NeoCOAST, the first neoadjuvant platform trial of novel immuno-oncology combinations, demonstrates that combined therapies improve responses and immunologic cell activation compared to durvalumab (anti–PD-L1) monotherapy in resectable non–small cell lung cancer.
Abstract
Neoadjuvant chemoimmunotherapy improves pathologic complete response rate and event-free survival in patients with resectable non–small cell lung cancer (NSCLC) versus chemotherapy alone. NeoCOAST was the first randomized, multidrug platform trial to examine novel neoadjuvant immuno-oncology combinations for patients with resectable NSCLC, using major pathologic response (MPR) rate as the primary endpoint. Eighty-three patients received a single cycle of treatment: 26 received durvalumab (anti–PD-L1) monotherapy, 21 received durvalumab plus oleclumab (anti-CD73), 20 received durvalumab plus monalizumab (anti-NKG2A), and 16 received durvalumab plus danvatirsen (anti-STAT3 antisense oligonucleotide). MPR rates were higher for patients in the combination arms versus durvalumab alone. Safety profiles for the combinations were similar to those of durvalumab alone. Multiplatform immune profiling suggested that improved MPR rates in the durvalumab plus oleclumab and durvalumab plus monalizumab arms were associated with enhanced effector immune infiltration of tumors, interferon responses and markers of tertiary lymphoid structure formation, and systemic functional immune cell activation.
Significance:
A neoadjuvant platform trial can rapidly generate clinical and translational data using candidate surrogate endpoints like MPR. In NeoCOAST, patients with resectable NSCLC had improved MPR rates after durvalumab plus oleclumab or monalizumab versus durvalumab alone and tumoral transcriptomic signatures indicative of augmented immune cell activation and function.
See related commentary by Cooper and Yu, p. 2306.
This article is featured in Selected Articles from This Issue, p. 2293
INTRODUCTION
The treatment landscape for patients with advanced non–small cell lung cancer (NSCLC) has evolved dramatically over the last decade, as evidenced by approvals for dozens of new targeted and immuno-oncology agents. Translation of these therapeutic advances to earlier-stage NSCLC has lagged behind despite historically poor outcomes in this setting (1). However, combinations of multiple agents in the perioperative setting are now under evaluation in clinical trials, with a major focus on neoadjuvant therapy (2).
Several studies have previously evaluated PD-(L)1 inhibitors as neoadjuvant monotherapy for patients with early-stage NSCLC, with major pathologic response (MPR) and pathologic complete response (pCR) rates ranging from 6.7% to 45.0% and 0% to 16.2%, respectively (3–10). The PD-1 inhibitor nivolumab plus platinum-based chemotherapy is currently approved in several jurisdictions, including the United States and Canada, for use as neoadjuvant treatment for select patients with resectable NSCLC based on the results of the phase III CheckMate 816 study (11–13). The combination of nivolumab and chemotherapy led to significant increases in the pCR rate (24.0% vs. 2.2%) and event-free survival (EFS; median 31.6 vs. 20.8 months) compared with chemotherapy alone in patients with resectable, stage IB–IIIA NSCLC [per American Joint Committee on Cancer (AJCC), 7th edition]. Moreover, patients with pCR had longer EFS compared with patients without pCR, and the combination did not increase the incidence of adverse events (AE) or impede the feasibility of surgery (13). These results were reproduced in the context of resectable, stage III NSCLC (AJCC 7th edition) in the phase II NADIM2 trial, in which complete resection, progression-free survival, and overall survival were all significantly increased by the addition of perioperative immunotherapy to neoadjuvant chemotherapy (14). Furthermore, in the LCMC3 study, two cycles of the PD-L1 inhibitor atezolizumab as neoadjuvant monotherapy led to an MPR rate of 20% and a pCR rate of 6% in patients with previously untreated, resectable, stage IB–IIIB NSCLC (AJCC 8th edition) who had surgery (8).
The impact of dual immune-checkpoint inhibitor therapy in patients with resectable NSCLC was assessed in the single-arm, phase II study from Reuss and colleagues (15) and the phase II randomized NEOSTAR study (5), both of which evaluated nivolumab alone and in combination with the anti-CTLA4 monoclonal antibody (mAb) ipilimumab for patients with previously untreated, resectable, stage I–IIIA NSCLC (AJCC 7th edition). In the study by Reuss and colleagues (15), the pCR rate in patients treated with nivolumab plus ipilimumab was 33% in the resected population, although the study was terminated early, partly due to 33% of patients experiencing grade 3–5 treatment-related AEs (TRAE; ref. 15). In the intent-to-treat (ITT) population of the NEOSTAR trial, 38% and 22% of patients had an MPR in the combination and monotherapy arms, respectively, whereas 29% and 9% had a pCR, respectively (5); the rate of grade 3 or higher TRAEs was 10% in the combination arm. Most recently, the ipilimumab–nivolumab arm of CheckMate 816, though discontinued early due to the changing treatment landscape in advanced lung cancer, reported a pCR rate of 20% in patients with stage IB–IIIA, resectable NSCLC (16). Overall, these results illustrate that multi–immune pathway modulation may be superior to monotherapy and support the evaluation of other immunomodulatory agents with novel mechanisms of action, in combination with anti–PD-(L)1 therapy, with the goal of augmenting the pathologic responses and improving survival outcomes, thereby increasing the cure rate of resectable NSCLC.
NeoCOAST (NCT03794544) is the first phase II, open-label, randomized, multicenter, multidrug platform, window-of-opportunity study of neoadjuvant therapy for patients with previously untreated, early-stage, resectable NSCLC. Patients received a single cycle of the anti–PD-L1 mAb durvalumab (17) alone or in combination with one of three novel immune-oncology agents—oleclumab, monalizumab, or danvatirsen—using MPR rate as the primary efficacy endpoint. The short-term administration of the study drugs in the neoadjuvant setting provided an opportunity to rapidly evaluate the activity, feasibility, safety, and immune modulation of novel immuno-oncology agents combined with durvalumab compared with durvalumab alone. These agents were chosen due to the potential for their mechanisms of action to be additive or synergistic with durvalumab (based on preliminary evidence of clinical activity in combination with durvalumab in solid tumors), having an established recommended dose in combination with durvalumab, and an acceptable safety profile (18–25). Oleclumab, an anti-CD73 human IgG1 mAb, selectively binds to and inhibits CD73; by inhibiting CD73 and reducing its cell-surface expression, oleclumab reduces extracellular adenosine production and promotes antitumor immunity (ref. 22; Supplementary Fig. S1A). In a phase I study, oleclumab combined with durvalumab had a manageable safety profile and showed promising antitumor activity in patients with EGFR-mutant NSCLC (20). The anti-NKG2A monalizumab is a first-in-class, humanized IgG4 mAb that specifically binds to and blocks the inhibitory receptor NKG2A from binding to HLA-E, thereby reducing inhibition of natural killer (NK) and CD8+ T cells and enhancing antitumor immunity (ref. 18; Supplementary Fig. S1B). In a phase Ib trial, monalizumab combined with durvalumab was shown to have manageable safety and demonstrated modest clinical activity in patients with recurrent ovarian cancer (19). Both oleclumab and monalizumab were assessed in the phase II COAST trial, which showed that durvalumab combined with oleclumab or monalizumab improved objective response rates (ORR) and prolonged progression-free survival in patients with unresectable, stage III NSCLC versus durvalumab alone, with no new or significant safety signals (23). The anti-STAT3 antisense oligonucleotide danvatirsen reduces expression of STAT3 by targeted downregulation of STAT3 mRNA, reverses a suppressive tumor microenvironment, and promotes proinflammatory gene expression changes (ref. 25; Supplementary Fig. S1C). Durvalumab in combination with danvatirsen has previously been shown to have a manageable safety profile in early-phase studies across multiple tumor types (21, 26), and demonstrated encouraging efficacy signals in patients with recurrent/metastatic head and neck squamous cell carcinoma (21). However, the clinical development of danvatirsen was discontinued by the study sponsor due to strategic portfolio prioritization after the rights for further development of the molecule were returned to the company from which it was licensed; as a result, only response and safety data are available and are reported for the durvalumab + danvatirsen treatment arm in the present study.
Here, we report the clinical efficacy and safety outcomes from all treatment arms, as well as translational correlates from the durvalumab monotherapy, durvalumab + oleclumab, and durvalumab + monalizumab treatment arms of the NeoCOAST trial.
RESULTS
Participants
Between March 20, 2019, and September 21, 2020, a total of 84 patients with stage IA3–IIIA NSCLC (per AJCC 8th edition) were enrolled in 17 study centers across seven countries (Canada, France, Italy, Portugal, Spain, Switzerland, and the United States). The study design is shown in Fig. 1A. Among 84 enrolled patients, 83 received study treatment. Twenty-seven patients were randomized to durvalumab monotherapy, 21 to durvalumab + oleclumab, 20 to durvalumab + monalizumab, and 16 to durvalumab + danvatirsen (Fig. 1B); treatment in each arm was administered over a 4-week period prior to the planned surgery. One patient randomized to the durvalumab monotherapy arm did not receive any study treatment due to an uncontrolled intercurrent illness prior to therapy initiation. Baseline disease characteristics were representative of the intended patient population and were overall well balanced across the four arms, although the durvalumab + monalizumab arm included a higher proportion of patients with stage IA3 disease at study entry (n = 6; 30.0%) and the durvalumab arm included a lower proportion of patients with stage IIIA disease (n = 2; 7.4%; Table 1). Representativeness of the study participants is described in Supplementary Table S1. Overall, 80 (95.2%) patients completed planned neoadjuvant therapy (durvalumab ± novel agent), of whom 78 (92.9%) completed the follow-up assessment on day 105: 26 (96.3%) in the durvalumab monotherapy arm, 18 (85.7%) in the durvalumab + oleclumab arm, 19 (95.0%) in the durvalumab + monalizumab arm, and 15 (93.8%) in the durvalumab + danvatirsen arm (Fig. 1B). At the time of the final data cutoff (September 15, 2021), all patients were off study.
Figure 1.
Study design and patient disposition. A, Eligible patients with resectable, early-stage [stage IA3 (>2 cm) to IIIA per AJCC staging, 8th edition) NSCLC were randomized to receive one 28-day cycle of durvalumab monotherapy or durvalumab in combination with oleclumab, monalizumab, or danvatirsen. Patients were stratified by lymph node involvement. Surgical resection was planned to occur between days 29 and 42 after the first dose of neoadjuvant therapy. After surgery, patients were followed for AEs up to day 105. The primary endpoint was the MPR rate, defined as the proportion of patients with ≤10% residual viable tumor cells in the surgical specimen (primary tumor and sampled lymph nodes at surgery). Tumor samples were collected, where possible, at screening (day −1 to day −21) and at surgery. Blood samples were collected, where possible, at screening (pretherapy), at baseline (day 1), at the end of neoadjuvant treatment (day 28), and at the end of study (day 105 ± 21 days). Stool samples were collected, where possible, at screening (within 21 days of the start of treatment) and on treatment (day 15 to day 28). B, Flow diagram depicts the disposition of patients through the phases of the study, from screening, neoadjuvant treatment, surgical resection, and study completion. The total numbers of patients in the ITT and as-treated populations, as well as reasons for discontinuations of treatment, are shown. Danva, danvatirsen; Durva, durvalumab; ECOG PS, Eastern Cooperative Oncology Group performance status; Mona, monalizumab; Ole, oleclumab; Q4W, once every 4 weeks; Q2W, once every 2 weeks; QW, every week. *The danvatirsen arm was stopped early, as the program was discontinued. †Patients who completed treatment with novel agent. ‡One patient did not receive all planned doses of danvatirsen but had surgery. ¶Death due to perioperative complications not considered related to treatment.
Table 1.
Baseline characteristics and demographics
| Durva | Durva + Ole | Durva + Mona | Durva + Danva | Total | |
|---|---|---|---|---|---|
| (n = 27) | (n = 21) | (n = 20) | (n = 16) | (N = 84) | |
| Median age (min, max) | 67 (51, 83) | 65 (52, 80) | 64.5 (54, 82) | 71.5 (56, 87) | 67.5 (51, 87) |
| Sex, n (%) | |||||
| Male | 14 (51.9) | 12 (57.1) | 14 (70.0) | 10 (62.5) | 50 (59.5) |
| Female | 13 (48.1) | 9 (42.9) | 6 (30.0) | 6 (37.5) | 34 (40.5) |
| ECOG PS a , n (%) | |||||
| 0 | 19 (73.1) | 12 (57.1) | 12 (60.0) | 10 (62.5) | 53 (63.1) |
| 1 | 7 (26.9) | 9 (42.9) | 8 (40.0) | 6 (37.5) | 30 (35.7) |
| Smoking, n (%) | |||||
| Never | 6 (22.2) | 1 (4.8) | 1 (50.0) | 1 (6.3) | 9 (10.7) |
| Current or former | 21 (77.8) | 20 (95.2) | 19 (95.0) | 15 (93.8) | 75 (89.3) |
| Stage, n (%) | |||||
| IA3 | 4 (14.8) | 1 (4.8) | 6 (30.0) | 1 (6.3) | 12 (14.3) |
| IB | 7 (25.9) | 4 (19.0) | 2 (10.0) | 1 (6.3) | 14 (16.7) |
| IIA | 3 (11.1) | 4 (19.0) | 1 (5.0) | 2 (12.5) | 10 (11.9) |
| IIB | 11 (40.7) | 7 (33.3) | 8 (40.0) | 7 (43.8) | 33 (39.3) |
| IIIA | 2 (7.4) | 5 (23.8) | 3 (15.0) | 5 (31.3) | 15 (17.9) |
| Histology, n (%) | |||||
| Adenocarcinoma | 18 (66.7) | 14 (66.7) | 11 (55.0) | 8 (50.0) | 51 (60.7) |
| Squamous cell carcinoma | 9 (33.3) | 7 (33.3) | 6 (30.0) | 4 (25.0) | 26 (31.0) |
| Large cell carcinoma | 0 | 0 | 2 (10.0) | 1 (6.3) | 3 (3.6) |
| Adenosquamous carcinoma | 0 | 0 | 0 | 1 (6.3) | 1 |
| Platen epithelial carcinoma | 0 | 0 | 0 | 1 (6.3) | 1 |
| Not specified | 0 | 0 | 1 (5.0) | 1 (6.3) | 2 |
Abbreviations: Danva, danvatirsen; Durva, durvalumab; ECOG PS, Eastern Cooperative Oncology Group performance status; Mona, monalizumab; Ole, oleclumab.
aECOG PS was not available for one patient in the durvalumab monotherapy arm.
Clinical Activity
All efficacy endpoints are reported for the ITT population, which comprised 84 patients across the four treatment arms. MPR rates were 11.1% [3/27; 95% confidence interval (CI), 2.4–29.2] in the durvalumab monotherapy arm, 19.0% (4/21; 95% CI, 5.4–41.9) in the durvalumab + oleclumab arm, 30.0% (6/20; 95% CI, 11.9–54.3) in the durvalumab + monalizumab arm, and 31.3% (5/16; 95% CI, 11.0–58.7) in the durvalumab + danvatirsen arm (Supplementary Table S2). Compared with durvalumab monotherapy, the proportion of patients with MPR was higher in the combination arms (Fig. 2A). pCR rates were 3.7% (1/27; 95% CI, 0.1–19.0) in the durvalumab monotherapy arm, 9.5% (2/21; 95% CI, 1.2–30.4) in the durvalumab + oleclumab arm, 10.0% (2/20; 95% CI, 1.2–31.7) in the durvalumab + monalizumab arm, and 12.5% (2/16; 95% CI, 1.6–38.3) in the durvalumab + danvatirsen arm (Supplementary Table S2).
Figure 2.
Pathologic regressions at surgery; genomic profiles of ITT population and correlates with pathologic responses. A, The magnitude of pathologic response is shown by percent residual viable tumor cells in resected tumor and nodal samples for all patients with available data (total N = 75; durvalumab monotherapy arm: n = 24; durvalumab + oleclumab arm: n = 18; durvalumab + monalizumab arm: n = 18; durvalumab + danvatirsen arm: n = 15), and annotated with histologic subtype, tumor mutational burden (TMB; mutations/megabase), and history of smoking. PD-L1 status (≥1% positive; <1% negative) from baseline tumor biopsies was determined by IHC (SP263) for all evaluable patients (n = 33). Presence of activating EGFR mutations or ALK fusions was determined by whole-exome sequencing (n = 34). B, Residual viable tumor cells (RVT) from resected tumor and nodal samples are reported as 0% to 100%, and MPR (RVT ≤10%) for n = 60 patients (durvalumab monotherapy: n = 24, durvalumab + oleclumab: n = 18; durvalumab + monalizumab: n = 18). Best response by Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST) is reported for n = 58 patients. Somatic tumor alterations identified in EGFR, KRAS, STK11, KEAP1, TP53, ALK, and RET genes are reported from tumor tissue for n = 35 patients. For patients with evaluable circulating tumor DNA (ctDNA) samples at baseline, each patient is identified as having detected or no detected ctDNA at baseline. For patients with detectable ctDNA at baseline (total N = 20; durvalumab monotherapy: n = 6; durvalumab + oleclumab: n = 7; durvalumab + monalizumab: n = 7), molecular response is depicted at end-of-treatment (day 28, n = 14) and follow-up (day 105, n = 15) time points for all patients with evaluable ctDNA at those time points and represented as complete molecular response [100% reduction in variant allele frequency (VAF) from baseline, also referred to as complete clearance], partial molecular response (≥50% reduction in VAF from baseline), or no molecular response (<50% reduction in VAF from baseline). BESTRESP, best response; FU, follow-up; MR, molecular response; WT, wild-type.
The MPR rate in the resected population was 22.2% (95% CI, 6.4–47.6) in the durvalumab + oleclumab arm, 33.3% (95% CI, 13.3–59.0) in the durvalumab + monalizumab arm, and 33.3% (95% CI, 11.8–61.6) in the durvalumab + danvatirsen arm compared with 12.5% (95% CI, 2.7–32.4) in the durvalumab monotherapy arm. We observed a lower median percentage of residual viable tumor (%RVT) in resected tumor and nodal samples from patients treated with durvalumab + oleclumab and durvalumab + danvatirsen (40.0% and 30.0%, respectively) compared with durvalumab alone (60.0%; Supplementary Fig. S2). Computed tomographic imaging was planned prior to neoadjuvant treatment and again after treatment (prior to surgery) for all patients; radiographic responses are summarized in Supplementary Table S2.
Safety and Toxicity
Safety endpoints are reported for the as-treated population (all patients who received at least one dose of any study treatment), which comprised 83 patients (Fig. 1B). The incidence and severity of treatment-emergent AEs (TEAE) were similar across all treatment arms (Table 2). TRAEs (as assessed by the investigator) occurred in nine (34.6%), 12 (57.1%), 10 (50.0%), and seven (43.8%) patients in the durvalumab monotherapy, durvalumab + oleclumab, durvalumab + monalizumab, and durvalumab + danvatirsen arms, respectively; grade ≥3 TRAEs occurred in one (4.8%) patient in the durvalumab + oleclumab arm (diabetic ketoacidosis) and one (6.3%) patient in the durvalumab + danvatirsen arm (procedural hemorrhage). Serious TRAEs occurred in three patients: one (3.8%) in the durvalumab monotherapy arm (immune-mediated arthritis), one (4.8%) in the durvalumab + oleclumab arm (diabetic ketoacidosis), and one (6.3%) in the durvalumab + danvatirsen arm (procedural hemorrhage). Three patients discontinued treatment due to AEs (not treatment-related): one (4.8%) in the durvalumab + oleclumab arm (grade 1 transient ischemic attack), one (5.0%) in the durvalumab + monalizumab arm (grade 2 COVID-19), and one (6.3%) in the durvalumab + danvatirsen arm (grade 2 hepatic enzyme increase). One death occurred 11 days after surgery due to a surgical AE of a bronchial anastomotic complication accompanied by postoperative COVID-19 infection in the durvalumab + danvatirsen arm, deemed by the investigators not to be related to treatment.
Table 2.
Safety summary (as-treated population)
| Incidence, n (%) | Durva | Durva + Ole | Durva + Mona | Durva + Danva |
|---|---|---|---|---|
| (n = 26) | (n = 21) | (n = 20) | (n = 16) | |
| Any TEAE | 18 (69.2) | 19 (90.5) | 15 (75.0) | 13 (81.3) |
| Grade ≥3 TEAEs | 5 (19.2) | 3 (14.3) | 2 (10.0) | 5 (31.3) |
| Serious TEAEs | 3 (11.5) | 2 (9.5) | 1 (5.0) | 5 (31.3) |
| Any TRAE | 9 (34.6) | 12 (57.1) | 10 (50.0) | 7 (43.8) |
| Grade ≥3 TRAEs | 0 | 1 (4.8) | 0 | 1 (6.3) |
| Serious TRAEsa | 1 (3.8) | 1 (4.8) | 0 | 1 (6.3) |
| AEs leading to treatment discontinuation | 0 | 1 (4.8) | 1 (5.0) | 1 (6.3) |
| Deathsb | 0 | 0 | 0 | 1 (6.3) |
Abbreviations: Danva, danvatirsen; Durva, durvalumab; Mona, monalizumab; Ole, oleclumab.
aSerious TRAEs included one patient with immune-mediated arthritis in the Durva arm; one patient with diabetic ketoacidosis in the Durva + Ole arm; and one patient with procedural hemorrhage in the Durva + Danva arm.
bDeath in the Durva + Danva arm was due to an AE of bronchial anastomosis complication, deemed not to be related to either study drug.
The most common TEAEs by preferred term with a frequency greater than 10% in any treatment arm are reported in Supplementary Table S3. Overall, 38 (45.8%) patients experienced TRAEs; the most frequently reported were fatigue (10.8%), and asthenia and pruritis (each 6.0%; Supplementary Table S4).
AEs of special or potential interest (AESI/AEPI) are summarized in Supplementary Table S5. Overall, 31 (37.3%) patients experienced an AESI/AEPI; there were no AESI/AEPIs with an outcome of death. Immune-mediated AEs (imAE) occurred in three patients: one (3.8%) in the durvalumab monotherapy arm (grade 2 immune-mediated arthritis and grade 2 musculoskeletal pain), one (4.8%) in the durvalumab + oleclumab arm (grade 3 diabetic ketoacidosis), and one (5.0%) in the durvalumab + monalizumab arm (grade 1 maculopapular rash). Serious imAEs occurred in one (3.8%) patient in the durvalumab monotherapy arm (grade 2 immune-mediated arthritis), and 1 (4.8%) patient in the durvalumab + oleclumab arm (grade 3 diabetic ketoacidosis). There were no imAEs with an outcome of death.
Surgical Resectability
The proportion of patients for whom surgery was feasible (i.e., able to proceed with surgery between day 29 and day 42 after week 1, day 1, as defined in the clinical study protocol) was similar between treatment arms. Overall, 76 (91.6%) patients received surgery, of whom 72 (86.7%) received surgery within 42 days (the protocol-defined time was not considered to be a delay). Resection rates in the durvalumab + oleclumab, durvalumab + monalizumab, durvalumab + danvatirsen, and durvalumab monotherapy arms were 81.0%, 90.0%, 93.8%, and 84.6%, respectively. The mean time to surgical resection from week 1, day 1 was 38.2 days. Overall, five (6.6%) patients’ surgery was delayed beyond day 42: one due to an AE of pneumonia, not considered to be treatment-related (57 days from week 1, day 1; durvalumab monotherapy arm), three patients had a scheduling delay (49 and 43 days from week 1, day 1 in the durvalumab monotherapy arm and 57 days from week 1, day 1 in the durvalumab + danvatirsen arm), and one patient was no longer a candidate for surgery following a second opinion (78 days from week 1, day 1; durvalumab + oleclumab arm). Of the seven treated patients unable to complete surgery, five had progressive tumors that were deemed no longer resectable by the treating physicians, one was lost to follow-up, and another had a serious AE (pneumonia, nontreatment-related) and was no longer eligible for surgery.
Association between MPR Rate and Pretherapy Clinical or Biomarker Characteristics
Overall, more patients with stage I or II disease had an MPR (Supplementary Table S6). MPRs were more frequent in patients with pretherapy tumor PD-L1 expression ≥1% versus those with <1% [overall, 35 (41.7%) patients had evaluable PD-L1 status], in both the durvalumab + oleclumab and durvalumab + monalizumab arms, but not in the durvalumab monotherapy or durvalumab + danvatirsen arms (no patients with an MPR in the durvalumab or durvalumab + danvatirsen arms were evaluable for baseline PD-L1; Supplementary Table S6). Among six patients with an MPR and evaluable PD-L1 expression, five had PD-L1–positive tumors (two patients in the durvalumab + oleclumab arm and three patients in the durvalumab + monalizumab arm; Supplementary Fig. S3A); one patient with an MPR had a PD-L1–negative but CD73-high (defined as ≥10% of tumor cells) tumor in the durvalumab + oleclumab arm (Supplementary Table S6). Increased CD8+ T-cell density before therapy was not associated with MPR in any arm (Supplementary Fig. S3B). Pretherapy density of NKG2A+ cells in the tumor area was not associated with MPR in any arm (Supplementary Table S6; Supplementary Fig. S3C). In the durvalumab + oleclumab arm, all patients with evaluable pretherapy tumoral CD73 IHC expression (n = 11) who also had an MPR were CD73-high (Supplementary Fig. S3D). In the same arm, high CD73 expression was numerically correlated with fewer viable tumor cells at surgery (Rho = -0.5, P = 0.14; Supplementary Fig. S3E). The opposite trend was observed with durvalumab monotherapy, where high CD73 expression at baseline was significantly correlated with a greater percentage of viable tumor at surgery (Rho = 0.85, P = 0.01). A patient with an MPR in the durvalumab + oleclumab arm demonstrated a pretherapy-to-surgery decrease in CD73+ tumor cells and an increase in NKG2A+ cells and CD8+ T cells in the tumor microenvironment (Supplementary Fig. S3F).
Somatic Mutational Profiling
Somatic tumor alterations were profiled by whole-exome sequencing (WES) from tumor and matched blood DNA. Notably, among patients who experienced an MPR, two had EGFR exon 21 L858R driver mutations (both in the durvalumab + oleclumab arm; stage 1B N0 and stage IIA N0). RET (CCDC6–RET) and ALK (ALK–EML4) gene fusions were observed in two patients without an MPR (one in the durvalumab monotherapy arm and one in the durvalumab + monalizumab arm, respectively). Additionally, KRAS alterations were observed in nine patients without an MPR (two G12A, four G12C, one G12D, and two Q61H), both with and without co-occurring STK11 or KEAP1 alterations (Fig. 2B). Tumor mutational burden ranged from 0.11 to 22.02 mutations per megabase and was not correlated with %RVT in the surgical specimen (Rho = 0.19, P = 0.30) in analyzed treatment groups.
Modulatory Transcriptomic Changes in Tumors from Pre– to Post–Neoadjuvant Treatment
RNA sequencing (RNA-seq) analysis of pretherapy and resected tumor tissues [from formalin-fixed, paraffin-embedded (FFPE) samples] from the durvalumab monotherapy, durvalumab + oleclumab, and durvalumab + monalizumab arms revealed treatment-related transcriptomic changes from pre- to posttreatment. A numerically greater number of differentially expressed genes (DEG; adj P < 0.1) in surgical tumor samples compared with pretherapy tumor samples were observed in the durvalumab + oleclumab and durvalumab + monalizumab arms, versus the durvalumab monotherapy arm (Supplementary Table S7; Fig. 3A). Expression of genes associated with NK cells (KLRC1, GNLY) and CD8 T cells (CD8A, GZMK) increased after treatment in all arms, with a greater magnitude in the durvalumab + oleclumab and durvalumab + monalizumab [1–5 log2 fold change (FC), adj P < 0.1] arms compared with durvalumab monotherapy [0–1 log2FC, not significant (n.s.); Fig. 3B]. A gene signature associated with tertiary lymphoid structure (TLS) formation was calculated from pretherapy and surgery tumor transcriptomes, and identified significant upregulation of TLS signature after treatment with durvalumab + monalizumab (P < 0.05; Supplementary Fig. S4). Gene set enrichment analyses (GSEA) revealed that programs associated with the interferon pathway, as well as inflammatory signatures, were significantly upregulated after treatment with durvalumab + oleclumab [normalized enrichment score (NES) >1.5; adj P < 0.05] and durvalumab + monalizumab (NES >1.5; adj P < 0.05; Fig. 3A). Pathway enrichment analyses supported enrichment of Th1/Th2 and antigen presentation pathways in these treatment arms. Moreover, pathways involved in T-cell receptor signaling were enriched in the durvalumab + oleclumab arm, and pathways involved in NK-cell and B-cell receptor signaling were enriched in the durvalumab + monalizumab arm. Immune deconvolution to estimate relative abundance and distinct subsets of immune cells in the tumor microenvironment revealed an increase in CD8 T-cell and B-cell signature scores following treatment with durvalumab + monalizumab (Supplementary Fig. S5). T cells, cytotoxic lymphocytes, and CD8 T cells were increased after treatment with durvalumab + oleclumab and durvalumab + monalizumab. Evaluation of individual genes related to TLS, chemokines, and immune cells suggested increased expression of markers associated with TLS after treatment with durvalumab + oleclumab and durvalumab + monalizumab (Fig. 3A).
Figure 3.
Treatment-related transcriptomic changes in tumor and peripheral blood. A, Left: differential gene expression between tumors collected at pretherapy and at surgery was assessed in all patients with evaluable paired tumor samples (total, n = 13 patients). Paired samples in each arm had the following MPR rates: durvalumab monotherapy: 0/4; durvalumab + oleclumab: 1/5; durvalumab + monalizumab: 2/4. Right: gene set enrichment analysis (GSEA) was used to identify gene sets and signatures significantly down- or upregulated from pretherapy to surgery on each treatment arm. The durvalumab monotherapy arm is not pictured, as no significant gene enrichment was observed. B, Patients with MPR are indicated in closed teal circle; patients without MPR are indicated in open gray circle. Top and top middle: mRNA from select genes associated with T cells, NK cells, and cytotoxicity is shown from pretreatment and surgery tumor tissue (n = 69 samples; n = 35 pretherapy, n = 34 surgery). Patients with paired samples (an evaluable sample from both pretreatment and surgery) are connected by a line. Bottom and bottom middle: mRNA from select genes associated with tumor and blood lymphocyte recruitment is shown from pretherapy to end-of-treatment peripheral blood collections (n = 120 samples; n = 65 pretherapy, n = 55 end-of-treatment). Y-axes units are all [Log2 (TPM + 0.01)], where TPM is transcripts per million. DN, down; Durva, durvalumab; IPA, Ingenuity Pathway Analysis; Mona, monalizumab; Ole, oleclumab.
Profiling of Protein Expression in Tumors
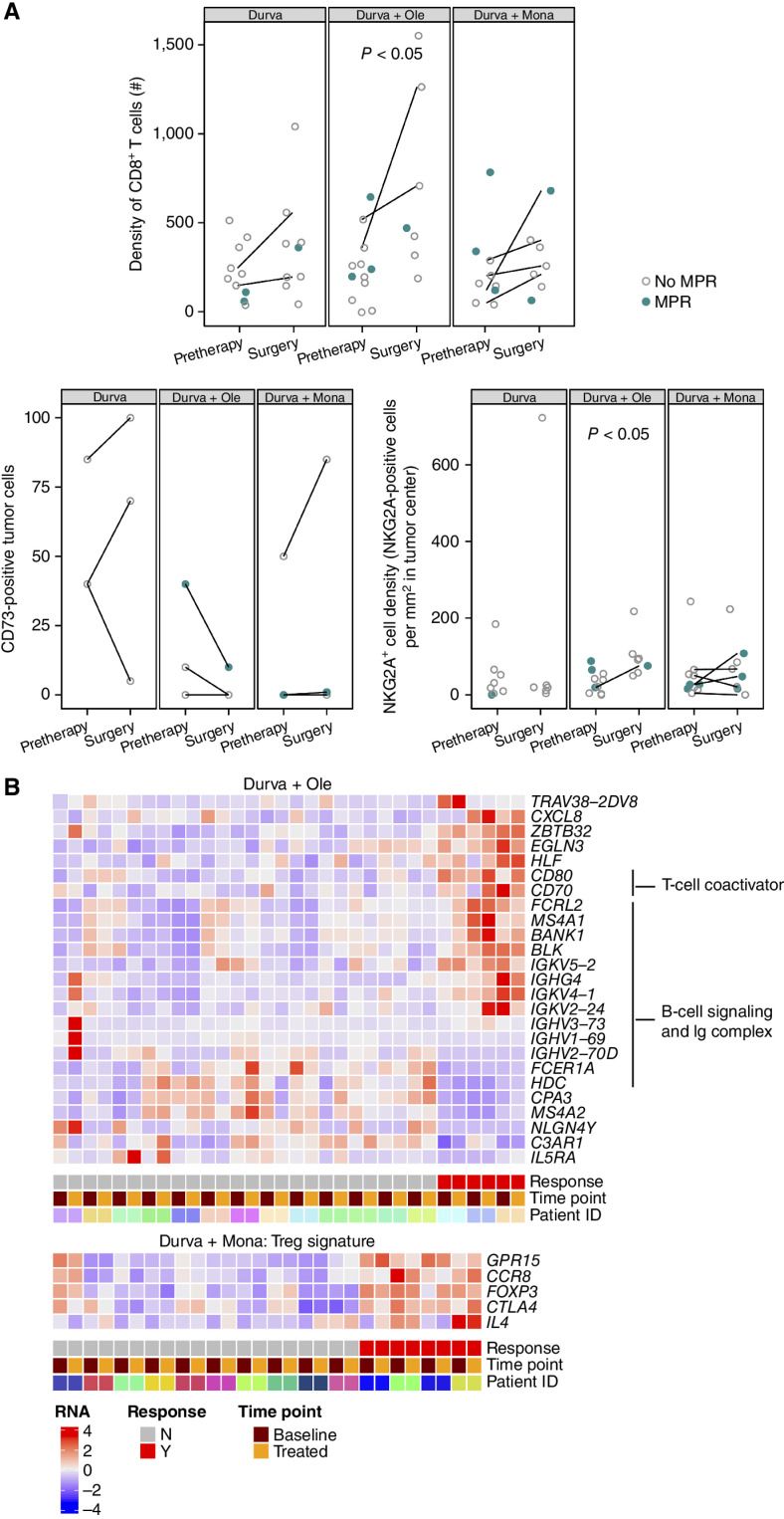
Pharmacodynamic changes, including increased CD8+ T cells in the tumor microenvironment, were observed in all treatment arms (FC: 1.9–2.1, P < 0.05 in the durvalumab + oleclumab arm, n.s. in all other arms; Fig. 4A). We also evaluated the expression of CD73 on tumor cells by IHC in paired baseline and resected tumors; this analysis revealed a decrease in the percentage of tumor cells expressing CD73 (at any intensity) after treatment with durvalumab + oleclumab, but not after treatment in the other arms (Fig. 4A). NKG2A+ cell density in the tumor area significantly increased after treatment with durvalumab + oleclumab (FC: 2.1, P < 0.05; Fig. 4A).
Figure 4.
Treatment-related changes in the tumor microenvironment by IHC; relationship of IHC and peripheral blood RNA biomarkers and MPR. A, IHC biomarkers are shown in all patients with evaluable tumor samples at pretherapy and surgery. Patients with MPR are indicated in closed teal circle; patients without MPR are indicated in open gray circle. Patients with paired samples (an evaluable sample from both pretreatment and surgery) are connected by a line. Top: CD8+ (SP239) T-cell density as number of positive cells/mm2 tumor area in all evaluable patients (n = 54 samples). Bottom left: Percentage of tumor cells positive for CD73 (D7F9A) at any intensity in paired cases only (n = 20 samples from 10 paired cases). Bottom right: NKG2A+ (AR9352) cell density as number of positive cells/mm2 tumor area (n = 49 samples). B, Heat map of selected genes that were significantly differentially expressed between patients with and without an MPR in analyses of patients with paired pretherapy and end-of-treatment samples (n = 54). These analyses identified numerous genes associated with T-cell coactivation, B-cell signaling, and Ig complex upregulated in the peripheral blood of patients with an MPR in the durvalumab + oleclumab arm (n = 16 paired cases). In the durvalumab + monalizumab arm, genes associated with regulatory T cells (Treg) are upregulated in patients with an MPR (n = 14 paired cases). Durva, durvalumab; Mona, monalizumab; Ole, oleclumab.
Genomic Profiling of Peripheral Blood Transcriptome and Stool Microbiome in Patients with and without an MPR
We performed total RNA-seq on whole blood from pre- and posttreatment samples and found significant gene expression changes between responders (MPR) and nonresponders (no MPR) in each treatment arm, controlling for time point. The number of DEGs in the durvalumab + oleclumab (n = 161) and durvalumab + monalizumab (n = 126) arms was much higher than in the durvalumab monotherapy arm [n = 31; adj P < 0.1 and abs(FC) ≥1.5]. Gene ontology enrichment analysis showed that DEGs in the durvalumab + oleclumab arm were enriched in immune genes (Fig. 4B), especially genes involved in the B-cell receptor signaling pathway (fold enrichment = 9.25, adj P = 0.02), and immunoglobulin complex (fold enrichment = 6.35, adj P = 0.018). Regulatory T-cell gene signatures were significantly upregulated in the durvalumab + monalizumab arm (Fig. 4B; adj P = 0.002). The DEGs that were found to be associated with MPR were unique to their respective treatment arms and not associated with MPR in other treatment arms.
We characterized the gut microbiome from stool samples at baseline from 53 patients (18, 14, and 12 in the durvalumab monotherapy, durvalumab + oleclumab, and durvalumab + monalizumab arms, respectively) and evaluated the alpha- and beta-diversity and the taxonomic abundances observed in patients with an MPR compared with those without. In all arms combined, a trend of greater richness in patients with an MPR was observed, but was not statistically significant (P = 0.09; Supplementary Fig. S6A), though we noted a significant difference in the richness at baseline between patients with an MPR and those without in the durvalumab + oleclumab arm (P = 0.041; Supplementary Fig. S6B). Comparison of the gut microbiome diversity by response status did not reveal apparent differences by MPR when comparing inverse Simpson alpha-diversity (P = 0.90; Supplementary Fig. S6C and S6D) or when ordinating beta-diversity measured by binary Jaccard index (P = 0.172; Supplementary Fig. S6E). However, differential enrichment analysis demonstrated that the relative abundance of several bacterial taxa, such as Phascolarctobacterium succinatutens, Streptococcus parasanguinis, and Lactobacillus paragasseri, was increased in responders, in addition to Acidaminococcaceae and Streptococcaceae families (Supplementary Fig. S6F). We observed no differences in Akkermansia muciniphila overall or by treatment arm. Similarly, we observed no obvious differences in other taxa that have been previously associated with immunotherapy response, including Ruminococcacceae, Faecalibacterium prausnitzii (27), or Bifidobacterium (28), likely due to the small sample size or other factors such as the type and length of neoadjuvant treatment, or geographic and dietary variation. In order to investigate geographic differences, we ordinated beta-diversity distances (binary Jaccard) using principal coordinate analysis, from which we observed a potential clustering effect between the U.S. and European regions, though the difference was not statistically significant (P = 0.18).
Circulating Tumor DNA at Baseline and on Study
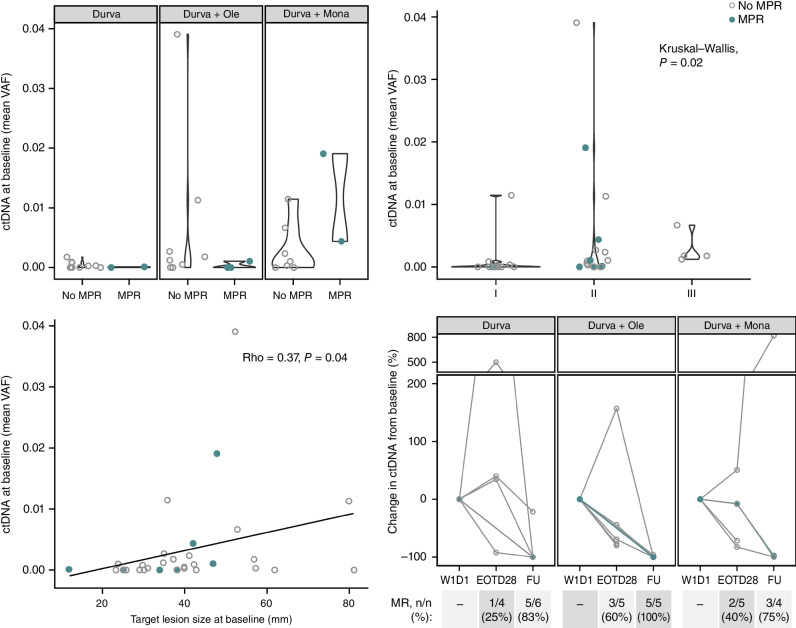
WES on blood and tumor was successful for 35 of 55 of the patients with available tumor tissue (11 baseline and 24 surgical samples); from these, the designing of a personalized panel for tracking somatic mutations in circulating tumor DNA (ctDNA) was successful for 33 of 35 patients. At cycle 1, day 1 predose, 60.6% (20/33) of evaluable patients had detectable ctDNA and were evaluable for molecular response on treatment, whereas 39.4% (13/33) had no detectable ctDNA (Supplementary Fig. S7A). The MPR rate for patients with no detectable ctDNA at baseline (3/13; 23%) was similar to that for patients with detectable ctDNA at baseline (4/20; 20%), and there was no difference in mean variant allele frequency (VAF) at baseline between individuals with an MPR compared with those without (Fig. 5, top left). Detectable ctDNA at baseline was associated with clinical disease stage at study entry; of the 33 patients who were evaluable for ctDNA at baseline, four of four with stage III disease had detectable ctDNA at cycle 1, day 1 compared with four of 12 with stage I and 12 of 17 with stage II disease (Fig. 5, top right). Mean plasma VAF was moderately correlated with tumor size at study entry (Rho = 0.37, P = 0.04; Fig. 5, bottom left), reflecting greater ctDNA burden in patients with larger tumors. For patients with detectable ctDNA at baseline, molecular response (≥50% reduction in VAF from baseline; ref. 29) and complete clearance (100% reduction in VAF from baseline) were evaluated after treatment and after surgery at follow-up. Molecular responses were seen in 25.0% (1/4), 60.0% (3/5), and 40.0% (2/5) of patients after treatment and 83.3% (5/6), 100% (5/5), and 75.0% (3/4) of patients after surgery in the durvalumab monotherapy, durvalumab + oleclumab, and durvalumab + monalizumab arms, respectively (Fig. 5, bottom right; Supplementary Fig. S7B). No complete clearances were observed after treatment, whereas 25% to 85% of patients per arm had complete clearance after surgery. Twenty-one percent (5/24) of patients had molecular residual disease (defined as detectable ctDNA after surgery), ranging from 10% to 60% per arm (Supplementary Fig. S7A). All patients who had an MPR and were evaluable for ctDNA were molecular responders after surgery (n = 3), including two patients with a pCR and complete clearance of ctDNA (Supplementary Fig. S7B; Fig. 2B).
Figure 5.
ctDNA dynamics as surrogate for response. Patients with MPR are indicated in closed teal circle; patients without MPR are indicated in open gray circle. Top left: mean VAF at baseline is compared between patients with an MPR and with no MPR across all arms. Top right: mean VAF at baseline associated across stage I, II, or III disease. Bottom left: mean VAF at baseline correlated with the sum of diameters among target lesions at baseline (mm) (total patients with ctDNA evaluable at baseline N = 33; durvalumab monotherapy arm: n = 13; durvalumab + oleclumab arm: n = 11; durvalumab + monalizumab arm: n = 9). Bottom right: for patients with detectable ctDNA at baseline (total N = 20; durvalumab monotherapy arm: n = 6; durvalumab + oleclumab arm: n = 7, durvalumab + monalizumab arm: n = 7), molecular response (≥50% reduction in VAF from baseline) is depicted at end of treatment (EOT; day 28) and follow-up (FU; day 105) time points for all patients with evaluable ctDNA at those time points (N are depicted above). W1D1, week 1, day 1.
DISCUSSION
Previous single-arm studies examining PD-(L)1 pathway blockade for the treatment of resectable NSCLC have evaluated PD-(L)1 inhibitors alone and/or in combination with CTLA4 inhibitors or with platinum-based chemotherapy. However, little is known regarding the clinical impact of neoadjuvant PD-(L)1 inhibition in combination with novel immuno-oncology agents for patients with resectable NSCLC. Here, we report the clinical outcomes and translational correlates of the NeoCOAST trial, the first randomized, multidrug platform study to evaluate one cycle of neoadjuvant durvalumab as monotherapy or in combination with oleclumab, monalizumab, or danvatirsen, using MPR rate as a primary endpoint, in this setting. We found that combining durvalumab with each of the three novel immunomodulatory agents resulted in greater MPR rates in the ITT population than those seen with a single dose of durvalumab monotherapy, without any new safety signals for durvalumab alone or in combination with the novel agents. Our correlative studies suggest that the improved MPR rates seen in the combination arms may be a result of enhanced immune cell activation and function, as evidenced by transcriptomic changes in the tumor microenvironment and peripheral blood. Between March 2019 and September 2020, despite the logistical challenges of this new type of neoadjuvant platform study for resectable lung cancer, all patients were recruited in a timely fashion with no safety signals and no impact on the conduct of curative intent surgery.
In our trial, the MPR and pCR rates after a single dose of neoadjuvant durvalumab (11.1% and 3.7%, respectively) were consistent with the rates reported from other studies of PD-(L)1 inhibitors in this treatment setting (3–10). The addition of oleclumab, monalizumab, or danvatirsen to durvalumab resulted in greater MPR (19.0%, 30%, and 31.3%, respectively) and pCR (9.5%, 10%, and 12.5%, respectively) rates than with durvalumab alone (MPR 11.1%; pCR 3.7%); however, the danvatirsen arm was discontinued early due to the sponsor's decision to terminate the program. The MPR rates generated by these novel combinations are broadly consistent with those from other chemotherapy-free combinatorial neoadjuvant immunotherapy trials (5, 15), whereas the percentage of patients with pCR was lower compared with previous studies of PD-1 or PD-1/CTLA4 blockade (5, 13, 15). It should be emphasized, however, that the rates of MPR and pCR achieved in the NeoCOAST trial were in response to a short 4-week course of neoadjuvant therapy. It remains to be determined how pCR rates would change in the context of a longer neoadjuvant therapeutic course with these agents.
No new safety signals were identified across the different arms in NeoCOAST. Although the incidence of TRAEs reported in the durvalumab + oleclumab and durvalumab + monalizumab arms (57.1% and 50.0%, respectively) appears to be higher compared with the durvalumab monotherapy arm (34.6%), the actual number of TRAEs reported was similar across the different arms. TRAEs were reported in 12 and 10 patients in the durvalumab + oleclumab and durvalumab + monalizumab arms, respectively, compared with nine in the durvalumab monotherapy arm.
Although our study revealed an association between baseline tumor PD-L1 expression in the durvalumab + oleclumab and durvalumab + monalizumab arms, we did note responses in some tumors lacking PD-L1 expression. This observation is consistent with results from other recent trials evaluating neoadjuvant immune-checkpoint inhibitors (5, 6). Similar findings were also reported in the unresectable setting as part of the COAST study, in which survival benefit was demonstrated with durvalumab + oleclumab and durvalumab + monalizumab compared with durvalumab alone in patients with unresectable, stage III NSCLC, irrespective of tumor PD-L1, CD73, NKG2A, or HLA-E expression (23).
The utility of CD73 as both a predictive and pharmacodynamic biomarker for oleclumab therapy warrants further study. Of the five patients with high CD73 expression in the durvalumab + oleclumab arm, three had an MPR. We noted that high baseline CD73 expression by IHC in tumor samples was associated with fewer viable tumor cells at surgery in the durvalumab + oleclumab arm, suggesting a potential predictive value of identifying tumors more likely to benefit from this combination. In contrast, in the durvalumab monotherapy arm, high baseline tumor CD73 expression was associated with greater viable tumor cells at surgery, consistent with the immunosuppressive effects of CD73 in the tumor microenvironment. Consistent with the mechanism of action of oleclumab, we observed that tumoral CD73 expression decreased in resected tumor samples compared with baseline specimens in the durvalumab + oleclumab arm, but not in other treatment arms. This observation on pharmacodynamic activity is similar to findings from a recently reported phase I study of oleclumab alone or combined with durvalumab in patients with advanced colorectal or pancreatic cancer (24), in which the tumoral CD73 decrease was accompanied by an increase in CD8+ T-cell infiltration. It is also worth noting that two patients with MPR in the durvalumab + oleclumab arm had tumor EGFR L858R activating mutations. A recent characterization of the immune landscape of EGFR-mutant NSCLC identified the CD73/adenosine pathway as a potential therapeutic vulnerability in this patient population and that CD73 blockade inhibited tumor growth in murine models of EGFR-mutant NSCLC (30); furthermore, oleclumab is now also being explored in an ongoing phase I/II trial (NCT03381274) in combination with an EGFR inhibitor in patients with EGFR-mutated NSCLC (31).
Previous studies of neoadjuvant combination immune-checkpoint inhibition have shown increased immune infiltrates with effector function in resected tumors compared with monotherapy (5). In the phase II randomized NEOSTAR trial, for example, there was enhanced tumor infiltration by effector, tissue-resident memory, and effector memory T cells after combined PD-1/CTLA4 blockade (5). A separate study reported abundant cytotoxic T cells in resected tumors responding to the same neoadjuvant combination treatment (15). In our study, we found that combining durvalumab with oleclumab or monalizumab enhanced NK- and CD8 T effector cell recruitment and function in resected tumors, compared with baseline samples. Our transcriptomic analyses performed in resected tumors and in blood after therapy supported augmented expression of gene signatures associated with cytotoxicity, TLS, and lymphocyte recruitment, possibly mediated by IFNγ-induced cytokines, in the durvalumab + oleclumab and durvalumab + monalizumab arms compared with the durvalumab monotherapy arm. These findings are consistent with previous studies in other cancers demonstrating increased tumor immunity upon NKG2A and PD-L1 inhibition through similar immune-mediated mechanisms in murine models and patients (18). Like the present NeoCOAST study, multiomic immune profiling in the phase II platform NEOSTAR study also underscored immune cell populations and phenotypes, including effector memory CD8+ T, B, and myeloid cells and markers of TLS, that were preferentially increased after neoadjuvant chemotherapy plus combined PD-1/CTLA4 blockade (32).
Recent evidence suggests that ctDNA clearance before surgery may be closely associated with pCR after neoadjuvant therapy with nivolumab plus chemotherapy (13). In NeoCOAST, we did not observe complete clearance before surgery in any patient who was evaluable for molecular response. Possibly, these findings were related to the short duration of neoadjuvant treatment in this study. Nevertheless, we did observe substantial reductions in ctDNA VAF, and these reductions were more common in patients with fewer viable tumor cells at surgery (Supplementary Fig. S7B). Moreover, the number of patients with no detected ctDNA increased progressively from pretreatment to posttreatment and postsurgery follow-up, supporting a link between reduction in ctDNA and greater pathologic regression (Supplementary Fig. S7A). Notably, surgery was the most effective intervention to result in the clearance of ctDNA.
In the gut microbiome, we observed a trend of greater richness in patients with an MPR compared with those without and a significant differential abundance of several bacteria taxa. The association between microbiome and response was not as profound as reported in other studies (27, 28, 33), which may be influenced by the global geography of the NeoCOAST patient population; the microbial composition would likely be less homogeneous than if the patients were restricted to a smaller geographic area.
In conclusion, the primary goal of this platform study was to seek preliminary signals in order to rapidly identify promising neoadjuvant combinatorial strategies to be tested in subsequent, larger studies. We integrated multiomics translational studies in the NeoCOAST trial to identify potential mechanisms of response to therapy and streamline the next generation of neoadjuvant immunotherapy–based clinical trials. However, the study was underpowered to draw conclusive biomarker correlations in the various treatment cohorts given the complexity of the assays and availability of baseline tissue. Nevertheless, the concept that such efforts are feasible in the context of early-stage lung cancer within a multicenter design is provocative and promising, suggesting that additional investigations of this kind are required to drive further progress for these patients. In this study, MPR rates in all combination arms were numerically higher than with durvalumab alone, without new safety signals, and translational correlates revealed enhanced tumor and systemic immune effector activation and function as well as ctDNA dynamic changes suggestive of VAF reduction in patients with greater tumor pathologic regression. Further to the results of the CheckMate 816 study, in which neoadjuvant nivolumab plus chemotherapy was shown to improve pCR and EFS in patients with early-stage, resectable NSCLC (13), recently presented data from planned interim analyses of the phase III AEGEAN study have shown that perioperative durvalumab combined with neoadjuvant chemotherapy led to a statistically significant and clinically meaningful improvement in EFS and pCR rate, versus neoadjuvant chemotherapy alone followed by surgery, for patients with resectable, early-stage NSCLC (34). Similarly, recently published findings from a prespecified interim analysis of the phase III Keynote-671 study showed that neoadjuvant pembrolizumab (a PD-1 inhibitor) plus chemotherapy followed by resection and adjuvant single-agent pembrolizumab demonstrated a statistically significant and clinically meaningful improvement in EFS compared with neoadjuvant placebo plus chemotherapy followed by adjuvant placebo in patients with early-stage NSCLC (35). Based on results from NeoCOAST and CheckMate 816, the global, phase II, randomized NeoCOAST-2 study has been initiated (NCT05061550), which is evaluating durvalumab + oleclumab and chemotherapy, durvalumab + monalizumab and chemotherapy, the novel bispecific PD-1/CTLA4 inhibitor volrustomig + chemotherapy, the novel antibody–drug conjugate datopotamab deruxtecan + durvalumab and chemotherapy, or the first-in-class antileukemia inhibitory factor mAb AZD0171 + durvalumab and chemotherapy as neoadjuvant treatment, followed by surgery and adjuvant durvalumab + oleclumab, durvalumab + monalizumab, volrustomig alone, durvalumab alone, or AZD0171 + durvalumab, respectively, in patients with resectable, stage IIA–IIIB NSCLC (AJCC 8th edition; ref. 36). The co–primary endpoints of the study are pCR and safety and feasibility; collection of baseline and post–neoadjuvant therapy tumor samples is mandated for translational analyses (36).
METHODS
Study Design, Hypotheses, and Endpoints
This is a phase II, open-label, multicenter, randomized, multidrug platform study of neoadjuvant durvalumab alone or in combination with oleclumab, monalizumab, or danvatirsen in patients with resectable, early-stage [stage IA3 (>2 cm) to IIIA] NSCLC. Eligible patients were randomized to receive either one 28-day cycle of durvalumab monotherapy, or durvalumab in combination with oleclumab, monalizumab, or danvatirsen. Patients were stratified by lymph node involvement. Surgical resection was planned to occur between days 29 and 42 after the first dose of neoadjuvant therapy. After surgery, patients were followed up to day 105 (Fig. 1A).
This study was designed to seek preliminary efficacy signals by calculating the MPR rates and their CIs of durvalumab monotherapy or in combination with oleclumab, monalizumab, or danvatirsen. It was not statistically powered to make definitive conclusions for any hypotheses tested. The primary hypothesis tested was that neoadjuvant durvalumab alone or in combination, administered over a 28-day treatment period to patients with resectable, early-stage NSCLC, would lead to a pathologic response within the resected tumor specimen. The secondary hypothesis tested was that neoadjuvant durvalumab alone or in combination would demonstrate an acceptable safety profile and would not result in delayed surgery in patients with resectable, early-stage NSCLC.
The primary endpoint of the MPR rate was defined as the proportion of patients with ≤10% residual viable tumor cells in the surgical specimen and sampled lymph nodes at surgery. MPR was assessed locally based on previously described criteria (37). The secondary endpoint of pCR was defined as the proportion of patients with no residual viable tumor cells in the resected tumor specimen or sampled lymph nodes at surgery; local review of the resected specimens was used for assessment of pCR. Additional secondary endpoints included the feasibility of planned surgery, safety and tolerability, pharmacokinetics, and immunogenicity of durvalumab alone and in combination. As part of the safety evaluation, AESIs, AEPIs, and imAEs were assessed in all treatment arms (full definitions are included in the Supplementary Materials). Exploratory endpoints included best overall response (BOR) and ORR per Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST v1.1; ref. 38), evaluation of tumor and blood biomarkers, ctDNA dynamics and microbiome composition, and functional/metabolic profile.
Eligible patients had previously untreated, resectable, stage IA3 to IIIA (for patients with N2 disease, only those with a single nodal station ≤3 cm were eligible) NSCLC according to the 8th edition of AJCC staging classification, an Eastern Cooperative Oncology Group performance status of 0 or 1, and adequate organ and marrow function. Key exclusion criteria included mixed small cell/non–small cell histology; requirement of pneumonectomy (as assessed by the surgeon prior to enrollment) to obtain potentially curative resection of primary tumor; prior treatment with PD-(L)1 or CTLA4 inhibitors; active or prior documented autoimmune or inflammatory disorders; history of active primary immunodeficiency; active infections; and uncontrolled intercurrent illness.
Study Oversight, Ethical Approval, and Ethical Standards
All patients enrolled in this study provided written informed consent. Safety reviews of all enrolled patients were conducted at least twice a year by a Safety Review Committee. The trial was conducted according to the principles of the Declaration of Helsinki and the International Council for Harmonisation Good Clinical Practice guidelines. Independent ethics committees or Institutional Review Boards at each participating center approved the protocol. Data were collected and analyzed by the investigators, and all authors approved and agreed to submit the final manuscript for publication. The authors vouch for the accuracy and completeness of the data and the fidelity of the trial to the study protocol.
Interventions
Eligible patients were randomized 1:1:1:1 to receive durvalumab 1,500 mg every 4 weeks (Q4W) alone or in combination with oleclumab 3,000 mg every 2 weeks (Q2W), monalizumab 750 mg Q2W, or danvatirsen 200 mg every week (including a 7-day lead-in period of danvatirsen 200 mg on days 1, 3, and 5: week 0). Patients were enrolled and randomized using a central system (an interactive voice/Web response system, IXRS). As part of the screening procedures, patients underwent clinical radiographic staging, which included PET-CT, contrast CT, and brain magnetic resonance imaging. Surgical resection was planned to occur within 14 days after the last dose of neoadjuvant therapy. Surgical resection of the primary tumor and mediastinal lymph nodes was performed based on surgeons’ discretion and institutional standards.
Pathologic Assessment
Pathologic samples were collected at baseline (day −21 to −1) and at surgical resection (days 29 to 42). For baseline samples, archival biopsies collected within ≤6 months of study entry were acceptable; otherwise, a new (fresh) biopsy was required. Pathologic tumor response (MPR and pCR) in resected specimens was assessed locally according to Pataer and colleagues (37).
Statistical Methods and Sample Size Justification
Clinical data are presented mainly by descriptive statistics; 95% CIs were calculated for key endpoints at times for understanding the precision of the point estimates. However, no formal statistical inferences are made about clinical data. This study was designed to obtain preliminary clinical efficacy, safety, pharmacokinetic, and immunogenicity data on durvalumab in combination with novel agents compared with durvalumab monotherapy. It was not designed to make definitive power and type one error considerations for a hypothesis test. The sample size was therefore determined so that 95% CIs for comparing efficacy signals between durvalumab monotherapy and combination therapy arms were a reasonable width, in line with the purpose of this study, per the sponsor's discretion. At the time of trial design, the MPR rate for durvalumab monotherapy arm was assumed to be 30%, based on a weighted average of available data, including the pilot study of neoadjuvant nivolumab in patients with resectable, early-stage NSCLC, which reported an MPR rate of 45% (9/20; ref. 6) and the LCMC3 trial of neoadjuvant atezolizumab in patients with resectable NSCLC, which reported an MPR rate of 21% (4/19; ref. 39). Owing to the small sample size, we calculated exact CIs when assessing clinical efficacy signals.
Tissue, Blood, and Fecal Microbiome Sampling
Tumor specimens were collected (when available) pretreatment as fresh tumor biopsies, or archival if collected within 6 months of study entry, and were mandatory for approximately 50% of patients. Tumor specimens were collected (when available) at surgery for evaluation of exploratory biomarkers after treatment. Whole-blood samples for mRNA sequencing were collected pretreatment (week 1, day 1) and after treatment (day 28). Peripheral blood samples for ctDNA were collected pretreatment, at the end of treatment (day 28), and at follow-up (day 105 ± 21 days). Stool samples for microbiome (when available) were collected pretreatment, within 21 days of the start of treatment, and after treatment at any time after week 3, day 1 through end of treatment (day 28; Fig. 1A).
Pathologic Assessment of PD-L1 and Other Proteins
Pretreatment and on-treatment tumor tissue, when available, was analyzed by hematoxylin and eosin, IHC, and multiplex immunofluorescence. FFPE tumor sections (4-μm-thick) were stained and analyzed as follows: PD-L1 (Ventana, SP263) analyzed using tumor center scoring; CD73 (Cell Signaling Technology, D7F9A) was scored by a pathologist for tumor membrane percent of positivity (% positive tumor cells); NKG2A (Abcam, AR9352) and CD8 (Ventana, SP239) were scored using in-house software image analysis (Computational Pathology, AstraZeneca) using density scoring (number of positive cells/mm2 area). For the number of samples available for correlative analyses, please refer to individual figure legends.
RNA-seq from Tumor Tissue
mRNA was extracted from pretreatment and on-treatment FFPE tumor tissue, when available, using the HudsonAlpha Discovery FFPE Tissue Extraction Method. Libraries were constructed from 250 pg to 10 ng RNA input using Takara RNA amplification and rRNA-reduced library construction kit, and sequenced on an Illumina NovaSeq 6000 at a depth of 200 million base reads (100 million paired-end). Data were generated based on 69 samples (35 baseline and 34 on-treatment) for three arms (durvalumab monotherapy: 12 baseline, 14 on-treatment; durvalumab + oleclumab: 13 baseline, 11 on-treatment; durvalumab + monalizumab: 10 baseline, nine on-treatment). The numbers of available paired sample sets (patients with both baseline and on-treatment samples, n = 13 total) in each treatment arm were as follows: four in the durvalumab monotherapy arm; five in the durvalumab + oleclumab arm; and four in the durvalumab + monalizumab arm.
The RNA-seq pipeline implemented in bcbio-nextgen (version 1.2.7; https://bcbio-nextgen.readthedocs.org/en/latest/) was used for quality control and gene expression quantification. Reads were aligned to the UCSC build GRCh38 Homo Sapiens genome using STAR (version 2 6.1; ref. 40). Alignments were evaluated for evenness of coverage, rRNA content, genomic context of alignments, and complexity using a combination of FastQC, Qualimap, and custom tools (41). Transcripts per million (TPM) measurements per isoform were generated by alignment-based quantification using Salmon (version 1.4.0; ref. 42) and used to estimate the abundance of genes. The aggregated gene counts were used for differential gene expression analyses with DESeq2 (43).
Individual hypotheses on differences between pretherapy and surgery samples were tested for genes or gene signatures related to TLS, effector cells, chemokines, and other features using Wilcoxon-rank sum test. GSEA was performed using R package “fGSEA” (bioRxiv 2021.02.01.060012) using hallmark gene sets from MSigDB (44). Enrichment P values were calculated as described in bioRxiv 2021.02.01.060012, and P values were adjusted using Benjamini–Hochberg methods. Pathway enrichment analysis was conducted using Ingenuity Pathway Analysis (QIAGEN; https://www.qiagen.com/us/products/discovery- and-translational-research/next-generation-sequencing/informatics-and-data/interpretationcontent-databases/ingenuity-pathway-analysis). MCP-Counter tool (45) was used to estimate immune cell abundances for each sample and condition using immune gene signatures within MCP-Counter.
RNA-seq from Whole Blood
Whole blood was collected in PaxGene RNA tubes and processed with TruSeq Stranded Total RNA with Illumina Ribo-Zero plus rRNA Depletion + Globin Reduction RNA Library Preparation. Libraries were sequenced on Illumina PE100 base reads at a depth of 100 million. Data were generated on 120 samples from 66 patients in the durvalumab monotherapy, durvalumab + oleclumab, and durvalumab + monalizumab arms. Fifty-four patients had both pretreatment and on-treatment data, 11 patients had only pretreatment data, and one patient had only on-treatment data. Quality control and gene expression quantification were carried out in the same way as for samples from tumor tissue, as described previously.
TPM measurements aggregated per gene were used for GSVAs (package GSVA, Bioconductor, https://bioconductor.org/packages/release/bioc/manuals/GSVA/man/GSVA.pdf). The values were first normalized using log2, and the minimal (0.5) value was appended to zero. The GSVA method was used with a minimum of five genes in the signature and at least 80% overlap of genes in the signature. Differential gene expression was performed using DESeq2, and differential gene signature analysis was performed using the LIMMA package with Benjamini–Hochberg correction.
WES and ctDNA Analysis
Tumor mutation profiling was performed by WES on FFPE tumor tissue and matched normal blood samples (whole blood or peripheral blood mononuclear cells). When available, the surgical resection tissue was used (n = 35), and in all other cases, pretreatment tumor tissue was used (n = 20; total N = 55). gDNA was extracted from both, and WES was performed to greater than 180× coverage of tumor tissue and 50× coverage on matched normal blood. A personalized, tumor-informed Signatera 16-plex assay was developed for individual patients (n = 33) based on somatic tumor alterations and performed on plasma ctDNA from pretreatment, after treatment (day 28), and follow-up (day 205) time points. Moreover, somatic alterations, microsatellite instability, and tumor mutational burden were evaluated from tumor and matched normal tissue from 34 patients with WES data passing quality control thresholds using pipeline software bcbio-nextgen (https://zenodo.org/record/5781867#.Y43iay-l1cQ). Variant calling was performed using VarDict v1.7.0 (46), down to a VAF of 1% (before filtering and curation), and variant effects annotated by snpEff v4.3.1t.
Fecal Microbiome Sample Processing and Analysis
Whole metagenomic sequencing was conducted at Diversigen on samples from 38 patients (15 in the durvalumab arm, 12 in the durvalumab + oleclumab arm, and 11 in the durvalumab + monalizumab arm). A 100-mg sample was extracted with the MagAttract PowerSoil DNA EP kit (QIAGEN) using mechanical-based lysis with garnet beads. The extraction was automated for high-throughput on the Hamilton STARLet. Samples were subsequently quantified with the Quant-iT PicoGreen dsDNA Assay (Invitrogen), and libraries were prepared with a procedure adapted from the Illumina DNA Prep Kit (Illumina). Next, the libraries were quantified with the Quant-iT PicoGreen dsDNA assay (Invitrogen) and confirmed using the High Sensitivity NGS Fragment Analysis Kit (1 bp–6,000 bp; Agilent) on an Agilent Fragment Analyzer. Finally, libraries were sequenced on an Illumina NovaSeq 6000 using a paired-end 2 × 150 bp flow cell. KneadData was used to parse paired-end fastq files and remove reads that mapped to the human and phiX genomes using default parameters (https://github.com/biobakery/kneaddata). The filtered reads were subsequently processed using MetaPhlAn3 for taxonomic identification (47). Alpha-diversity (inverse Simpson) and richness were calculated after estimating the total reads per sample using the “-t rel_ab_with_stats” parameter of MetaPhlAn3. Beta-diversity distances were estimated using the binary Jaccard matrix and ordinated using principal coordinate analysis. Differential enrichment analysis to identify candidate taxa was done using LEfSe (48).
Data Availability Statement
Data underlying the findings described in this article may be obtained in accordance with AstraZeneca's data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data for studies directly listed on Vivli can be requested through Vivli at www.vivli.org. Data for studies not listed on Vivli could be requested through Vivli at https://vivli.org/members/enquiries-about-studies-not-listed-on-the-vivli-platform/. The AstraZeneca Vivli member page is also available, outlining further details: https://vivli.org/ourmember/astrazeneca/.
Supplementary Material
Additional details on safety endpoints. Supplementary tables: 1, Representativeness of study participants; 2, Efficacy summary; 3, Adverse events occurring in ≥10% of patients; 4, Treatment-related adverse events occurring in ≥5% of patients; 5, Adverse events of special or potential interest; 6, Baseline biomarker association with response. Supplementary figures: 1, Mechanism of action for each study drug; 2, Violin plots with median % residual viable tumor cells; 3, Additional immunohistochemistry biomarker to major pathological response; 4, Calculation of gene signatures associated with tertiary lymphoid structure (TLS) formation; 5, Changes in immune cell populations by deconvolution; 6, Stool microbiome richness and diversity; 7, ctDNA dynamics.
Supplementary Table 7. Tumor RNA sequencing. (File too large to be included in main supplementary file).
Acknowledgments
The authors thank the patients, their families and caregivers, and all the investigators involved in this study. This study (NCT03794544) was funded by AstraZeneca. The authors also thank the AstraZeneca Oncology Data Science Platform and Omics Data Operation teams for supporting RNA-seq and WES data preprocessing. Medical writing support for the development of this manuscript, under the direction of the authors, was provided by Connor Keating of Ashfield MedComms (Manchester, UK), an Inizio company, and funded by AstraZeneca.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T. Cascone reports personal fees and other support (institutional contracted support) from MedImmune/AstraZeneca during the conduct of the study, as well as grants, personal fees, and other support from Bristol Myers Squibb, personal fees from Regeneron, Merck & Co., Pfizer, Genentech, Arrowhead Pharmaceuticals, Roche, Medscape, OncLive, PeerView, Clinical Care Options, and The Mark Foundation for Cancer Research, personal fees and other support from Physicians’ Education Resource and IDEOlogy Health, and other support from the Parker Institute for Cancer Immunotherapy, the Society for Immunotherapy of Cancer, the International Association for the Study of Lung Cancer, Dava Oncology, and EMD Serono outside the submitted work. G. Kar reports other support from AstraZeneca during the conduct of the study. J.D. Spicer reports grants and personal fees from AstraZeneca during the conduct of the study; grants and personal fees from Bristol Myers Squibb, Merck, Roche, and Protalix Biotherapeutics, grants from CLS Therapeutics and Novartis, and personal fees from Eisai outside the submitted work. R. García-Campelo reports personal fees from AstraZeneca during the conduct of the study, as well as grants and personal fees from MSD, Merck, Sanofi, and Pfizer, and personal fees from Roche, Takeda, Amgen, Organon, and Boehringer Ingelheim outside the submitted work. W. Weder reports advisory board participation for AstraZeneca. D.B. Daniel reports grants and other support from the Sarah Cannon Research Institue during the conduct of the study, as well as grants from AstraZeneca, G1 Therapeutics, Bristol Myers Squibb, Genentech, Guardant Health, Janssen, Merch, AbbVie, Novartis, ARMO BIoSciences, Lilly, Daiichi Sankyo, Roche, and EQRx outside the submitted work. D.R. Spigel reports grants from MedImmune during the conduct of the study, as well as grants and other support from AbbVie, AstraZeneca, Bristol Myers Squibb, BeiGene, Roche/Genentech, GSK, Ipsen, Jazz Pharmaceuticals, Lilly, Lyell Immunopharma, Monte Rosa Therapeutics, Novartis, and Novocure, grants from Aeglea Biotherapeutics, Agios, Arcus, Arrys Therapeutics, Ascendis Pharma, Asher Biotherapeutics, Astellas, Bayer, BIND Therapeutics, BioNTech, Blueprint Medicines, Boehringer Ingelheim, Calithera, Celgene, Celldex, Clovis, Cyteir Therapeutics, Daiichi Sankyo, Eisai, Elevation Oncology, Ellipses Pharma, EMD Serono, Endeavor, Erasca, Evelo Biosciences, Faeth Therapeutics, Foundation Bio, FujiFilm Pharmaceuticals, G1 Therapeutics, Gilead Sciences, GRAIL, Hutchinson MediPharma, ImClone Systems, Incyte, Janssen, Janux Therapeutics, Kronos Bio, Loxo Oncology, MacroGenics, MedImmune, Merck, Millennium Pharmaceuticals, Moderna, Molecular Partners, Nektar, Neon Therapeutics, Oncologie, Peloton Therapeutics, Pfizer, PTC Therapeutics, PureTech Health, Razor Genomics, Repare Therapeutics, Rgenix, Seagen, Shenzhen Chipscreen Biosciences, Strata Oncology, Stemline Therapeutics, Synthekine, Taiho, Takeda Pharmaceuticals, Tango Therapeutics, Tarveda, Tesaro, Tizona Therapeutics, Transgene, UT Southwestern, Verastem, and the Zai Laboratory, and other support from Amgen, Regeneron Pharmaceuticals, and Sanofi-Aventis outside the submitted work. J. Mazieres reports grants from AstraZeneca during the conduct of the study, as well as grants from Roche, Bristol Myers Squibb, MSD, Pfizer, Pierre Fabre, and AbbVie outside the submitted work. J. Oliveira reports grants from AstraZeneca outside the submitted work. E.H. Yau reports grants from NextCure outside the submitted work. A.I. Spira reports grants from LAM Therapeutics, Regeneron, Roche, AstraZeneca, Boehringer Ingelheim, Astellas Pharma, MedImmune, Novartis, Incyte, AbbVie, Ignyta, Takeda, Macrogenics, CytomX Therapeutics, Astex Pharmaceuticals, Bristol Myers Squibb, Loxo, Arch Therapeutics, Gritstone, Plexxikon, Amgen, Daiichi Sankyo, ADCT, Janssen Oncology, Mirati Therapeutics, Rubius, Synthekine, Mersana, Blueprint Medicines, Alkermes, Revolution Medicines, Medikine, Black Diamond Therapeutics, BluPrint Oncology, Nalo Therapeutics, Scorpion Therapeutics, ArriVent Biopharma, and Revolution Medicines during the conduct of the study, as well as personal fees from CytomX Therapeutics, AstraZeneca/MedImmune, Merck, Takeda, Amgen, Janssen Oncology, Novartis, Bristol Myers Squibb, Bayer, Incyte, Amgen, Novartis, Mirati Therapeutics, Gritstone Oncology, Jazz Pharmaceuticals, Takeda, Janssen Research and Development, Mersana, Gritstone Bio, Daiichi Sankyo/AstraZeneca, Regeneron, Lilly, Black Diamond Therapeutics, Sanofi, Array BioPharma, AstraZeneca/MedImmune, Merck, Bristol Myers Squibb, Blueprint Medicines, and Eli Lilly outside the submitted work. V. Anagnostou reports grants from AstraZeneca, Bristol Myers Squibb, Delfi Diagnostics, and Personal Genome Diagnostics, and personal fees from Neogenomics and AstraZeneca outside the submitted work, as well as patent 63/276,525 issued, patent 17/779,936 issued, patent 16/312,152 issued, patent 16/341,862 issued, patent 17/047,006 issued, and patent 17/598,690 issued. L.-Y. Cheng is an employee of AstraZeneca, who receives long-term incentives of stocks from AstraZeneca every year as part of the work benefits AstraZeneca gives to its employees. Y. Zheng reports personal fees from AstraZeneca during the conduct of the study. J. Rodriguez-Canales reports other support from AstraZeneca outside the submitted work. V. Gopalakrishnan reports other support from AstraZeneca during the conduct of the study, as well as US patent PCT/US17/53,717 issued and US patent WO2020106983A1 pending. B.R. Sellman reports other support from AstraZeneca during the conduct of the study. Y. Soo-Hoo reports other support from AstraZeneca during the conduct of the study; other support from AstraZeneca outside the submitted work; and is an employee of and shareholder in AstraZeneca. R. Kumar reports other support from AstraZeneca during the conduct of the study, as well as patent for anti-CD73 antibody pending. L. McGrath reports personal fees from AstraZeneca during the conduct of the study; personal fees from Jounce Therapeutics outside the submitted work; and patent 11692038 issued to Gilead Sciences. P.M. Forde reports grants and other support from AstraZeneca during the conduct of the study; other support from Ascendis, Curevac, G1, Genelux, Gritstone, Merck, Janssen, F-star, Sanofi, Amgen, Fosun, Teva, Synthekine, Flame, Iteos, and Tavotek, grants and other support from Bristol Myers Squibb, Novartis, and Regeneron, and grants from BioNTech and Nextpoint outside the submitted work; and patent PCT/US2022/079403 pending. No disclosures were reported by the other authors.
Authors’ Contributions
T. Cascone: Conceptualization, investigation, writing–original draft. G. Kar: Data curation, methodology, writing–original draft. J.D. Spicer: Conceptualization, investigation, writing–original draft. R. García-Campelo: Investigation, writing–review and editing. W. Weder: Writing–review and editing. D.B. Daniel: Investigation, writing–review and editing. D.R. Spigel: Investigation, writing–review and editing. M. Hussein: Writing–review and editing. J. Mazieres: Writing–review and editing. J. Oliveira: Writing–review and editing. E.H. Yau: Writing–review and editing. A.I. Spira: Writing–review and editing. V. Anagnostou: Writing–review and editing. R. Mager: Formal analysis, project administration, writing–review and editing. O. Hamid: Formal analysis, project administration, writing–review and editing. L.-Y. Cheng: Formal analysis, writing–review and editing. Y. Zheng: Formal analysis, project administration, writing–review and editing. J. Blando: Formal analysis, project administration, writing–review and editing. T.H. Tan: Formal analysis, project administration, writing–review and editing. M. Surace: Formal analysis, project administration, writing–review and editing. J. Rodriguez-Canales: Formal analysis, project administration, writing–review and editing. V. Gopalakrishnan: Data curation, formal analysis, project administration, writing–review and editing. B.R. Sellman: Formal analysis, project administration, writing–review and editing. I. Grenga: Conceptualization, supervision, writing–original draft, project administration. Y. Soo-Hoo: Conceptualization, supervision, project administration, writing–review and editing. R. Kumar: Conceptualization, supervision, writing–original draft, project administration. L. McGrath: Conceptualization, data curation, formal analysis, supervision, writing–original draft, project administration. P.M. Forde: Conceptualization, investigation, writing–original draft.
References
- 1. Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552–9. [DOI] [PubMed] [Google Scholar]
- 2. Wu D, Huang H, Zhang M, Li Z, Wang S, Yu Y, et al. The global landscape of neoadjuvant and adjuvant anti–PD-1/PD-L1 clinical trials. J Hematol Oncol 2022;15:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altorki NK, McGraw TE, Borczuk AC, Saxena A, Port JL, Stiles BM, et al. Neoadjuvant durvalumab with or without stereotactic body radiotherapy in patients with early-stage non-small-cell lung cancer: a single-centre, randomised phase 2 trial. Lancet Oncol 2021;22:824–35. [DOI] [PubMed] [Google Scholar]
- 4. Besse B, Adam J, Cozic N, Chaput-Gras N, Planchard D, Mezquita L, et al. 1215O-SC neoadjuvant atezolizumab (A) for resectable non-small cell lung cancer (NSCLC): results from the phase II PRINCEPS trial. Ann Oncol 2020;31:S794–S5. [Google Scholar]
- 5. Cascone T, William WN Jr, Weissferdt A, Leung CH, Lin HY, Pataer A, et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in operable non-small cell lung cancer: the phase 2 randomized NEOSTAR trial. Nat Med 2021;27:504–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forde PM, Chaft JE, Smith KN, Anagnostou V, Cottrell TR, Hellmann MD, et al. Neoadjuvant PD-1 blockade in resectable lung cancer. N Engl J Med 2018;378:1976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao S, Li N, Gao S, Xue Q, Ying J, Wang S, et al. Neoadjuvant PD-1 inhibitor (sintilimab) in NSCLC. J Thorac Oncol 2020;15:816–26. [DOI] [PubMed] [Google Scholar]
- 8. Chaft JE, Oezkan F, Kris MG, Bunn PA, Wistuba II, Kwiatkowski DJ, et al. Neoadjuvant atezolizumab for resectable non-small cell lung cancer: an open-label, single-arm phase II trial. Nat Med 2022;28:2155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tong BC, Gu L, Wang X, Wigle DA, Phillips JD, Harpole DH Jr, et al. Perioperative outcomes of pulmonary resection after neoadjuvant pembrolizumab in patients with non-small cell lung cancer. J Thorac Cardiovasc Surg 2022;163:427–36. [DOI] [PubMed] [Google Scholar]
- 10. Wislez M, Mazieres J, Lavole A, Zalcman G, Carre O, Egenod T, et al. Neoadjuvant durvalumab for resectable non-small-cell lung cancer (NSCLC): results from a multicenter study (IFCT-1601 IONESCO). J Immunother Cancer 2022;10:e005636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. BMS. Canadian Product Monograph [cited 2023 March 09]. Available from: https://pdf.hres.ca/dpd_pm/00068871.PDF.
- 12. US Food and Drug Administration. OPDIVO (Nivolumab) Label [cited 2023 Mar 09]. Available from:https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/125554s119lbl.pdf.
- 13. Forde PM, Spicer J, Lu S, Provencio M, Mitsudomi T, Awad MM, et al. Neoadjuvant nivolumab plus chemotherapy in resectable lung cancer. N Engl J Med 2022;386:1973–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Provencio M, Nadal E, González-Larriba JL, Martínez-Martí A, Bernabé R, Bosch-Barrera J, et al. Perioperative nivolumab and chemotherapy in stage III non-small-cell lung cancer. N Engl J Med 2023;389:504–13. [DOI] [PubMed] [Google Scholar]
- 15. Reuss JE, Anagnostou V, Cottrell TR, Smith KN, Verde F, Zahurak M, et al. Neoadjuvant nivolumab plus ipilimumab in resectable non-small cell lung cancer. J Immunother Cancer 2020;8:e001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Forde PM, Spicer J, Lu S, Provencio M, Mitsudomi T, Awad MM, et al. Nivolumab + platinum-doublet chemotherapy vs chemotherapy as neoadjuvant treatment for resectable (IB–IIIA) non-small cell lung cancer in the phase 3 CheckMate 816 trial [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2021; 2021 Apr 10–15 and May 17–21. Philadelphia (PA): AACR; Cancer Res 2021;81(13_Suppl):Abstract nr CT003 (unpublished oral presentation).
- 17. Stewart R, Morrow M, Hammond SA, Mulgrew K, Marcus D, Poon E, et al. Identification and characterization of MEDI4736, an antagonistic anti-PD-L1 monoclonal antibody. Cancer Immunol Res 2015;3:1052–62. [DOI] [PubMed] [Google Scholar]
- 18. Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell 2018;175:1731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Banerjee S, Oaknin A, Sanchez-Simon I, Salgado AC, Patel SP, Oza A, et al. 518 Phase 1B trial of monalizumab (NKG2A inhibitor) plus durvalumab: safety and efficacy in patients with metastatic ovarian, cervical, and microsatellite-stable endometrial cancers. Int J Gynecol Cancer 2020;30:A86–A87. [Google Scholar]
- 20. Bendell JC, LoRusso P, Overman MJ, Noonan AM, Kim D-W, Strickler J, et al. Safety and efficacy of the anti-CD73 monoclonal antibody (mAb) oleclumab ± durvalumab in patients (pts) with advanced colorectal cancer (CRC), pancreatic ductal adenocarcinoma (PDAC), or EGFR-mutant non-small cell lung cancer (EGFRm NSCLC). J Clin Oncol 39, 2021. (suppl 15; abstr 9047). [Google Scholar]
- 21. Cohen EEW, Harrington KJ, Hong DS, Mesia R, Brana I, Perez Segura P, et al. A phase Ib/II study (SCORES) of durvalumab (D) plus danvatirsen (DAN; AZD9150) or AZD5069 (CX2i) in advanced solid malignancies and recurrent/metastatic head and neck squamous cell carcinoma (RM-HNSCC): Updated results. Ann Oncol 2018;29 Suppl 8:VIII372. [Google Scholar]
- 22. Hay CM, Sult E, Huang Q, Mulgrew K, Fuhrmann SR, McGlinchey KA, et al. Targeting CD73 in the tumor microenvironment with MEDI9447. Oncoimmunology 2016;5:e1208875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herbst RS, Majem M, Barlesi F, Carcereny E, Chu Q, Monnet I, et al. COAST: an open-label, phase II, multidrug platform study of durvalumab alone or in combination with oleclumab or monalizumab in patients with unresectable, stage III non-small-cell lung cancer. J Clin Oncol 2022;40:3383–93. [DOI] [PubMed] [Google Scholar]
- 24. Bendell J, LoRusso P, Overman M, Noonan AM, Kim D-W, Strickler JH, et al. First-in-human study of oleclumab, a potent and selective anti-CD73 monoclonal antibody, alone or in combination with durvalumab in patients with advanced solid tumors. Cancer Immunol Immunother 2023;72:2443–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Proia TA, Singh M, Woessner R, Carnevalli L, Bommakanti G, Magiera L, et al. STAT3 antisense oligonucleotide remodels the suppressive tumor microenvironment to enhance immune activation in combination with anti-PD-L1. Clin Cancer Res 2020;26:6335–49. [DOI] [PubMed] [Google Scholar]
- 26. Ribrag V, Lee ST, Rizzieri D, Dyer MJS, Fayad L, Kurzrock R, et al. A phase 1b study to evaluate the safety and efficacy of durvalumab in combination with tremelimumab or danvatirsen in patients with relapsed or refractory diffuse large B-cell lymphoma. Clin Lymphoma Myeloma Leuk 2021;21:309–17. [DOI] [PubMed] [Google Scholar]
- 27. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goldberg SB, Narayan A, Kole AJ, Decker RH, Teysir J, Carriero NJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res 2018;24:1872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le X, Negrao MV, Reuben A, Federico L, Diao L, McGrail D, et al. Characterization of the immune landscape of EGFR-mutant NSCLC identifies CD73/adenosine pathway as a potential therapeutic target. J Thorac Oncol 2021;16:583–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim DW, Kim SW, Camidge DR, Shu CA, Marrone KA, Le X, et al. CD73 inhibitor oleclumab plus osimertinib in previously treated patients with advanced T790M-negative EGFR-mutated NSCLC: A brief report. J Thorac Oncol 2023;18:650–6. [DOI] [PubMed] [Google Scholar]
- 32. Cascone T, Leung CH, Weissferdt A, Pataer A, Carter BW, Godoy MCB, et al. Neoadjuvant chemotherapy plus nivolumab with or without ipilimumab in operable non-small cell lung cancer: the phase 2 platform NEOSTAR trial. Nat Med 2023;29:593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oh B, Boyle F, Pavlakis N, Clarke S, Eade T, Hruby G, et al. The gut microbiome and cancer immunotherapy: can we use the gut microbiome as a predictive biomarker for clinical response in cancer immunotherapy? Cancers 2021;13:4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heymach JV, Harpole DH Jr, Mitsudomi T, Taube JM, Galffy G, Hochmair M, et al. AEGEAN: a phase 3 trial of neoadjuvant durvalumab + chemotherapy followed by adjuvant durvalumab in patients with resectable NSCLC [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2023; Part 2 (Clinical Trials and Late-Breaking Research); 2023 Apr 14–19; Orlando, FL. Philadelphia (PA): AACR; Cancer Res 2023;83(8_Suppl):Abstract nr CT005. [Google Scholar]
- 35. Wakelee H, Liberman M, Kato T, Tsuboi M, Lee S-H, Gao S, et al. Perioperative pembrolizumab for early-stage non–small-cell lung cancer. N Engl J Med 2023;389:491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guisier F, Bennouna J, Spira AI, Kim D-W, Shim BY, Sater HA, et al. NeoCOAST-2: a phase 2 study of neoadjuvant durvalumab plus novel immunotherapies (IO) and chemotherapy (CT) or MEDI5752 (volrustomig) plus CT, followed by surgery and adjuvant durvalumab plus novel IO or volrustomig alone in patients with resectable non-small-cell lung cancer (NSCLC). J Clin Oncol 41, 2023. (suppl 16; abstr TPS8604). [Google Scholar]
- 37. Pataer A, Kalhor N, Correa AM, Raso MG, Erasmus JJ, Kim ES, et al. Histopathologic response criteria predict survival of patients with resected lung cancer after neoadjuvant chemotherapy. J Thorac Oncol 2012;7:825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 39. Rusch V, Chaft JE, Johnson B, Wistuba IV, Kris MG, Lee JM, et al. Neoadjuvant atezolizumab in resectable non-small cell lung cancer (NSCLC): initial results from a multicenter study (LCMC3). J Clin Oncol 36, 2018. (suppl; abstr 8541). [Google Scholar]
- 40. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Garcia-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Gotz S, Tarazona S, et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 2012;28:2678–9. [DOI] [PubMed] [Google Scholar]
- 42. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 2016;44:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beghini F, McIver LJ, Blanco-Miguez A, Dubois L, Asnicar F, Maharjan S, et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 2021;10:e65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional details on safety endpoints. Supplementary tables: 1, Representativeness of study participants; 2, Efficacy summary; 3, Adverse events occurring in ≥10% of patients; 4, Treatment-related adverse events occurring in ≥5% of patients; 5, Adverse events of special or potential interest; 6, Baseline biomarker association with response. Supplementary figures: 1, Mechanism of action for each study drug; 2, Violin plots with median % residual viable tumor cells; 3, Additional immunohistochemistry biomarker to major pathological response; 4, Calculation of gene signatures associated with tertiary lymphoid structure (TLS) formation; 5, Changes in immune cell populations by deconvolution; 6, Stool microbiome richness and diversity; 7, ctDNA dynamics.
Supplementary Table 7. Tumor RNA sequencing. (File too large to be included in main supplementary file).
Data Availability Statement
Data underlying the findings described in this article may be obtained in accordance with AstraZeneca's data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data for studies directly listed on Vivli can be requested through Vivli at www.vivli.org. Data for studies not listed on Vivli could be requested through Vivli at https://vivli.org/members/enquiries-about-studies-not-listed-on-the-vivli-platform/. The AstraZeneca Vivli member page is also available, outlining further details: https://vivli.org/ourmember/astrazeneca/.



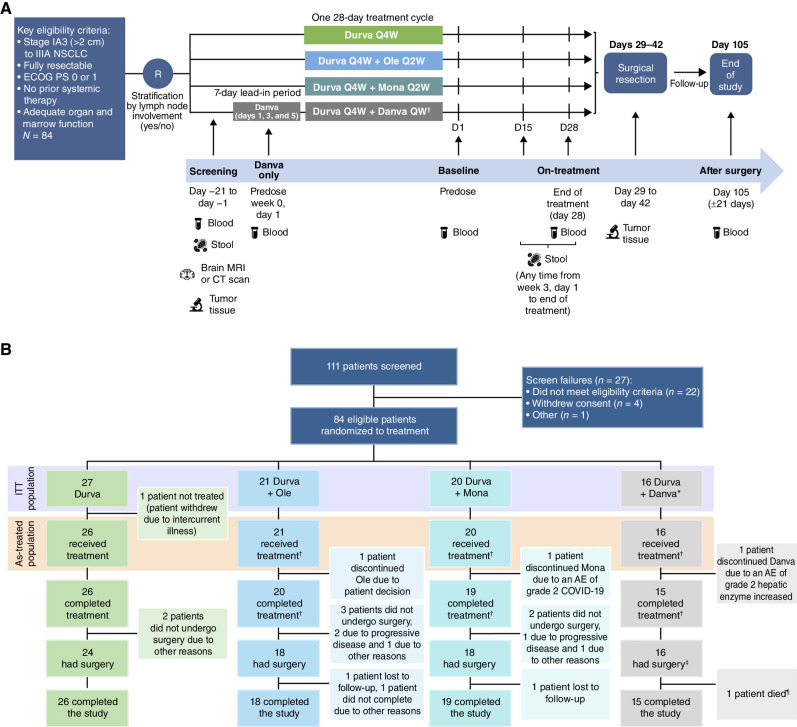
![Figure 2. Pathologic regressions at surgery; genomic profiles of ITT population and correlates with pathologic responses. A, The magnitude of pathologic response is shown by percent residual viable tumor cells in resected tumor and nodal samples for all patients with available data (total N = 75; durvalumab monotherapy arm: n = 24; durvalumab + oleclumab arm: n = 18; durvalumab + monalizumab arm: n = 18; durvalumab + danvatirsen arm: n = 15), and annotated with histologic subtype, tumor mutational burden (TMB; mutations/megabase), and history of smoking. PD-L1 status (≥1% positive; <1% negative) from baseline tumor biopsies was determined by IHC (SP263) for all evaluable patients (n = 33). Presence of activating EGFR mutations or ALK fusions was determined by whole-exome sequencing (n = 34). B, Residual viable tumor cells (RVT) from resected tumor and nodal samples are reported as 0% to 100%, and MPR (RVT ≤10%) for n = 60 patients (durvalumab monotherapy: n = 24, durvalumab + oleclumab: n = 18; durvalumab + monalizumab: n = 18). Best response by Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST) is reported for n = 58 patients. Somatic tumor alterations identified in EGFR, KRAS, STK11, KEAP1, TP53, ALK, and RET genes are reported from tumor tissue for n = 35 patients. For patients with evaluable circulating tumor DNA (ctDNA) samples at baseline, each patient is identified as having detected or no detected ctDNA at baseline. For patients with detectable ctDNA at baseline (total N = 20; durvalumab monotherapy: n = 6; durvalumab + oleclumab: n = 7; durvalumab + monalizumab: n = 7), molecular response is depicted at end-of-treatment (day 28, n = 14) and follow-up (day 105, n = 15) time points for all patients with evaluable ctDNA at those time points and represented as complete molecular response [100% reduction in variant allele frequency (VAF) from baseline, also referred to as complete clearance], partial molecular response (≥50% reduction in VAF from baseline), or no molecular response (<50% reduction in VAF from baseline). BESTRESP, best response; FU, follow-up; MR, molecular response; WT, wild-type.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/6bf3/10618740/cb4db42ba0d5/2394fig2.jpg)
![Figure 3. Treatment-related transcriptomic changes in tumor and peripheral blood. A, Left: differential gene expression between tumors collected at pretherapy and at surgery was assessed in all patients with evaluable paired tumor samples (total, n = 13 patients). Paired samples in each arm had the following MPR rates: durvalumab monotherapy: 0/4; durvalumab + oleclumab: 1/5; durvalumab + monalizumab: 2/4. Right: gene set enrichment analysis (GSEA) was used to identify gene sets and signatures significantly down- or upregulated from pretherapy to surgery on each treatment arm. The durvalumab monotherapy arm is not pictured, as no significant gene enrichment was observed. B, Patients with MPR are indicated in closed teal circle; patients without MPR are indicated in open gray circle. Top and top middle: mRNA from select genes associated with T cells, NK cells, and cytotoxicity is shown from pretreatment and surgery tumor tissue (n = 69 samples; n = 35 pretherapy, n = 34 surgery). Patients with paired samples (an evaluable sample from both pretreatment and surgery) are connected by a line. Bottom and bottom middle: mRNA from select genes associated with tumor and blood lymphocyte recruitment is shown from pretherapy to end-of-treatment peripheral blood collections (n = 120 samples; n = 65 pretherapy, n = 55 end-of-treatment). Y-axes units are all [Log2 (TPM + 0.01)], where TPM is transcripts per million. DN, down; Durva, durvalumab; IPA, Ingenuity Pathway Analysis; Mona, monalizumab; Ole, oleclumab.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/6bf3/10618740/df6d114ca665/2394fig3.jpg)