/group). Consistent with the significantly enhanced efficacy seen among patient samples ex vivo, tumor burden (assessed by CD45-positive cells in spleens) after 3 weeks of treatment was reduced in Ven + ruxolitinib-treated animals relative to either single-agent or vehicle, a result concordant with previous observations on a xenograft model using an AML cell line (ref. 21; Fig. 5A).
Figure 5.
Patterns of sensitivity and resistance for the type 3 combination Ven + ruxolitinib. A, Patient-derived xenograft evaluation of Ven + ruxolitinib in vivo. Mononuclear cells from a newly diagnosed AML patient were injected into NSGS mice, allowed to engraft to ∼1% human CD33 chimerism, and cohorts (n = 4/group) were treated with venetoclax (Ven; 50 mg/kg), ruxolitinib (30 mg/kg), or Ven + ruxolitinib (30/50 mg/kg) daily for 3 weeks. Animals were sacrificed and splenic disease burdens were compared using one-way ANOVA. *, P < 0.05; **, P < 0.01. B, Volcano plot of differential gene expression (DE) by RNA-seq in OCI-AML2 cells following treatment with Ven alone or in combination with ruxolitinib in triplicate for 24 hours. Genes highlighted in blue and orange indicate those with significantly decreased and increased expression, respectively, in cells treated with the combination compared with single-agent Ven. C, Predicted protein interaction network between drug targets of the combination (highlighted in red) and genes downregulated upon Ven + ruxolitinib treatment compared with Ven alone in OCI-AML2 cells. D, Principal component analysis of CRISPR/Cas9 screen sequencing data obtained from YUSA library-transduced OCI-AML2 cells at days 7, 14, and 21 after treatment with DMSO, Ven, or Ven + ruxolitinib. Clusters representing Ven-treated and Ven + ruxolitinib–treated cells are highlighted in purple and blue, respectively. Duplicate samples were evaluated for each condition. E, Volcano plot of changes in sgRNA expression (log2 fold change) for Ven-treated cells relative to DMSO control cells at day 21. Select genes highlighted in blue in the left tail showed unique sgRNA-sensitizing effects for Ven treatment but not Ven + ruxolitinib. F, Volcano plot of changes in sgRNA expression (log2 fold change) for Ven + ruxolitinib-treated cells relative to DMSO control cells at day 21. Select genes highlighted in orange in the right tail showed sgRNA enrichment consistent with unique resistance to Ven + ruxolitinibruxolitinib treatment but not Ven alone. On both CRISPR volcano plots (C–D), sgRNAs for select genes previously identified in Ven resistance CRISPR screens (BAX, PMAIP1, and TP53) are highlighted in red in each plot's resistant (positive) tail. Select unique Ven-sensitizing gene hits (G) and Ven + ruxolitinib resistance gene hits (H) are listed.
To dissect mechanisms underlying the enhanced efficacy of Ven + ruxolitinib, we first profiled multiple AML cell lines to identify a representative model. In line with the broad efficacy seen in ex vivo patient samples, Ven + ruxolitinib showed synergistic inhibition in the majority of eight lines tested, with contrastingly decreased efficacy in two non-AML leukemic cell lines (Supplementary Fig. S3). The most potent combination sensitivity was observed for OCI-AML2 cells, despite having little to no sensitivity to ruxolitinib alone. RNA-seq of OCI-AML2 cells after treatment with venetoclax or Ven + ruxolitinib revealed 41 differentially expressed genes (Fig. 5B; Supplementary Table S7). The majority of the genes downregulated by the combination compared with venetoclax alone associate with the principal drug targets (Fig. 5C), some of which have known roles in differentiation (22–24).
OCI-AML2 cells were also analyzed using genome-wide CRISPR-Cas9 screens following treatment with DMSO, venetoclax, or the Ven + ruxolitinib combination. The ruxolitinib-only screen was not performed, given the lack of sensitivity of these cells to single-agent ruxolitinib. Principal component analysis indicated discrete clusters separating DMSO controls, venetoclax-only treatment, and Ven + ruxolitinib treatment by both days 14 and 21 of treatment (Fig. 5D). Comparison of profiles for venetoclax and combination-treated cells to DMSO controls revealed discrete patterns for sensitivity and resistance (Fig. 5E–H). Many sgRNAs were depleted upon venetoclax but not combination treatment, suggesting a role for knockdown of these genes in further sensitization to venetoclax. Notably, this included many genes that are direct regulators of JAK–STAT signaling, such as TYK2, a JAK family kinase and a direct target of ruxolitinib (Fig. 5E and G). Other venetoclax-sensitizing hits included cytokine pathways identified from RNA-seq data and transcriptional regulators that govern cell differentiation programs. With respect to genes involved in resistance, among the sgRNAs most significantly enriched in both venetoclax- and Ven + ruxolitinib-treated cells were those targeting TP53, BAX, and PMAIP1, an expected result in concordance with our previous findings (25). sgRNAs with enrichment in the combination-treated cells but not those treated with venetoclax alone were more varied, with some of these targeting genes with roles as negative regulators of JAK–STAT signaling and others suggesting immediate resensitizing drug combinations (Fig. 5F and H).
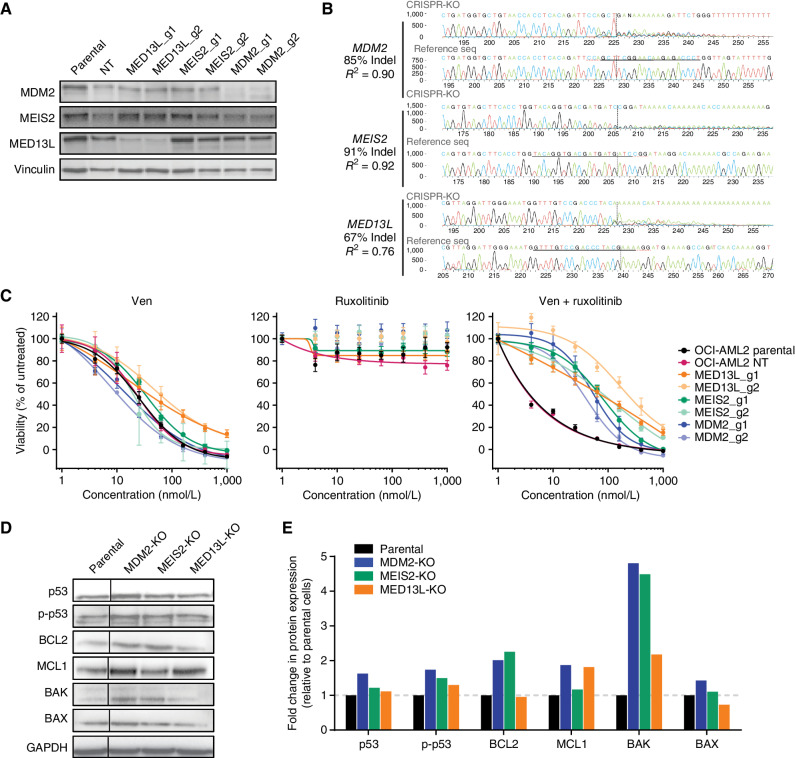
We evaluated three candidate effector genes—MED13L, MEIS2, and MDM2—for validation of genes and pathways conferring unique resistance to Ven + ruxolitinib. Single-cell OCI-AML2 knockouts were formed with two independent guides for candidate genes (Supplementary Table S8). Guide effectiveness evaluated by immunoblot analysis indicated near-complete loss of MED13L and MDM2, whereas robust reduction of MEIS2 levels was not apparent (Fig. 6A), despite parallel evidence of near complete allelic knockout at the DNA level for all three genes via TIDE analysis comparison with parental OCI-AML2 control cells (Fig. 6B). Consistent with screen results, knockout of these genes promoted resistance to the Ven + ruxolitinib combination and restored sensitivity to a level similar to that seen with venetoclax alone, but did not affect response to single-agent venetoclax (Fig. 6C). Furthermore, loss of MDM2, but not MED13 L or MEIS2, induced resistance to the MDM2 inhibitor idasanutlin despite all lines remaining similarly sensitive to venetoclax alone (Supplementary Fig. S4). Immunoblot analysis showed levels of total and phosphorylated TP53 to be elevated to varying degrees in knockout lines, most prominently for MDM2-KO. Levels of BCL2 and related apoptotic family members MCL1, BAK, and BAX were also elevated to varying extents in cells with Ven + ruxolitinib resistance gene knockouts compared with parental cells (Fig. 6D and E).
Figure 6.
Validation of CRISPR screen hits for resistance to the Ven + ruxolitinib combination. A, Immunoblot analysis of OCI-AML2 cells transduced with two independent guide RNAs for genes identified in the Ven + ruxoltinib CRISPR/Cas9 resistance screen. Cells were lysed, subjected to SDS-PAGE, transferred to PVDF membranes, and probed for levels of MED13L, MDM2, and MEIS2; vinculin was used as a protein loading control. Parental denotes untransduced OCI-AML2 cells; NT, nontargeting control guide. B, TIDE sequencing comparisons of genomic loci for MED13L, MDM2, and MEIS2 relative to parental OCI-AML2 controls following transduction and CRISPR modification. C, Dose-response curves for OCI-AML2 cells transduced as in B with two independent guide RNAs for MED13L, MDM2, and MEIS2 and tested for sensitivity to Ven, ruxolitinib, and Ven + ruxolitinib. Data points represent the mean normalized cell viability ± SEM for three replicates. Parental denotes untransduced OCI-AML2 cells; NT, nontargeting control guide. D, Immunoblot analysis of indicated proteins in OCI-AML2 parental and CRISPR knockout (KO) lines for MED13L, MDM2, or MEIS2. The vertical line denotes the cropping of nonadjacent lanes to juxtapose samples for ease of comparison. E, Quantification by densitometry of protein levels detected by immunoblot analysis in D. Band intensities were first blanked and normalized to loading control vinculin levels within lanes, then normalized to levels in parental OCI-AML2 cells and shown as fold change in expression.
DISCUSSION
Although its current indication is limited to newly diagnosed elderly patients with AML or patients with comorbidity concerns, the combination of venetoclax plus a hypomethylating agent (HMA) has improved the rate and depth of remission. Composite complete response rates range from 66% to 74% among these patients, with median overall survival between 14.7 and 16.4 months, representing an improvement over HMA alone (9, 26, 27). This highlights the importance of both defining the underlying features of patients who best respond to treatment and also identifying alternative combination strategies to manage emergent resistance to venetoclax plus hypomethylating agent therapy (28, 29).
A direct comparison of the activity of single-agent venetoclax versus Ven + azacytidine has not been evaluated in a randomized trial setting; however, in the first-line setting, azacytidine has been compared with Ven + azacytidine, where CR/CRi rates of 28% for azacytidine alone and 66% for Ven + azacytidine were reported (9). In the relapsed/refractory setting, venetoclax showed a 19% overall response rate (30). Venetoclax has been tested as a single agent in newly diagnosed AML, with single-agent run-in administered for 7 days prior to administration of 5 + 2 (31). The venetoclax run-in resulted in significant and impressive reductions of bone marrow blasts over the 7-day period, especially within molecular subgroups with known strong responses to Ven + azacytidine (e.g., NPM1 and IDH1/2). Venetoclax has also been combined with low-dose Ara-C (LDAC), exhibiting equivalent activity to Ven + azacytidine (32). These data show that venetoclax can exhibit clinical activity in a variety of different combination scenarios, which argues for the exploration of more diverse venetoclax doublet and triplet combinations. In our ex vivo data, the activity of Ven + azacytidine aligned with expected responses in genetic subgroups and with cell-surface immunophenotypes. In addition, clinical responses in the cases where both ex vivo and clinical data were available showed predictivity of the ex vivo result. Overall, these data suggest that our ex vivo assay provides a rapid readout for the prediction of potentially useful venetoclax-based combinations, and the findings of this data set may be useful for the development of novel venetoclax-combination regimens.
Analysis of clinical responses to Ven + azacytidine indicated some molecularly based stratifications, such as stronger response rates in patients harboring IDH1/2 or NPM1 variants and poorer responses in patients with TP53 or NRAS/KRAS mutations (9, 10, 33). Here, we show that ex vivo drug-sensitivity screening data from primary AML patient samples recapitulate these observations for Ven + azacytidine and implicate potential new markers for response stratification such as decreased sensitivity in patient samples harboring the CBFB-MYH11 (inv(16) or t(16;16)) rearrangement. Patients with this rearrangement also generally present with myelomonocytic differentiation and abnormal eosinophils (34). The bimodal distribution seen for some combinations upon alignment with individual mutations suggests that incorporation of a second clinical or cell state feature may better discern optimal patient subtypes for a given combination.
Tumor cell state exerts a critical influence on venetoclax efficacy with preferential selectivity for more primitive, less differentiated leukemic cells compared with more mature, monocytic counterparts (12–15). To this end, classification schemes based on gene-expression scores that capture variations in leukemic hierarchy composition represent powerful biomarkers of response to targeted agents (19, 20). However, stratifying patients with AML on the basis of tumor cell state expression signatures will require further development and resources before its clinical implementation in routine care is realized. Nevertheless, our results provide the context for aligning ex vivo drug combination sensitivities, including Ven + azacytidine, with both transcriptomic gene signatures of cell state and clinical features as part of current standard care. We demonstrate that patient attributes obtained from clinical labs, such as differential blood counts (e.g., % monocytes) and immunophenotyping (e.g., CD14 and CD64), represent sufficient surrogate biomarkers to predict sensitivity to Ven + azacytidine. Additional stratification of sensitivity to Ven + azacytidine was evident upon combining genetic and cell state markers, such as IDH1/2 mutation status and CD64 expression. Furthermore, our findings suggest many novel partner drugs may offer superior, enhanced efficacy when combined with venetoclax for patients who exhibit monocytic disease, thereby targeting AML tumors with greater selectivity.
Three patterns of venetoclax partner combination sensitivities emerged from our analysis, ranging from Ven + azacytidine-like (type 1) to differentiated selectivity (type 2) to broad efficacy irrespective of cell state (type 3). Each type may have utility, depending on specific patient characteristics. We note primary leukemic cells isolated from patient samples are viable in these assay conditions but do not proliferate. Consequently, drugs whose activity requires cycling cells will have reduced sensitivity, which may provide an underestimation of the utility of the combination. In addition, drugs whose mechanism of action requires transcriptomic rewiring occurring over many cycles may similarly have reduced sensitivity. For example, the ex vivo sensitivity profile for Ven + azacytidine may reflect a mechanism of azacytidine activity that likely requires more than 3 days of exposure to observe its activity. It is noteworthy that patients undergoing azacytidine therapy require 3 to 4 cycles of dosing before clinical benefit is achieved (15, 35). The Ven + azacytidine combination showed enhanced activity in 47% of the matched samples tested ex vivo (Fig. 1B), as measured by a combination ratio (defined as the combination AUC value divided by that of the most potent single-agent). Consistent with previous clinical observations showing Ven + azacytidine sensitivity on primitive but not mature tumor cells (15), we found the ex vivo activity profile of Ven + azacytidine coincides with primitive clinical features. We identified many venetoclax-inclusive combinations with greater potency and enhanced efficacy and these may require sequential or altered dosing regimens to accommodate toxicity and resistance constraints as has been done for Ven + ibrutinib in chronic lymphocytic leukemia (36, 37) or their addition as a third agent to the Ven + azacytidine backbone (e.g., clinicaltrials.gov: NCT03471260 and NCT04140487). Consistent with the activity of FLT3 inhibitors previously reported as single agents in AML (38, 39), the combination of quizartinib and sorafenib with venetoclax demonstrated increased ex vivo sensitivity in patient samples harboring FLT3-ITD mutations (Fig. 4D). Moreover, multiple type 2 combinations, including Ven + quizartinib, showed associations with sensitivity in TP53-mutated samples, which may reflect a subset that harbor mutant TP53 yet feature a more mature cell state phenotype.
Our CRISPR-Cas9 genome screen of single-agent venetoclax revealed sensitizing hits in multiple JAK–STAT pathway genes, suggesting a heightened dependence of venetoclax-treated cells on an actionable pathway targeted by ruxolitinib. Many of the resistance hits in the Ven + ruxolitinib CRISPR-Cas9 screen are in regulatory or transcription factor coding genes, suggesting reprogramming at a global transcriptional level is required to evade combination sensitivity. Protein interaction network analysis (String-db.org) predicted associations for CRISPR hits for MED13L and MDM2 with the molecular targets of the Ven + ruxolitinib combination (BCL2 and JAKs), with TP53 implicated as a common interaction. Enhanced levels of TP53 were detected in all three CRISPR-KO lines consistent with the preservation of venetoclax sensitivity in the TP53-KO cell lines (25), while compromising ruxolitinib activity in the combination by transcriptional reprogramming via loss of MDM2, MED13L, or MEIS2. MDM2 has a well-established role in activating TP53, and inhibitors of MDM2 are dependent on wild-type TP53 (40). Prior studies using colon cancer cells have shown that loss of MED13L altered levels of super-enhancer-regulated gene expression (41); in AML cells, inhibition of Mediator complex kinases led to increased expression of super-enhancer-regulated genes (42). MEIS2 has essential roles in hematopoietic differentiation (43) and proliferation, self-renewal, and AML disease progression (44). We also observed varying levels of increased MCL1 for all three CRISPR-KO lines, and this was most pronounced in MDM2-KO cells. As inhibition of JAK–STAT signaling diminishes levels of MCL1 (45), this represents a potential resistance mechanism involving rescue via MCL1 levels. Direct inhibition of MCL1 combined with venetoclax has been shown to overcome resistance to venetoclax in AML (46). Underlying the efficacy of Ven + ruxolitinib may be the depletion of MCL1 via JAK–STAT inhibition thereby enhancing sensitivity to BCL2 inhibition.
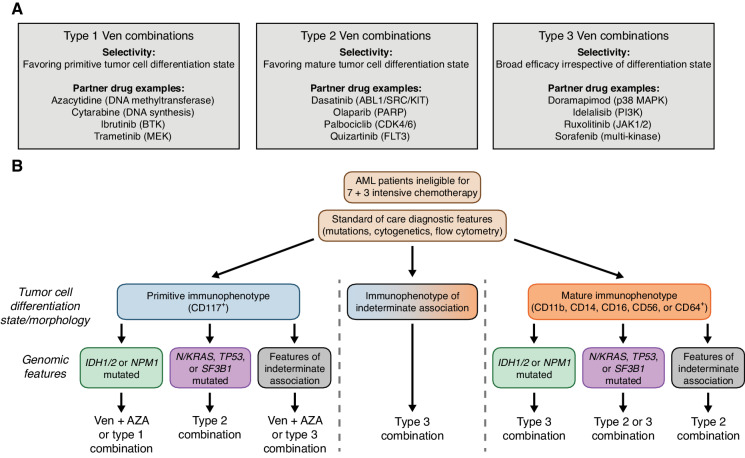
Building on the success of Ven + azacytidine begins with a detailed understanding of which patients are most likely to benefit from its use. We show that such stratification sensitivity is associated with both transcriptomic signatures of tumor cell state and select clinical and genetic subtypes. Mapping ex vivo drug sensitivity data to clinical features opens opportunities for developing additional venetoclax partner combinations that may benefit patients who lack sensitivity to Ven + azacytidine, affording the possibility of defining a taxonomy of AML based on drug sensitivities informed by the readily available set of clinical and genetic features obtained as part of standard care (Fig. 7A and B). For a cancer as heterogeneous as AML, the ability to use all available clinical and genetic patient features, along with evolving diagnostic tools, to guide venetoclax-combination treatment selection may enable improved depth and duration of response in patients.
Figure 7.
Prioritizing venetoclax-based combination therapy for AML. A, Categories of venetoclax-inclusive combination selectivity from this study. Select combination partner drugs representing FDA-approved agents and their canonical targets are shown. Complete annotations for all tested combinations in this study, along with their type 1, 2, and 3 designations, are provided in Supplementary Table S1. B, Decision scheme for optimizing therapeutic application of Ven + azacytidine and other venetoclax-combination strategies based on standard clinical immunophenotype/differentiation state and mutation features. Ex vivo combination sensitivity data were expressed as z-scores relative to the mean AUC value for each combination to account for differences in overall potency. Scaled sensitivity data were grouped by combination type and then further aligned based on the presence of the indicated clinical immunophenotype and genomic features to optimize patient sample subgroup activity. Given Ven + azacytidine is the standard of care for patients who are ineligible for 7 + 3 chemotherapy, it may be that alternate type combinations or partner drugs added to a backbone of Ven + azacytidine as a triplet may offer improved responses.
METHODS
Study Approval
Clinical specimens used in this study were collected following written informed consent from patients according to a protocol approved by the Institutional Review Board at Oregon Health and Science University (IRB# 9570; 4422) based on the recommendations of the Belmont Report. MNCs were isolated from blood or bone marrow drawn from 433 patients diagnosed with AML. MNCs were isolated by Ficoll gradient separation and used immediately in drug-sensitivity assays or for the preparation of DNA and RNA. In vivo murine xenograft studies were performed in accordance with an approved protocol from the Oregon Health and Science University Institutional Animal Care and Use Committee (IACUC).
Ex Vivo Inhibitor Assay
Small-molecule inhibitors were purchased from LC Laboratories or Selleck Chemicals or obtained from a corporate partner, reconstituted in DMSO, and stored at −80°C. Inhibitors (single agents or equimolar combinations of two single agents) were evaluated in a concentration series ranging from 10 μmol/L to 0.0137 μmol/L. Freshly isolated MNCs from primary patient bone marrow, peripheral blood, or leukapheresis specimens were seeded into prepared drug plates at 1E4 cells per well. At the time of seeding, cell viability was >90%. The median percentage of blasts in the bone marrow of patient samples surveyed was 72.5% (range, 0.5%–98%). Under these culture conditions, cells maintain viability but do not proliferate. Cell viability was assessed after 3 days using an MTS reagent (CellTiter96; Promega). A probit regression curve was fit to the [0,100]-bounded cell viability percentages for log10-transformed drug concentrations in the seven-dose-point series to derive IC50 and AUC. Ex vivo sensitivity data were normalized to untreated MNCs for each patient sample analyzed but were not further adjusted based on percentages of blasts detected in the clinical labs. For comparisons of sensitivity with nonleukemic blast cells, stromal cell lines HS-5 and HS-27 were plated at 1,000 cells/well in 384-well plates and tested on the same set of venetoclax combinations in three independent experiments conducted in triplicate. Fresh MNCs isolated from nonleukemic bone marrow obtained as a surgical discard from three different individuals (OHSU Department of Orthopedics) were plated at 1E4/well, also in triplicate. For some inhibitors and combinations, matched sensitivity data were available from AML patient sample 22-00044.
Univariate Analysis of Clinical and Genetic Features
Disease-specific panels of clinical, prognostic, genetic, cytogenetic, and flow cytometry-detected immunophenotype characteristics were manually curated from patient electronic medical records. Genetic characterization of patient samples included results of a clinical deep-sequencing panel of genes commonly mutated in hematologic malignancies. Associations between ex vivo inhibitor single-agent or combination efficacy (as assessed by AUC) were evaluated by the Mann–Whitney U test for categorical patient- or disease-based features and by Spearman rank correlation for continuous features. For pairwise feature-based stratification of drug combination sensitivity, AUC values for all 26 venetoclax combinations were binned into four subgroups for all possible pairs of categorical features from a panel of 42 mutated genes, 7 recurrent cytogenetic rearrangements, and 9 surface antigens. Feature pair breakdowns of combination AUC with at least 3 samples in all four possible subgroups were evaluated by the Kruskal–Wallis test across subgroups. Benjamini–Hochberg FDR correction of P values was applied for all tests within a given combination. For select highlighted comparisons, pairwise Mann–Whitney tests were performed between subgroups, with P values adjusted for family-wise error rate (FWER) by the Hochberg method.
Training versus Test Set Stratification
Available samples were divided into independent training and test subgroups of samples from the full sample cohort after stratifying on five factors: availability of RNA-seq data, availability of immunophenotyping data, collection year of the sample, FLT3-ITD status, and percentage of peripheral blood monocytes. Balance between the sample sets was evaluated for a panel of clinical and genetic features by the Mann–Whitney U test for categorical variables and Spearman rank correlation for continuous variables.
Correlation of Drug-Sensitivity with Gene Signatures of Tumor Cell State
Gene-level RNA-seq counts were generated as described (47) and normalized using conditional quantile normalization (48). Cell states, defined using six transcriptomic signatures for hematopoietic cell differentiation states (17), were deconvoluted and scored from bulk RNA-seq as described (19, 20). Drug sensitivity was dissected with respect to transcriptomic signatures for hematopoietic cell differentiation states (17) for 21 venetoclax combinations and their respective 22 single agents with gene expression and ex vivo drug data. Pearson correlation coefficients were estimated for each cell type score (as computed in ref. 20) versus AUC comparison, with P values corrected for FWER (Hochberg method) across the six cell states for each single agent or combination.
Multivariable Ridge Regression for Assessing Relative Strength of Feature-Sensitivity Associations
Ridge regression modeling of the square root of AUC for specific venetoclax-inclusive combinations was applied to newly diagnosed AML patient samples to quantify the association between individual patient features and combination efficacy while adjusting for other features. This form of penalized regression was chosen over ordinary least squares regression for its ability to better accommodate collinearity. The shrinkage parameter was chosen as the value that minimized mean squared error upon applying 5-fold cross-validation to the training set samples. To improve the visual display, ridge regression coefficient estimates were bounded between −0.5 and 0.5 in the heat map representations. Feature sets (i.e., model predictors) were unique to the extent of missing data for some features and were not considered if >5% missing. Only those combinations screened on at least 30 training set samples and 20 test set samples were included, resulting in 22 combinations. The heat map of ridge regression estimates (one model per venetoclax combination) allows for the identification of combinations with the highest and lowest ex vivo efficacy for a given patient attribute.
Xenograft Generation and In Vivo Drug Testing
MNCs from a male patient with newly diagnosed AML (FLT3-ITD and mutated NPM1, DNMT3A, SOCS1, and TET2) were injected into tail veins of NSGS mice (1 × 106 MNCs per mouse) and allowed to engraft until the peripheral blood showed approximately 1% human CD33 chimerism. Engraftment was detected using human CD33 and human CD45 antibodies by flow cytometry, with values ranging from 0.9% to 9.6%. Cohorts (n = 4/group) were then treated with vehicle, venetoclax (50 mg/kg), ruxolitinib (30 mg/kg), or Ven + ruxolitinib (30/50 mg/kg) daily for three weeks. Animals were sacrificed and splenic disease burdens were compared using one-way ANOVA with Tukey post-test to correct for multiple comparisons. Data were not further adjusted based on the level of engraftment. Spleens were analyzed normalizing the number of human cells to the spleen weight (in mg). The data represent a single study conducted with four mice in each treatment cohort, as this was the minimum needed for statistical power.
Cell Lines
Human AML cell lines (OCI-AML2, OCI-AML3, CMK, HL-60, Marimo, MOLM14, U-937, and UT-7), non-AML leukemic cell lines (K562 and RCH-ACV), HEK 293T/17 cells, and stromal cell lines (HS-5 and HS-27) were purchased from ATCC. Cell lines were authenticated by the DNA sequencing core at OHSU and assessed for Mycoplasma negativity using the MycoAlert Detection kit (Lonza).
RNA-Seq of OCI-AML2 Cells
OCI-AML2 parental cells treated with Ven + ruxolitinib (0.1 μmol/L and 1 μmol/L, respectively) or venetoclax alone (0.5 μmol/L) in triplicate for 24 hours and processed for RNA isolation (RNeasy Mini Kit, Qiagen Inc.). RNA quality was assessed using an Agilent Bioanalyzer. Libraries were prepared using the TruSeq Stranded mRNA Kit (Illumina Inc.) and sequenced on a NovaSeq 6000 (Illumina Inc.). Fastq files were assembled using Bcl2Fastq (Illumina Inc.). Reads were aligned to Ensembl v75 gene models (GRCh37 build of the human genome) using Kallisto (v0.46.2). Gene-level counts were produced using tximport v1.18.0 (49) with the “lengthScaledTPM” option. Differential expression was performed using the limma-trend v3.46.0 methodology (50). The predicted protein interaction network between drug targets and genes downregulated upon Ven + ruxolitinib treatment was derived using open-source software available at string-db.org.
Lentivirus Production and Transduction
HEK 293T/17 cells were transfected at low passage using Lipofectamine-2000 (Life Technologies Inc.) with single transfer vectors in combination with packaging plasmids, psPAX2 (Addgene, #12260) and VSVG (Life Technologies Inc.). Viral supernatants were collected, filtered through 0.45-μm filters, and used for transduction as previously described (25). Cells were used for downstream assays following 2 to 3 weeks after transduction.
CRISPR-Cas9 Screening and Single Gene Inactivation by Individual sgRNAs
Venetoclax and Ven + ruxolitinib–resistant Cas9-expressing cells were generated using lentiCas9-Blast (Addgene, #52962). Loss-of-function screens were performed using a human genome-wide sgRNA library (ref. 51; Addgene #67989), as described (25). Cells (5E6) were collected to ensure library representation and overall cultures were expanded to 120 million. Transduced cells (3E7) were separated in individual flasks and treated with DMSO, venetoclax (500 nmol/L), or Ven + ruxolitinib (100 nmol/L and 1 μmol/L, respectively) in duplicate, for 14 to 21 days. Cells (2-3E7) were collected at each time point from each sample to ensure 300× representation of the library. CRISPR PCR-amplified barcode libraries were generated as previously described (25). Inactivation of individual genes was performed with sgRNAs cloned into plentiCRISPRv2 (Addgene, #52961).
Immunoblotting
Lysates from OCI-AML2 parental and CRISPR-derived knockout cell lines (50 μg) were separated on precast 4% to 15% Tris-Glycine gradient gels (Bio-Rad), transferred onto Immobilon-P 37TPVDF37T membranes (Millipore Inc.). Following incubation with primary antibodies for MED13 L (Abcam#8783), MDM2 (Abcam#259265), MEIS2 (Abcam#174270), p53 (Abcam#26), phospho-p53 (Cell Signaling Technology #82530), BCL2 (Cell Signaling Technology #4222), MCL1 (Abcam#5453), BAX (Cell Signaling Technology #41162), BAK (Cell Signaling Technology #12105), or vinculin (Cell Signaling Technology #13901), the membranes were probed with species-specific HRP-conjugated IgG antibodies, coated with luminescent substrate and imaged with the Bio-Rad Gel Doc Imaging System. Protein densitometry was performed using ImageJ software (NIH) by normalizing the blanked intensity of each band of interest in a given lane to corresponding levels of loading control protein and expressing it as a fold change relative to levels in parental OCI-AML2 cells.
Statistical Analysis
For comparisons of matched drug combination and single-agent data for a given venetoclax combination, AUC values were compared by the Nemenyi test. Univariate comparisons of ex vivo drug sensitivity with respect to binary categorical variables (e.g., positive vs. negative for a given feature) were performed using a Mann–Whitney test. Differences in median values and their corresponding 95% confidence intervals were computed using the Hodges–Lehmann method. Differences in drug sensitivity across multiple subgroups defined by the status of two categorical variables were screened by the Kruskal–Wallis test. For categorical bivariate feature comparisons, P values were adjusted by the Benjamini–Hochberg step-up method to correct for all feature pairs compared for a given combination; select comparisons of multiple subgroups with FDR-adjusted P < 0.05 were subsequently analyzed by pairwise Mann–Whitney U test between subgroups with FWER-based correction of P values by Hochberg method. Continuous variable features (e.g., % monocytes in PB) were compared with combination ex vivo AUC values by Spearman rank correlation. Correlations of single-agent or combination AUC with tumor cell state gene-expression signatures were performed by Pearson correlation followed by FWER adjustment of P values across the six cell states tested for each combination. Drug sensitivity and feature correlation heat maps were generated using the pHeatmap R package (RRID:SCR_016418) using unsupervised pairwise average linkage clustering by either the Euclidean or Canberra method. For in vivo xenograft studies, tumor burden (as measured by splenic abundance of human CD45 + cells) was compared between treated arms (4 animals per group) by one-way ANOVA with Tukey post-test for multiple comparisons. For genome-wide CRISPR-Cas9 knockout screens, sgRNA-level enrichment statistics were summarized at the gene level by selecting the sgRNA (among a pool of 5 targeting the same gene) with the middle P value as a representative. Robust rank aggregation was performed after ranking genes by their mid P value (52). For in vitro drug profiling of select CRISPR gene knockout cell lines, all treatment conditions were tested in triplicate and fit by four-parameter nonlinear regression curves.
Data Availability
Patient sample RNA-sequencing data used in this study were previously submitted to dbGaP and Genomic Data Commons as part of the Beat AML Cohort and are publicly available. The dbGaP study ID is 30641 and the accession ID is phs001657.v2.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001657.v2.p1). Single-agent and drug combination ex vivo data (AUC values) along with associated specimen clinical annotations for the full cohort of patients analyzed in this study are available in Supplementary Table S9. CRISPR-Cas9 data are available at Gene Expression Omnibus (GSE216087).
Supplementary Material
Novel VEN combo partner drugs, targets, and status
Comparison of feature balance between training and test cohorts
Correlations of cell state gene sig with drug sensitivity
Associations with mutations, immunophenotype, and cytogenetics
Ridge regression coefficients
Pairwise feature KW test screen
Cell line DE genes for VEN+Ruxolitinib
Validation target sgRNA and TIDE primer reagent sequences
Full ex vivo AUC values and clinical annotations
Comparison of ex vivo sensitivity to venetoclax combinations with respect to their constituent single-agent drugs.
Screening of venetoclax combinations and respective single agents against non-leukemic human stromal and bone marrow mononuclear cells
Sensitivity and synergy of the Ven+Ruxolitinib combination in AML cell lines
Sensitivity of OCI-AML2 CRISPR/Cas9-KO cell line models to venetoclax or the MDM2 antagonist idasanutlin
Acknowledgments
The authors thank the patients for the generous use of their tissue samples and the Massively Parallel Sequencing Shared Resource at Oregon Health and Science University for technical support. This work was supported by the Drug Sensitivity and Resistance Network, NIH, NCI grant U54CA224019 (J.W. Tyner and B.J. Druker) and the Cancer Target Discovery and Development Network grant U01CA217862 (B.J. Druker). This work was additionally supported by NCI award R01CA262758 (J.W. Tyner and S.E. Kurtz), the V Foundation for Cancer Research (J.W. Tyner), the Gabrielle's Angel Foundation for Cancer Research (J.W. Tyner), the Anna Fuller Fund (J.W. Tyner), the Mark Foundation for Cancer Research (J.W. Tyner), and the Silver Family Foundation (J.W. Tyner). We thank Dr. Curt Civin (University of Maryland) for suggesting the inclusion of artemisinin and David McCoy (OCRTI, OHSU) for providing access to clinical immunophenotype data.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
NoteSupplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
D. Bottomly reports grants from NIH NCI 2U54CA224019 during the conduct of the study. S.K. McWeeney reports grants from NIH NCI 2U54CA224019-05 during the conduct of the study. J. Minnier reports grants from NIH during the conduct of the study. J.N. Saultz reports grants from IKENA Oncology and personal fees from the Rigel Advisory Board outside the submitted work. B.J. Druker reports grants from NIH/NCI and other support from HHMI during the conduct of the study; personal fees from Amgen, Aptose Biosciences, Iterion Therapeutics, Blueprint Medicines, Cepheid, GRAIL, Enliven Therapeutics, Nemucore Medical Innovations, VB Therapeutics, Vincerx Pharma, RUNX1 Research Program, Adela, DNA Seq, RUNX1 Research Program, and other support from MCED Consortium, Beat AML LLC, Astra-Zeneca outside the submitted work; in addition, B.J. Druker has a patent for Treatment of Gastrointestinal Stromal Tumors issued, licensed, and with royalties paid from Novartis. J.W. Tyner reports grants from Acerta, grants from Agios, Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Kronos, Meryx, Petra, Schrodinger, Seattle Genetics, Syors, Takeda, Tolero, and other support from Recludix outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
C.A. Eide: Conceptualization, data curation, formal analysis, writing–original draft, writing–review and editing. S.E. Kurtz: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, writing–original draft, writing–review and editing. A. Kaempf: Formal analysis, Methodology, writing–review and editing. N. Long: Data curation, methodology, writing–review and editing. S.K. Joshi: Formal analysis, validation, writing–review and editing. T. Nechiporuk: Formal analysis, Methodology. A. Huang: Validation, writing–review and editing. C.A. Dibb: Validation. A. Taylor: Validation. D. Bottomly: Formal analysis, methodology. S.K. McWeeney: Formal analysis, methodology. J. Minnier: Supervision, Methodology. C.A. Lachowiez: Formal analysis, validation, writing–review and editing. J.N. Saultz: Resources, data curation, supervision, funding acquisition. R.T. Swords: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. A. Agarwal: Data curation, formal analysis, validation, methodology, writing–review and editing. B.H. Chang: Resources, data curation, formal analysis, supervision, funding acquisition, validation, writing–review and editing. B.J. Druker: Resources, data curation, supervision, funding acquisition, methodology. J.W. Tyner: Conceptualization, resources, supervision, funding acquisition, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Kantarjian H, Ravandi F, O'Brien S, Cortes J, Faderl S, Garcia-Manero G, et al. Intensive chemotherapy does not benefit most older patients (age 70 years or older) with acute myeloid leukemia. Blood 2010;116:4422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pettit K, Odenike O. Defining and treating older adults with acute myeloid leukemia who are ineligible for intensive therapies. Front Oncol 2015;5:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dombret H, Seymour JF, Butrym A, Wierzbowska A, Selleslag D, Jang JH, et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood 2015;126:291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fenaux P, Mufti GJ, Hellström-Lindberg E, Santini V, Gattermann N, Sanz G, et al. Azacitidine prolongs overall survival and reduces infections and hospitalizations in patients with WHO-defined acute myeloid leukaemia compared with conventional care regimens: an update. Ecancermedicalscience 2008;2:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Craddock CF, Houlton AE, Quek LS, Ferguson P, Gbandi E, Roberts C, et al. Outcome of azacitidine therapy in acute myeloid leukemia is not improved by concurrent vorinostat therapy but is predicted by a diagnostic molecular signature. Clin Cancer Res 2017;23:6430–40. [DOI] [PubMed] [Google Scholar]
- 7. Del Gaizo Moore V, Letai A. BH3 profiling–measuring integrated function of the mitochondrial apoptotic pathway to predict cell fate decisions. Cancer Lett 2013;332:202–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 2013;19:202–8. [DOI] [PubMed] [Google Scholar]
- 9. DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AH, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med 2020;383:617–29. [DOI] [PubMed] [Google Scholar]
- 10. DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 2020;135:791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stahl M, Menghrajani K, Derkach A, Chan A, Xiao W, Glass J, et al. Clinical and molecular predictors of response and survival following venetoclax therapy in relapsed/refractory AML. Blood Adv 2021;5:1552–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bisaillon R, Moison C, Thiollier C, Krosl J, Bordeleau ME, Lehnertz B, et al. Genetic characterization of ABT-199 sensitivity in human AML. Leukemia 2020;34:63–74. [DOI] [PubMed] [Google Scholar]
- 13. Kuusanmäki H, Leppä AM, Pölönen P, Kontro M, Dufva O, Deb D, et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica 2020;105:708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang H, Nakauchi Y, Köhnke T, Stafford M, Bottomly D, Thomas R, et al. Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat Cancer 2020;1:826–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pei S, Pollyea DA, Gustafson A, Stevens BM, Minhajuddin M, Fu R, et al. Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov 2020;10:536–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. [DOI] [PubMed] [Google Scholar]
- 17. van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 2019;176:1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med 2018;24:1859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng AGX, Bansal S, Jin L, Mitchell A, Chen WC, Abbas HA, et al. A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia. Nat Med 2022;28:1212–23. [DOI] [PubMed] [Google Scholar]
- 20. Bottomly D, Long N, Schultz AR, Kurtz SE, Tognon CE, Johnson K, et al. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell 2022;40:850–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karjalainen R, Pemovska T, Popa M, Liu M, Javarappa KK, Majumder MM, et al. JAK1/2 and BCL2 inhibitors synergize to counteract bone marrow stromal cell-induced protection of AML. Blood 2017;130:789–802. [DOI] [PubMed] [Google Scholar]
- 22. Gingras MC, Margolin JF. Differential expression of multiple unexpected genes during U937 cell and macrophage differentiation detected by suppressive subtractive hybridization. Exp Hematol 2000;28:65–76. [DOI] [PubMed] [Google Scholar]
- 23. Pietras EM, Mirantes-Barbeito C, Fong S, Loeffler D, Kovtonyuk LV, Zhang S, et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol 2016;18:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spilsbury K, O'Mara MA, Wu WM, Rowe PB, Symonds G, Takayama Y. Isolation of a novel macrophage-specific gene by differential cDNA analysis. Blood 1995;85:1620–9. [PubMed] [Google Scholar]
- 25. Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D'Alessandro A, et al. The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov 2019;9:910–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pollyea DA, Pratz K, Letai A, Jonas BA, Wei AH, Pullarkat V, et al. Venetoclax with azacitidine or decitabine in patients with newly diagnosed acute myeloid leukemia: long-term follow-up from a phase 1b study. Am J Hematol 2021;96:208–17. [DOI] [PubMed] [Google Scholar]
- 27. Pratz KW, Jonas BA, Pullarkat VA, Thirman MJ, Garcia JS, Fiedler W, et al. Long-term follow-up of the phase 3 viale: a clinical trial of venetoclax plus azacitidine for patients with untreated acute myeloid leukemia ineligible for intensive chemotherapy. Blood 2022;140( Suppl 1):529–31.35951346 [Google Scholar]
- 28. Ilyas R, Johnson IM, McCullough K, Al-Kali A, Alkhateeb HB, et al. Outcome of patients with acute myeloid leukemia following failure of frontline venetoclax plus hypomethylating agent therapy. Haematologica 2023 Mar 2. doi: 10.3324/haematol.2022.282677 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 29. Maiti A, Rausch CR, Cortes JE, Pemmaraju N, Daver NG, Ravandi F, et al. Outcomes of relapsed or refractory acute myeloid leukemia after frontline hypomethylating agent and venetoclax regimens. Haematologica 2021;106:894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, et al. Efficacy and biological correlates of response in a phase II study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov 2016;6:1106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chua CC, Roberts AW, Reynolds J, Fong CY, Ting SB, Salmon JM, et al. Chemotherapy and venetoclax in elderly acute myeloid leukemia trial (CAVEAT): a phase Ib dose-escalation study of venetoclax combined with modified intensive chemotherapy. J Clin Oncol 2020;38:3506–17. [DOI] [PubMed] [Google Scholar]
- 32. Wei AH, Strickland SA Jr, Hou JZ, Fiedler W, Lin TL, Walter RB, et al. Venetoclax combined with low-dose cytarabine for previously untreated patients with acute myeloid leukemia: results from a phase Ib/II study. J Clin Oncol 2019;37:1277–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 2019;133:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu PP, Hajra A, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood 1995;85:2289–302. [PubMed] [Google Scholar]
- 35. Silverman LR, Fenaux P, Mufti GJ, Santini V, Hellström-Lindberg E, Gattermann N, et al. Continued azacitidine therapy beyond time of first response improves quality of response in patients with higher-risk myelodysplastic syndromes. Cancer 2011;117:2697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hillmen P, Rawstron AC, Brock K, Muñoz-Vicente S, Yates FJ, Bishop R, et al. Ibrutinib plus venetoclax in relapsed/refractory chronic lymphocytic leukemia: the CLARITY study. J Clin Oncol 2019;37:2722–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wierda WG, Allan JN, Siddiqi T, Kipps TJ, Opat S, Tedeschi A, et al. Ibrutinib plus venetoclax for first-line treatment of chronic lymphocytic leukemia: primary analysis results from the minimal residual disease cohort of the randomized phase II CAPTIVATE study. J Clin Oncol 2021;39:3853–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cortes J, Perl AE, Dohner H, Kantarjian H, Martinelli G, Kovacsovics T, et al. Quizartinib, an FLT3 inhibitor, as monotherapy in patients with relapsed or refractory acute myeloid leukaemia: an open-label, multicentre, single-arm, phase 2 trial. Lancet Oncol 2018;19:889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ravandi F, Alattar ML, Grunwald MR, Rudek MA, Rajkhowa T, Richie MA, et al. Phase 2 study of azacytidine plus sorafenib in patients with acute myeloid leukemia and FLT-3 internal tandem duplication mutation. Blood 2013;121:4655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, et al. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood 2005;106:3150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuuluvainen E, Domenech-Moreno E, Niemela EH, Makela TP. Depletion of mediator kinase module subunits represses superenhancer-associated genes in colon cancer cells. Mol Cell Biol 2018;38:e00573–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 2015;526:273–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang M, Wang H, Wen Y, Chen X, Liu X, Gao J, et al. MEIS2 regulates endothelial to hematopoietic transition of human embryonic stem cells by targeting TAL1. Stem Cell Res Ther 2018;9:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lai CK, Norddahl GL, Maetzig T, Rosten P, Lohr T, Sanchez Milde L, et al. Meis2 as a critical player in MN1-induced leukemia. Blood Cancer J 2017;7:e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res 2002;8:945–54. [PubMed] [Google Scholar]
- 46. Ramsey HE, Fischer MA, Lee T, Gorska AE, Arrate MP, Fuller L, et al. A novel MCL1 inhibitor combined with venetoclax rescues venetoclax-resistant acute myelogenous leukemia. Cancer Discov 2018;8:1566–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 2012;13:204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep 2016;17:1193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 2012;28:573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Novel VEN combo partner drugs, targets, and status
Comparison of feature balance between training and test cohorts
Correlations of cell state gene sig with drug sensitivity
Associations with mutations, immunophenotype, and cytogenetics
Ridge regression coefficients
Pairwise feature KW test screen
Cell line DE genes for VEN+Ruxolitinib
Validation target sgRNA and TIDE primer reagent sequences
Full ex vivo AUC values and clinical annotations
Comparison of ex vivo sensitivity to venetoclax combinations with respect to their constituent single-agent drugs.
Screening of venetoclax combinations and respective single agents against non-leukemic human stromal and bone marrow mononuclear cells
Sensitivity and synergy of the Ven+Ruxolitinib combination in AML cell lines
Sensitivity of OCI-AML2 CRISPR/Cas9-KO cell line models to venetoclax or the MDM2 antagonist idasanutlin
Data Availability Statement
Patient sample RNA-sequencing data used in this study were previously submitted to dbGaP and Genomic Data Commons as part of the Beat AML Cohort and are publicly available. The dbGaP study ID is 30641 and the accession ID is phs001657.v2.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001657.v2.p1). Single-agent and drug combination ex vivo data (AUC values) along with associated specimen clinical annotations for the full cohort of patients analyzed in this study are available in Supplementary Table S9. CRISPR-Cas9 data are available at Gene Expression Omnibus (GSE216087).



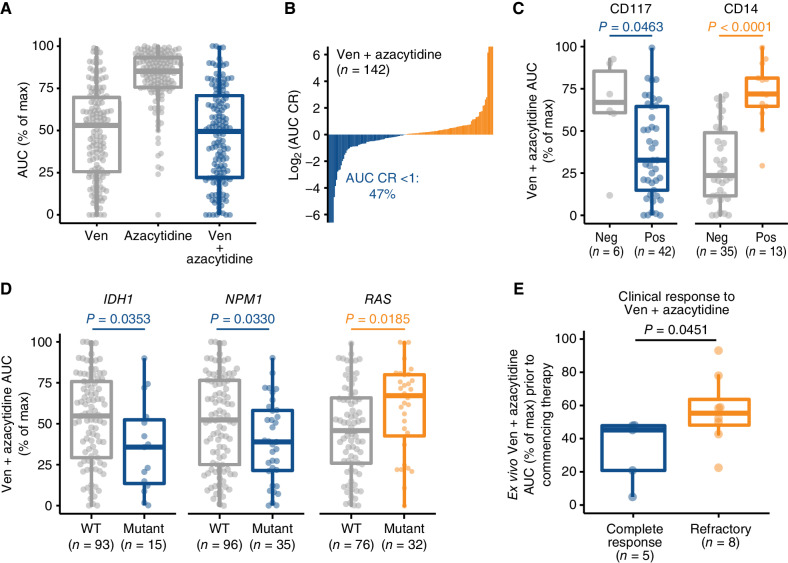
![Figure 2. Novel partners with venetoclax augment the sensitivity of Ven + azacytidine. A, Comparisons of overall ex vivo potency (as measured by median AUC) and enhanced efficacy [median AUC combination ratio (CR)] for venetoclax (Ven) combined with azacytidine and 30 novel drug partners among primary AML patient specimens. Numbers of evaluable samples and fractions exhibiting the highest single-agent (HSA) synergy for each combination are shown as point size and color, respectively. Note that absolute numbers of samples tested with a particular combination vary as they were added to drug panels over time. Ven + azacytidine is highlighted in red for reference comparison purposes. B, Heat map of matched ex vivo sensitivity data [AUC % of maximum (max)] for 95 AML patient samples tested with Ven + azacytidine, a subset of novel Ven combinations, and their respective single agents. C–E, Comparisons of Ven combination sensitivities with respect to AML type (C), ELN 2017 risk (D), and peripheral blood (PB) monocyte percentage from differential blood counts obtained at the time of sample collection (E). For categorical variables (de novo/secondary diagnosis and ELN 2017 risk) are compared by Mann–Whitney test; points indicate the difference of median AUC and bars indicate the 95% confidence interval around this estimate. Negative and positive differences in median values reflect greater sensitivity and resistance, respectively, in patient samples with secondary AML or adverse risk. Percent monocytes are correlated with combination AUC by Spearman rank test, where negative and positive Spearman r values denote correlation with sensitivity and resistance, respectively. Blue and orange coloring represent statistically significant associations with sensitivity and resistance, respectively; gray color indicates the comparison was not statistically significant. Secondary AML denotes instances in which a patient's AML transformed from one of multiple disease states.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/88fd/10618724/0ea50934f976/452fig2.jpg)
![Figure 3. Varying venetoclax partner drugs alters associations of sensitivity with AML tumor cell state. A, Concordance of drug-sensitivity cell state Pearson correlations for training and test sample cohorts. Colors indicate significant P values [after adjustment for family-wise error rate (FWER)] for both cohorts (purple), training set only (green), and test set only (orange). B, Clustered heat map of Pearson correlation r values for venetoclax (Ven) combination sensitivities (left) and respective single-agent sensitivities (not clustered, right) with respect to tumor cell state gene-expression signatures. Negative (blue) and positive (orange) correlation values correspond to sensitivity and resistance, respectively. C, Sensitivity of Ven + azacytidine and selected Ven combinations with respect to individual patient sample cell state. Dots represent individual patient specimens across the cell state spectrum defined by monocyte-like versus progenitor-like expression scores. Ex vivo drug sensitivity (AUC) is denoted in the color scale hue of each dot. Max, maximum.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/88fd/10618724/276d2b283597/452fig3.jpg)