Development of a diagnostic model for early colorectal cancer based on epigenetic analysis of PBMCs supports the utility of altered DNA methylation in immune cells for cancer diagnosis.
Abstract
An effective blood-based method for the diagnosis of colorectal cancer has not yet been developed. Molecular alterations of immune cells occur early in tumorigenesis, providing the theoretical underpinning for early cancer diagnosis based on immune cell profiling. Therefore, we aimed to develop an effective detection method based on peripheral blood mononuclear cells (PBMC) to improve the diagnosis of colorectal cancer. Analysis of the genome-wide methylation landscape of PBMCs from patients with colorectal cancer and healthy controls by microarray, pyrosequencing, and targeted bisulfite sequencing revealed five DNA methylation markers for colorectal cancer diagnosis, especially early-stage colorectal cancer. A single-tube multiple methylation–specific quantitative PCR assay (multi-msqPCR) for simultaneous detection of five methylation markers was established, which allowed quantitative analysis of samples with as little as 0.1% PBMC DNA and had better discriminative performance than single-molecule detection. Then, a colorectal cancer diagnostic model (CDM) based on methylation markers and the multi-msqPCR method was constructed that achieved high accuracy for early-stage colorectal cancer (AUC = 0.91; sensitivity = 81.18%; specificity = 89.39%), which was improved compared with CEA (AUC = 0.79). The CDM also enabled a high degree of discrimination for advanced adenoma cases (AUC = 0.85; sensitivity = 63.04%). Follow-up data also demonstrated that the CDM could identify colorectal cancer potential up to 2 years before currently used diagnostic methods. In conclusion, the approach constructed in this study based on PBMC-derived DNA methylation markers and a multi-msqPCR method is a promising and easily implementable diagnostic method for early-stage colorectal cancer.
Significance:
Development of a diagnostic model for early colorectal cancer based on epigenetic analysis of PBMCs supports the utility of altered DNA methylation in immune cells for cancer diagnosis.
Introduction
According to GLOBOCAN 2020 database, colorectal cancer ranks third in incidence and second in mortality worldwide (1). It is well accepted that patients with early-stage colorectal cancer (stages I and II) have a better 5-year relative survival rate than those at advanced stages (stages III and IV; ref. 2). Unfortunately, 60% to 70% of patients with colorectal cancer are diagnosed at advanced stages, limiting their curative outcomes (3). In addition, young patients with colorectal cancer typically exhibit more advanced disease and adverse pathologic features, likely leading to poorer survival outcomes (4). Detection in earlier stages of tumors or asymptomatic patients is, therefore, of paramount importance for reducing the incidence and mortality of these patients. Colonoscopy and fecal immunochemical test (FIT) have been recommended as the first-tier options for colorectal cancer screening in the population ages 50 to 74 years, according to the American College of Gastroenterology (ACG) Clinical Guidelines (5). However, the invasive nature and high costs of colonoscopy create a barrier to participation (6). FIT may improve participation rates, while its sensitivity and specificity are suboptimal for colorectal cancer detection (7, 8). Therefore, developing specific, sensitive, and noninvasive biomarkers for colorectal cancer diagnosis is urgently necessary.
It is clear that colorectal cancer is closely related to immunity, and immune cells play a pivotal role in colorectal cancer by dynamically responding to variations in the tumor and protecting the host against tumor development (9). Peripheral blood mononuclear cells (PBMC) consisting of circulating monocytes, T cells, B cells, and natural killer (NK) cells, can be easily obtained from routinely collected blood and closely reflect the immune response status (10). In recent years, reports of PBMCs as surrogate markers for various diseases, such as inflammatory diseases like rheumatoid arthritis and malignant diseases like renal cell carcinoma, have gradually emerged (11, 12). However, although many efforts have been made, the roles of PBMCs in the tumorigenesis of colorectal cancer remain largely unexplored.
Abnormal alterations of DNA methylation have been recognized as the most dominant phenomenon occurring during the initiation and progression of multiple cancer types, including colorectal cancer (13). Regardless of the biological consequences of aberrant DNA methylation, this epigenetic alteration can be regarded as a molecular signature and one of the first detectable neoplastic changes for tumor diseases (14, 15). Several studies on identifying cancer-specific DNA methylation biomarkers have been reported in recent years (16). Regarding colorectal cancer, several aberrantly methylated genes, such as SEPT9, SFRP2, SDC2, ADHFE1, and IKZF1, are associated with colorectal cancer in various samples and suggested as potential markers (17–20). However, the potential value of aberrant DNA methylation in PBMCs for the early diagnosis of colorectal cancer has seldom been investigated.
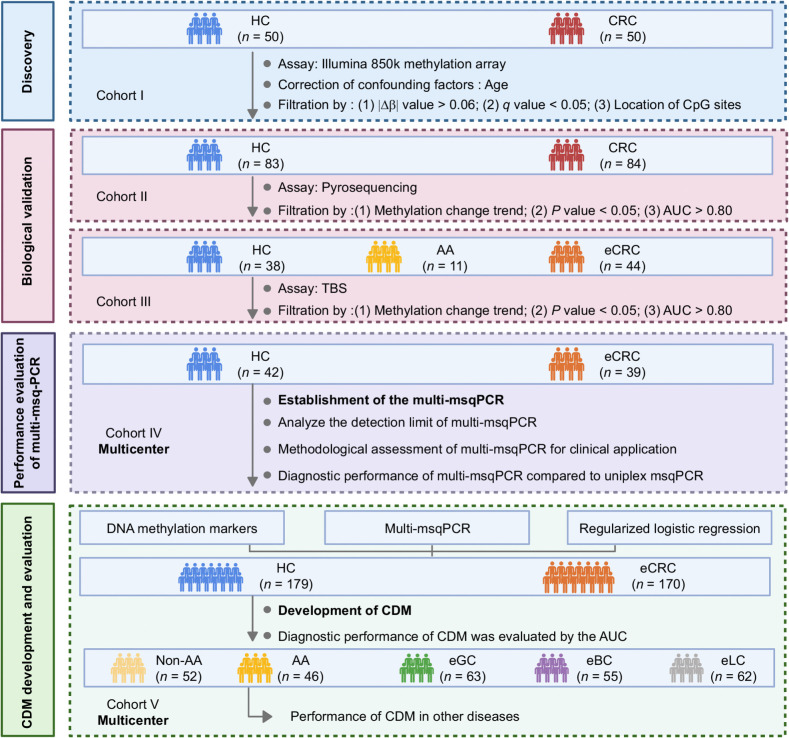
Accordingly, our current study had four consecutive and independent objectives. First, we performed the latest HM850K methylation array to reveal the DNA methylation landscape of PBMCs in colorectal cancer and identify DNA methylation markers capable of discriminating colorectal cancer from healthy controls (Discovery). Second, candidate methylation markers were biologically validated via two different methods in two independent cohorts (biological validation). Subsequently, a multiple methylation-specific quantitative PCR assay (multi-msqPCR) was developed to quantify methylation levels of selected methylation markers (performance evaluation of multi-msqPCR). Finally, a colorectal cancer diagnostic model (CDM) based on DNA methylation markers and multi-msqPCR technology was generated, and its feasibility for identifying early-stage colorectal cancer cases was evaluated using the area under the receiver operating characteristic curve (AUC) analysis (CDM development and evaluation).
Materials and Methods
Biospecimen sources
Total of 1,068 participants were originally enrolled in this analysis from 10 hospitals of China between May 2020 and June 2022. Participants in cohorts I to III were recruited from the Second Hospital of Shandong University and Qilu Hospital of Shandong University. Participants in cohorts IV and V were recruited from the Second Hospital of Shandong University, Qilu Hospital of Shandong University, the 960 Hospital of the PLA Joint Logistic Support Force, the First Hospital of Dalian Medical University, the First Affiliated Hospital of Anhui Medical University, Affiliated Tumor Hospital of Nantong University, Nanjing First Hospital, Guangdong Provincial People's Hospital, Lanzhou University Second Hospital, the Seventh Affiliated Hospital of Sun Yat-Sen University. The age of all participants ranged from 23 to 82 years; the gender ratio was roughly equal between male and female participants. Clinical and demographic parameters on patient samples are summarized in Table 1 and Supplementary Table S1, and the differences in age and sex between subgroups of each cohort were calculated by statistical methods. The final protocol, any amendments, and informed consent documentation were reviewed and approved by the ethical committees of each hospital participating centers. All participants can understand and sign written informed consent in the study.
Table 1.
Main clinical characteristics of participants in study.
| Age | Gender | TNM stage (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Mean ± SD | Range | P | Male | Female | P | I–II | III–IV | |
| Discovery (cohort I) | HC (n = 50) | 53 ± 13 | 31–82 | 0.013 | 20 (40%) | 30 (60%) | 0.420 | / | / |
| CRC (n = 50) | 59 ± 12 | 26–76 | 24 (48%) | 26 (52%) | 33 (66%) | 17 (34%) | |||
| Biological validation I (cohort II) | HC (n = 83) | 61 ± 7 | 45–81 | 0.537 | 40 (48%) | 43(52%) | 0.316 | / | / |
| CRC (n = 84) | 62 ± 10 | 39–81 | 47 (56%) | 37 (44%) | 43 (51%) | 41 (49%) | |||
| Biological validation II (cohort III) | HC (n = 38) | 59 ± 9 | 35–75 | 0.194 | 16 (42%) | 22 (58%) | 0.853 | / | / |
| AA (n = 11) | 62 ± 6 | 51–68 | 4 (36%) | 7 (64%) | / | / | |||
| eCRC (n = 44) | 62 ± 8 | 45–77 | 20 (45%) | 24 (55%) | 44 (100%) | 0 (0%) | |||
| Performance evaluation of multi-msqPCR (cohort IV) | HC (n = 42) | 62 ± 8 | 42–79 | 0.677 | 24 (57%) | 18 (43%) | 0.766 | / | / |
| eCRC (n = 39) | 61 ± 9 | 40–79 | 21 (54%) | 18 (46%) | 39 (100%) | 0 (0%) | |||
| CDM development and evaluation (cohort V) | HC (n = 179) | 57 ± 9 | 35–79 | 0.181 | 87 (49%) | 92 (51%) | 0.236a | / | / |
| Non-AA (n = 52) | 58 ± 12 | 23–76 | 33 (63%) | 19 (37%) | / | / | |||
| AA (n = 46) | 58 ± 8 | 36–72 | 24 (52%) | 22 (48%) | / | / | |||
| eCRC (n = 170) | 59 ± 12 | 30–79 | 100 (59%) | 70 (41%) | 170 (100%) | 0 (0%) | |||
| eGC (n = 63) | 58 ± 10 | 33–77 | 39 (62%) | 24 (38%) | 63 (100%) | 0 (0%) | |||
| eBC (n = 55) | 57 ± 11 | 34–78 | 0 (0%) | 55 (100%) | 55 (100%) | 0 (0%) | |||
| eLC (n = 62) | 59 ± 9 | 33–72 | 34 (55%) | 28 (45%) | 62 (100%) | 0 (0%) | |||
Abbreviations: AA, advanced adenomas; CDM, CRC diagnostic model; CRC, colorectal cancer; eBC, early-stage breast cancer; eCRC, early-stage colorectal cancer; eGC, early-stage gastric cancer; eLC, early-stage lung cancer; HC, healthy controls; Non-AA, non–advanced adenomatous polyps.
aBreast cancer was excluded in the statistical analysis of gender differences between groups.
All cases included in this study were first required to exclude pathogenic factors that affected the methylation status of PBMCs (such as inflammatory bowel disease, acute and chronic infections, autoimmune diseases, and hematologic diseases, etc.). Then, healthy controls were recruited from gastroenterology clinic, and further inclusion criteria were as follows: (i) no significant abnormalities were found in colonoscopy; (ii) no obvious abnormality in blood routine; (iii) no disease of the intestinal system, (iv) imaging and histology confirmed no cancer. Similarly, non–advanced adenomatous polyps and advanced adenoma need to be confirmed by colonoscopy and pathologic examination when necessary, participants in both groups were also required to exclude other intestinal disorders. Non–advanced adenoma includes hyperplastic polyps, inflammatory polyps and other polyps with a diameter of less than 10 mm, which generally do not have the possibility of cancer. Advanced adenoma was defined as those with a diameter of >10 mm or with villous components or with high-grade dysplasia. Colorectal cancer cases were selected based on the further inclusion criteria: (i) primary diagnosis of colorectal cancer; (ii) confirmed by pathologic examination; (iii) without other colorectal lesions; (iv) without extra-colorectal malignancies; (v) did not receive any treatment, including surgery, chemotherapy, or radiotherapy. Participants with other tumors, including early-stage gastric cancer, early-stage breast cancer, and early-stage lung cancer, are required to meet the following criteria: (i) a single type of early-stage tumor was pathologically confirmed; (ii) no intestinal lesions; (iii) has not received any form of treatment.
Prior to colonoscopy, 2 mL venous blood with EDTA-2K anticoagulant and 200 μL of serum after coagulation centrifugation were collected from all subjects on an empty stomach in the morning. PBMCs were separated from peripheral blood using Histopaque-1077 (Sigma-Aldrich) within 30 minutes after blood collection. Precipitation of PBMCs and serum samples were stored in −80°C refrigerator and mailed to the Second Hospital of Shandong University (Jinan, P.R. China) via sufficient dry ice for follow-up processing.
DNA extraction and bisulfite conversion
DNA extraction, bisulfite conversion, and DNA methylation detection of PBMCs were performed by professionally trained staff from the Second Hospital of Shandong University. Samples from each cohort were processed at the same time. DNA was extracted from PBMCs using DNeasy Blood & Tissue Kit (Qiagen Sciences Inc.) following the manufacturer's instructions, and the concentration was determined by NanoDrop ND-1000 (Thermo Fisher Scientific) spectrophotometer. According to the manufacturer's instructions, isolated DNA from PBMCs that passed the preliminary quality control steps was then processed for bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research).
Assay techniques
Illumina 850K methylation analysis
For the discovery stage, DNA methylation status was assessed with Infinium MethylationEPIC BeadChip (Illumina), which covers more than 850,000 CpG sites. Approximately 250 ng of bisulfite-converted DNA was applied to the array and analyzed, and SNP-associated probes and those corresponding to the sex chromosomes were reserved. The thresholds given by Illumina were used for the quality check of the raw data of each probe and data normalization. β value was used to represent DNA methylation level, and it ranged from 0 to 1. The Gend Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using an R package clusterProfiler.
Pyrosequencing
DNA methylation levels of candidate CpG sites were first analyzed by pyrosequencing. Primers for pyrosequencing assays were designed using the PyroMark Assay Design software (Supplementary Table S2). Then, PCR amplification of bisulfite-converted DNA was performed using SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology) in a 10-μL reaction system. Pyrosequencing reactions were performed in a PyroMark Q96 ID pyrosequencer (Qiagen Sciences Inc.), and the pyrosequencing data were analyzed by PyroMark Q96 software according to the manufacturer's instructions.
Targeted bisulfite sequencing
PCR primer sets for targeted bisulfite sequencing (TBS) were carefully designed using Methylation Primer (Supplementary Table S3). Agarose electrophoresis was performed after PCR, and amplicons of the targeted DNA sequences were separated and purified through the QIAquick Gel Extraction Kit (Qiagen Sciences Inc.). After libraries were quantified and pooled together from different samples, sequencing was performed on the Illumina NextSeq platform following the manufacturer's protocol. In addition, BSseeker2, a commonly used tool for analyzing the bisulfite sequencing results, was performed in our study to map bisulfite-treated reads and methylation calling.
Multi-msqPCR
Amplification primers and probes for bisulfite-converted DNA sequences were designed using MethPrimer (University of California San Francisco, San Francisco, CA; Supplementary Table S4). The PCR products for CpG sites were TA-cloned into the pGEM-T Easy vector (Promega) for cloning and sequencing to ensure specific amplification. For multi-msqPCR reactions, three hypermethylated CpG sites, two hypomethylated CpG sites, and a reference gene (ACTB) were analyzed together in a single tube using FAM, CY5, and VIC-based probes, respectively. PCR amplification of bisulfite-converted DNA was performed using KAPA Probe Fast qPCR Master Mix Kit (Roche) in a 10-μL reaction system containing 5 μL of 2× Master Mix, 0.25 μmol/L of each target primer, 0.06 μmol/L of each ACTB primer, 0.1 μmol/L of each target probe, 0.05 μmol/L of ACTB probe, 0.2 μL ROX Low, and 1 μL of bisulfite-converted DNA (DNA concentration is about 50 ng/μL). The two-round PCR was performed as follows: 95°C/3 minutes, followed by 40 cycles of 95°C/3 seconds and 60°C/30 seconds, and then 72 °C/30 seconds. Methylation levels (ΔCq = Cq ACTB – Cq biomarker) were calculated from the single-tube reactions of each sample, and a higher ΔCq value represents a higher methylation level. Bisulfite-converted DNA samples were tested in random order, and the experimental operators were blinded to the grouping of patients.
CEA assay
CEA was quantitated from serum samples using the Carcinoembryonic Antigen (CEA) Assay Kit (Roche Diagnostics) on the Roche Cobas e801 analyzer by professional operators from the Second Hospital of Shandong University.
Statistical analysis
In each cohort, age between the two groups was compared using Mann–Whitney U test and the age among three or more groups was compared using Kruskal–Wallis tests; sex distribution was compared using χ2 test.
Discovery
The Illumina 850 K methylation dataset were analyzed using the ChAMP package in R. Because patients with colorectal cancer were significantly older than healthy controls in the discovery stage, we used the logistic regression to explore the association between CpG sites and colorectal cancer, by including age as a covariate to adjust for potential confounding bias. Then, the P values were corrected for multiple hypothesis testing using the Benjamini–Hochberg procedure to obtain a q value. CpG sites with |Δβ|≥0.06 and q value ≤ 0.05 were defined as differentially methylated positions (DMP). The partial least squares-discriminant analysis (PLS-DA), a multivariate dimensionality reduction tool (21), was performed to identify discriminant DNA methylation features of colorectal cancer cases versus healthy controls using R package ropls.
Biological validation I &II
The Mann–Whitney U test was used to compare the methylation levels of CpG sites in the case and healthy control groups, as well as between different clinical feature subgroups of colorectal cancer cases (e.g., age, sex, location, tumor size, and stage). The clinical performance of CpG sites were visualized by the receiver operating characteristic curve (ROC) and evaluated by AUC calculation.
Performance evaluation of multi-msqPCR
The Mann–Whitney U test was used to compare the methylation levels of CpG sites in the case and healthy control groups. AUC was performed to evaluated the clinical performance of each CpG site and multiple CpG sites. The method of DeLong and colleagues (22) was used to compare AUCs and measure significance of differences.
CDM development and evaluation
We used regularized logistic regression to build a colorectal cancer diagnostic classifier, using the glmnet package (23). The variables input for each sample in the analyses included ΔCq(multiple hypermethylated sites) and ΔCq(multiple hypomethylated sites). The function cv.glmnet returned a sequence of lambdas (λs) and models. Ten-fold cross-validation was performed to select the optimal hyperparameter associated with the minimum mean squared value. We then derived a formula based on the selected λ and extracted variables with their corresponding coefficients. This formula was 1/[1+EXP (−(1.967375+1.512778*ΔCq(multiple hypermethylated sites)−1.956483 *ΔCq(multiple hypomethylated sites))]. This formula was performed to calculate the diagnostic probability value of early-stage colorectal cancer, the cut-off value was decided by ROC curve and Youden index based on the CDM development dataset (170 early-stage colorectal cancer cases and 179 healthy controls). The clinical performance of CDM was visualized by ROC and evaluated by AUC calculation. The Mann–Whitney U test was used to compare CDM scores between cases and controls. The statistical significance of the difference between the AUCs were compared with the method of DeLong and colleagues (22).
Sample size considerations
The required sample size was determined using PASS software, and the primary outcome was the AUC for each marker, as calculated from the methylation level of each marker with colorectal cancer in comparison with healthy controls (24). For the confirmation of each CpG sites or combined CpG sites in independent PBMC samples, assuming an estimated AUC of 0.80 (25) or greater was deemed the lower bound of acceptable accuracy from a null value of 0.5 with 90% power. A minimum of 17 cases and healthy controls were required with a case:control ratio of 1:1 to achieve this difference at a two-sided significance level of 0.05, and participants enrolled in all cohort were sufficient.
Data and materials availability
The Illumina 850 K methylation data from the discovery phase is publicly available in the Gene Expression Omnibus (GSE240324). All data associated with this study are present in the article and the Supplementary Data. All other raw data generated in this study are available upon request from the corresponding author. All resources generated in this study are available under a completed material transfer agreement at the request of the corresponding author.
Ethical statement
The corresponding author, on behalf of all authors, jointly and severally, certifies that their institution has approved the protocol for any investigation involving humans and that all experimentation was conducted in conformity with ethical and humane principles of research.
Results
Participants and clinical characteristics
A total of 1,068 participants were included in this study. In the discovery set, 50 colorectal cancer cases and 50 healthy controls confirmed by colonoscopy were included in cohort I. The mean age of these colorectal cancer cases was 59 years (range 26–76 years) and significantly higher than in healthy controls (P = 0.013). There were no significant differences between groups in sex (P = 0.420). Patients in the biological validation stage were divided into a cohort II comprising 84 colorectal cancer cases (51% were in the early-stage) and 83 healthy controls and a cohort III with 44 early-stage colorectal cancer cases, 11 advanced adenoma cases, and 38healthy controls. Within both cohort, there were no significant differences between groups in age (cohort II, P = 0.537, cohort III, P = 0.194) and sex (cohort II, P = 0.316, cohort III, P = 0.853). In the performance evaluation of multi-msqPCR stage, a cohort IV including 39 early-stage colorectal cancer cases and 42 healthy controls was used, clinical parameters of age (P = 0.677) and sex (P = 0.766) did not significantly differ by case/control status. In the CDM development and evaluation stage, a total of 627 cases were enrolled as cohort V. These individuals comprised 179 healthy controls, 52 Non–advanced adenoma, 46 advanced adenoma, 170 early- stage colorectal cancer, and 180 other types of tumors (including 63 early-stage gastric cancer, 55 early-stage breast cancer, and 62 early-stage lung cancer), there were no significant differences in age (P = 0.181) and sex (P = 0.236) between the different case and control groups. The detailed clinical features of participants are summarized in Table 1 and Supplementary Table S1.
Genome-wide DNA methylation analysis reveals the DNA methylation landscape of PBMCs in colorectal cancer
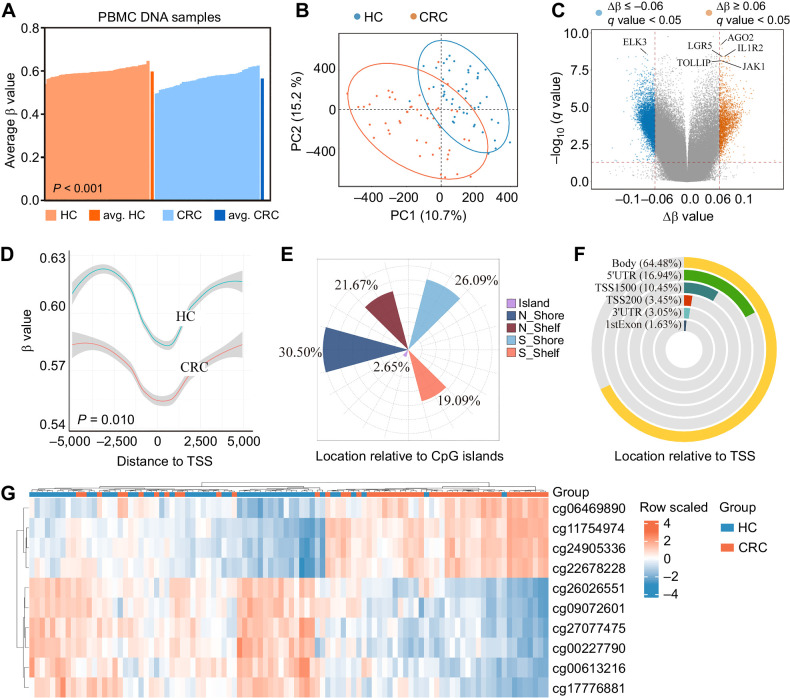
The study design and implementation are summarized in Fig. 1. To comprehensively delineate colorectal cancer–specific DNA methylation patterns in PBMCs, we first performed genome-wide Illumina 850K microarray in 50 colorectal cancer cases and 50 healthy controls from cohort I. Figure 2A shows the overall methylation level of each sample, which was consistent with the previous report that immune cells are globally hypomethylated in colorectal cancer (P < 0.001; ref. 26). PLS-DA was performed and plotted on the basis of the DNA methylation status of CpG sites, and there were differences between colorectal cancer cases and healthy controls (Fig. 2B). DMPs were defined as CpG sites with an absolute |Δβ|≥ 0.06 and a q value < 0.05. There were 8,155 DMPs between colorectal cancer and healthy controls, among which, 2,085 were significantly hypermethylated, and 6,070 were significantly hypomethylated in patients with colorectal cancer. A volcano plot was generated to visualize the DMPs, and representative genes related to immunity [such as IL1R2 (27, 28) and TOLLIP (29)] were identified (Fig. 2C). As CpG sites located within 0–2 kb upstream or downstream from the transcription start site (TSS) normally regulate gene expression in PBMCs, we explored the global methylation changes of DMPs in patients with colorectal cancer. On the basis of the gene location annotation, we investigated a total of 1,819 DMPs were close to TSS, which showed overall hypomethylation trend in colorectal cancer cases (P = 0.010, Fig. 2D). Classification of DMPs according to their location relative to CpG islands (CpGI) revealed that the proportion of DMPs in N_Shore (0–2 kb upstream from CpGIs) was the largest compared with other regions (Fig. 2E). Figure 2F shows the insights into the functional genomic regions of DMPs, with a majority of DMPs located in the gene bodies, whereas fewer DMPs were aligned to the regulatory regions.
Figure 1.
Overall workflow of the study design. AA, advanced adenoma; CDM, CRC diagnostic model; CRC, colorectal cancer; eCRC, early-stage colorectal cancer; HC, healthy controls; non-AA, non–advanced adenomatous polyps.
Figure 2.
DNA methylation landscape of colorectal cancer delineated using Illumina 850K DNA methylation array in the discovery stage. A, Average methylation levels of all CpG sites in PBMCs DNA that passed quality control for individual healthy controls (HC; orange), average of all healthy controls (avg. healthy controls; dark orange), individual colorectal cancer (blue), and average of all colorectal cancer (avg. colorectal cancer; dark blue), P values were determined by the Mann–Whitney U test. B, PLS-DA performed using all CpG sites. Samples from colorectal cancer and healthy controls are shown in different colors; blue dots, healthy controls; orange dots, colorectal cancer. C, Volcano plot representing DMPs:|Δβ (methylation average in colorectal cancer cases - methylation average in healthy controls) | > 0.06 and q value < 0.05 (q value means Benjamini–Hochberg Padj value. Red, significantly hypermethylated CpG sites; blue, hypomethylated CpG sites in colorectal cancer cases. Representative genes related to immunity were annotated. D, The global methylation change of DMPs close to TSS in PBMCs DNA of colorectal cancer cases was hypomethylated. P values were determined by the Kolmogorov–Smirnov test. E, Classification of differentially methylated CpG sites according to CpG islands. F, Classification of differentially methylated CpG sites according to their location relative to their genomic distribution. G, Hierarchical clustering of ten DMPs from the discovery set analysis of Illumina 850K data. CRC, colorectal cancer; PC, principal component.
As shown in Supplementary Fig. S1A, GO analysis indicate that DMPs were associated with functions essential for immune response (e.g., response to IL15, positive regulation of type 2 immune response, leukocyte aggregation, IL15-mediated signaling pathway). KEGG enrichment also showed that DMPs are associated with functions essential for immune response such as T-cell receptor signaling pathway, NK cell–mediated cytotoxicity, B-cell receptor signaling pathway (Supplementary Fig. S1B). Our results indicated that DMPs in PBMCs were involved in immunologic response during colorectal cancer progression, providing a theoretical basis for their use as potential markers for colorectal cancer diagnosis.
Identification of DMPs in PBMCs to distinguish colorectal cancer cases from healthy controls (biological validation phase I)
Taking into account criteria, such as |Δβ|, q value, location, and specific primer design, 13 CpG sites were initially selected (see more details in Supplementary Methods). But cg10187233, cg08214069, and cg25975690 were excluded due to unsuccessful primer design for pyrosequencing, and eventually 10 CpG sites (cg06469890, cg11754974, cg24905336, cg22678228, cg26026551, cg09072601, cg27077475, cg00227790, cg00613216, and cg17776881) were selected for further studies (Fig. 2G; Supplementary Table S5), the AUC of these CpG sites ranged from 0.81 to 0.88 (Supplementary Table S6).
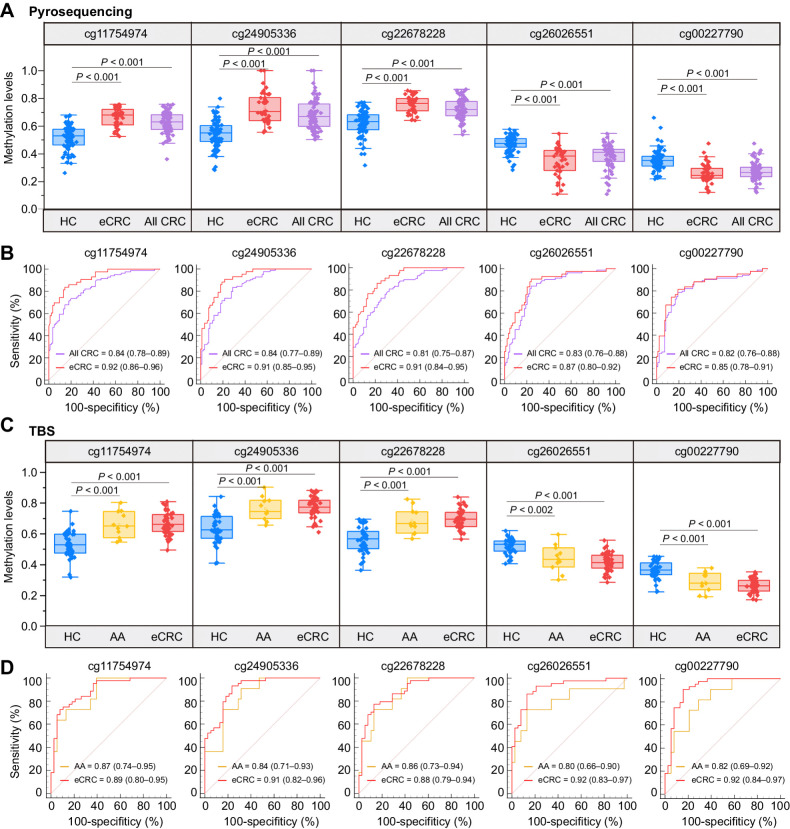
Therefore, methylation levels of these candidate CpG sites were further evaluated via pyrosequencing assay in cohort II. According to the following three principles: (i) the methylation change trend was consistent with microarray results; (ii) P value < 0.05; (iii) AUC > 0.80. Five CpG sites (cg11754974, cg24905336, cg22678228, cg26026551, cg00227790) met all the criteria and advanced to the next round of validation (see more details in Supplementary Data; Supplementary Fig. S2A and S2B). The AUC for individual CpG sites was greater than 0.80, with the sensitivity ranging from 67.86% to 84.52% (Fig. 3A and B; Supplementary Table S7). Meanwhile, we found there was an obvious feature that methylation levels of all five CpG sites were significantly different between early-stage colorectal cancer (stage I/II) and advanced colorectal cancer(stage III/IV) cases (Supplementary Table S8). Figure 3A shows that there were statistically significant differences in each individual CpG site between early-stage colorectal cancer cases and healthy controls. AUC, sensitivity, and specificity of these individual markers for early-stage colorectal cancer diagnosis were also calculated, where AUC ranged from 0.85 to 0.92, and sensitivity ranged from 81.40% to 90.70% (Fig. 3B; Supplementary Table S7). Our data indicated that these CpG sites have the potential to distinguish early-stage colorectal cancer from healthy controls.
Figure 3.
Validation of candidate DNA methylation biomarkers for colorectal cancer diagnosis. A, In biological validation phase I, pyrosequencing was performed in 84 all-stage colorectal cancer cases (including 43 early-stage colorectal cancer cases) and 83 healthy controls to analyze methylation levels of selected markers, of which, cg11754974, cg24905336, and cg22678228 were significantly hypermethylated, and cg26026551 and cg00227790 were significantly hypomethylated, P values were determined by the Mann–Whitney U test. B, ROC and the associated AUCs illustrate the diagnostic potential of five DMPs individually in different groups (purple, all-stage colorectal cancer vs. healthy controls; red, early-stage colorectal cancer versus healthy controls) using pyrosequencing. C, In biological validation phase II, methylation levels of five candidate CpG sites were quantified by the TBS in 38 healthy controls, 11 advanced adenoma cases, and 44 early-stage colorectal cancer cases. P values were determined by the Mann–Whitney U test. D, ROC curves display the 5 DMPs classification performance for distinguishing eCRC and advanced adenoma cases against healthy controls using TBS (red, early-stage colorectal cancer vs. healthy controls; golden, advanced adenoma vs. healthy controls). AA, advanced adenomas; CRC, colorectal cancer; eCRC, early-stage colorectal cancer; HC, healthy controls.
To uncover which types of cells contributed to the overall PBMC methylation variances, we separated T cells, B cells, and the T/B-lymphocyte–depleted cells (including monocytes, and NK cells) from 11 PBMC samples, including five colorectal cancers and six healthy controls. The differential methylation trends of five CpG sites in T cells, B cells, and the T/B-lymphocyte–depleted cells were consistent with PBMCs (Supplementary Fig. S3A).
Identification of DMPs in PBMCs to distinguish early-stage colorectal cancer cases from healthy controls (biological validation phase II)
Colorectal cancer develops through a stepwise accumulation of genetic and epigenetic alterations in precancerous lesions, and the detection of precancerous and early-stage colorectal cancer during the screening is essential for the prevention of colorectal cancer (30). Therefore, we decided to investigate the diagnostic efficacy of methylation changes in early-stage colorectal cancer and precancerous lesions in our following experiment. Biological validation was performed in additional independent DNA samples from 44 early-stage colorectal cancer cases, 11 advanced adenoma cases, and 38 healthy controls (cohort III) through another detection method, TBS assay. Figure 3C shows that significant methylation level differences of five candidate CpG sites were validated in early-stage colorectal cancer cases compared with healthy controls. Moreover, moderate differences in methylation levels of individual CpG sites were also observed in advanced adenoma cases, indicating that the DNA methylation was altered in the precursor lesions of colorectal development. On the basis of the data obtained from TBS, the AUC, diagnostic sensitivity, and specificity of early-stage colorectal cancer were calculated, and the results showed that AUC ranged from 0.88 to 0.92, sensitivity ranged from 72.73% to 93.18%, and specificity ranged from 76.32% to 92.11% (Fig. 3D; Supplementary Table S9). Besides, the performance of the five markers for distinguishing between advanced adenoma and healthy control samples was assessed, with the AUC, sensitivity, and specificity of selected markers ranging from 0.80 to 0.87, from 72.73% to 100.00%, and from 60.53% to 86.84%, respectively. Taken together, the abovementioned results validated the diagnostic value of the five methylation sites, especially in early-stage colorectal cancer cases.
Performance evaluation of multi-msqPCR
The above strategies for detecting methylation levels relied on sequencing, which might not be suitable for clinical translation and certainly not generally affordable. We developed a single-tube multi-msqPCR for clinical application to apply our DNA methylation markers to early-stage colorectal cancer diagnosis more broadly. This assay contained three different types of fluorophore probes. FAM fluorophores were used to label all three hypermethylated CpG sites, CY5 was used to label both two hypomethylated CpG sites, and VIC was used for the ACTB control assay. Therefore, the multi-msqPCR assay measure the total methylation of hypermethylated and hypomethylated CpG sites, rather than the methylation level of individual CpG sites (Supplementary Fig. S4). That is, Cq(multiple hypermethylated sites) represents the cumulative methylation levels of cg11754974, cg24905336, cg22678228, and Cq(multiple hypomethylated sites) represents the cumulative methylation levels of cg26026551 and cg00227790. The multiplex CpG sites produced a higher ΔCq value than corresponding individual CpG sites, confirming that multi-msqPCR could achieve fluorescence signal accumulation (Fig. 4A). PBMC DNA samples extracted from 39 early-stage colorectal cancer cases and 42 healthy controls (cohort IV) were used to further evaluate the performance of the multi-msqPCR. Supplementary Figure S5A reveals that the methylation difference between the early-stage colorectal cancer cases and healthy controls (ΔΔCq) was significantly enlarged by multiple detections of the three hypermethylated sites than in the uniplex-msqPCR assay (all P values < 0.001; Supplementary Table S10). Similar results were also observed when detecting two hypomethylated sites (Supplementary Table S10). The AUC of multiple hypermethylated sites and multiple hypomethylated sites for early-stage colorectal cancer was 0.89 [95% confidence interval (95% CI), 0.81–0.95] and 0.87 (95% CI, 0.78–0.94), respectively (Supplementary Fig. S5B), which were higher than the corresponding individual CpG site detection (P values are shown in Supplementary Table S11), indicating that multi-msqPCR could discriminate early-stage colorectal cancer cases from healthy controls better than uniplex-msqPCR assay.
Figure 4.
Performance evaluation of multi-msqPCR. A, Comparison between the multi-msqPCR and uniplex-msqPCR assays. The multi-msqPCR assay produced higher ΔCq values than uniplex-msqPCR assays [6.13 (ΔCq multiple hypermethylated sites) versus 1.44 (ΔCq cg11754974), 1.97 (ΔCq cg24905336), 0.47 (ΔCq cg22678228; left) and 4.43 (ΔCq multiple hypomethylated sites) versus 2.18 (ΔCq cg26026551), 1.79 (ΔCq cg00227790; right)]. B, Assessment of the analytic sensitivity of the multi-msqPCR assay. Primer probe sets for multiple hypermethylated sites (left) and multiple hypomethylated sites (right) were tested for two technical replicates with fold dilutions of 50 ng bisulfite-converted DNA. The multi-msqPCR assay detected tumor DNA signals with as little as 0.1%. ΔCq = (CqACTB− Cqbiomarker).
To evaluate the clinical applicability of multi-msqPCR, we tested the primer probe sets using clinical PBMCs DNA samples diluted to 50%, 25%, 10%, 1%, and 0.1% with water to determine the detection limit of the multi-msqPCR assay. All individual primer probe sets for CpG sites were able to quantitatively detect PBMCs DNA at 1% diluted clinical samples, while only the multi-msqPCR assay could detect tumor DNA at as low as 0.1% diluted clinical samples (Fig. 4B; Supplementary Fig. S6A–S6E). Because stability is an essential prerequisite for biomarkers, we performed two experiments to verify the stability of multi-msqPCR in our study. The coefficient of variation (CV), including intra-assay variation and inter-assay variation, was less than 5%, indicating that variation between the detection results of multi-msqPCR was clinically acceptable (Supplementary Fig. S6F). Second, converted DNA samples were stored at − 80°C for 0, 30, 60, and 90 days. Results indicated that storage for 90 days did not affect the methylation level of candidate five methylation markers (Supplementary Fig. S6G). Collectively, our data indicated that the established multi-msqPCR was an ideal clinical test.
Development and evaluation of the CDM for early-stage colorectal cancer diagnosis
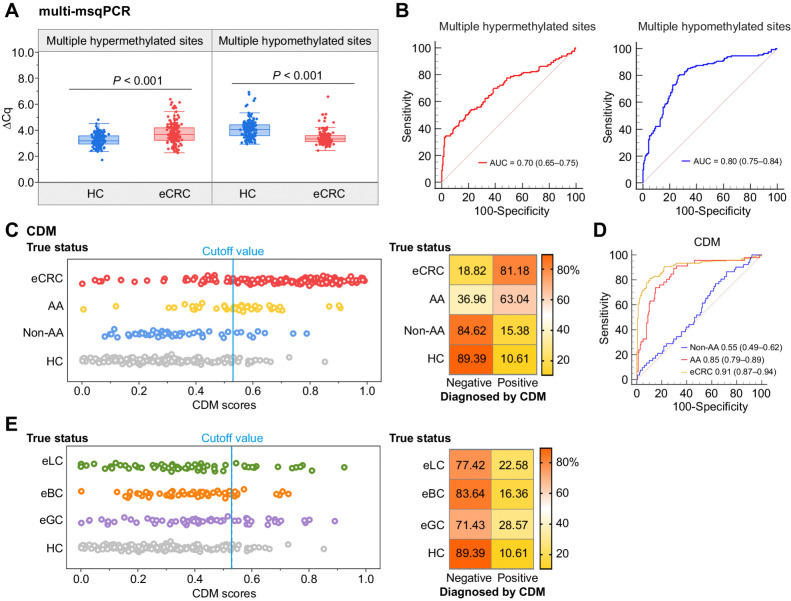
To further assess the performance of multi-msqPCR for clinical application, we recruited a cohort V including 170 early-stage colorectal cancer cases and 179 healthy controls from multiple hospitals in China. The methylation level of multiple hypermethylated sites was significantly higher, and the multiple hypomethylated sites was significantly lower in early-stage colorectal cancer cases compared with healthy controls (Fig. 5A). The AUC value of multiple hypermethylated sites and multiple hypomethylated sites for discrimination between early-stage colorectal cancer cases and healthy controls was 0.70 (95% CI, 0.65–0.75) and 0.80 (95% CI, 0.75–0.84), respectively (Fig. 5B). Considering not all of the CpG sites revealed aberrant DNA methylation in all early-stage colorectal cancer samples, we combined their DNA methylation levels into a regularized logistic regression to obtain a higher dimensionality diagnostic model. CDM, which could discern healthy controls and early-stage colorectal cancer samples and ranging from 0 to 1. As shown in Fig. 5C, distribution plots illustrated CDM score discrimination for early-stage colorectal cancer cases. The performance of CDM for early-stage colorectal cancer reached an AUC value of 0.91 (95% CI, 0.87–0.94), which yielded a sensitivity of 81.18% (95% CI, 74.5–86.8%) and a specificity of 89.39% (95% CI, 83.9–93.5%) using a cut-off value of 0.5252 determined by Youden's index, corresponding to an accuracy of 85.39% (Fig. 5C and D; Supplementary Table S12). ACG Clinical Guidelines recommend colonoscopy and FIT for at-risk groups aged 50–74 years (5). However, there is no consensus on colorectal cancer screening in low- and moderate-risk groups under the age of 50 (young patients with colorectal cancer). CDM assay performance was evaluated in all young participants in cohort V (including 35 young patients with early-stage colorectal cancer cases and 39 young healthy controls, there were no significant differences in age (P = 0.116) and sex (P = 0.103) between two groups). The CDM scores for young patients with early-stage colorectal cancer cases were significantly higher than young healthy controls (P < 0.001, Supplementary Fig. S7A). Notably, the CDM could discriminate the samples of early young patients with colorectal cancer cases from young healthy controls with an AUC of 0.93 (95% CI, 0.85–0.98), a sensitivity of 77.14%, and a specificity of 92.31% (Supplementary Fig. S7A and S7B). In general, our results show the advantages of using CDM as a noninvasive tool to diagnose early-stage colorectal cancer, which would improve the diagnostic rate of colorectal cancer.
Figure 5.
Development of CDM for early-stage colorectal cancer diagnosis. A, Methylation levels of PBMCs DNA as quantified by the multi-msqPCR assay in 349 individuals (170 early-stage colorectal cancer and 179 healthy controls) from multiclinical sites, where a higher ΔCq value represents a higher methylation level. P values between different groups were determined by the Mann–Whitney U test. B, The ROCs demonstrate the performance of the multi-msqPCR assay for distinguishing early-stage colorectal cancer cases from healthy controls in multicenter cohort. C, A CDM score of 0.5252 was used as the cut-off value. Samples were classified as negative or positive according to the CDM score and the detection percentages are shown in the corresponding confusion matrix on the right. D, The ROCs of the CDM for the detection of early-stage colorectal cancer, advanced adenoma, and non–advanced adenoma cases. E, The multi-msqPCR assay was performed in 180 patients with different types of cancer (62 early-stage lung cancer cases, 55 early-stage breast cancer, and 63 early-stage gastric cancer). All samples were classified as negative or positive according to the cut-off value of 0.5252 and the classification results are shown in the corresponding confusion matrix on the right. AA, advanced adenomas; CDM, CRC diagnostic model; CRC, colorectal cancer; eGC, early-stage gastric cancer; eBC, early-stage breast cancer; eLC, early-stage lung cancer HC, healthy controls; non-AA, non-advanced adenomatous polyps.
To provide maximum benefit, a colorectal cancer diagnostic tool need to detect precursor lesions in the colorectum, but not benign lesions. The CDM classifier was then locked and applied to 52 non–advanced adenoma and 46 advanced adenoma cases in cohort V. Compared with healthy controls, CDM scores were higher in advanced adenoma cases (P < 0.001) but not significantly different from non– advanced adenoma cases (P = 0.243; Fig. 5C; Supplementary Table S13). Positive and negative classifications of CDM were also determined using the cut-off value of 0.5252, 84.62% of 52 non- advanced adenoma cases were successfully identified as negative. The CDM detected advanced adenoma cases with 63.04% (95% CI, 47.5%–76.8%) sensitivity, AUC was 0.85 (95% CI, 0.79–0.89; Fig. 5D; Supplementary Table S12). Our data suggested that CDM was able to distinguish early-stage colorectal cancer from the normal colorectal epithelium and benign lesions. We also applicated CDM in patients with other common clinical cancer types, including 63 early-stage gastric cancer, 55 early-stage breast cancer, and 62 early-stage lung cancer cases in cohort V. According to the determined cut-off value of 0.5252, 28.57% of early-stage gastric cancer, 16.36% of early-stage breast cancer, and 22.58% of early-stage lung cancer were incorrectly identified as positive (Fig. 5E; Supplementary Table S12). Together, these results preliminarily confirmed the clinical applicability of CDM in the diagnosis of early-stage colorectal cancer.
Diagnostic performance of CDM compared with conventional methods
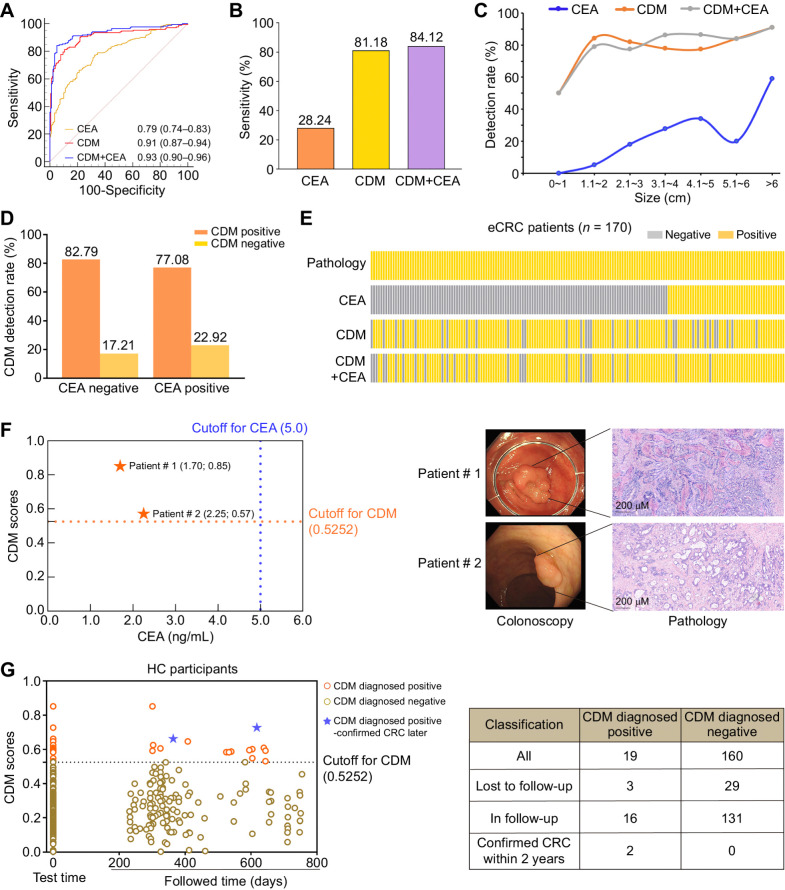
The single marker CEA assay is a routine method used for blood-based colorectal cancer screening. The CEA test was synchronously conducted on participants from cohort V. The superior performance of CDM was observed in comparison with CEA (AUC = 0.79, P < 0.001 vs. CDM). At the threshold of 5 ng/mL, the sensitivity of CEA for early-stage colorectal cancer was only 28.24%. The accuracy was higher when the CDM assay and CEA were combined, with a sensitivity of 84.12% (95% CI, 77.7%–89.3%), a specificity of 94.97% (95% CI, 90.7%–97.7), and an AUC of 0.93 (95% CI, 0.90–0.96; Fig. 6A and B). The above data suggested that these markers captured a broader spectrum of epigenetic heterogeneity of colorectal cancer than individual markers alone (AUC for CDM+CEA = 0.93, P = 0.004 vs. CDM, P < 0.001 vs. CEA). Our results indicated that detection rates of all three detection methods showed an increasing trend with the tumor size of colorectal cancer (Fig. 6C). In our current study, more than two-thirds (122/170) of cases with early-stage colorectal cancer were CEA negative. We thus investigated the performance of CDM in CEA-negative early-stage colorectal cancer cases, of which, 101 (82.79%) cases were CDM positive (Fig. 6D). Our data indicated that CDM was a preferable noninvasive biomarker for the detection of early-stage colorectal cancer when compared with CEA (Fig. 6E).
Figure 6.
Performance evaluation of CDM in detecting early-stage colorectal cancer. A, The ROCs of CDM, CEA, or both for colorectal cancer versus healthy controls. B, The sensitivity of the CDM, CEA, or both in early-stage colorectal cancer cases were calculated by determined cut-off values (cut-off value for CDM = 0.5252; cut-off value for CEA = 5 ng/mL; cut-off value for CDM+CEA = 0.5776). C, Detection rates of CDM, CEA, or both in patients with early-stage colorectal cancer overall increased with tumor size. D, CDM detection rate in CEA-negative and CEA-positive early-stage colorectal cancer cases. E, For patients with early-stage colorectal cancer in cohort V, the real disease status and the disease status diagnosed by CDM and CEA, or both are shown in a waffle chart. Each column represents an individual participant. Gray, patients diagnosed as negative according to corresponding method; yellow, positive. F, Example of two patients with tumor detected by CDM but missed by CEA, who were later diagnosed by pathology. The magnifications of the images in pathology were ×20. G, Scatter plots of follow-up results of healthy control participants in cohort V diagnosed by CDM at the time of enrollment. CDM, CRC diagnostic model; CRC, colorectal cancer; eCRC, early-stage colorectal cancer; HC, healthy controls.
Surprisingly, the potential utility of CDM is highlighted by two cases that were diagnosed by CDM but missed by CEA and colonoscopy. Poor bowel preparation leads to inadequate examination and missed diagnosis at the first examination. But the next colonoscopy confirmed colonic epithelial tumor of low malignant potential by pathology (Fig. 6F). These data suggested the advantage of CDM in the detection of early-stage colorectal cancer.
CDM may indicate the possibility of colorectal cancer up to 2 years
DNA methylation alterations enables early detection of cancer. Xu and colleagues demonstrated that DNA methylation–based blood test can detect tumors up to four year earlier before conventional diagnosis (31).We therefore conducted a follow-up study of 179 healthy controls in cohort V. According to current follow-up data, among 19 healthy controls diagnosed positive by CDM at enrollment, 3 were lost to follow-up, and 2 participants were confirmed as colorectal cancer within 2 years of follow-up. Moreover, among the 160 CDM-negative patients at enrollment, 29 were lost to follow-up, and no colorectal cancer was confirmed within 2 years (Fig. 6G). Our finding suggested that the CDM assay had potential in prediagnosis of colorectal cancer in advance, and further large-scale investigations should be conducted.
Discussion
Aberrant DNA methylation of tumor-associated genes may present at the early stage of malignant transformation, indicating its potential as a reliable discriminatory marker for cancer detection (32). In addition, an accurate, noninvasive diagnostic test for both early-stage colorectal cancer and advanced precancerous lesions throughout the colorectum would be transformative. In the currentstudy, we demonstrated that five clinically applicable DNA methylation biomarkers could identify patients with colorectal cancer. Next, a CDM assay with high analytic sensitivity for candidate biomarkers was established for the early detection of colorectal cancer, which showed satisfactory diagnostic value for early-stage of colorectal cancer and precancerous lesions.
Despite considerable efforts, there has been limited clinical success in developing valuable, easily applicable, and noninvasive diagnostic approaches for early screening and detection of colorectal cancer. In 2014, a large, prospective trial including 7,941 patients has been conducted by Church and colleagues to assess the accuracy of methylated SEPT9 DNA for colorectal cancer screening using a commercially available assay. Sensitivity and specificity for all stages of colorectal cancer are 48.2% and 91.5%, respectively. Concerning different tumor stages, combined sensitivity for stages I–II is only 44.7%, and combined sensitivity for stages I–III is 45.1% (33). Other noninvasive approaches, such as fecal testing, also tend not to detect early-stage colorectal cancer with adequate sensitivity and specificity (34). Therefore, scholars are flocking to identify biomarkers for tumor diagnosis based on blood samples. To the best of our knowledge, ours was the first comprehensive whole-methylome discovery effort to identify highly discriminant markers for colorectal cancer from PBMC samples. In the discovery phase performed with Infinium Methylation EPIC array, thousands of DMPs for colorectal cancer were discovered, and a majority of the differentially methylated CpG sites were hypomethylated, which was consistent with the previous study that global DNA methylation in PBMCs is lower in participants who develop cancer during the follow-up period (35). Because candidate CpG sites in PBMCs showed more pronounced methylation differences in early stages colorectal cancer, the phase II biological validation study was biased towards early-stage colorectal cancer to best reflect the intended surveillance population. Carrying forward, five CpG sites were eventually selected as candidate DMPs for early diagnosis of colorectal cancer. We separated T cells, B cells, and the T/B-lymphocyte–depleted cells to uncover which types of cells contributed to the overall PBMC methylation variances. The results indicated that T/B cells depleted preparations are more similar to the PBMCs than T and B cells separately. Because the main cellular components of T/B cells depleted preparations are NK cells, monocytes and dendritic cells, which related to the rapid and powerful antitumor function of immune cells such as NK cells in innate immunity, play an important role in tumor progression (36).
Sequencing is required to interrogate tumor DNA methylation markers in previous studies (31, 37, 38), making potential applications of diagnostics tests too expensive and time-consuming for all subjects. One important finding of our study was the development of a multi-msqPCR technique for the clinical application that allowed simultaneous quantification of five validated DNA methylation markers in a single-tube qPCR assay. This approach could analyze up to 384 samples at a time and provide clinical reports within 6.5–7 hours. Furthermore, based on multitargeted PCR of specific markers, our CDM could avoid the high sequencing cost and shorten detection time, making it a more routine and cost-effective application. A clinical study with stage I/II colorectal cancer was performed and verified the technical advantages of CDM, showing a very good and reproducible clinical performance in detecting methylation. Several blood-based methylation marker candidates have been previously reported for the early detection of colorectal cancer, such as TMEFF2 and SEPT9, with AUC values of only 0.72 and 0.80 for diagnosing colorectal cancer (17). In this study, CDM showed a better performance for early detection of colorectal cancer with a higher AUC of 0.91. Effective detection of advanced adenoma cases is of paramount importance (39). Our study clearly demonstrated that the detection rate of CDM assay for advanced adenoma was 63.04% in cohort V. Such a value was much higher compared with another proposed blood methylation marker, SEPT9, of which, the sensitivity is only 11.2% (33), which was a major strength of this study. Given this performance characteristic, the CDM classifier had the potential to improve the general screening detection of advanced adenoma throughout the development of colorectal cancer. We further validated the diagnostic ability of CDM in cohort V consisting of 180 participants with other types of early-stage cancer, our results demonstrated that CDM had certain false-positive for other tumors. It is worth mentioning that the DNA methylation changes are more pronounced in the early stage, while the specimens of other tumor types were in early stages, which means that the false-positive of CDM in other tumors would be further reduced if all stages were included. More types of cancer and more participants will be included in the further to validate the specificity of CDM.
More importantly, we compared the diagnostic efficiency of CDM with CEA in cohort V, the most commonly used blood-based colorectal cancer biomarker worldwide. The results showed that CEA performed less well than CDM, only identifying 48 of 170 (28.24%) participants with early-stage colorectal cancer. The performance of CEA in our study was similar to previous literature reports, in which colonoscopy is the reference standard. CDM also demonstrated diagnostic value in CEA-negative early-stage colorectal cancer, and a combination of CDM with CEA increased the diagnostic accuracy for early-stage colorectal cancer compared with CDM or CEA alone. The noninvasive screening strategy CDM in our study might improve screening adherence and increase participation rates.
Despite these encouraging preliminary results, several potential limitations need to be emphasized in our current study. First, the sample size of the clinical validation cohort was not sufficiently large enough. A multicenter cohort with a large-scale sample size and more comparable cancer types should ideally be included in the future. We anticipated that there was an opportunity to further obtain more high-quality data for training and then improve the diagnostic performance. Second, lesion size measurements were not always available, and cases with multiple lesions were classified according to their largest or most advanced diseases. Finally, the quality of colonoscopy should be controlled in future cohort studies to avoid the omission of relevant tumors due to the inexperience of clinicians, which might adversely affect estimates of CDM assay specificity.
Collectively, we developed a new, promising and efficient diagnostic method based on five DNA methylation biomarkers and multi-msqPCR technology, CDM, which could diagnosis early-stage colorectal cancer and precancerous lesions with higher accuracy and sensitivity than conventional modalities. Additional research should be done to evaluate whether CDM can be further improved to increase the benefit to patients with early-stage colorectal cancer.
Supplementary Material
Supplementary methods
Supplementary tables
Supplementary figures
Acknowledgments
The authors thank all of the physicians at the multicenter hospitals for facilitating the collection of blood samples. The authors also thank Professor Tao Zhang, Department of Biostatistics, School of Public Health, Shandong University, and Professor Yongyue Wei, Peking University Health Science Center, for their humble support in the experimental design and statistical disorders of this subject. This work was supported by grants from Shandong University Clinical Research Project (2020SDUCRCA002), the Key Research and Development Program of Shandong Province (2021ZLGX02, 2020CXGC011304, and 2022CXGC020507), the National Natural Science Foundation of China (81972007, 82122041, 82002228, and 82130067), Taishan Scholars Climbing Program of Shandong Province (NO.tspd20210323), Young Taishan Scholars Program of Shandong Province (NO.tsqn201909176 and NO.tsqn202211323), Outstanding Young and Middle-aged Scholar of Shandong University, Shandong Provincial Natural Science Foundation (ZR2020QH280), and Tumor Biomarker Innovation Team Foundation of Jinan City (2021GXRC020).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
Y. Xie: Software, formal analysis. P. Li: Conceptualization, software, funding acquisition, writing–original draft. D. Sun: Methodology, writing–original draft. Q. Qi: Software, writing–original draft, writing–review and editing. S. Ma: Formal analysis, methodology. Y. Zhao: Writing-original draft. S. Zhang: Resources. T. Wang: Resources, software. J. Wang: Data curation, software. S. Li: Sample collection. T. Gong: Sample collection. H. Xu: Sample collection. M. Xiong: Sample collection. G. Li: Sample collection. C. You: Sample collection. Z. Luo: Sample collection. J. Li: Conceptualization, resources, writing–review and editing. C. Wang: Formal analysis, supervision, funding acquisition, validation, project administration. L. Du: Funding acquisition, validation, investigation, project administration.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet 2005;365:153–65. [DOI] [PubMed] [Google Scholar]
- 3. Bray C, Bell LN, Liang H, Collins D, Yale SH. Colorectal cancer screening. WMJ 2017;116:27–33. [PubMed] [Google Scholar]
- 4. Demb J, Liu L, Murphy CC, Doubeni CA, Martinez ME, Gupta S. Young-onset colorectal cancer risk among individuals with iron-deficiency anaemia and haematochezia. Gut 2020;70:1529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shaukat A, Kahi CJ, Burke CA, Rabeneck L, Sauer BG, Rex DK. ACG clinical guidelines: colorectal cancer screening 2021. Am J Gastroenterol 2021;116:458–79. [DOI] [PubMed] [Google Scholar]
- 6. Chen H, Li N, Ren J, Feng X, Lyu Z, Wei L, et al. Participation and yield of a population-based colorectal cancer screening programme in China. Gut 2019;68:1450–7. [DOI] [PubMed] [Google Scholar]
- 7. Quintero E, Castells A, Bujanda L, Cubiella J, Salas D, Lanas A, et al. Colonoscopy versus fecal immunochemical testing in colorectal-cancer screening. N Engl J Med 2012;366:697–706. [DOI] [PubMed] [Google Scholar]
- 8. Imperiale TF, Gruber RN, Stump TE, Emmett TW, Monahan PO. Performance characteristics of fecal immunochemical tests for colorectal cancer and advanced adenomatous polyps: a systematic review and meta-analysis. Ann Intern Med 2019;170:319–29. [DOI] [PubMed] [Google Scholar]
- 9. Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 2017;17:97–111. [DOI] [PubMed] [Google Scholar]
- 10. Sookoian S, Gemma C, Fernandez Gianotti T, Burgueno A, Alvarez A, Gonzalez CD, et al. Effects of rotating shift work on biomarkers of metabolic syndrome and inflammation. J Intern Med 2007;261:285–92. [DOI] [PubMed] [Google Scholar]
- 11. Edwards CJ, Feldman JL, Beech J, Shields KM, Stover JA, Trepicchio WL, et al. Molecular profile of peripheral blood mononuclear cells from patients with rheumatoid arthritis. Mol Med 2007;13:40–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Twine NC, Stover JA, Marshall B, Dukart G, Hidalgo M, Stadler W, et al. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res 2003;63:6069–75. [PubMed] [Google Scholar]
- 13. Werner RJ, Kelly AD, Issa JJ. Epigenetics and precision oncology. Cancer J 2017;23:262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol 2016;8:a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barault L, Amatu A, Siravegna G, Ponzetti A, Moran S, Cassingena A, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 2018;67:1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414–23. [DOI] [PubMed] [Google Scholar]
- 18. Bartak BK, Kalmar A, Peterfia B, Patai AV, Galamb O, Valcz G, et al. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics 2017;12:751–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan J, Li J, Guo S, Tao C, Zhang H, Wang W, et al. Genome-wide DNA methylation profiles of low- and high-grade adenoma reveals potential biomarkers for early detection of colorectal carcinoma. Clin Epigenetics 2020;12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Symonds EL, Pedersen SK, Baker RT, Murray DH, Gaur S, Cole SR, et al. A blood test for methylated BCAT1 and IKZF1 vs. a fecal immunochemical test for detection of colorectal neoplasia. Clin Transl Gastroenterol 2016;7:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vogelstein JT, Bridgeford EW, Tang M, Zheng D, Douville C, Burns R, et al. Supervised dimensionality reduction for big data. Nat Commun 2021;12:2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–45. [PubMed] [Google Scholar]
- 23. Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox's proportional hazards model via coordinate descent. J Stat Softw 2011;39:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kisiel JB, Raimondo M, Taylor WR, Yab TC, Mahoney DW, Sun Z, et al. New DNA methylation markers for pancreatic cancer: discovery, tissue validation, and pilot testing in pancreatic juice. Clin Cancer Res 2015;21:4473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 2010;5:1315–6. [DOI] [PubMed] [Google Scholar]
- 26. Yang R, Cheng S, Luo N, Gao R, Yu K, Kang B, et al. Distinct epigenetic features of tumor-reactive CD8+ T cells in colorectal cancer patients revealed by genome-wide DNA methylation analysis. Genome Biol 2019;21:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang L, Qiang J, Yang X, Wang D, Rehman AU, He X, et al. IL1R2 blockade suppresses breast tumorigenesis and progression by impairing USP15-dependent BMI1 stability. Adv Sci (Weinh) 2020;7:1901728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boraschi D, Italiani P, Weil S, Martin MU. The family of the interleukin-1 receptors. Immunol Rev 2018;281:197–232. [DOI] [PubMed] [Google Scholar]
- 29. Liu X, Ren X, Zhou L, Liu K, Deng L, Qing Q, et al. Tollip orchestrates macrophage polarization to alleviate intestinal mucosal inflammation. J Crohns Colitis 2022;16:1151–67. [DOI] [PubMed] [Google Scholar]
- 30. He X, Wu K, Ogino S, Giovannucci EL, Chan AT, Song M. Association between risk factors for colorectal cancer and risk of serrated polyps and conventional adenomas. Gastroenterology 2018;155:355–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med 2020;12:eaax7533. [DOI] [PubMed] [Google Scholar]
- 32. Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer 2004;4:707–17. [DOI] [PubMed] [Google Scholar]
- 33. Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hellebrekers DM, Lentjes MH, van den Bosch SM, Melotte V, Wouters KA, Daenen KL, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res 2009;15:3990–7. [DOI] [PubMed] [Google Scholar]
- 35. Friso S, Udali S, Guarini P, Pellegrini C, Pattini P, Moruzzi S, et al. Global DNA hypomethylation in peripheral blood mononuclear cells as a biomarker of cancer risk. Cancer Epidemiol Biomarkers Prev 2013;22:348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu SY, Fu T, Jiang YZ, Shao ZM. Natural killer cells in cancer biology and therapy. Mol Cancer 2020;19:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clarke MA, Gradissimo A, Schiffman M, Lam J, Sollecito CC, Fetterman B, et al. Human papillomavirus DNA methylation as a biomarker for cervical precancer: consistency across 12 genotypes and potential impact on management of HPV-positive women. Clin Cancer Res 2018;24:2194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bozic T, Kuo CC, Hapala J, Franzen J, Eipel M, Platzbecker U, et al. Investigation of measurable residual disease in acute myeloid leukemia by DNA methylation patterns. Leukemia 2022;36:80–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 2012;142:248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods
Supplementary tables
Supplementary figures



![Figure 4. Performance evaluation of multi-msqPCR. A, Comparison between the multi-msqPCR and uniplex-msqPCR assays. The multi-msqPCR assay produced higher ΔCq values than uniplex-msqPCR assays [6.13 (ΔCq multiple hypermethylated sites) versus 1.44 (ΔCq cg11754974), 1.97 (ΔCq cg24905336), 0.47 (ΔCq cg22678228; left) and 4.43 (ΔCq multiple hypomethylated sites) versus 2.18 (ΔCq cg26026551), 1.79 (ΔCq cg00227790; right)]. B, Assessment of the analytic sensitivity of the multi-msqPCR assay. Primer probe sets for multiple hypermethylated sites (left) and multiple hypomethylated sites (right) were tested for two technical replicates with fold dilutions of 50 ng bisulfite-converted DNA. The multi-msqPCR assay detected tumor DNA signals with as little as 0.1%. ΔCq = (CqACTB− Cqbiomarker).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8dd9/10618739/2b5a52f42d30/3636fig4.jpg)