A mass spectrometry–based immunopeptidomics characterizes the antigenic landscape of primary leukemic stem and progenitor cells in acute myeloid leukemia, identifying prime targets for the development of T cell–based immunotherapies targeting leukemia-repopulating cells.
Abstract
Therapy-resistant leukemia stem and progenitor cells (LSC) are a main cause of acute myeloid leukemia (AML) relapse. LSC-targeting therapies may thus improve outcome of patients with AML. Here we demonstrate that LSCs present HLA-restricted antigens that induce T-cell responses allowing for immune surveillance of AML. Using a mass spectrometry–based immunopeptidomics approach, we characterized the antigenic landscape of patient LSCs and identified AML- and AML/LSC-associated HLA-presented antigens absent from normal tissues comprising nonmutated peptides, cryptic neoepitopes, and neoepitopes of common AML driver mutations of NPM1 and IDH2. Functional relevance of shared AML/LSC antigens is illustrated by presence of their cognizant memory T cells in patients. Antigen-specific T-cell recognition and HLA class II immunopeptidome diversity correlated with clinical outcome. Together, these antigens shared among AML and LSCs represent prime targets for T cell–based therapies with potential of eliminating residual LSCs in patients with AML.
Significance:
The elimination of therapy-resistant leukemia stem and progenitor cells (LSC) remains a major challenge in the treatment of AML. This study identifies and functionally validates LSC-associated HLA class I and HLA class II–presented antigens, paving the way to the development of LSC-directed T cell–based immunotherapeutic approaches for patients with AML.
See related commentary by Ritz, p. 430 .
This article is featured in Selected Articles from This Issue, p. 419
INTRODUCTION
A major challenge in treatment of acute myeloid leukemia (AML) is the elimination of leukemia stem and progenitor cells (LSC). LSCs represent therapy-resistant cells that persist after treatment and are considered a main cause of relapse (1, 2). The latter occurs in a large proportion of patients despite the treatment advances in recent years and initially high remission rates resulting in the still very high mortality of AML (3).
Evidence that LSCs can be targeted and eliminated by the immune system arises from graft-versus-leukemia effects after allogeneic stem cell transplantation (4). However, immunotherapeutic approaches such as immune checkpoint inhibition, bispecific antibodies, and chimeric antigen receptor T cells that revolutionized treatment of other malignant diseases in recent years (5–8), showed only limited success in AML so far (9). One reason might be that these therapies are directed against bulk AML cells and do not specifically target LSCs. Thus, the identification of novel LSC-specific immune targets is of paramount importance to develop therapies with the potential to eradicate those cells. The target structures for such T cell–mediated immune responses are tumor-associated antigenic peptides that are presented on the surface of cancer cells by human leukocyte antigen (HLA) molecules. Hematopoietic stem and progenitor cells (HSPC), in health and disease, constitutively present antigens not only on HLA class I but also via HLA class II molecules allowing for direct interaction with antigen-specific CD4+ T cells, suggesting an immune surveillance mechanism that may effectively suppress leukemia onset upon LSC transformation (10). Promiscuous HLA class II–presented peptides, which bind to multiple HLA alleles (11, 12), might represent universally applicable targets for T cell–based immunotherapies to eliminate LSCs in AML. However, a systematic characterization of the immunopeptidome, that is, the entirety of naturally presented HLA-restricted peptides, and definition of the specific antigens that allow for LSC-directed immune surveillance is lacking. Mass spectrometry–based analyses of the immunopeptidome from different tumor entities enabled the identification of different groups of naturally presented tumor antigens: (i) neoepitopes from tumor-specific mutations (13, 14), (ii) cryptic neoepitopes originating from noncoding regions such as 5′ and 3′ untranslated region (UTR), noncoding RNAs (ncRNA), intronic and intergenic regions, or shifted reading frames in annotated protein coding regions (off-frame; refs. 15–20) and (iii) nonmutated tumor-associated antigens arising through differential gene expression or protein processing in tumor cells (21–25).
Here, we characterized the antigenic landscape of LSCs and AML bulk cells using mass spectrometry–based immunopeptidomics. We identified AML/LSC-associated (defined as shared among AML bulk cells and LSCs) HLA-restricted peptides that mediate immune surveillance in AML and constitute broadly applicable antigens for T cell–based immunotherapeutic approaches to specifically target LSCs in patients with AML.
RESULTS
Mass Spectrometry–Based Immunopeptidomics Uncovers the Antigenic Landscape of Primary LSCs
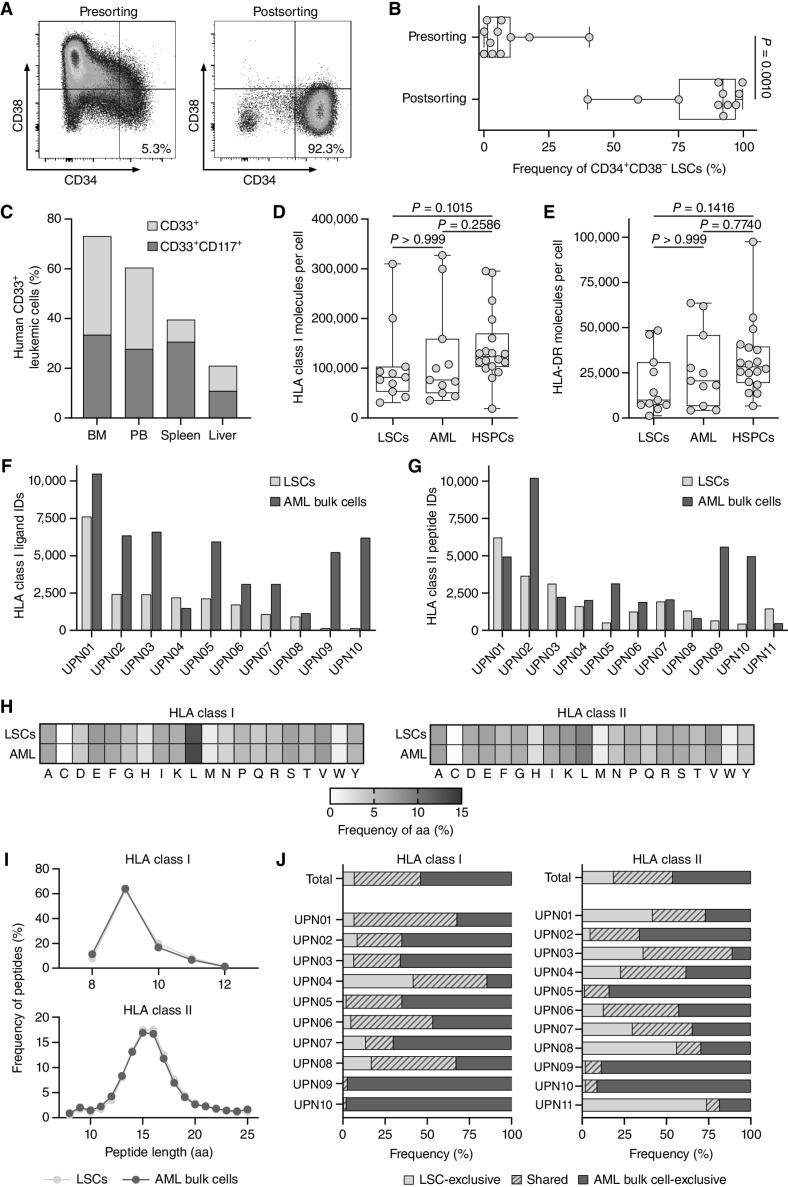
To investigate the immunopeptidomic landscape of primary LSCs (phenotypically defined as CD34+CD38− cells throughout this study), we isolated them from peripheral blood mononuclear cells (PBMC) of patients with AML. We screened samples of 26 patients with AML for the presence of CD34+CD38− LSCs (26) revealing a median LSC frequency of 0.2% (Supplementary Table S1; Supplementary Fig. S1), selecting 11 samples for sorting. LSC enrichment resulted in a median frequency of 92.1% CD34+CD38− cells postsorting (Fig. 1A and B; Supplementary Table S1). Stemness features of the sorted LSCs were validated by transplantation assays in NOD/SCID/IL2Rγnull (NSG) mice as reflected by in vivo leukemic engraftment of human CD33+ and CD33+CD117+ cells in the bone marrow, peripheral blood, spleen, and liver (Fig. 1C).
Figure 1.
Immunopeptidome analysis of enriched primary CD34+CD38− LSCs. A, Representative flow cytometry analysis of CD34+CD38− LSC frequencies pre- and postenrichment using magnetic-activated cell sorting (MACS) with the markers CD34 and CD38. B, Frequencies of CD34+CD38− LSCs in primary AML patient samples (n = 11) pre- and postsorting determined by flow cytometry. C,In vivo leukemic engraftment of LSCs (UPN01) in NOD/SCID/IL2Rγnull mice (n = 4). Flow cytometry–based analysis of the frequency of human CD33+ and CD33+CD117+ leukemic cells in the bone marrow, peripheral blood, spleen, and liver of NOD/SCID/IL2Rγnull mice 31 weeks after intrafemoral transplantation of 6 × 105 human CD34+CD38− LSCs. D and E, Surface expression of HLA class I (D) and HLA-DR molecules (E) determined by flow cytometry on AML patient-derived CD34+CD38− LSCs (n = 11) and CD34+CD38+ AML cells (n = 11) and on HV-derived CD34+ HSPCs (n = 18, thereof n = 7 hematopoietic stem cell apheresis from G-CSF mobilized blood donations of patients with nonhematologic malignancies). F and G, Number of mass spectrometric identified HLA class I- (F) and HLA class II-presented peptides (G) on LSCs and corresponding AML bulk cells (n = 10 for HLA class I, n = 11 for HLA class II). H, Amino acid distribution within the LSC- and AML bulk cell-derived HLA class I (left) and HLA class II (right) immunopeptidomes based on the unique peptide identifications in each cohort. I, Peptide length distribution of HLA class I ligands (top) and HLA class II peptides (bottom) in the immunopeptidome of LSCs and corresponding AML bulk samples. J, Overlap analysis of HLA class I ligand (left) and HLA class II peptide (right) identifications of LSCs and corresponding AML bulk samples on patient-individual and cohort-wide level. In B, D, and E, data points represent individual samples. Boxes represent median and 25th to 75th percentiles, whiskers are minimum to maximum. C, Data are presented as bar graphs with mean. B, Paired Wilcoxon signed rank test. D and E, Kruskal–Wallis test. Abbreviations: BM, bone marrow; PB, peripheral blood; ID, identification; UPN, uniform patient number; aa, amino acid.
As T cell—based immunotherapy requires sufficient HLA expression on target cells, we quantified HLA surface expression of CD34+CD38− LSCs and CD34+CD38+ AML cells as well as CD34+ HSPCs using PBMCs from patients with AML and healthy volunteers (HV), respectively. HLA surface expression on LSCs was comparable with CD34+CD38+ AML cells and CD34+ HSPCs for HLA class I and slightly decreased for HLA-DR (Fig. 1D and E).
Mass spectrometry–based analysis of the naturally presented HLA-restricted peptides, the so-called immunopeptidome, of LSC and corresponding bulk AML samples revealed a total of 16,342 and 32,961 unique HLA class I (comprising HLA-A, -B, and -C) ligands, respectively (Fig. 1F). 16,638 and 25,128 different HLA class II- (comprising HLA-DR, -DP, and -DQ) presented peptides were identified for LSCs and corresponding bulk AML samples, respectively (Fig. 1G).
HLA class I- and HLA class II-presented peptides of LSCs and corresponding AML bulk cells showed comparable specificities in terms of amino acid compositions and peptide length distributions (Fig. 1H and I; Supplementary Fig. S2A and S2B). The amino acid composition of the LSC- and AML bulk cell-presented peptides, that is, the abundance of certain amino acids, revealed no differences (Fig. 1H), even with respect to specific positions within the peptides (Supplementary Fig. S2A and S2B). The peptide length distribution is also comparable between the cell types, with the majority of HLA class I- and HLA class II–restricted peptides being 9 and 15 to 16 amino acids long, respectively (Fig. 1I). LSC- and corresponding AML bulk cell–derived immunopeptidomes revealed an overlap of HLA-presented peptides with 39.4% and 35.1% shared HLA class I and HLA class II peptides, respectively. 6.8% and 18.7% of the total identified HLA class I- and HLA class II-restricted peptides, respectively, showed exclusive presentation on LSCs (Fig. 1J).
Together, these data demonstrate that LSCs present HLA class I- and HLA class II–restricted antigens comparable but not identical to AML bulk cells highlighting the importance to integrate LSC-specific targets when selecting AML-associated antigens for immunotherapeutic approaches.
Comparative Immunopeptidome Profiling Identifies AML/LSC-Associated HLA Class I–Restricted Peptides
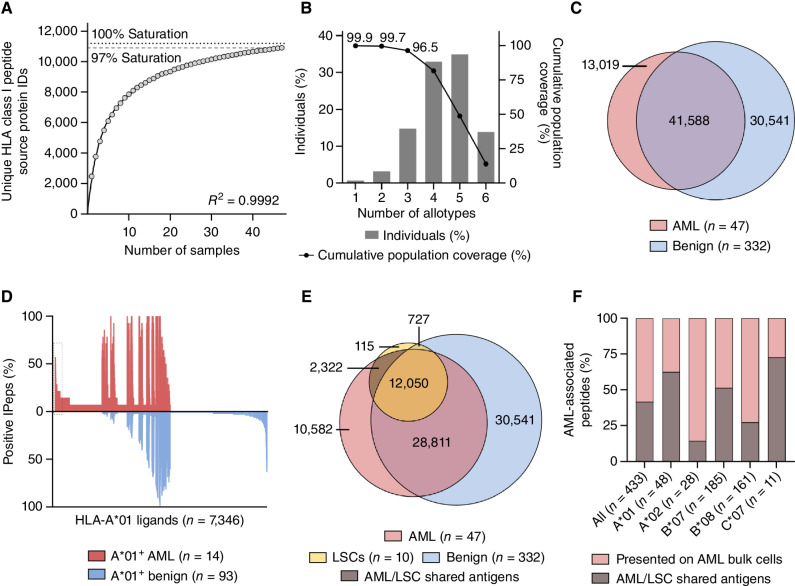
To explore antigens shared between LSCs (sorted CD34+CD38−) and bulk AML cells (unsorted PBMCs) that might be suitable for the dual immunotherapeutic targeting of AML bulk cells and LSCs (termed AML/LSC antigens), we comprehensively mapped the HLA class I immunopeptidome of 47 primary AML samples (Supplementary Tables S2 and S3) including the above-described samples sorted for LSCs. We identified a total of 72,042 unique HLA class I ligands that are derived from 10,609 source proteins (Supplementary Table S3). Thereby, 97% coverage of the estimated maximal attainable number of distinct source proteins was obtained (Fig. 2A). The AML dataset comprised 48 different HLA class I allotypes. 99.9% of the world population carry at least one of these allotypes (Fig. 2B). Curated AML/LSC immunopeptidomes were compared with a benign immunopeptidome dataset (25, 27), which comprises among others PBMCs, CD34+-enriched HSPCs, and various solid organ tissues and contains 72,129 unique HLA class I ligands (Fig. 2C), to identify AML/LSC-associated antigens absent from benign tissues. The distribution of HLA allotypes in the AML cohort was comparable with the benign immunopeptidome cohort (Supplementary Fig. S3A). For the identification of broadly applicable AML-associated antigens, we aimed for the selection of target antigens that not only fulfill the criterion of AML exclusivity, but also exhibit a high prevalence within the AML cohort and are presented by common HLA class I allotypes. Therefore, allotype-specific comparative profiling of AML and benign immunopeptidome datasets were performed for the common allotypes HLA-A*01 (30% frequency in AML cohort), -A*02 (49%), -B*07 (26%), -B*08 (23%), and -C*07 (55%, Supplementary Fig. S3A), which achieve 71% coverage within the world population (28). This revealed, respectively, 48, 28, 185, 161, and 11 AML-exclusive antigens with high allotype-specific frequencies ranging between 20% and 58% within the AML cohort (Fig. 2D; Supplementary S3B and S3C). 99.5% (433/435) of AML antigens were not presented on PBMC samples obtained from patients with AML in molecular remission (Supplementary Table S3), thereby further confirming AML specificity (Supplementary Table S4). AML-associated peptides showed different intensity ranks in the patients’ specific immunopeptidomes most ranking within the second and third quartiles of the intensity distribution (Supplementary Fig. S4).
Figure 2.
Comparative immunopeptidome profiling identifies AML/LSC-associated HLA class I antigen targets. A, Saturation analysis of HLA class I-restricted peptide source proteins of the AML cohort. The mean number of unique source proteins (y-axis) for a given cohort size (number of samples, x-axis) has been calculated by 1,000 random samplings from the entirety of AML immunopeptidomes. The number of unique source protein identifications (y-axis) is shown as a function of cumulative immunopeptidome analysis of AML samples (n = 47, x-axis). Exponential regression was used to extrapolate the maximum attainable coverage of different source proteins (dotted line, 11,193 proteins). The dashed line depicts the HLA-restricted peptide source proteome coverage achieved in the AML cohort (10,906 proteins). B, HLA class I allotype population coverage within the AML cohort compared with the world population (www.iedb.org). The frequencies of individuals within the world population carrying up to six HLA allotypes (x-axis) of the AML dataset are indicated as gray bars on the left y-axis. The cumulative percentage of population coverage is depicted as black dots on the right y-axis. C, Overlap analysis of HLA class I ligand identifications of primary AML samples (n = 47, curated immunopeptidome data) and benign samples (n = 332). D, Allotype-specific comparative immunopeptidome profiling based on the frequency of HLA-A*01–restricted peptide presentation in HLA-A*01–positive AML (n = 14) and benign samples (n = 93). Frequencies of positive immunopeptidomes for the respective HLA ligand (x-axis) are indicated on the y-axis. HLA-A*01 ligands (n = 7,346) are depicted on the x-axis, sorted according to the frequency of AML and benign samples presenting the respective ligand. The box on the left highlights the subset of AML-associated antigens showing AML-exclusive, high frequent presentation. E, Overlap analysis of HLA class I ligand identifications of LSC samples (n = 10) with AML bulk (n = 47) and benign samples including CD34+-enriched HSPCs (n = 332). F, Proportion of all HLA class I and of HLA class I allotype-specific high frequent AML-associated peptides that are also presented on LSCs (AML/LSC shared antigens). N indicates the number of peptides. Abbreviations: ID, identification; IPep, immunopeptidome.
By comparing the AML bulk and benign datasets to the immunopeptidomics data of the LSC-sorted samples, we identified 2,322 AML/LSC shared antigens, absent on any benign tissue including CD34+-enriched HSPCs (Fig. 2E; Supplementary Table S5). Within these AML/LSC-exclusive antigens, 181 peptides represented high frequent AML-associated targets identified for the common HLA allotypes HLA-A*01 (30/48, 62.5% of A*01 targets are shared on LSCs), A*02 (4/28, 14.3%), B*07 (95/185, 51.4%), B*08 (44/161, 27.3%), and C*07 (8/11, 72.7%, Fig. 2F; Supplementary Table S4). In total, 41.8% (181/433) of the high frequent AML-exclusive antigen targets were shared by LSCs (Fig. 2F; Supplementary Table S4) and thus constitute promising targets for the dual T cell–based targeting of AML bulk cells and LSCs.
Identification of AML/LSC-Associated HLA Class II–Restricted Antigens
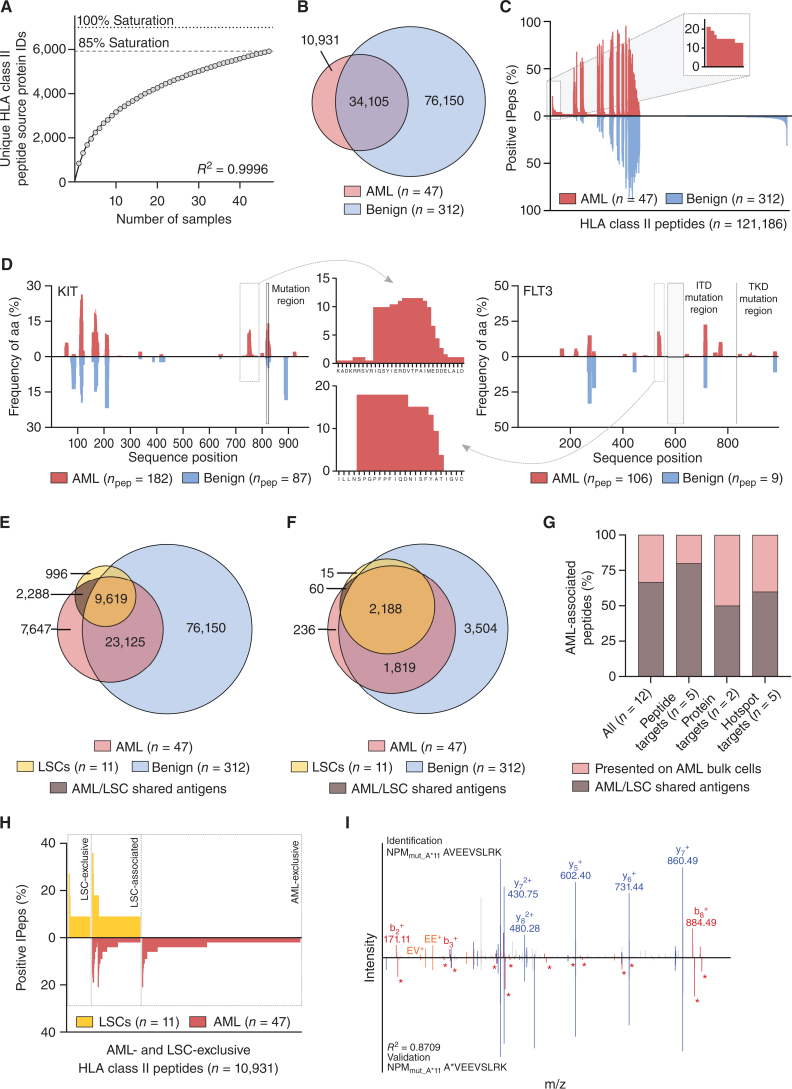
Because CD4+ T cells also play important direct and indirect roles in anticancer immunity, we aim to identify HLA class II–presented AML and AML/LSC antigens. A total of 61,205 unique HLA class II peptides originating from 5,922 source proteins and obtaining 85% of the estimated maximum attainable source protein coverage were identified by mass spectrometry–based immunopeptidomics (Supplementary Tables S2 and S3; Fig. 3A). Utilizing a previously established immunopeptidome profiling platform (25), we delineated three groups of AML-associated antigens: (i) peptide targets, (ii) protein targets, and (iii) hotspot targets (Supplementary Table S6). Overlap analysis and comparative profiling with a benign dataset on peptide level revealed 10,931 AML-exclusive HLA class II–restricted peptides (Fig. 3B) of which 5 were found in at least 15% of samples without detection of any length variants on benign samples (Fig. 3C; Supplementary Fig. S5A; Supplementary Table S6). HLA peptide source protein profiling revealed 311 AML-exclusive proteins, of which CCL23 and RRS1 show frequent (17% and 15% of AML samples) and significant AML-associated presentation with 7 and 2 proteotypic peptides, that is, uniquely derived from the respective proteins, respectively (Supplementary Fig. S5B–S5D; Supplementary Table S6). Hotspots of antigen presentation reflect distinct regions of proteins, which are more prone to produce HLA peptides. These hotspots depend on proteasomal cleavage, peptide processing, and HLA-binding rules (29). In particular for HLA class II–presented antigens, which are characterized by the presentation of different length variants in distinct patients, hotspot analysis represents a suitable tool for antigen identification. We here identified 5 AML-associated hotspots by peptide clustering, based on mapping identified peptides to their positions within the source protein, with representation frequencies of at least 15% within KIT, FLT3, AP2B1, HPRT, and IL1AP (Fig. 3D; Supplementary Fig. S5E; Supplementary Table S6).
Figure 3.
Identification of AML/LSC-associated HLA class II antigen targets by comparative immunopeptidome profiling. A, Saturation analysis of HLA class II–restricted peptide source proteins of the AML cohort. Therefore, the mean number of unique source proteins (y-axis) for a given cohort size (number of samples, x-axis) has been calculated by 1,000 random samplings from the entirety of AML immunopeptidomes. The number of unique source protein identifications (y-axis) shown as a function of cumulative immunopeptidome analysis of AML samples (n = 47, x-axis). Exponential regression was used to extrapolate the maximum attainable number of different source protein identifications (dotted lines, 6,992 proteins). The dashed line depicts the HLA-restricted peptide source proteome coverage achieved in the AML cohort (5,922 proteins). B, Overlap analysis of HLA class II peptide identifications of primary AML (n = 47, curated immunopeptidome data) and benign samples (n = 312). C, Comparative HLA class II immunopeptidome profiling based on the frequency of HLA-restricted presentation in AML and benign immunopeptidomes. Frequencies of positive immunopeptidomes for the respective HLA peptide (x-axis) are indicated on the y-axis. HLA class II peptides (n = 121,186) are depicted on the x-axis, sorted according to the frequency of AML and benign samples presenting the respective peptide. The box on the left and its magnification highlight the subset of AML-associated antigens showing AML-exclusive, high frequent presentation. D, Hotspot analysis of the proteins KIT and FLT3 by HLA class II-presented peptide clustering, respectively. Identified peptides were mapped to their amino acid positions within the source protein. Representation frequencies of amino acid counts within each cohort for the respective amino acid position (x-axis) are indicated on the y-axis. The boxes and their magnifications highlight the identified hotspots with the respective amino acids on the x-axis. Protein regions commonly mutated in AML are marked with light gray boxes. E and F, Overlap analysis of HLA class II peptide (E) and source protein (F) identifications of LSC samples (n = 11) with AML (n = 47) and benign samples (n = 312). Overlap analysis, comparative profiling, and hotspot analysis were performed with curated AML immunopeptidome data. G, Analysis of the proportion of AML-associated HLA class II–presented peptide, protein, and hotspot targets that are presented on both LSCs and AML bulk cells (AML/LSC shared antigens). H, Comparative profiling of AML- and LSC-exclusive HLA class II peptide identifications (not identified on benign tissue samples) based on the frequency of HLA-restricted presentation in the immunopeptidomes of LSC (n = 11) and AML (n = 47) samples. Frequencies of positive immunopeptidomes for the respective HLA peptide (x-axis) are indicated on the y-axis. HLA class II peptides (n = 10,931) are depicted on the x-axis, sorted according to the frequency of LSC and ANL samples presenting the respective peptide. The boxes mark subsets of LSC-exclusive antigens, LSC-associated antigens showing presentation on both LSCs and AML bulk cells (AML/LSC shared antigens), and AML-exclusive antigens. I, Mass spectrometry–based neoantigen validation using an isotope-labeled synthetic peptide. The experimentally eluted peptide NPMmut_A*11 (identification, above the x-axis) was validated with the corresponding synthetic peptide (validation, mirrored on the x-axis). Identified b-, y- and internal ions are marked in red, blue and orange, respectively. Ions containing the isotopic labeled amino acid are marked with an asterisk. Abbreviations: ID, identification; aa, amino acid; ITD, internal tandem duplication; TKD, tyrosine kinase domain; IPep, immunopeptidome; npep, number of peptides.
AML-associated peptide, protein, and hotspot targets were screened for their presentation on LSC samples (Fig. 3E and F; Supplementary Table S7), which identified 66.7% (8/12) as AML/LSC shared antigens (Fig. 3G; Supplementary Table S6) and thus prime targets for dual T cell–based targeting of LSCs and AML bulk cells. Of note, one high frequent LSC-exclusive peptide (on 27% of LSC samples) as well as 4 LSC-associated peptides and 3 LSC-associated protein targets (on 27% of LSC samples with dual presentation on AML bulk cells) were identified in the HLA class II immunopeptidome data, which constitute further highly interesting candidates for LSC immune targeting (Fig. 3H; Supplementary Fig. S5F; Supplementary Table S6). The abundance of AML- and AML/LSC-associated antigens differ in the patient-individual immunopeptidomes with the majority ranking in the second and third quartiles of the intensity distribution (Supplementary Fig. S6).
Mutation-Derived and Cryptic Neoantigens Are Represented in the AML Immunopeptidome
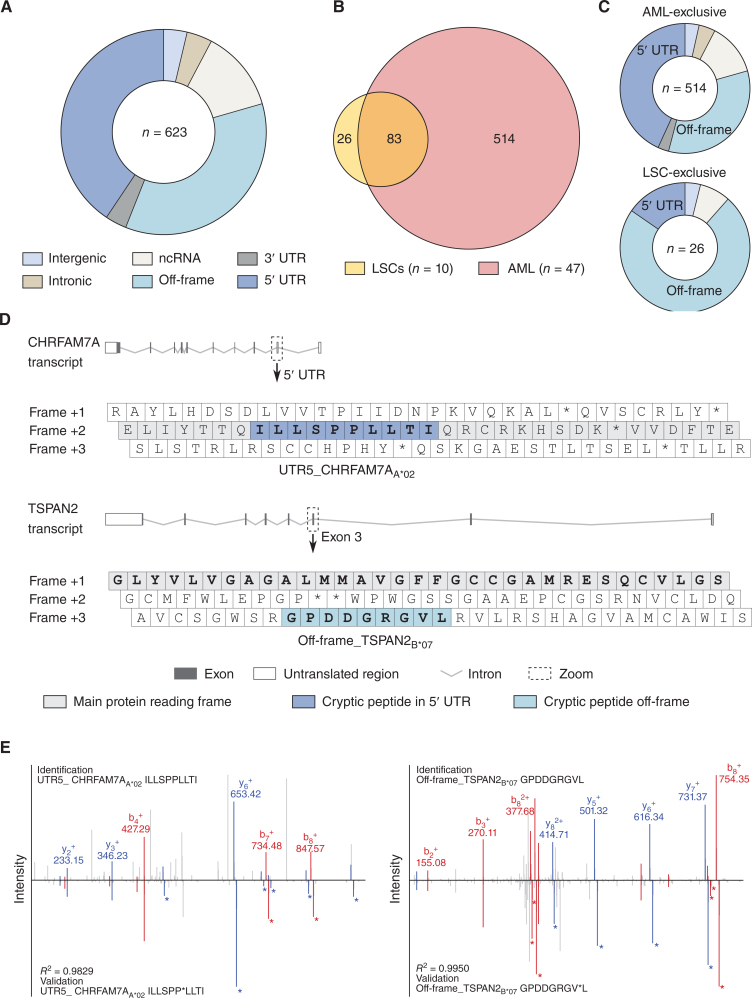
The HLA class I and HLA class II AML/LSC immunopeptidome datasets were further screened for naturally presented, mutation-derived neoepitopes from common AML-specific mutations (195 mutations within 23 genes representing 107 different mutation sites; Supplementary Table S8). Two naturally presented NPM1 mutation–derived HLA-A*11- and HLA-A*03- (NPMmut_A*11 AVEEVSLRK, NPMmut_A*03 LAVEEVSLR) and one IDH2 R140Q mutation–derived HLA class II- (IDH2mut_II KLKKMWKSPNGTIQNILGGTVF) restricted neoepitope(s) were identified on AML bulk cells, but not on LSCs, respectively. The three neoepitopes were identified in 100% (2/2), 33% (1/3), and 100% (1/1) of HLA-matched samples with the appropriate mutation profile, respectively. Mass spectrometric identification was validated by comparative measurement of synthetic peptides and verification of the respective mutations for each patient (Fig. 3I; Supplementary Figs. S7 and S8). In total, 96% (22/23) of mutation-bearing proteins are represented by HLA peptides that are identified in AML and/or benign immunopeptidomes. In contrast, only 19% (20/107) of the specific mutation sites within these proteins were directly covered by wild-type peptides, as most recurrent AML-specific mutations are located in “dark spots” of the immunopeptidome, defined as protein regions without any detectable HLA-presented peptides (Fig. 3D). Besides classical neoepitopes derived from AML-specific mutations, 623 cryptic AML-associated HLA class I peptides derived from noncanonical gene products were identified in the AML immunopeptidomes using the established algorithm Peptide-PRISM developed for the detection of cryptic peptides (30). AML-associated cryptic peptides were mainly derived from 5′ UTR and off-frame regions (Fig. 4A). 109 of these peptides were identified on LSC samples with 26 LSC-exclusive peptides (Fig. 4B) mainly originating from off-frame regions (Fig. 4C and D). High frequent AML/LSC-associated cryptic neoepitopes were validated using isotope-labeled synthetic peptides (Fig. 4E).
Figure 4.
Cryptic peptides are presented in AML- and LSC-derived immunopeptidomes. A, Distribution of identified AML-associated cryptic HLA class I-presented peptides (n = 623) among the respective genomic categories. B, Overlap analysis of cryptic peptide identifications of LSC samples (n = 10) with AML (n = 47). C, Distribution of AML-exclusive (n = 514, top) and LSC-exclusive (n = 26, bottom) cryptic peptides among the respective genomic categories. D, Examples for 5′ UTR- (top, UTR5_ CHRFAM7AA*02) and off-frame–derived (bottom, off-frame_TSPAN2B*07) cryptic HLA peptides. The symbols depict the transcripts of CHRFAM7A and TSPAN2. The zoom box highlights the 5′ UTR and exon 3 with the three reading frames including the respective cryptic peptides. E, Spectral validation of the cryptic peptides UTR5_ CHRFAM7AA*02 (left) and Off-frame_TSPAN2B*07 (right). Comparison of fragment spectra (m/z on x-axis) of cryptic peptides eluted from primary samples (identification) to their corresponding isotope-labeled synthetic peptides (validation, mirrored on the x-axis). Identified b- and y-ions are marked in red and blue, respectively. Ions containing isotope-labeled amino acids are marked with asterisks. Abbreviations: ncRNA, noncoding RNA; UTR, untranslated region.
Preexisting and De Novo Inducible AML/LSC Antigen-Specific T-cell Responses in Patients with AML and HVs
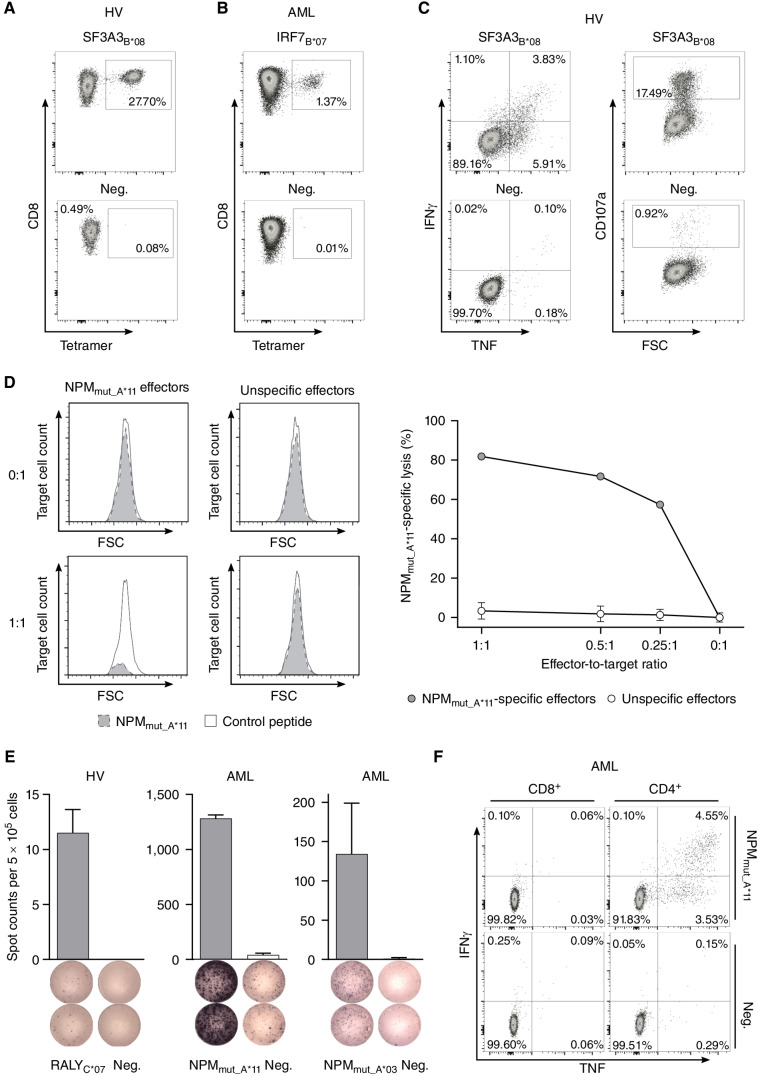
To evaluate whether the immunopeptidome-defined antigens comprising neoepitopes, cryptic peptides and AML/LSC-associated antigens, mediate T-cell effector functions in vitro and in vivo, two cohorts of patients with AML and HVs (Supplementary Tables S9 and S10) were investigated for antigen-specific T cells and corresponding immune responses (Table 1; for spectra validation see Supplementary Figs. S7 and S8). Artificial antigen-presenting cell (aAPC)-based in vitro priming experiments were used as an in vitro model for the feasibility of vaccination-induced de novo T-cell priming of naïve T cells and ELISpot assays were utilized for the detection of preexisting memory T-cell responses demonstrating an in vivo peptide-specific T-cell activation and active antitumor immune response. Using in vitro priming of naïve CD8+ T cells of HLA-matched HVs, de novo induction, and effective expansion of antigen-specific CD8+ T cells was observed for 14 of 15 HLA class I peptides including neoepitopes and cryptic peptides in 66% to 100% of analyzed HV samples (Fig. 5A; Table 1; Supplementary Fig. S9A). De novo induction of antigen-specific T cells was also achieved using samples of patients with AML without preexisting immune responses (Fig. 5B; Supplementary Fig. S9A). Antigen-specific T cells showed multifunctionality with IFNγ and TNF cytokine production as well as upregulation of the degranulation marker CD107a upon peptide stimulation (Fig. 5C; Supplementary Fig. S9B). NPMmut_A*11-directed CD8+ T cells specifically lysed NPMmut_A*11-loaded autologous cells in vitro with up to 82% target cell lysis compared with unspecific effector cells at various effector-to-target ratios (Fig. 5D).
Table 1.
AML- and AML/LSC-associated peptides.
| HLA class I–presented antigens | |||||||
|---|---|---|---|---|---|---|---|
| Memory T-cell responses | In vitro T-cell priming | ||||||
| ARP2A*01 | DIDTRSEFY | ARP2 | A*01 | 0/13 (0%) | 0/12 (0%) | 2/2 (100%) | n.a. |
| MYNNA*01 | FSEYFGAIY | MYNN | A*01 | 0/11 (0%) | 0/12 (0%) | 2/2 (100%) | TNF+ IFNγ+ CD107a+ |
| CEBPA A*01 | YLDGRLEPLY | CEBPA | A*01 | 0/11 (0%) | 0/12 (0%) | 2/2 (100%) | TNF+ IFNγ+ |
| CCNA1A*02 | SLLEADPFL | CCNA1 | A*02 | 0/16 (0%) | 0/14 (0%) | 4/4 (100%) | TNF+ IFNγ+ CD107a+ |
| DOCK8A*02 | IILDALPQL | DOCK8 | A*02 | 0/15 (0%) | 0/14 (0%) | 4/4 (100%) | TNF+ IFNγ+ CD107a+ |
| RUSF1A*02 | ILNDVAMFL | RUSF1 | A*02 | 0/14 (0%) | 0/15 (0%) | 4/4 (100%) | TNF+ IFNγ+ CD107a+ |
| STT3BB*07 | APESKHKSSL | STT3B | B*07 | 1/11 (9%) | 0/15 (0%) | 2/3 (66%) | n.a. |
| IRF7B*07 | APGLHLEL | IRF7 | B*07 | 0/12 (0%) | 0/16 (0%) | 5/5 (100%) | TNF+ IFNγ+ CD107a+ |
| OST48B*07 | APTIVGKSSL | OST48 | B*07 | 0/10 (0%) | 0/12 (0%) | 0/7 (0%) | — |
| ABCF2B*08 | DLDTRVAL | ABCF2 | B*08 | 0/11 (0%) | 0/13 (0%) | 2/3 (66%) | n.a. |
| TLE1B*08 | APTPRIKAEL | TLE1 | B*08/B*07 | 0/11 (0%) | 0/13 (0%) | 3/3 (100%) | n.a. |
| SF3A3B*08 | EGYGRYLDL | SF3A3 | B*08/B*14 | 0/11 (0%) | 0/11 (0%) | 1/1 (100%) | TNF+ IFNγ+ CD107a+ |
| UBE3BC*07 | SRPPLLGF | UBE3B | C*07 | 1/15 (7%) | 0/14 (0%) | n.a. | n.a. |
| CSN3C*07 | AYHELAQVY | CSN3 | C*07 | 0/15 (0%) | 0/14 (0%) | n.a. | — |
| RALYC*07 | IYSGYIFDY | RALY | C*07 | 1/15 (7%) | 1/13 (8%) | n.a. | TNF+ IFNγ+ |
| NPMmut_A*11 | AVEEVSLRK | NPMmut type A | A*11 | 1/13 (8%) | 0/9 (0%) | 3/3 (100%) | TNF+ IFNγ+ CD107a+ |
| NPMmut_A*03 | LAVEEVSLR | NPMmut type A | A*03 | 3/10 (30%) | 0/9 (0%) | n.a. | n.a. |
| UTR5_ CHRFAM7AA*02 | ILLSPPLLTI | UTR5 CHRFAM7A | A*02 | 0/12 (0%) | 0/23 (0%) | 1/1 (100%) | TNF+ IFNγ+ |
| Off-frame_TSPAN2B*07 | GPDDGRGVL | Off-frame TSPAN2 | B*07 | 0/7 (0%) | 0/23 (0%) | 2/2 (100%) | TNF+ IFNγ+ CD107a+ |
| HLA class II–presented antigens | |||||||
|---|---|---|---|---|---|---|---|
| Memory T-cell response | |||||||
| Peptide ID | Target class | Peptide sequence | Source protein | AML | HVs | Functionality of peptide-specific CD4+ T cells | |
| GALT7II | Peptide target | GNQLFRINEANQLMQ | GALT7 | 4/35 (11%) | 3/15 (20%) | TNF+ IFNγ+ CD107a+ | |
| CLC11II | Peptide target | DRQQMEALTRYLRAAL | CLC11 | 1/37 (3%) | 1/15 (7%) | n.a. | |
| APOBII | Peptide target | LGQEVALNANTKNQKIR | APOB | 1/37 (3%) | 1/15 (7%) | TNF+ IFNγ+ | |
| IL1APII_1 | Peptide target | NGRTFHLTRTLTVK | IL1AP | 3/33 (9%) | 5/15 (33%) | TNF+ IFNγ+ | |
| CCL23II | Protein target | SKPGVIFLTKKGRRF | CCL23 | 0/34 (0%) | 5/20 (25%) | TNF+ IFNγ+ | |
| FLT3II | Hotspot target | SPGPFPFIQDNISFYA | FLT3 | 5/33 (15%) | 1/14 (7%) | TNF+ IFNγ+ | |
| IL1APII_2 | Hotspot target | LDTMRQIQVFEDEPAR | IL1AP | 2/34 (6%) | 0/14 (0%) | TNF+ IFNγ+ | |
| HPRTII | Hotspot target | VVGYALDYNEYFRDL | HPRT | 4/31 (13%) | 1/14 (7%) | TNF+ IFNγ+ | |
| KITII | Hotspot target | IGSYIERDVTPAIM | KIT | 0/32 (0%) | 0/14 (0%) | — | |
| LTV1II | LSC-exclusive peptide target | PHRKKKPFIEKKKAVSFHLVHR | LTV1 | 0/31 (0%) | 2/14 (14%) | TNF+ IFNγ+ | |
| PPGBII | LSC-associated peptide target | KHLHYWFVESQKDPEN | PPGB | 1/31 (3%) | 0/14 (0%) | TNF+ IFNγ+ | |
| ITALII | LSC-associated peptide target | ETLHKFASKPASEFVK | ITAL | 2/31 (6%) | 0/14 (0%) | n.a. | |
| TACTII | LSC-associated protein target | DRVKLGTDYRLHLSPV | TACT (CD96) | 1/31 (3%) | 1/14 (7%) | TNF+ IFNγ+ | |
| G6PC3II | LSC-associated protein target | ERPEWIHVDSRPF | G6PC3 | 0/32 (0%) | 0/14 (0%) | — | |
| IDH2mut_II | Neoepitope | KLKKMWKSPNGTIQNILGGTVF | IDH2 R140Q | 0/31 (0%) | 6/25 (24%) | TNF+ IFNγ+ | |
NOTE: Immunogenicity analysis of AML- and AML/LSC-associated peptides. Preexisting memory T-cell responses in AML patient and HV samples were investigated by IFNγ ELISpot assays after peptide-specific 12-day in vitro expansion. In vitro T-cell priming of peptide-specific T cells was conducted using aAPCs. Functionality of peptide-specific T cells was analyzed by intracellular cytokine (IFNγ, TNF) and degranulation marker (CD107a) staining.
Abbreviations: ID, identification; n.a., not available.
Figure 5.
Immunogenicity analysis of HLA class I–restricted targets. A and B,De novo induction of peptide-specific CD8+ T cells using aAPC-based in vitro priming with PBMC samples of HVs (A) and patients with AML (B). Representative example of flow cytometry–based tetramer staining. Graphs show single, viable cells stained for CD8 and PE-conjugated tetramer of indicated specificity. The negative control depicts the tetramer staining of T cells from the same donor primed with an HLA-matched control peptide. Results for all analyzed HVs and patients with AML are shown in Supplementary Fig. S9A. C, Flow cytometry–based functional characterization of peptide-specific CD8+ T cells after in vitro aAPC-based priming by intracellular cytokine (IFNγ, TNF) and degranulation marker (CD107a) staining. Representative example after stimulation with the peptide SF3A3B*08 (top) compared with an HLA-matched control peptide (bottom). Graphs show single, viable CD8+ cells stained for IFNγ and TNF (left) as well as CD107a (right). Results for all analyzed HVs and patients with AML are shown in Supplementary Fig. S9B. D, Specific cell lysis of NPMmut_A*11-loaded autologous target cells in comparison to HLA-matched control peptide-loaded target cells by NPMmut_A*11-specific CD8+ T cells compared with no cell lysis by unspecific effector cells derived from in vitro–primed cells of an HV analyzed by flow cytometry–based VITAL assay. Unspecific effectors were evaluated in three independent replicates and results are shown as mean with SD for the three replicates. E and F, Detection of preexisting HLA class I peptide-specific T-cell responses by IFNγ ELISpot assay (E) and intracellular cytokine staining (F) after 12-day in vitro expansion using peripheral blood mononuclear cell samples of HVs and patients with AML. Representative examples are depicted. Data are presented as bar graphs with mean and SD of duplicates. Graphs show single, viable cells stained for CD8 (left) and CD4 (right) and the cytokines IFNγ and TNF. Data for all analyzed samples are shown in Supplementary Fig. S10. Abbreviations: neg., HLA-matched negative control peptide; FSC, forward scatter.
IFNγ ELISpot assays after peptide-specific 12-day in vitro expansion revealed preexisting HLA class I peptide–specific memory T-cell responses in up to 30% and 8% of HLA-matched AML patient and HV samples, respectively, targeting 26.3% (5/19) of HLA class I–restricted peptides (Fig. 5E; Table 1), with immune responses mainly mediated by CD4+ T cells (Fig. 5F; Supplementary Fig. S10). No preexisting memory T-cell responses targeting cryptic peptides (UTR5_ CHRFAM7AA*02, Off-frame_TSPAN2B*07) were observed in patients with AML or HVs, whereas neoepitope-specific memory responses were detectable frequently in AML patient samples (up to 30%, Table 1). Moreover, strong preexisting CD4+ T cell–mediated immune responses targeting HLA class II–restricted peptides (86.7%; 13/15) were observed using IFNγ ELISpot assays after 12-day in vitro expansion in up to 15% and 33% of patients with AML and HVs, respectively (Fig. 6A–C; Supplementary Fig. S10, Table 1). The FLT3-derived peptide FLT3II (SPGPFPFIQDNISFYA) elicited an additional CD8+ T cell–mediated immune response (Supplementary Fig. S11A and S11B). In silico prediction with the respective patient's HLA allotype revealed four potential HLA class I–restricted peptides embedded in the long HLA class II sequence (Supplementary Fig. S11C).
Figure 6.
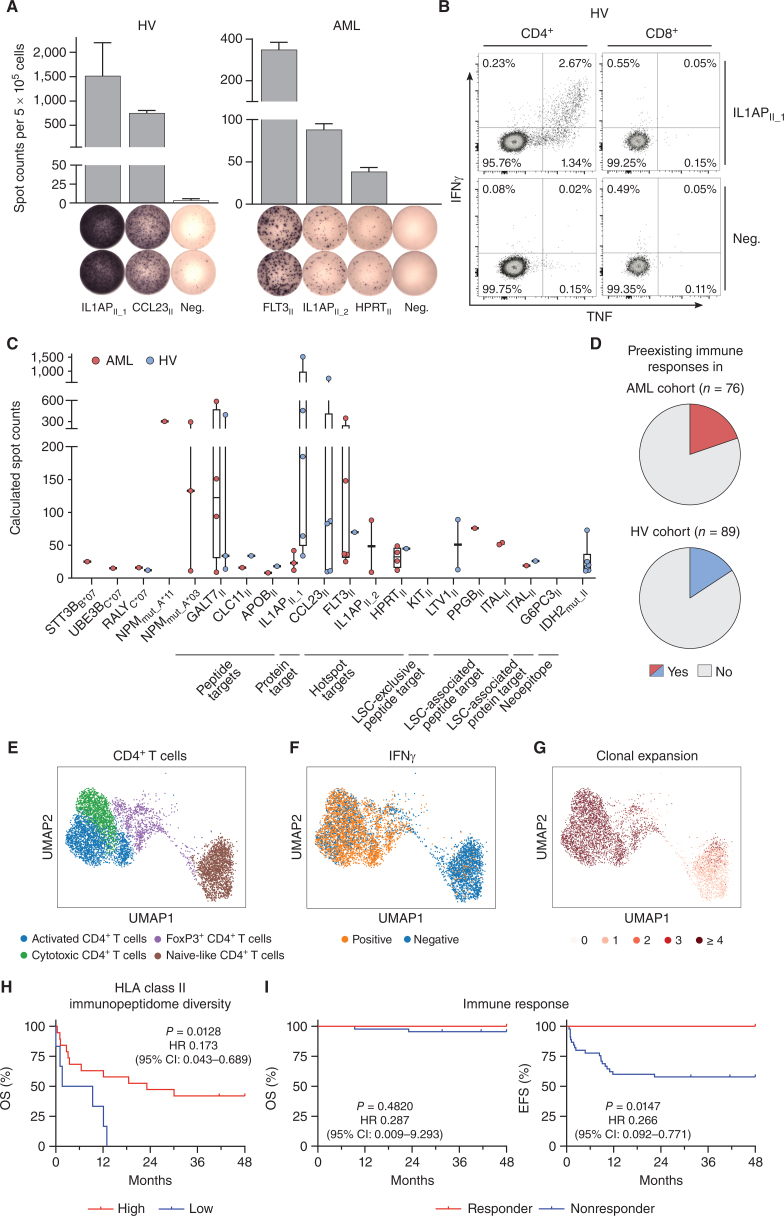
Immunogenicity analysis of HLA class II–restricted targets and impact of immunopeptidome diversity and peptide-specific immune responses on AML patient outcome. A and B, Detection of preexisting HLA class II peptide-specific T-cell responses by IFNγ ELISpot assay (A) and intracellular cytokine staining (B) after 12-day in vitro expansion using PBMC samples of HVs and patients with AML. Representative examples are depicted. Data are presented as bar graphs with mean and SD of duplicates. Graphs show single, viable cells stained for CD4 (left) and CD8 (right) and the cytokines IFNγ and TNF. Intracellular cytokine staining results for all analyzed samples are shown in Supplementary Fig. S10. C, Intensity of T-cell responses in terms of calculated spot counts in IFNγ ELISpot assays after 12-day stimulation against the respective AML- and AML/LSC-associated HLA class I and HLA class II–restricted antigenic peptide using PBMCs of AML patients and HVs. Dots represent data from individual donors. Data is shown for donors with T-cell responses. Boxes represent median and 25th to 75th percentiles, whiskers are minimum to maximum. D, Pie charts depicting the recognition rate (individuals with preexisting T-cell responses/tested individuals) of AML- and AML/LSC-associated peptides in PBMC samples of patients with AML and HVs as assessed by IFNγ ELISpot assay after 12-day in vitro expansion. Patients with AML and HVs were judged to possess preexisting immune responses when a positive T-cell response against at least one of the AML- and AML/LSC-associated peptides were observed in IFNγ ELISpot assays after peptide-specific 12-day in vitro expansion. T-cell responses were considered positive when >10 spots/500,000 cells were counted and the mean spot counts was at least three-fold higher than the negative control. E–G, scRNAseq analysis of CD4+ T cells of HLA class II AML/LSC peptide-stimulated and IFNγ secretion–based sorted PBMCs of patients with AML (n = 3) after 12-day in vitro expansion. E, Uniform Manifold Approximation and Projection (UMAP) plot showing the different subclusters of CD4+ T cells. Colors represent cell type classification. F, UMAP plot depicting the Boolean discretization of IFNγ+ CD4+ T cells. G, UMAP plot showing the TCR clonality of sequenced CD4+ T cells. Color code indicates the number of clonotypes per cell. H, Impact of HLA class II–restricted immunopeptidome diversity in terms of unique AML-exclusive HLA class II–restricted peptide presentation on overall survival (OS) of patients with AML (n = 25 total, n = 19 high, n = 6 low). Immunopeptidome diversity was classified as low and high according to the median peptide yields in the AML immunopeptidome cohort. I, Retrospective correlation analysis revealing the impact of preexisting antigen-specific T-cell responses as assessed by IFNγ ELISpot assay after 12-day in vitro expansion against HLA class II–restricted AML/LSC-associated peptides on OS (left) and event-free survival (EFS, right) of patients with AML (n = 56 total, n = 11 responder, n = 45 nonresponder). Patients with AML were classified as responders when they showed a positive T-cell response against one or multiple of the HLA class II–restricted AML/LSC-associated peptides in IFNγ ELISpot assays after peptide-specific 12-day in vitro expansion. T-cell responses were considered positive when >10 spots/500,000 cells were counted and the mean spot counts was at least three-fold higher than the negative control. Kaplan–Meier analysis, log-rank test. Abbreviations: CI, confidence interval; neg., negative peptide; OS, overall survival; EFS, event-free survival.
Overall, 20% (15/76) of patients with AML and 16% (14/89) of HVs showed preexisting immune responses targeting the AML and AML/LSC antigens (Fig. 6D).
HLA Class II AML/LSC-Specific T-cell Responses Are Mediated by Clonally Expanded Th1 Memory CD4+ T Cells
We further characterized T-cell responses to HLA class II AML/LSC peptides by single-cell RNA sequencing (scRNA-seq) in combination with T-cell receptor (TCR) V(D)J sequencing (TCR-seq) analysis of peptide-stimulated and IFNγ secretion-based sorted CD4+ T cells. IFNγ+ antigen-specific CD4+ T cells showed an activated and cytotoxic effector memory phenotype with expression of Th1-specific TNF and TBX21 (encoding T-bet), the cytotoxicity markers GZMB (encoding granzyme B), and PRF1 (encoding Perforin-1), paired with an absence of the naïve or central memory markers SELL (encoding CD62L) and CCR7, the exhaustion marker TOX, and the Th2 cytokines IL4 and IL13 (Fig. 6E and F; Supplementary Fig. S12A and S12B). Single-cell TCR sequencing showed a high clonality of the IFNγ+ antigen-specific activated and cytotoxic CD4+ T cells (Fig. 6G; Supplementary Fig. S12C). A comparable phenotype of AML/LSC peptide-stimulated CD4+ T cells was shown in an ex vivo performed multicolor flow cytometry assay with a Th1-directed CD45RO+CD62L− effector memory T-cell population (Supplementary Fig. S12D and S12E).
Presentation and Immune Recognition of AML and AML/LSC Antigens Associates with Improved Survival of Patients with AML
Finally, the association of immunopeptidome-defined antigen presentation and corresponding peptide-specific immune recognition with clinical characteristics, disease control, and outcome of patients with AML was investigated. Presentation of AML-exclusive antigens, in terms of uniquely presented peptides per AML sample, that is, the diversity of the immunopeptidome, did not differ significantly according to demographics and AML disease characteristics including age, sex, ELN risk classification, karyotype, FLT3-ITD, and NPM1 mutation (AML immunopeptidome cohort; Supplementary Table S11), neither did these variables themselves affect patient survival (Supplementary Table S12). Whereas the diversity of the HLA class I AML-exclusive immunopeptidome did not show an impact on event-free (EFS) and overall (OS) survival in a retrospective analysis of the AML immunopeptidome cohort (Supplementary Fig. S13A; Supplementary Table S13), diverse presentation of HLA class II–restricted AML-exclusive antigens was associated with significantly longer OS (Fig. 6H; Supplementary Fig. S13B) suggesting a central role of these antigens for immune control in AML. This is further underscored by a correlation of preexisting memory T-cell responses with patient survival revealing improved EFS in a retrospective analysis for patients with AML showing spontaneous memory CD4+ T-cell responses assessed by IFNγ ELISpot assays after 12-day in vitro expansion targeting the HLA class II–restricted AML/LSC-associated antigens as compared with patients without AML/LSC-specific immune responses (Fig. 6I; Supplementary Table S14). Demographics such as sex and age did not affect patient survival (Supplementary Table S15). Within the patient group, OS was better in patients that underwent hematopoietic stem cell transplantation (Supplementary Table S15). A trend for improved EFS could also be observed in a correlation of preexisting memory T-cell responses with patient survival for AML patients after allogeneic stem cell transplantation (Supplementary Table S14) showing HLA class II–restricted AML/LSC-associated antigen-specific T-cell responses in ex vivo IFNγ ELISpot assays (Supplementary Fig. S13C).
DISCUSSION
In this study, we demonstrate that primary AML progenitor cells present HLA-restricted cancer antigens that induce T-cell responses and presumably mediate immune surveillance in human AML. These antigens comprise cancer-associated cryptic neoepitopes as well as unmutated self-antigens. Of note, neoantigens derived from cancer-specific mutations, described as main specificities of T-cell responses induced by immune checkpoint inhibition in solid tumors with high mutational burden (31, 32), were only identified for single AML-specific mutations, limited to specific HLA allotypes and only identified on AML bulk cells but not specifically on LSCs. The sensitivity of shotgun mass spectrometric discovery approaches, even in the context of immense technical improvements in the last decades, is limited as the immunopeptidome is a highly dynamic, rich, and complex assembly of peptides. Therefore, low-level presentation of these neoepitopes on LSCs cannot be excluded. The low frequent detection of neoepitopes on AML bulk cells is in line with several reports that show a distorted correlation of mRNA expression and limited or even lacking immunopeptidome presentation for mutated and unmutated tumor antigens (13, 33–35). This further highlights the immunopeptidome as independent complex layer formed by the antigen presentation machinery that does not necessarily mirror the transcriptome or proteome calling for direct and unbiased methods of HLA-restricted antigen identification as realized by our mass spectrometry–based immunopeptidomics approach. However, it is a well-known issue that immunoprecipitation-based HLA peptide isolation cannot distinguish between HLA-restricted peptides presented on the cell surface from intracellular HLA:peptide complexes that might never reach the cell surface and are thus not suitable candidates for immunotherapy approaches (36, 37). Therefore, potential tumor antigen candidates have to be validated and their cell surface presentation have to be proven prior to their clinical application. The prove of peptide immunogenicity, in particular, the detection of preexisting memory T-cell responses in patients, which we were able to detect for the majority of the analyzed AML/LSC-associated antigens, validate the surface presentation, and T-cell activation of the respective antigens.
Besides mutation-derived neoepitopes, noncanonical cryptic peptides were recently suggested as potential tumor antigens (15–19). Such peptides arise through rapid degradation (38) of noncoding translation products from novel open-reading frames (ORF), 5′ and 3′ UTRs, ncRNA, intronic and intergenic regions, or shifted reading frames in annotated protein-coding regions (off-frame; refs. 18–20, 39–42). Recent large-scale pan-cancer screens (15–17) revealed the average proportion of noncanonical peptides within the immunopeptidome to roughly 1% to 3%. Interestingly, some noncanonical peptides were confirmed to be shared across different individuals as well as tumor entities, whereas others are tumor-specific. We identified 623 AML-associated noncanonical cryptic peptides accounting to 0.9% of the total AML-derived immunopeptidome. The main sources of noncanonical peptides in our study were 5′ UTR and off-frames, which is in line with other studies (16, 17, 20). 5′ UTRs, translated through non-AUG start codons in upstream ORFs (42), contributed to 41% of noncanonical peptides. Exonic regions translated in alternative frames were the source for 35% of noncanonical peptides, which might arise from initiation codon readthrough (43), novel ORFs (40), or ribosomal slippage (39) during translation. Immune responses targeting cryptic neoepitopes were reported infrequent. Chong and colleagues screened more than 500 noncanonical peptides; however, immune recognition was only detected for a single peptide (15). In line, we could not detect any preexisting T-cell responses in patients with AML or HVs targeting cryptic peptides.
AML arises from molecular alterations at the HSPC level. Alike in healthy hematopoiesis, LSCs are the origin of clonal growth and therefore considered responsible for maintenance of the leukemic population (44, 45). Given the central role of LSCs in development and pathogenesis of leukemic disease (1, 2), therapeutic elimination of this population is central to prevent relapses. However, the biological similarity between LSCs and normal HSPCs as well as the specific biological properties of LSCs hamper the development of therapeutic strategies for their effective destruction (46). With regard to antigen presentation, we observed no significant differences in HLA surface expression between HV-derived HSPCs and patient-derived LSCs and bulk AML cells, in particular no loss or significant downregulation previously postulated as mechanism by which AML cells escape immune surveillance (47, 48). HLA ligands presented by LSCs and AML bulk cells showed similar features in terms of physiochemical properties. Comparative immunopeptidome profiling delineated a distinct population of LSC-exclusive antigens not presented on benign HSPCs. Within the entirety of AML-exclusive antigens, a relevant proportion of peptides were shared between LSCs and bulk AML cells.
Beyond CD8+ T cells, CD4+ T cells also play a central role in the development and maintenance of effective antitumor immunity (49). Previous reports showed that spontaneous and vaccine-induced tumor-specific T-cell responses are predominantly mediated by CD4+ T cells (24, 50–53). Furthermore, HLA class II–restricted tumor antigen presentation and antigen-specific CD4+ T-cell responses correlate with clinical outcome of patients with cancer (54, 55). HSPCs present MHC class II–restricted antigens that interact with antigen-specific CD4+ T cells in mice (10). This immune surveillance mechanism effectively eliminates transformed progenitor cells, thereby preventing leukemia onset. We detected highly frequent spontaneous memory T cells targeting HLA class II–restricted antigens in patients with AML and HVs and showed that this antigen-specific T-cell recognition and HLA class II immunopeptidome diversity mediate immune surveillance and impact clinical outcome in patients with AML. This observation, which has to be validated in future prospective larger cohort studies, provides first evidence for the impact of immunopeptidome diversity and peptide-specific T-cell responses for patient survival and elucidates the pathophysiologic role of HLA class II antigen presentation and underscores its relevance for immune control of malignant disease (47, 56). The latter is based on (i) activation of CD4+ T cells that mediate direct and indirect effector functions in anticancer immunity, (ii) additional direct activation of CD8+ T cells by embedded HLA class I T-cell epitopes, and (iii) the promiscuous binding to multiple different HLA allotypes, which qualifies these AML/LSC antigens as highly promising novel targets for broadly applicable immunotherapeutic approaches like vaccines, adoptive T-cell transfer, or TCR-engineered therapies.
Together, our results unraveled the immunopeptidomic landscape of LSCs and delineated AML/LSC-associated antigens that mediate immune surveillance in AML and may facilitate the development of novel LSC-directed immunotherapeutic approaches for patients with AML. We are currently preparing a multicentric, open-label, phase I clinical study that will address two central issues of former peptide-based vaccination trials (57–60) by selection of a personalized multipeptide vaccine from a peptide warehouse comprising our naturally presented AML/LSC-associated antigens and adjuvating the vaccine with the novel TLR1/2 agonist XS15 (61) that enables the induction of superior, long-lasting T-cell responses and thus will provide an in vivo evaluation of the AML/LSC antigens directly in humans.
METHODS
Patients and Blood Samples
For immunopeptidome analysis, PBMCs or bone marrow mononuclear cells from patients with AML (Supplementary Table S3) at the time of diagnosis, at relapse (n = 52), or in molecular remission (n = 8) as well as hematopoietic stem cell apheresis from G-CSF–mobilized blood donations (n = 8) of HVs (n = 1) and patients with nonhematologic malignancies (n = 7) were collected at the Departments of Hematology and Oncology in Tübingen and Dresden, Germany as well as at the Department of Medicine, Divisions of Hematology and Medical Oncology at the San Francisco University of California. For T cell—based assays, PBMCs from HVs (Supplementary Table S10, n = 92) and patients with AML (Supplementary Table S9, n = 78) after allogenic stem cell transplantation or in complete remission at different time points after standard treatment were collected. Cells were isolated by density gradient centrifugation and stored at −80°C. Clinical and survival data were collected within a follow-up phase of up to 48 months after the date of diagnosis. Written informed consent was obtained in accordance with the Declaration of Helsinki protocol. The study was performed according to the guidelines of the local ethics committees (373/2011B02, 454/2016B02, EK 20805217). HLA typing was carried out by the Department of Hematology and Oncology, Tübingen, Germany. Patient and HV demographic and clinical characteristics are provided in Supplementary Tables S3, S9, and S10.
HLA Surface Quantification
HLA surface expression was determined using the QIFIKIT bead-based quantification flow cytometric assay (Dako, catalog no. K007811–8) according to the manufacturer's instructions. In brief, cells were stained either with the pan-HLA class I-specific W6/32 mAb, the HLA-DR–specific L243 mAb (produced in-house), or IgG isotype control (BioLegend, catalog no. 400202, RRID: AB_2927399, clone MOPC-173), respectively. Polyclonal goat FITC anti-mouse antibody (Dako, catalog no. F047902, RRID: AB_578665) was used as a secondary antibody. After washing with normal mouse serum (eBioscience, catalog no. 24–5544–94) surface marker staining was performed using PE/Cy7 anti-human CD38 (BioLegend, catalog no. 356608, RRID: AB_2561903, clone HB-7), APC anti-human CD34 (BD Biosciences, catalog no. 555824, RRID: AB_398614, clone 581), and Pacific Blue anti-human CD45 (BD Biosciences, catalog no. 642275, RRID: AB_1645755, clone 2D1) antibodies. Aqua fluorescent reactive dye (Invitrogen, catalog no. L34957) was used as viability marker. Analyses were performed on a FACS Canto II cytometer (BD Biosciences). Only cell populations with ≥100 cells were analyzed for their HLA surface expression.
LSC Enrichment
Enrichment of LSCs from AML samples were either performed by fluorescence-activated cell sorting (FACS) at the Institute for Stem Cell Biology and Regenerative Medicine, Stanford, CA (UPN3–8, UPN11), or by magnetic-activated cell sorting (MACS) at the Institute for Cell Biology, Department of Immunology, University of Tübingen, Tübingen, Germany (UPN01, UPN02, UPN09, UPN10). For FACS, PBMCs were stained with APC anti-human CD34 (BD Biosciences, catalog no. 340441, RRID: AB_400514, clone 8G12), PE/Cy7 anti-human CD38, and PerCP/Cy5.5 anti-human CD3 (BioLegend, catalog no. 300328, RRID: AB_1575008, clone HIT3a), CD19 (BioLegend, catalog no. 302229, RRID: AB_2275547, clone HIB19), CD20 (BioLegend, catalog no. 302325, RRID: AB_893285, clone 2H7), and CD56 (BioLegend, catalog no. 318321, RRID: AB_893391, clone HCD56) mAbs and sorted on a FACSAria II or FACSAria III (BD Biosciences). MACS was performed with the human CD34 MultiSort (Miltenyi Biotec, catalog no. 130–056–701) and CD38 MicroBead Kits (Miltenyi Biotec, catalog no. 130–092–263). Sorted cells were stained with PE/Cy7 anti-human CD38, APC anti-human CD34, and Pacific Blue anti-human CD45 mAbs to determine the purity. Aqua fluorescent reactive dye was used as viability marker. Analyses were performed on a FACSCanto II cytometer. CD34+ HSPCs were magnetically enriched (CD34 MicroBead Kit, Miltenyi Biotec, catalog no. 130–046–702) from hematopoietic stem cell apheresis from G-CSF–mobilized blood donations of HVs and patients with nonhematologic malignancies.
Mice and Xenotransplantation Assays
Xenotransplantation assays were performed at the Department of Biomedicine, University of Basel and University Hospital Basel, Switzerland. NOD.Cg-Prkdcscid IL2rgtmWjl/Sz mice (NSG, The Jackson Laboratory, strain # 005557) were maintained under pathogen-free conditions according to the Swiss federal and state regulations. All animal experiments were approved by the Veterinäramt Basel-Stadt (24981). Xenotransplantation assays were performed as previously described (62). In brief, 6 × 105 primary human sorted CD34+CD38− LSCs were transplanted via intrafemoral injection into 8-week-old female NSG mice (n = 4). Engraftment was monitored as previously described (62) via routine bone marrow punctures or assessment of peripheral blood. Engraftment was defined as ≥1% human leukemic cells in murine peripheral blood or bone marrow as analyzed by multicolor flow cytometry using antibodies against human leukemic antigens. The panel includes fluorescent antibodies against human CD33 (BD Biosciences, catalog no. 555450, RRID: AB_395843, clone WM53), CD34 (BD Biosciences, catalog no. 340441, RRID: AB_400514, clone 8G12), CD133 (BD Biosciences, catalog no. 566595, RRID: AB_2739755, clone 293C3), CD117 (BD Biosciences, catalog no. 339195, RRID: AB_647418), CD45 (BD Biosciences, catalog no. 561865, RRID: AB_10896120, clone HI30), CD14 (eBiosciences, catalog no. 17–0149–42, RRID: AB_10669167, clone 61D3), CD13 (eBiosciences, catalog no. 12–0138–42, RRID: AB_10853031, clone WM15), CD3 (BioLegend, catalog no. 317318, RRID: AB_1937212, clone OKT3), and CD19 (BioLegend, catalog no. 302208, RRID: AB_314238, clone HIB19). All mice underwent final bone marrow, peripheral blood, and organ assessment by multicolor flow cytometry.
Isolation of HLA Ligands
HLA class I and HLA class II molecules were isolated from snap-frozen cell pellets by standard immunoaffinity purification (63) using the pan-HLA class I–specific W6/32, the pan-HLA class II–specific Tü-39, and the HLA-DR–specific L243 mAbs (produced in-house) cross-linked to CNBr-activated Sepharose (Sigma-Aldrich) to extract HLA ligands. Cells were lysed in lysis buffer [CHAPS (Panreac AppliChem), cOmplete protease inhibitor cocktail tablet (Roche) in PBS] for 1 hour on a shaker at 4°C, sonicated, and centrifuged (45 minutes, 4,000 rpm) and incubated again for 1 hour. Lysates were cleared by sterile filtration (5-μm filter unit; Merck Millipore) and cyclically passed through a column-based setup overnight at 4°C. Columns were washed with PBS (30 minutes) and ddH2O (1 hour). Peptides were eluted by 0.2% trifluoroacetic acid (TFA), isolated by ultrafiltration (Amicon filter units; Merck Millipore), lyophilized, and desalted using ZipTip pipette tips with C18 resin (Merck).
Mass Spectrometric Data Acquisition
For the mass spectrometric analysis (64), peptides were loaded on a 75 μm × 2 cm PepMap Nanotrap Column (Thermo Fisher Scientific) at a flow rate of 4 μL/minute for 10 minutes. Subsequent separation was performed by nanoflow high-performance liquid chromatography (RSLCnano, Thermo Fisher Scientific) using a 50 μm × 25 cm PepMap rapid separation column (Thermo Fisher Scientific, particle size of 2 μm) and a linear gradient ranging from 2.4% to 32.0% acetonitrile at an flow rate of 0.3 μL/minute over the course of 90 minutes. Eluting peptides were analyzed in technical replicates in an online-coupled Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectron spray ion source using a data dependent acquisition mode using a top speed collisional-induced dissociation (CID, normalized collision energy 35%, HLA class I peptides) or higher-energy collisional dissociation (HCD, normalized collision energy 30%, HLA class II peptides) fragmentation method. MS1 and MS2 spectra were detected in the Orbitrap with a resolution of 120,000 and 30,000, respectively. The maximum injection time was set to 50 ms and 150 ms for MS1 and MS2, respectively. The dynamic exclusion was set to 7 and 10 seconds for HLA class I and HLA class II, respectively. Mass range for HLA class I peptide analysis was set to 400–650 m/z with charge states 2+ and 3+ selected for fragmentation. For HLA class II peptide analysis, mass range was limited to 400–1,000 m/z with charge states 2+ to 5+ selected for fragmentation.
Data Processing
Data processing was performed as described previously (64). In brief, the SEQUEST HT search engine (University of Washington, Seattle, WA; ref. 65) was used to search the human proteome as comprised in the Swiss-Prot database (20,279 reviewed protein sequences, September 27, 2013) without enzymatic restriction. Precursor mass tolerance was set to 5 ppm, and fragment mass tolerance to 0.02 Da. Oxidized methionine was allowed as a dynamic modification. The FDR was estimated using the Percolator algorithm (66) and limited to 5% for HLA class I and 1% for HLA class II. Peptide lengths were limited to 8 to 12 amino acids for HLA class I and to 8 to 25 amino acids for HLA class II. Protein inference was disabled, allowing for multiple protein annotations of peptides. HLA class I annotation was performed using NetMHCpan 4.0 (67, 68) and SYFPEITHI (69) annotating peptides with percentile rank below 2% and ≥60% of the maximal score, respectively. Comparative profiling approaches, that is, overlap analysis and frequency-based comparisons of AML- and benign-derived immunopeptidomes, were performed with curated AML and LSC immunopeptidome data excluding peptides that are presented on only one single sample with PSM counts ≤3 (“one hit wonders”).
Screening for Neoepitopes
For neoepitope screening, we used a non-patient–individual mutFASTA, which includes the TOP100 recurrent AML-associated missense mutations specified in the COSMIC database (https://cancer.sanger.ac.uk/cosmic; ref. 70) supplemented with the most common NPM1 frame-shift mutations (type A, B, C, D, and E; ref. 71) as well as FLT3-ITD (72) and FLT3-TKD (73–77) mutations (Supplementary Table S8). Data processing of AML immunopeptidomics data with the mutFASTA were performed as described above. To minimize false positive identifications, more stringent filter criteria with XCorr ≥1 and ΔCn ≥0.2 were applied. After manual spectrum validation, candidate neoepitopes were produced as isotope-labeled synthetic peptides and used for spectral comparison and validation.
Peptide Synthesis and Spectrum Validation
Peptides were produced by the peptide synthesizer Liberty Blue (CEM) using the 9-fluorenylmethyl-oxycarbonyl/tert-butyl strategy (78). Spectrum validation of the experimentally eluted peptides was performed by computing the similarity of the spectra with corresponding isotope-labeled synthetic peptides measured in a complex matrix. The spectral correlation was calculated between the MS/MS spectra of the eluted and the synthetic peptide (79).
Identification of Cryptic Peptides
Cryptic HLA class I peptides were identified using Peptide-PRISM as described recently (30). De novo peptide sequencing was performed with PEAKS X (Bioinformatics Solutions Inc; ref. 80). Raw data refinement was performed with the following settings: (i) merge options: no merge; (ii) precursor options: corrected; (iii) charge options: no correction; (iv) filter options: no filter; (v) process: true; (vi) default: true; and (vii) associate chimera: yes. De novo sequencing was performed with parent mass error tolerance set to 10 ppm. Fragment mass error tolerance was set to 0.15 Da, and enzyme was set to none. The following variable modifications have been used: oxidation (M), pyro-Glu from Q (N-term Q), and carbamidomethylation (C). A maximum of three variable posttranslational modifications were allowed per peptide. Up to 10 de novo sequencing candidates were reported for each identified fragment ion mass spectrum, with their corresponding average local confidence score. Because we applied the chimeric spectra option of PEAKS X, two or more TOP10 candidate lists could be assigned to a single fragment ion spectrum. Two tables (“all de novo candidates” and “de novo peptides”) were exported from PEAKS for further analysis. All de novo sequence candidates were matched against the six-frame translated human genome (hg38) and the three-frame translated human transcriptome (ENSEMBL 90) using Peptide-PRISM. Results were filtered to 10% FDR for each category (CDS, UTR5, OffFrame, ncRNA, UTR3, intronic, and intergenic). NetMHCpan 4.0 was used to predict binding affinities for all identified HLA class I peptides for all HLA alleles of the corresponding sample. Shown AML-associated cryptic peptides are AML-exclusive peptides never identified on any benign tissue sample.
Amplification of Peptide-specific T Cells and IFNγ ELISpot Assay
PBMCs from patients with AML and HVs were pulsed with 1 μg/mL (HLA class I) or 5 μg/mL (HLA class II) per peptide and cultured for 12 days adding 20 U/mL IL2 (Novartis) on days 3, 5, and 7 (64, 81). Peptide-stimulated PBMCs were analyzed by ELISpot assay on day 12 (82). Spots were counted using an ImmunoSpot S5 analyzer (CTL) and T-cell responses were considered positive when >10 spots/500,000 cells were counted and the mean spot count was at least three-fold higher than the mean spot count of the negative control. The intensity of T-cell responses is depicted as calculated spot counts, which were calculated as the mean spot count of duplicates normalized to 5 × 105 cells minus the normalized mean spot count of the respective negative control.
Refolding
Biotinylated HLA–peptide complexes were manufactured as described previously (83) and tetramerized using PE-conjugated streptavidin (Invitrogen) at a 4:1 molar ratio.
Induction of Peptide-specific CD8+ T Cells with aAPCs
Priming of peptide-specific CTLs was conducted using aAPCs as described before (23, 84). In detail, 800,000 streptavidin-coated microspheres (5.6-μm diameter, Bangs Laboratories) were loaded with 200 ng biotinylated peptide–HLA complexes and 600 ng biotinylated anti-human CD28 antibody (clone 9.3, in-house production). MACS-sorted CD8+ T cells (CD8 Microbeads, Miltenyi Biotec, catalog no. 130–045–201) were cultured with 4.8 U/μL IL2 (R+D) and 1.25 ng/mL IL7 (PromoKine). Weekly stimulation with aAPCs (200,000 aAPCs per 1 × 106 CD8+ T cells) and 5 ng/mL IL12 (PromoKine) was performed four times. Induction of peptide-specific T cells was analyzed by tetramer staining.
Cytokine and Tetramer Staining
The frequency and functionality of peptide-specific CD8+ T cells was analyzed by tetramer and ICS as described previously (82, 85). For ICS, cells were pulsed with 10 μg/mL of individual peptide and incubated with 10 μg/mL Brefeldin A (Sigma-Aldrich) and 10 μg/mL GolgiStop (BD Biosciences) for 12-16 hours. Staining was performed using Cytofix/Cytoperm (BD Biosciences), PE/Cy7 anti-human CD8 (BioLegend, catalog no. 344711, RRID: AB_2044007, clone SK1), APC/Cy7 anti-human CD4 (BioLegend, catalog no. 300518, RRID: AB_314086, clone RPA-T4), Pacific Blue anti-human TNF (BioLegend, catalog no. 502920, RRID: AB_528965, clone MAb11), FITC anti-human CD107a (BioLegend, catalog no. 328606, RRID: AB_1186036, clone H4A3), and PE anti-human IFNγ (BioLegend, catalog no. 506507, RRID: AB_315440, clone B27) mAbs. Aqua fluorescent reactive dye (Invitrogen, catalog no. L34957) was used as viability marker. PMA and ionomycin (Sigma-Aldrich) served as positive control. The following peptides were used as negative control peptides: GSEELRSLY, POL_HV1BR, HLA-A*01; YLLPAIVHI, DDX5_HUMAN, HLA-A*02; RLRPGGKKK, GAG_HV1BR, HLA-A*03; TPGPGVRYPL, NEF_HV1BR, HLA-B*07; DIAARNVL, FAK1_HUMAN, HLA-B*08; ASEDYVAPPK, MKX_HUMAN, HLA-A*11; ETVITVDTKAAGKGK, FLNA_HUMAN, HLA class II. The frequency of peptide-specific CD8+ T cells after aAPC-based priming was determined by PE/Cy7 anti-human CD8 mAb and HLA:peptide tetramer-PE staining. Aqua fluorescent reactive dye (Invitrogen, catalog no. L34957) was used as viability marker. Cells of the same donor primed with an HLA-matched control peptide were used as negative control. The priming was considered successful if the frequency of peptide-specific CD8+ T cells was >0.1% of CD8+ T cells within the viable single cell population and at least three-fold higher than the frequency of peptide-specific CD8+ T cells in the negative control. The frequency of tetramer+, IFNγ+, TNF+, and CD107a+ T cells is depicted as calculated frequency, which is the frequency in the test well minus the frequency of the respective negative control. The same evaluation criteria were applied for ICS results. Samples were analyzed on a FACSCanto II cytometer.
Cytotoxicity Assay
Cytolytic capacity of peptide-specific CD8+ T cells was analyzed using the flow cytometry-based VITAL assay as described before (86, 87). Autologous CD8-depleted PBMCs were loaded with the test peptide or an HLA-matched control peptide and labeled with CFSE (Invitrogen) or FarRed (Invitrogen), respectively. Effector cells were added in the indicated effector-to-target ratios. Specific lysis of peptide-loaded target cells was calculated relative to control targets.
In-depth Phenotyping by Multicolor Flow Cytometry
For multicolor flow cytometry–based phenotyping of peptide-specific CD4+ T-cell responses, cells were stimulated with the pool of HLA class II–restricted AML/LSC-associated peptides (10 μg/mL of each peptide) and incubated with 10 μg/mL Brefeldin A and 10 μg/mL GolgiStop for 12-16 hours. Staining was performed using Cytofix/Cytoperm, PE/Cy7 anti-human CD8 (BioLegend, catalog no. 344711, RRID: AB_2044007, clone SK1), APC/Cy7 anti-human CD4 (BioLegend, catalog no. 300518, RRID: AB_314086, clone RPA-T4), Pacific Blue anti-human TNF (BioLegend, catalog no. 502920, RRID: AB_528965, clone MAb11), PE anti-human IFNγ (BioLegend, catalog no. 506507, RRID: AB_315440, clone B27), APC anti-human CD45RO (BioLegend, catalog no. 304210, RRID: AB_314426), PE-Dazzle 594 anti-human IL4 (BioLegend, catalog no. 500832, RRID: AB_2564036), and Brilliant Violet 650 anti-human CD62 L (BioLegend, catalog no. 304831, RRID: AB_2561461) mAbs. Zombie Aqua (BioLegend, catalog no. 423101) was used as viability marker. PMA and ionomycin served as positive control. The peptide ETVITVDTKAAGKGK (FLNA_HUMAN) was used as negative control. Samples were analyzed on a LSR Fortessa cytometer (BD Biosciences).
Single-Cell Immune Profiling
In vitro amplified and HLA class II peptide pool-stimulated memory T cells of patients with AML were enriched by the IFNγ Secretion Assay – Cell Enrichment and Detection Kit (PE) (Miltenyi Biotec, catalog no. 130–054–201) and prepared according to the 10 × Genomics cell preparation protocol. Single cells were partitioned into Gel Beads-in-Emulsion (GEMs) together with 10 × barcoded Gel Beads and reverse transcriptase enzymatic reaction using the Chromium X instrument (10X Genomics). Single-cell gene expression libraries and single-cell TCR (VDJ) libraries were then prepared using the Chromium Next GEM Single Cell 5′Kit v2 (10X Genomics, catalog no. PN-1000263), the Library Construction Kit (10X Genomics, catalog no. PN-1000190), and the Chromium Single Cell Human TCR Amplification Kit (10X Genomics, catalog no. PN-1000252) according to the manufacturer's instructions. Libraries were pooled and sequenced on a NOVASEQ 6000 (Illumina) at 37,773, 80,897, and 46,925 mean reads per cell, respectively. Samples were demultiplexed using bcl-convert version 3.9.3 (Illumina). Barcode processing, alignment, VDJ annotation, and single-cell 5′gene counting were performed using Cell Ranger Software version 7.1.0 (10X Genomics). Further data processing, visualization, and analysis were performed using scanpy version 1.9.1 and scirpy version 0.12.2 (88, 89) for all samples combined. Cells with unique gene counts <500 and without VDJ sequence associated, as well as cells with >20% of mitochondrial genes, were removed from the analysis. Data was log-normalized to a size factor of 10,000. Only highly variable genes were considered for dimensional reduction. Effect of total counts was regressed out and counts were scaled to unit variance and zero mean for each gene. The dimensionality reduction was done using principal component analysis (PCA). The neighborhood graph and UMAP embedding were computed using the UMAP algorithm (90) for 30 neighbors and the first 60 principal components (n_neighbors = 30, n_PC = 60). Unsupervised clustering was performed using the Leiden algorithm (91) resulting in 15 clusters with 14,697 cells in total. CD3+ clusters were selected for downstream analysis yielding 9 clusters with 3,058 cells (UPN114), 1,826 cells (UPN120) and 5,144 cells (UPN82), respectively. CD4 expressing clusters with 1,676 cells (UPN114), 616 cells (UPN120), 3,814 cells (UPN82) were then extracted from the previous clustering for further downstream analysis.
Software and Statistical Analysis
Overlap analysis was performed using BioVenn (92). The population coverage of HLA allotypes was calculated by the IEDB population coverage tool (www.iedb.org; ref. 28). For saturation analysis, the mean number of unique source proteins for a given cohort size (number of samples) has been calculated by 1,000 random samplings from the entirety of AML immunopeptidomes, that is, the number of source proteins of randomly picked samples was summed up for each cohort size and this process was repeated 1,000 times before calculating the average. An in-house Python script was used for the calculation of FDRs of AML-associated peptides at different presentation frequencies (64). Hotspot analysis (hotspot length ≥8 amino acids) of HLA class II immunopeptidomes was performed using an in-house R script that maps identified peptides according to their sequence onto its source protein and calculates representation frequencies of single amino acid positions within the respective cohorts. Flow cytometric data was analyzed using FlowJo 10.0.8 (Treestar). For survival analysis investigating the impact of the immunopeptidome diversity, peptide yields of AML-exclusive peptides were normalized to the cell number applied for immunopeptidome analysis. OS and EFS were depicted for low and high immunopeptidome diversity according to the median peptide yields in the AML immunopeptidome cohort and calculated by Kaplan–Meier method. The log-rank test was performed to test the difference of survival between the groups. For survival analysis investigating the impact of preexisting antigen-specific immune responses against HLA class II-restricted AML- and LSC-associated peptides as detected by IFNγ ELISpot assays patients were dichotomized into the group of responders showing a peptide-specific T-cell response and nonresponders without any detectable peptide-specific T-cell response. All figures and statistical analyses were generated using GraphPad Prism 9.4.1 (GraphPad Software). Data are displayed as mean with SD, box plots as median with 25th or 75th quantiles and min/max whiskers. Continuous data were tested for distribution and individual groups were tested by use of two-sided χ2 test, unpaired t test, unpaired Mann–Whitney U test, Kruskal–Wallis test, or paired Wilcoxon signed rank test, all performed as two-sided tests. If applicable adjustment for multiple testing was done. P values of <0.05 were considered statistically significant.
Data Availability
The mass spectrometry immunopeptidomics data generated in this study has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE (93) partner repository with the dataset identifier PXD038691. The mass spectrometry immunopeptidomics raw data (.raw files) can be viewed by the Thermo Xcalibur Qual Browser or other .raw file viewers. The processed search engine output (.msf files) can be viewed by the Proteome Discoverer software. The scRNA-seq data generated in this study have been deposited in the NCBI's Gene Expression Omnibus database with the dataset identifier GSE235080.
Supplementary Material
The Supplementary Tables file includes Tables S1 to S15. Table S1: Frequencies of CD34+CD38- LSCs within primary AML patient samples. Table S2: Immunopeptidomics cohort overview. Table S3: Patient characteristics of AML immunopeptidomics cohort. Table S4: AML- and AML/LSC-associated HLA class I targets. Table S5: AML/LSC shared HLA class I-presented antigens. Table S6: AML- and AML/LSC-associated HLA class II peptide, protein, and hotspot targets. Table S7: AML/LSC shared HLA class II-presented peptide targets. Table S8: Recurrent AML mutations included in neoepitope screening. Table S9: Characteristics of AML patient samples used for T cell-based assays. Table S10: Characteristics of HV samples used for T cell-based assays. Table S11: Immunopeptidome diversity according to demographics and disease characteristics. Table S12: Impact of demographics and disease characteristics on patient outcome in the immunopeptidome survival analysis group. Table S13: Patient characteristics of immunopeptidome survival analysis group. Table S14: Patient characteristics of immune response survival analysis group. Table S15: Impact of demographics and hematopoietic stem cell transplantation on patient outcome in the immune response survival group.
The Supplementary Figures file includes Figures S1 to S13. Figure S1: Gating strategy for the evaluation of LSC frequencies pre and post enrichment. Figure S2: Amino acid distribution within LSC and AML bulk immunopeptidomes. Figure S3: HLA class I immunopeptidomics. Figure S4: Abundance of AML- and AML/LSC-associated HLA class I ligands within the immunopeptidome. Figure S5: Identification of AML- and AML/LSC-associated HLA class II antigen targets by comparative immunopeptidome profiling. Figure S6: Abundance of AML- and AML/LSC-associated HLA class II peptides within the immunopeptidome. Figure S7: Spectral validation of HLA class I-restricted AML- and AML/LSC-associated peptides. Figure S8: Spectral validation of HLA class II-restricted AML- and AML/LSC-associated peptides. Figure S9: Magnitude and functionality of in vitro primed CD8+ T cells. Figure S10: Characterization of memory T cell responses detected in ELISpot assays. Figure S11: Further characterization of HLA class II-restricted antigens. Figure S12: In-depth characterization and phenotyping of HLA class II AML/LSC-specific CD4+ T cells by single-cell RNA sequencing and multi-color flow cytometry. Figure S13: Impact of immunopeptidome diversity and peptide-specific immune responses on patient survival.
Acknowledgments
We thank Ulrike Schmidt, Claudia Falkenburger, Beate Pömmerl, and Ulrich Wulle for technical support. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, WA 4608/1–2, to J.S. Walz; CRC1479 Project ID 441891347-P03, to N. Köhler, CRC/TRR167-Project ID 259373024-Z01, to M. Boerries and B. Appiah; CRC 1160-Project ID 256073931-Z02, to M. Boerries; CRC1479 Project ID 441891347-S1, to M. Boerries), the Deutsche Forschungsgemeinschaft under Germany's Excellence Strategy (EXC2180-390900677, to J.S. Walz and H.-G. Rammensee and CIBSS – EXC 2189 project ID 390939984, to N. Kohler), the Federal Ministry of Education and Research (03ZU1111LB, to M. Schmitz; MIRACUM-FKZ 01ZZ1801B and PM4Onco-FKZ 01ZZ2322A, to M. Boerries), the German Cancer Consortium (DKTK, to C. Lengerke, H.-G. Rammensee, and J.S. Walz), the Ernst Jung Prize for Medicine (to H.-G. Rammensee), the Landesforschungspreis of Baden-Württemberg (to H.-G. Rammensee), the Wilhelm Sander Stiftung (2016.177.3, to J.S. Walz), the José Carreras Leukämie-Stiftung (DJCLS 05 R/2017, to J.S. Walz), the Deutsche Krebshilfe (German Cancer Aid, 70114948, to J.S. Walz), the Swiss National Science Foundation (310030_179239, to C. Lengerke), the European Research Council (HemStem Consolidator Grant, to C. Lengerke), the Else Kröner-Fresenius-Stiftung (2019_A74, to N. Kohler), and the Fortüne Program of the University of Tübingen (2451–0-0 and 2581–0-0, to J.S. Walz and M. Roerden).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
NoteSupplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
A. Nelde reports a patent for EP22206337.2 pending. H. Schuster reports personal fees from Immatics Biotechnologies GmbH outside the submitted work; and is a currently employed by Immatics. J.S. Heitmann reports other support from Synimmune GmbH outside the submitted work. R. Majeti reports personal fees and other support from Kodikaz Therapeutic Solutions, Orbital Therapeutics, and Pheast Therapeutics; personal fees from 858 Therapeutics; and other support from Myelogene outside the submitted work. H.-G. Rammensee reports grants from DKTK, EXC2180, Ernst-Jung-Preis, and grants from Landesforschungspreis Baden-Württemberg during the conduct of the study; in addition, H.-G. Rammensee has a patent for “AML associated peptides” pending. J.S. Walz reports grants from German Research Foundation, German Research Foundation under Germany's Excellence Strategy, German Cancer Consortium, Wilhelm Sander Stiftung, grants from José Carreras Leukämie-Stiftung, German Cancer Aid, and grants from Fortüne Program of the University of Tübingen during the conduct of the study; in addition, J.S. Walz has a patent for EP22206337.2 pending. No disclosures were reported by the other authors.
Authors’ Contributions
A. Nelde: Conceptualization, data curation, formal analysis, investigation, visualization, writing–original draft, project administration, writing–review and editing. H. Schuster: Conceptualization, formal analysis, investigation, writing–review and editing. J.S. Heitmann: Resources, data curation, formal analysis, writing–review and editing. J. Bauer: Investigation, writing–review and editing. Y. Maringer: Investigation, writing–review and editing. M. Zwick: Investigation, writing–review and editing. J.-P. Volkmer: Investigation, writing–review and editing. J.Y. Chen: Investigation, writing–review and editing. A.M. Paczulla Stanger: Formal analysis, investigation, writing–review and editing. A. Lehmann: Formal analysis, visualization, writing–review and editing. B. Appiah: Formal analysis, funding acquisition, writing–review and editing. M. Märklin: Investigation, writing–review and editing. E. Rücker-Braun: Resources, writing–review and editing. H.R. Salih: Resources, writing–review and editing. M. Roerden: Resources, data curation, funding acquisition, writing–review and editing. S.M. Schroeder: Resources, data curation, writing–review and editing. M.-F. Häring: Resources, writing–review and editing. A. Schlosser: Formal analysis, writing–review and editing. J. Schetelig: Resources, writing–review and editing. M. Schmitz: Resources, funding acquisition, writing–review and editing. M. Boerries: Formal analysis, funding acquisition, writing–review and editing. N. Köhler: Formal analysis, funding acquisition, investigation, writing–review and editing. C. Lengerke: Resources, funding acquisition, investigation, writing–review and editing. R. Majeti: Investigation, writing–review and editing. I.L. Weissman: Resources, investigation, writing–review and editing. H.-G. Rammensee: Formal analysis, funding acquisition, writing–review and editing. J.S. Walz: Conceptualization, resources, formal analysis, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 2017;547:104–8. [DOI] [PubMed] [Google Scholar]
- 2. Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, Van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011;17:1086–93. [DOI] [PubMed] [Google Scholar]
- 3. Kantarjian H, Kadia T, DiNardo C, Daver N, Borthakur G, Jabbour E, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J 2021;11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kolb HJ. Graft-versus-leukemia effects of transplantation and donor lymphocytes. Blood 2008;112:4371–83. [DOI] [PubMed] [Google Scholar]
- 5. Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 2014;371:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Von Stackelberg A, Locatelli F, Zugmaier G, Handgretinger R, Trippett TM, Rizzari C, et al. Phase I/Phase II study of blinatumomab in pediatric patients with relapsed/refractory acute lymphoblastic leukemia. J Clin Oncol 2016;34:4381–9. [DOI] [PubMed] [Google Scholar]
- 9. Daver N, Alotaibi AS, Bücklein V, Subklewe M. T-cell-based immunotherapy of acute myeloid leukemia: current concepts and future developments. Leukemia 2021;35:1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hernández-Malmierca P, Vonficht D, Schnell A, Uckelmann HJ, Bollhagen A, Mahmoud MAA, et al. Antigen presentation safeguards the integrity of the hematopoietic stem cell pool. Cell Stem Cell 2022;29:760–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu Y, Petroni GR, Olson WC, Czarkowski A, Smolkin ME, Grosh WW, et al. Immunologic hierarchy, class II MHC promiscuity, and epitope spreading of a melanoma helper peptide vaccine. Cancer Immunol Immunother 2014;63:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol 1989;19:2237–42. [DOI] [PubMed] [Google Scholar]
- 13. Bassani-Sternberg M, Bräunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 2016;7:13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bauer J, Köhler N, Maringer Y, Bucher P, Bilich T, Zwick M, et al. The oncogenic fusion protein DNAJB1-PRKACA can be specifically targeted by peptide-based immunotherapy in fibrolamellar hepatocellular carcinoma. Nat Commun 2022;13:6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chong C, Müller M, Pak HS, Harnett D, Huber F, Grun D, et al. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat Commun 2020;11:1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ouspenskaia T, Law T, Clauser KR, Klaeger S, Sarkizova S, Aguet F, et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat Biotechnol 2021;40:209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bedran G, Gasser H-C, Weke K, Wang T, Bedran D, Laird A, et al. The immunopeptidome from a genomic perspective: Establishing the non-canonical landscape of MHC class I-associated peptides. Cancer Immunol Res 2023;11:747–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nelde A, Flötotto L, Jürgens L, Szymik L, Hubert E, Bauer J, et al. Upstream open reading frames regulate translation of cancer-associated transcripts and encode HLA-presented immunogenic tumor antigens. Cell Mol Life Sci 2022;79:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jürgens L, Wethmar K. The emerging role of uORF-Encoded uPeptides and HLA uLigands in cellular and tumor biology. Cancers 2022;14:6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marcu A, Schlosser A, Keupp A, Trautwein N, Johann P, Wölfl M, et al. Natural and cryptic peptides dominate the immunopeptidome of atypical teratoid rhabdoid tumors. J Immunother Cancer 2021;9:e003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Teck AT, Urban S, Quass P, Nelde A, Schuster H, Letsch A, et al. Cancer testis antigen Cyclin A1 harbors several HLA-A*02:01-restricted T cell epitopes, which are presented and recognized in vivo. Cancer Immunol Immunother 2020;69:1217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walz S, Stickel JS, Kowalewski DJ, Schuster H, Weisel K, Backert L, et al. The antigenic landscape of multiple myeloma: mass spectrometry (re)defines targets for T-cell-based immunotherapy. Blood 2015;126:1203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schuster H, Peper JK, Bösmüller HC, Rohle K, Backert L, Bilich T, et al. The immunopeptidomic landscape of ovarian carcinomas. Proc Natl Acad Sci U S A 2017;114:E9942–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nelde A, Maringer Y, Bilich T, Salih HR, Roerden M, Heitmann JS, et al. Immunopeptidomics-guided warehouse design for peptide-based immunotherapy in chronic lymphocytic leukemia. Front Immunol 2021;12:705974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bilich T, Nelde A, Bichmann L, Roerden M, Salih HR, Kowalewski DJ, et al. The HLA ligandome landscape of chronic myeloid leukemia delineates novel T-cell epitopes for immunotherapy. Blood 2019;133:550–65. [DOI] [PubMed] [Google Scholar]
- 26. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–7. [DOI] [PubMed] [Google Scholar]
- 27. Marcu A, Bichmann L, Kuchenbecker L, Kowalewski DJ, Freudenmann LK, Backert L, et al. HLA Ligand Atlas: A benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J Immunother Cancer 2021;9:e002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform 2006;7:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Müller M, Gfeller D, Coukos G, Bassani-Sternberg M. Hotspots” of antigen presentation revealed by human leukocyte antigen ligandomics for neoantigen prioritization. Front Immunol 2017;8:1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Erhard F, Dölken L, Schilling B, Schlosser A. Identification of the cryptic HLA-I immunopeptidome. Cancer Immunol Res 2020;8:1018–26. [DOI] [PubMed] [Google Scholar]
- 31. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014;515:572–6. [DOI] [PubMed] [Google Scholar]
- 34. Freudenmann LK, Marcu A, Stevanovi S. Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry. Immunology 2018;154:331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol 2013;31:e439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuznetsov A, Voronina A, Govorun V, Arapidi G. Critical review of existing MHC I immunopeptidome isolation methods. Molecules 2020;25:5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sturm T, Sautter B, Wörner TP, Stevanovi S, Rammensee H-G, Planz O, et al. Mild acid elution and MHC immunoaffinity chromatography reveal similar albeit not identical profiles of the HLA class I immunopeptidome. J Proteome Res 2021;20:289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malabat C, Feuerbach F, Ma L, Saveanu C, Jacquier A. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. eLife 2015;4:e06722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zook MB, Howard MT, Sinnathamby G, Atkins JF, Eisenlohr LC. Epitopes derived by incidental translational frameshifting give rise to a protective CTL response. J Immunol 2006;176:6928–34. [DOI] [PubMed] [Google Scholar]
- 40. Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MAS, Brocard M, et al. Extensive translation of small open reading frames revealed by poly-ribo-seq. eLife 2014;3:e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smart AC, Margolis CA, Pimentel H, He MX, Miao D, Adeegbe D, et al. Intron retention is a source of neoepitopes in cancer. Nat Biotechnol 2018;36:1056–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, et al. Translation from the 5’ untranslated region shapes the integrated stress response. Science 2016;351:aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bullock TNJ, Eisenlohr LC. Ribosomal scanning past the primary initiation codon as a mechanism for expression of CTL epitopes encoded in alternative reading frames. J Exp Med 1996;184:1319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Löwenberg B, Terpstra W. Maturation hierarchy of leukemic stem cells. Stem Cells 1998;16:85–8. [DOI] [PubMed] [Google Scholar]
- 45. Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med 1999;341:1051–62. [DOI] [PubMed] [Google Scholar]
- 46. Jordan CT. Unique molecular and cellular features of acute myelogenous leukemia stem cells. Leukemia 2002;16:559–62. [DOI] [PubMed] [Google Scholar]
- 47. Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJ, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med 2018;379:2330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vago L, Perna SK, Zanussi M, Mazzi B, Barlassina C, Stanghellini MTL, et al. Loss of Mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med 2009;361:478–88. [DOI] [PubMed] [Google Scholar]
- 49. Ostrand-Rosenberg S. CD4+ T lymphocytes: a critical component of antitumor immunity. Cancer Invest 2005;23:413–9. [PubMed] [Google Scholar]
- 50. Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547:217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Löwer M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547:222–6. [DOI] [PubMed] [Google Scholar]
- 52. Laheurte C, Dosset M, Vernerey D, Boullerot L, Gaugler B, Gravelin E, et al. Distinct prognostic value of circulating anti-telomerase CD4+ Th1 immunity and exhausted PD-1+/TIM-3+ T cells in lung cancer. Br J Cancer 2019;121:405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsuji T, Matsuzaki J, Ritter E, Miliotto A, Ritter G, Odunsi K, et al. Split T cell tolerance against a self/tumor antigen: spontaneous CD4+ but Not CD8+ T cell responses against p53 in cancer patients and healthy donors. PLoS One 2011;6:e23651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tegeler CM, Scheid J, Rammensee HG, Salih HR, Walz JS, Heitmann JS, et al. HLA-DR presentation of the tumor antigen MSLN associates with clinical outcome of ovarian cancer patients. Cancers 2022;14:2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Godet Y, Fabre E, Dosset M, Lamuraglia M, Levionnois E, Ravel P, et al. Analysis of spontaneous tumor-specific CD4 T-cell immunity in lung cancer using promiscuous HLA-DR telomerase-derived epitopes: potential synergistic effect with chemotherapy response. Clin Cancer Res 2012;18:2943–53. [DOI] [PubMed] [Google Scholar]
- 56. Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci G, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med 2019;25:603–11. [DOI] [PubMed] [Google Scholar]
- 57. Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanovi S, Gouttefangeas C, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 2019;565:240–5. [DOI] [PubMed] [Google Scholar]
- 58. Walter S, Weinschenk T, Stenzl A, Zdrojowy R, Pluzanska A, Szczylik C, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med 2012;18:1254–61. [DOI] [PubMed] [Google Scholar]
- 59. Yoshimura K, Minami T, Nozawa M, Kimura T, Egawa S, Fujimoto H, et al. A Phase 2 randomized controlled trial of personalized peptide vaccine immunotherapy with low-dose dexamethasone versus dexamethasone alone in chemotherapy-naive castration-resistant prostate cancer. Eur Urol 2016;70:35–41. [DOI] [PubMed] [Google Scholar]
- 60. Hubbard JM, Cremolini C, Graham RP, Moretto R, Mitchell JL, Wessling J, et al. Evaluation of safety, immunogenicity, and preliminary efficacy of PolyPEPI1018 off-the-shelf vaccine with fluoropyrimidine/bevacizumab maintenance therapy in metastatic colorectal cancer (mCRC) patients. J Clin Oncol 8, 2020. (suppl 15; abstr 4048). [Google Scholar]
- 61. Rammensee H-G, Wiesmüller K-H, Chandran PA, Zelba H, Rusch E, Gouttefangeas C, et al. A new synthetic toll-like receptor 1/2 ligand is an efficient adjuvant for peptide vaccination in a human volunteer. J Immunother Cancer 2019;7:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Paczulla AM, Dirnhofer S, Konantz M, Medinger M, Salih HR, Rothfelder K, et al. Long-term observation reveals high-frequency engraftment of human acute myeloid leukemia in immunodeficient mice. Haematologica 2017;102:854–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nelde A, Kowalewski DJ, Stevanovi S. Purification and identification of naturally presented MHC class I and II ligands. Methods Mol Biol 2019;1988:123–36. [DOI] [PubMed] [Google Scholar]
- 64. Kowalewski DJ, Schuster H, Backert L, Berlin C, Kahn S, Kanz L, et al. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci U S A 2015;112:E116–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 1994;5:976–89. [DOI] [PubMed] [Google Scholar]
- 66. Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 2007;4:923–5. [DOI] [PubMed] [Google Scholar]
- 67. Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med 2016;8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009;61:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rammensee H-G, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 1999;50:213–9. [DOI] [PubMed] [Google Scholar]
- 70. Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 2017;45:D777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 2005;352:254–66. [DOI] [PubMed] [Google Scholar]
- 72. Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 2012;485:260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Opatz S, Polzer H, Herold T, Konstandin NP, Ksienzyk B, Zellmeier E, et al. Exome sequencing identifies recurring FLT3 N676K mutations in core-binding factor leukemia. Blood 2013;122:1761–9. [DOI] [PubMed] [Google Scholar]
- 74. Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S. Prognostic relevance of FLT3-TKD mutations in AML: the combination matters–an analysis of 3082 patients. Blood 2008;111:2527–37. [DOI] [PubMed] [Google Scholar]
- 75. Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002;99:4326–35. [DOI] [PubMed] [Google Scholar]
- 76. Vempati S, Reindl C, Kaza SK, Kern R, Malamoussi T, Dugas M, et al. Arginine 595 is duplicated in patients with acute leukemias carrying internal tandem duplications of FLT3 and modulates its transforming potential. Blood 2007;110:686–94. [DOI] [PubMed] [Google Scholar]
- 77. Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 2001;97:2434–9. [DOI] [PubMed] [Google Scholar]
- 78. Sturm T, Leinders-Zufall T, Maek B, Walzer M, Jung S, Pömmerl B, et al. Mouse urinary peptides provide a molecular basis for genotype discrimination by nasal sensory neurons. Nat Commun 2013;4:1616. [DOI] [PubMed] [Google Scholar]
- 79. Toprak UH, Gillet LC, Maiolica A, Navarro P, Leitner A, Aebersold R. Conserved peptide fragmentation as a benchmarking tool for mass spectrometers and a discriminating feature for targeted proteomics. Mol Cell Proteomics 2014;13:2056–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics 2012;11:M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Berlin C, Kowalewski DJ, Schuster H, Mirza N, Walz S, Handel M, et al. Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. Leukemia 2015;29:647–59. [DOI] [PubMed] [Google Scholar]
- 82. Widenmeyer M, Griesemann H, Stevanovi S, Feyerabend S, Klein R, Attig S, et al. Promiscuous survivin peptide induces robust CD4+ T-cell responses in the majority of vaccinated cancer patients. Int J Cancer 2012;131:140–9. [DOI] [PubMed] [Google Scholar]
- 83. Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 1996;274:94–6. [PubMed] [Google Scholar]
- 84. Walter S, Herrgen L, Schoor O, Jung G, Wernet D, Bühring HJ, et al. Cutting edge: predetermined avidity of human CD8 T cells expanded on calibrated MHC/anti-CD28-coated microspheres. J Immunol 2003;171:4974–8. [DOI] [PubMed] [Google Scholar]
- 85. Neumann A, Hörzer H, Hillen N, Klingel K, Schmid-Horch B, Bühring HJ, et al. Identification of HLA ligands and T-cell epitopes for immunotherapy of lung cancer. Cancer Immunol Immunother 2013;62:1485–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nelde A, Walz JS, Kowalewski DJ, Schuster H, Wolz OO, Peper JK, et al. HLA class I-restricted MYD88 L265P-derived peptides as specific targets for lymphoma immunotherapy. Oncoimmunology 2017;126:2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hermans IF, Silk JD, Yang J, Palmowski MJ, Gileadi U, McCarthy C, et al. The VITAL assay: a versatile fluorometric technique for assessing CTL- and NKT-mediated cytotoxicity against multiple targets in vitro and in vivo. J Immunol Methods 2004;285:25–40. [DOI] [PubMed] [Google Scholar]
- 88. Sturm G, Szabo T, Fotakis G, Haider1 M, Rieder D, Trajanoski Z, et al. Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics 2020;36:4817–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wolf FA, Angerer P, Theis FJ. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nolet CJ, Lafargue V, Raff E, Nanditale T, Oates T, Zedlewski J, et al. Bringing UMAP closer to the speed of light with GPU acceleration. In:Proceedings of the AAAI Conference on Artificial Intelligence, 35(1), 418–26. 10.1609/aaai.v35i1.16118. [DOI] [Google Scholar]
- 91. Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 2019;26:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 2008;9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res 2019;47:D442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary Tables file includes Tables S1 to S15. Table S1: Frequencies of CD34+CD38- LSCs within primary AML patient samples. Table S2: Immunopeptidomics cohort overview. Table S3: Patient characteristics of AML immunopeptidomics cohort. Table S4: AML- and AML/LSC-associated HLA class I targets. Table S5: AML/LSC shared HLA class I-presented antigens. Table S6: AML- and AML/LSC-associated HLA class II peptide, protein, and hotspot targets. Table S7: AML/LSC shared HLA class II-presented peptide targets. Table S8: Recurrent AML mutations included in neoepitope screening. Table S9: Characteristics of AML patient samples used for T cell-based assays. Table S10: Characteristics of HV samples used for T cell-based assays. Table S11: Immunopeptidome diversity according to demographics and disease characteristics. Table S12: Impact of demographics and disease characteristics on patient outcome in the immunopeptidome survival analysis group. Table S13: Patient characteristics of immunopeptidome survival analysis group. Table S14: Patient characteristics of immune response survival analysis group. Table S15: Impact of demographics and hematopoietic stem cell transplantation on patient outcome in the immune response survival group.
The Supplementary Figures file includes Figures S1 to S13. Figure S1: Gating strategy for the evaluation of LSC frequencies pre and post enrichment. Figure S2: Amino acid distribution within LSC and AML bulk immunopeptidomes. Figure S3: HLA class I immunopeptidomics. Figure S4: Abundance of AML- and AML/LSC-associated HLA class I ligands within the immunopeptidome. Figure S5: Identification of AML- and AML/LSC-associated HLA class II antigen targets by comparative immunopeptidome profiling. Figure S6: Abundance of AML- and AML/LSC-associated HLA class II peptides within the immunopeptidome. Figure S7: Spectral validation of HLA class I-restricted AML- and AML/LSC-associated peptides. Figure S8: Spectral validation of HLA class II-restricted AML- and AML/LSC-associated peptides. Figure S9: Magnitude and functionality of in vitro primed CD8+ T cells. Figure S10: Characterization of memory T cell responses detected in ELISpot assays. Figure S11: Further characterization of HLA class II-restricted antigens. Figure S12: In-depth characterization and phenotyping of HLA class II AML/LSC-specific CD4+ T cells by single-cell RNA sequencing and multi-color flow cytometry. Figure S13: Impact of immunopeptidome diversity and peptide-specific immune responses on patient survival.
Data Availability Statement
The mass spectrometry immunopeptidomics data generated in this study has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE (93) partner repository with the dataset identifier PXD038691. The mass spectrometry immunopeptidomics raw data (.raw files) can be viewed by the Thermo Xcalibur Qual Browser or other .raw file viewers. The processed search engine output (.msf files) can be viewed by the Proteome Discoverer software. The scRNA-seq data generated in this study have been deposited in the NCBI's Gene Expression Omnibus database with the dataset identifier GSE235080.