Oncogenesis arises from hijacking a subset of transposable elements active in pluripotent stem cells into regulatory elements for lineage-specific transcription factors, such as the androgen receptor in prostate cancer.
Abstract
Transposable elements hold regulatory functions that impact cell fate determination by controlling gene expression. However, little is known about the transcriptional machinery engaged at transposable elements in pluripotent and mature versus oncogenic cell states. Through positional analysis over repetitive DNA sequences of H3K27ac chromatin immunoprecipitation sequencing data from 32 normal cell states, we report pluripotent/stem and mature cell state–specific “regulatory transposable elements.” Pluripotent/stem elements are binding sites for pluripotency factors (e.g., NANOG, SOX2, OCT4). Mature cell elements are docking sites for lineage-specific transcription factors, including AR and FOXA1 in prostate epithelium. Expanding the analysis to prostate tumors, we identify a subset of regulatory transposable elements shared with pluripotent/stem cells, including Tigger3a. Using chromatin editing technology, we show how such elements promote prostate cancer growth by regulating AR transcriptional activity. Collectively, our results suggest that oncogenesis arises from lineage-specific transcription factors hijacking pluripotent/stem cell regulatory transposable elements.
Significance:
We show that oncogenesis relies on co-opting transposable elements from pluripotent stem cells as regulatory elements altering the recruitment of lineage-specific transcription factors. We further discover how co-option is dependent on active chromatin states with important implications for developing treatment options against drivers of oncogenesis across the repetitive DNA.
This article is featured in Selected Articles from This Issue, p. 2293
INTRODUCTION
During normal development, pluripotent stem cells progressively lose plasticity as they commit to one of many diverse mature cell states (1). Such cell fate determination is ensured by chromatin variants, corresponding to segments of the genome that change chromatin state during cellular differentiation (2–4). Chromatin variants identify distinct classes of DNA elements contributing to cellular identity, inclusive of transcribed genes, their regulatory elements, and anchors of chromatin interactions (5–12). In pluripotent stem cells, chromatin variants at regulatory elements in nonrepetitive DNA reveal binding sites for pluripotent transcription factors, namely NANOG, OCT4 (POU5F1), and SOX2, while chromatin variants from somatic cell states relate to lineage-specific transcription factors (2, 3, 13–16). Similarly, chromatin variants from transcribed regions capture cell state–specific gene expression patterns (17, 18). Collectively, finding and characterizing chromatin variants across cell states reveal the regulatory processes that impact cell fate determination.
Oncogenesis is also governed by chromatin variants (8, 19–24). For instance, prostate cancer is characterized by chromatin variants that reveal regulatory elements serving as binding sites for pro-oncogenic transcription factors including the androgen receptor (AR), FOXA1, and HOXB13 (25–29). These chromatin variants are also enriched for mutations and risk variants of prostate cancer (28, 30–34). From recent advances in genome analysis, chromatin variants are revealing new insights across repetitive DNA sequences that constitute over half of the human genome (4). For example, chromatin variants repress the expression of endogenous retroviral sequences in prostate and other cancer types to prevent the activation of the viral mimicry response (35–37). Chromatin variants are also reported at repeat DNA that reveal the co-option of transposable elements (TE) as “regulatory TEs” promoting oncogene overexpression (38–40). While the latter aligns with promoter, enhancer, or anchor of chromatin interaction functions for TEs in normal development (41), whether such regulatory TEs rely on a transcriptional machinery shared or not with nonrepetitive DNA sequences is unknown. In this study, we identified and characterized repetitive DNA serving as regulatory TEs across normal and prostate cancer cell states to report a role for AR and FOXA1 at TEs active in prostatic epithelium compared with pluripotent stem cells, and a switch in primary prostate tumors for AR toward a subset of regulatory TEs active in pluripotent stem cells that contribute to oncogenic growth.
RESULTS
Pluripotent Stem Cells to Mature Cell and Tissue States Harbor Distinct Regulatory TEs
Pluripotent stem cells give rise to diverse somatic tissue states. To systematically evaluate the enrichment for TE families with regulatory properties in pluripotent stem cells and somatic tissue states, we looked for enrichment of TE families within H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) data generated in 339 individual samples from 32 different cell and tissue states (ENCODE project released data; Supplementary Fig. S1A; Supplementary Table S1). H3K27ac is a feature of chromatin states that typifies active regulatory elements and is characteristic of chromatin variants defining cell state identity (5, 7, 42). Of all repetitive DNA, we focused on 971 phylogenetically defined families of TEs (43), excluding simple repeats, satellites, short nuclear RNA (snRNA)/rRNA, and repeats of unknown subfamilies from our analysis. Using ChromVAR adapted to TE families (bioRxiv 2021.02.16.431334), we compared pluripotent stem cells to each individual cell/tissue state. We observed a higher number of TE families enriched within H3K27ac chromatin variants in pluripotent stem cells when compared with any of the non-extraembryonic mature cell or tissue states (Fig. 1A; Supplementary Tables S2 and S3). We also found 92 TE families enriched only within H3K27ac chromatin variants from a single mature cell or tissue state (Fig. 1B; Supplementary Table S4). In comparison, 23 TE families were defined as “regulatory TEs” based on being uniquely enriched in H3K27ac chromatin variants of pluripotent stem cells, across all pairwise comparisons between pluripotent stem cells with mature cell/tissue states (Fig. 1B; Supplementary Table S4). By grouping TE families into TE superfamilies according to Repbase (see Methods), we observed that regulatory TE families found in mature cell and tissue states are not restricted to a single superfamily but rather members of ERV, LINE, solo LTR, SINE, and Transposon superfamilies (Fig. 1C). For instance, adrenal glands harbor chromatin variants enriched for TEs from the MER65A family, part of the ERV1 superfamily (q = 5.86e-07, Fig. 1D). Similarly, TEs of the L1MC3 family, part of the LINE1 superfamily, are enriched in the chromatin variants from keratinocytes (q = 0.006, Fig. 1D). Finally, TEs of the AluSq4 family, part of the SINE1 superfamily, are preferentially found in chromatin variants from neutrophils (q = 0.004, Fig. 1D). In contrast, the 23 regulatory TE families in pluripotent stem cells belong solely to the ERV1 superfamily (Fig. 1C), exemplified by the LTR7 TE family (q ≤0.0085, Fig. 1D; Supplementary Table S4), known to control expression of the pluripotent stem cell–specific ERVH noncoding RNA (44, 45). Collectively, from assessing the enrichment of TE families over regulatory elements identified from H3K27ac ChIP-seq, we discovered families of regulatory TEs specific to pluripotent stem versus mature cell and tissue states, whereby pluripotent stem cell specificity is skewed toward TEs of the ERV1 superfamily.
Figure 1.
The landscape of TE families in active regulatory elements of pluripotent stem cells (PSC) and somatic tissues. A, Bar plot showcasing the number of TE families differentially enriched between PSCs and each individual tissue state. Note that the number of TE families enriched is always higher compared with somatic tissues. GastroS, gastric sphincter; Mem, memory; Mon, monocyte; Nervs, nerves and neural connections. B, Frequency plot of TE families enriched across the different number of somatic tissues and consistently enriched in PSCs. Note that TE families are mostly enriched in one somatic tissue state and a set of 23 TE families is always enriched in PSCs. C, Comparison of the number of PSC- and tissue-specific TE families. Colors correspond to the tissue state showing enrichment of a given number of TE families (left) or to TE superfamilies (right). Note that PSC-specific TE families are populated only by ERV1. D, Examples of tissue state–specific TE families. Box plots show differential deviation Z-scores in PSCs and in all somatic tissue states. E, Number of PSC-specific regulatory TE families (rTE) bound by pluripotency factors NANOG, SOX2, POU5F1 (OCT4), and KLF4.
Mature Cell States Are Defined by Regulatory TEs for Lineage-Specific Transcription Factors
We next assessed whether regulatory TEs could serve as docking sites for transcription factors. Using the ReMap atlas of 1,185 transcription factor cistromes (46), we specifically measured the propensity of transcription factors to bind to families of regulatory TEs from pluripotent stem cell state. From the regulatory TE families specific to pluripotent stem cells, we found an enrichment for the cistrome of the pluripotency factors NANOG, SOX2, OCT4 (POU5F1), and KLF4 (Fig. 1E), in agreement with previous reports (47, 48). Next, we specifically focused on prostate tissue, where transformation into localized prostate cancer and subsequent progression to advanced stages of the disease are typified by an initial expansion of chromatin variants across nonrepetitive DNA sequences related to lineage-specific transcription factors (25, 26, 29), followed by a transition toward chromatin variants linked to primitive cell states (28). Comparing H3K27ac ChIP-seq data from benign prostate (n = 13) and pluripotent stem cell (n = 25) samples, regardless of all other somatic tissues, identified 97 regulatory TE families enriched in benign prostate samples and 315 regulatory TE families enriched in pluripotent stem cells (q ≤0.01, Fig. 2A–C; Supplementary Table S5). Regulatory TE families from benign prostate belong to the LINE5, SINE, ERV2, and solo-LTRs superfamilies (Fig. 2C). Regulatory TE families from pluripotent stem cells, compared with benign prostate, are members of the ERV1, Transposon and SINE1, and LINE1 superfamilies (Fig. 2C).
Figure 2.
Distinct TE families populate the active regulatory elements of pluripotent stem cells (PSC) and benign prostate tissue. A, Heat map displaying the deviation (Dev) Z-scores of TE families differentially enriched in H3K27ac-positive chromatin between PSCs and benign prostate (rows, q-value ≤0.01). B, Volcano plot showing median difference in deviation Z-scores for each TE family enriched in H3K27ac-positive chromatin between PSCs and benign prostate tissue state vs. the −log10 q-value for that difference. The gray dashed line corresponds to −log10(q-value) = 2 (q-value = 0.01). The number of TE families enriched in H3K27ac-positive chromatin in PSCs or benign prostate is reported at the top. n.s., not significant. C, Direct comparison of PSCs vs. benign prostate. Y-axis shows all TE superfamilies with enriched TE families in H3K27ac-positive chromatin; individual points denote distinct TE families. The LINE1, ERV2, LTR (no ERVs), and Transposon families are enriched in benign prostate, whereas the ERV1, ERV3, and SINE1 families are enriched in PSCs (left). Number of TE families enriched in H3K27ac-positive chromatin in PSCs or benign prostate, divided by TE superfamily (right). TE superfamilies ordered from most highly enriched in H3K27ac-positive chromatin in PSCs to benign prostate. D and E, Frequency of transcription factor (TF) cistromes enriched at regulatory TE families (rTE) enriched in PSCs (D) and benign prostate (E). The top 5% most frequently enriched transcription factor cistromes are shown. Transcription factors associated with benign prostate development are indicated in red in F and G. F and G, Transcription factor cistromes enriched over L1PA6 and LTR5B TE families. Every dot corresponds to a transcription factor cistrome considered in the analysis. Red dashed lines correspond to −log10(q-value) = 1.3 (q-value = 0.05) and OR = 1 thresholds; q-values correspond to FDR-corrected Fisher exact test. H, Individual AR (left) or FOXA1 (right) cistromes enriched over TE families enriched in benign prostate tissue state and with enrichment of the AR or FOXA1 cistrome (according to Fig. 1D). Box plots showing enrichment GIGGLE scores of individual AR cistromes profiled in cell lines derived from prostate, mammary, or other tissue state. I, Enrichment of AR and FOXA1 DNA recognition sequences within L1PA6 and LTR5B TE families. Bars represent −log10(q-value) for each TE family for transcription factor DNA recognition sequences. The red dashed line corresponds to −log10(q-value) = 1.3 (q-value = 0.05) threshold; q-values correspond to Benjamini-corrected P values.
We next examined which transcription factors could bind to the 97 regulatory TE families from benign prostate as opposed to the 315 from pluripotent stem cells. Using the ReMap atlas of transcription factor cistromes, we discovered that CHD7, NANOG, SOX2, OCT4 (POU5F1), TEAD4, GATA6, and TRIM28 were among the top 5% of transcription factors prone to bind to the largest number of regulatory TE families in pluripotent stem cells (Fig. 2D; Supplementary Table S6). In comparison, the top 5% of transcription factors in benign prostate included CTCF, AR, XBP1, FOXA1, RELA, GRHL3, and hypoxia-inducible factor-1α (HIF1A; Fig. 2E; Supplementary Table S7). We specifically reported enrichment of the top 5% transcription factor cistromes for at least 17 of the 97 regulatory TE families from benign prostate (Fig. 2E). Noteworthy, compared with other mature cell and tissue states originating from the endoderm germ layer and pluripotent stem cells, the AR cistrome was uniquely enriched across the regulatory TE families in benign prostate, together with ERG, STAG1, TFAP2C, FOXA2, NFYA, and ZFX (Fig. 2E; Supplementary Fig. S1B–S1D). The FOXA1 cistrome was found to preferentially occupy regulatory TE families in prostate, liver, and pluripotent stem cells. This observation is in agreement with previous studies reporting essential FOXA1 functions in liver development and pluripotent stem cell identity (refs. 49, 50; Fig. 2D and E; Supplementary Fig. S1B–S1D; Supplementary Tables S8–S10).
The specific enrichment of AR and FOXA1 cistromes at benign prostate regulatory TE families was prominent at elements from the L1PA6 and LTR5B families [L1PA6: AR q = 0.011, LogOR (lgOR) = 1.79 and FOXA1 q = 2.94e-05, lgOR = 2.16; LTR5B: AR q = 5.68e-06, lgOR = 2.57 and FOXA1: q = 3.23e-07, lgOR = 2.84; Fig. 2F and G]. We further validated these observations by computing the cistrome enrichment over regulatory TEs using an independent approach relying on the GIGGLE tool (51). Our analysis reveals GIGGLE scores for the enrichment of AR and/or FOXA1 cistromes across 18 and 20 of the 97 regulatory TE families from benign prostate, respectively (Fig. 2H; Supplementary Tables S11 and S12), most notable for the L1PA6 and LTR5B regulatory TE families (Fig. 2H; Supplementary Tables S11 and S12). In parallel, sequence analysis identified enrichment for the DNA recognition motifs for AR and FOXA1, namely the androgen responsive elements (ARE) and the forkhead motifs (FKH), respectively, among motifs present within L1PA6 and LTR5B elements (Fig. 2I; Supplementary Fig. S1E). Collectively, these results suggest that mature benign prostate harbors regulatory TE families that can be bound by the lineage-specific transcription factors AR and FOXA1 (52, 53), revealing that regulatory elements in repetitive DNA relate to the same transcriptional machinery active over nonrepetitive DNA (25, 26, 29).
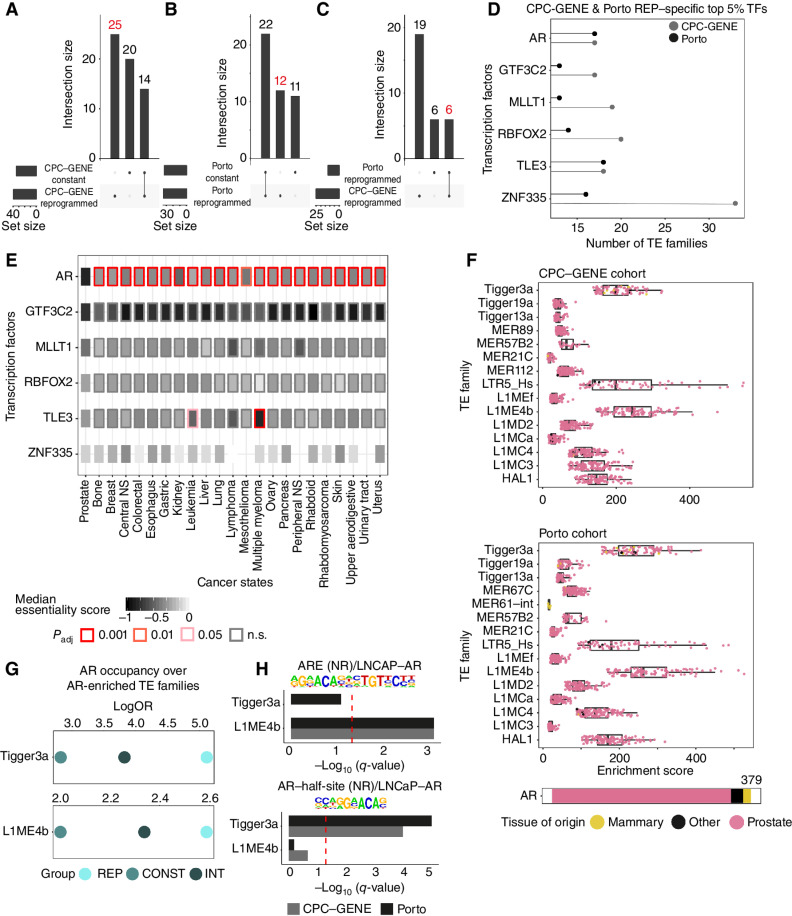
Regulatory TEs Define a Reprogrammed and Constant Prostate Cancer Subtype
Cell state transition leading normal cells to transform into cancer cells is accompanied by gains and losses of chromatin variants (23). In localized prostate cancer, chromatin variants found at nonrepetitive DNA sequences enable lineage-specific transcription factors, such as AR and FOXA1, to drive oncogene expression (25, 26, 29). We therefore assessed how transformation to localized prostate cancer could impact regulatory properties of TEs. Using H3K27ac ChIP-seq data from two independent cohorts of localized prostate tumors [CPC-GENE (25, 27, 54), n = 48, and Porto, n = 92 (26); Supplementary Fig. S1A] to score the enrichment of TE families in regulatory elements, we revealed two subgroups of prostate tumors based on unsupervised hierarchical clustering (Fig. 3A and B). We labeled a subgroup “constant” because it showed no significant enrichment for regulatory TE families (Fig. 3C and D; Supplementary Tables S13 and S14). We labeled the other subgroup “reprogrammed,” being composed of localized prostate tumors enriched for regulatory TE families (191 and 357 families for the CPC-GENE and Porto cohorts, respectively; Fig. 3C and D; Supplementary Tables S13 and S14). Comparing regulatory TE families found in the reprogrammed subgroup across both prostate tumor cohorts identified 186 shared regulatory TE families, corresponding to 97% and 52% of regulatory TE families from the CPC-GENE and Porto cohorts, respectively (Fig. 3E). Individual elements belonging to AluJb, HAL1, and Tigger3a families exemplify reprogrammed specific regulatory TEs (Supplementary Fig. S2A). A third subgroup of localized prostate tumors was also detected in the Porto cohort, being similar to the “reprogrammed” subgroup but with lower levels of enrichment score for regulatory TE families (Fig. 3B; Supplementary Table S15). Accordingly, this “intermediate” subgroup had 205 significantly enriched regulatory TE families, and all of them were in common with the “reprogrammed” subgroup (Supplementary Fig. S2B). We observed that 164 of the 186 (88%) regulatory TE families were shared with those found in pluripotent stem cells (164/315, 52%; Fig. 3F). Only 22 of the 186 (12%) regulatory TE families from the reprogrammed subgroup were shared with those found in the benign prostate (22/97, 23%; Fig. 3F). The 164 TE families shared with pluripotent stem cells showed a repartition into TE superfamilies very similar to pluripotent stem cells, with TE families mostly belonging to the ERV1, Transposon and SINE1, LINE1, and ERV3 superfamilies (Supplementary Fig. S2C). This suggests a high degree of similarity between prostate tumors of the reprogrammed subgroup with pluripotent stem cells. In agreement, our “reprogramming score” based on the 186 regulatory TE families shared between the CPC-GENE and Porto cohort prostate tumours (see Methods) is enriched in the reprogrammed prostate tumor subgroup, while the intermediate prostate tumor subgroup partially enriches and the constant prostate tumor subgroup is depleted for this signature (Fig. 3G and H). We also found pluripotent stem cells to rank high for the reprogramming score, while benign prostate samples were depleted for this score (Fig. 3I). Assessing the enrichment of TE families in accessible chromatin, using the Assay of Transposase Accessible Chromatin (ATAC) data previously generated from prostate adenocarcinoma (PRAD) samples [The Cancer Genome Atlas (TCGA) cohort; ref. 55; Supplementary Fig. S1A)], stratified tumors into two subgroups (Supplementary Fig. S2D; Supplementary Table S16). One subgroup of PRAD tumors enriched for 139 regulatory TE families, while the second subgroup was deprived of any enrichment (Supplementary Fig. S2E; Supplementary Table S16). We labeled the first TCGA subgroup as “reprogrammed” because 108 of the 139 (78%) regulatory TE families from these tumors were shared with the reprogrammed tumors from the CPC-GENE and Porto cohorts (108/186: 58%; Supplementary Fig. S2F). A total of 102 of the 108 regulatory TE families (102/108: 94%) were shared with pluripotent stem cells (102/315: 33%), while only six (6/108: 6%; 6/97: 6%) were shared with benign prostate (Supplementary Fig. S2G). Furthermore, the reprogrammed TCGA tumors were assigned a higher reprogramming score compared with the other TCGA tumors we labeled as “constant” (Supplementary Fig. S2H). We next investigated whether individual reprogrammed elements were involved in onco-exaptation, which has been previously linked to oncogene expression. A total of 13 individual elements from nine of the 164 regulatory TE families in reprogrammed prostate tumors (CPC-GENE and Porto) were shown to be onco-exapted in primary prostate tumors (ref. 39; Supplementary Table S17). These 13 individual elements correspond to two elements from the AluJb, AluSp, AluSx, and AluSz TE families, as well as one element from the AluJr, AluSc8, AluSx1, L1MB3, and MER41A TE families (Supplementary Table S17). While onco-exaptation was linked to oncogene expression (39), only seven of the 13 onco-exapted elements classify as regulatory elements in tumors from the CPC-GENE and Porto cohorts (four in CPC-GENE tumors and three in Porto tumors; Supplementary Fig. S2I). Collectively, our results show how localized prostate tumors differ from each other based on regulatory TE families by stratifying to constant or reprogrammed subgroups. We further show similarities from the families of TEs with regulatory properties between the reprogrammed subgroup and pluripotent stem cells, as opposed to benign prostate tissue.
Figure 3.
TE families are reprogrammed in a subset of patients with prostate cancer. A and B, Heat maps displaying the deviation (Dev) Z-scores of TE families differentially enriched in H3K27ac-positive chromatin between pluripotent stem cells and benign prostate tissue state across H3K27ac profiles of CPC-GENE patients (A; n = 48) or Porto patients (B; n = 92). BCR, biochemical recurrence; NE score, neuroendocrine score; Neg, negative; Pos, positive; REP score, reprogramming score. C and D, Volcano plots showing median difference in deviation Z-scores for each TE family enriched in H3K27ac-positive chromatin between reprogrammed (REP) and constant (CONST) patients (CPC-GENE patients in C, Porto patients in D) vs. the −log10 q-value for that difference. The gray dashed line corresponds to −log10(q-value) = 2 (q-value = 0.01). The number of TE families enriched in reprogrammed or constant patients is reported at the top. n.s., not significant. E, UpSet plot showing the intersection of TE families enriched in reprogrammed CPC-GENE and Porto patients. Note that the vast majority of TE families are commonly enriched. F, UpSet plot showing the intersection of TE families enriched in pluripotent stem cells (PSC), benign prostate, and reprogrammed prostate cancer patients. Note that 164 TE families are commonly enriched in patients with reprogrammed prostate cancer and PSCs, corresponding to reprogrammed TE families, while 22 TE families are shared between patients with prostate cancer and benign prostate. Red font highlights the set of TE families described in the corresponding results section (more precisely, 164 TE families common to prostate cancer patients and pluripotent stem cells and 22 TE families common to prostate cancer patients and benign prostate epithelium). G–I, Box plot displaying the reprogramming score in reprogrammed and constant CPC-GENE (G) and Porto patients [with intermediate (INT) patients; H], and in pluripotent stem cells and benign prostate (I). P value results of Wilcoxon test are showcased on the box plot. J, Breakdown of common prostate cancer genetic aberrations called from whole-genome sequencing data in each of the 48 CPC-GENE samples separated by TE-based clustering (left). Comparison of the frequency of genetic aberrations using Fisher exact test (right). The red dashed line corresponds to −log10(P) = 1.3 (P = 0.05) threshold. Note that no genetic aberration tested was found significantly different between reprogrammed and constant patients. NA, not available.
Stratifying prostate tumors from the CPC-GENE and Porto cohorts based on the H3K27ac ChIP-seq data over the nonrepetitive genome does not cluster tumors into the reprogrammed and constant subgroups (Supplementary Fig. S3A and S3B). Furthermore, we could not observe a relationship between the mutational status of prostate cancer driver genes, such as TP53, PTEN, SPOP, CHD1, RB1, NKX3-1, CDKN1B, and MYC, and the reprogrammed and constant subgroup stratification (Fig. 3J). This result also applied to the TMPRSS2–ERG translocation, previously linked to distinct H3K27ac signal distribution over nonrepetitive DNA in primary prostate tumors (refs. 25, 26; Fig. 3J; Supplementary Fig. S3C). Expanding this analysis to clinical (time to biochemical recurrence, Gleason score, age at diagnosis) and molecular features [AR expression, hypoxia score, genomic instability (percentage of genome altered score), pretreatment PSA, and neuroendocrine score] also failed to find any correlation with tumor stratification from regulatory TEs (Supplementary Fig. S3D–S3J; Supplementary Tables S18 and S19). However, we observed a higher AR activity score in reprogrammed patients compared with constant ones (Supplementary Fig. S3H). Collectively, these results suggest that a subset of tumors harbor a pluripotent stem cell–like biology based on regulatory TE families and that these account for a distinct mechanism controlling prostate cancer cell state identity compared with nonrepetitive regulatory elements.
Regulatory TEs in Prostate Tumors from the Reprogrammed Subgroup Are Binding Sites for the Lineage-Specific Transcription Factor AR
We next assessed how the 164 regulatory TE families from the “reprogrammed” subgroup of localized prostate tumors could impact transcriptional processes. We first used the ReMap atlas of transcription factor cistromes to find transcription factors prone to bind to the largest number of regulatory TE families from the reprogrammed subgroup. This identified 39 and 34 transcription factors from the CPC-GENE and Porto cohorts, respectively (Fig. 4A and B; Supplementary Tables S20 and S21). Repeating this analysis considering elements from the 164 regulatory TE families with H3K27ac ChIP-seq signal in prostate tumors of the “constant” subgroup identified 34 and 33 transcription factors in the CPC-GENE and Porto cohorts, respectively (Fig. 4A and B; Supplementary Tables S22 and S23). Fourteen and 22 were common to the two analyses (Fig. 4A and B), while 25 and 12 transcription factors were unique to the reprogrammed subgroup from the CPC-GENE and Porto cohorts, respectively, with six found in both cohorts (Fig. 4A–C; Supplementary Tables S20–S23). These include the AR, GTF3C2, MLLT1, RBFOX2, TLE3, and ZNF335 transcription factors (Fig. 4D). Although the 164 regulatory TE families from the reprogrammed subgroup were common to pluripotent stem cells, no pluripotent stem cell factors were identified from the cistrome enrichment analysis, arguing that regulatory TEs unique to pluripotent stem cells are biased to those relevant to the recruitment of pluripotent stem cell factors. In contrast, the 164 regulatory TE families acted as binding sites for prostate cancer oncogenic factors, such as AR, and other transcription factors that could be implicated in prostate cancer oncogenesis. Using the RNAi-based essentiality screen results from the Cancer Dependency Map (DepMap) project (56) revealed a negative median essentiality score for all six transcription factors in prostate cancer cell lines (Fig. 4E). Noteworthy, AR was the only transcription factor significantly more essential in prostate cancer cell lines compared with cell lines from other cancer states (Fig. 4E). The dependency on AR in tumors of the reprogrammed subgroup could not be justified from its expression levels (Supplementary Fig. S3J). From the GIGGLE analysis, we observed consistent high enrichment scores for AR cistromes generated mainly in samples originating from prostate, inclusive of cancer cells, across regulatory TE families identified from the ReMAP analysis from the CPC-GENE or Porto cohort (Fig. 4F). The Tigger3a, LTR5_Hs, and L1ME4b regulatory TE families had the highest median GIGGLE score in CPC-GENE or Porto cohort toward AR cistromes (Fig. 4F). Taking advantage of previously published matched H3K27ac and AR ChIP-seq data from localized prostate tumors of the Porto cohort (26), we validated our observations in vivo by detecting a higher enrichment of the AR cistrome in the reprogrammed subgroup over nine of the 16 TE families in reprogrammed compared with constant and intermediate subgroup samples, including the Tigger3a and L1ME4b TE families (Fig. 4G; Supplementary Fig. S4A). AR directly binds to a DNA homodimer known as ARE that consists of two 6-bp-long “half-sites,” spaced by 3 bp (57). AR also has strong binding capacities for the AR “half-site” (58); therefore, we investigated the enrichment of both ARE and AR half-sites within TE families enriched for the AR cistrome. DNA recognition motif analysis detected a significant enrichment for ARE and AR half-site motifs, respectively, within Tigger3a and L1ME4b regulatory TEs from the reprogrammed subgroup (Fig. 4H; Supplementary Fig. S4B). As a whole, these results suggest that regulatory TEs found in tumors from the reprogrammed subgroup can serve as docking sites for AR, supportive of an AR-dependent function over repetitive DNA sequences in a subset of localized prostate tumors.
Figure 4.
Reprogrammed TE families act as binding sites for AR. A and B, UpSet plot showing the intersection of transcription factor cistromes found in the top 5% most frequent cistromes enriched over reprogrammed TE families in reprogrammed or constant patients (CPC-GENE in A and Porto in B).Red font highlights the sets of transcription factor cistromes of interest (unique to reprogrammed CPC-GENE or Porto prostate cancer patients). C, UpSet plot showing the intersection of transcription factors commonly found in the top 5% most frequent cistromes enriched over reprogrammed TE families in the reprogrammed subgroup (CPC-GENE and Porto) but absent in the constant subgroup. Red font highlights the sets of transcription factor cistromes of interest (transcription factor cistromes common to reprogrammed CPC-GENE and Porto prostate cancer patients). The six factors labeled in red are then used for D and E. D, Frequency of transcription factor cistromes (TF) enriched at reprogrammed TE families in reprogrammed CPC-GENE and Porto patients. The top 5% most frequently enriched transcription factor cistromes specific to reprogrammed patients is shown. E, Overview of essentiality scores of TFs in D across all cancer types available in DepMap with more than five cell lines, based on RNAi data. The distribution of the essentiality scores of each transcription factor calculated in prostate cancer cell lines was compared with the distribution of the same transcription factor calculated in each other cancer state cell line. Rectangle inner color corresponds to median essentiality score, while border color corresponds to pairwise t test Benjamini–Hochberg–corrected P values (Padj). NS, nervous system; n.s., not significant. F, Individual AR cistromes enriched over reprogrammed TE families with enrichment of AR cistrome shown in B. Box plots showing enrichment GIGGLE scores of individual AR cistromes profiled in cell lines derived from prostate, mammary, or other tissue state in CPC-GENE (top) or Porto (bottom) reprogrammed patients. Note that the Tigger3a, L1ME4b, and LTR5_Hs TE families are the top three TE families for enrichment of AR cistromes. G, Enrichment of AR cistromes profiled in reprogrammed (REP), intermediate (INT), and constant (CONST) Porto patients at the Tigger3a and L1ME4b TE families. H, Enrichment of AR DNA recognition sequences within Tigger3a and L1ME4b TE families. Bars represent −log10(q-value) for CPC-GENE or Porto reprogrammed patients. The red dashed line corresponds to −log10(q-value) = 1.3 (q-value = 0.05) threshold; q-values correspond to Benjamini-corrected P values.
Regulatory TEs Essential for the Growth of Reprogrammed Subgroup Prostate Tumors
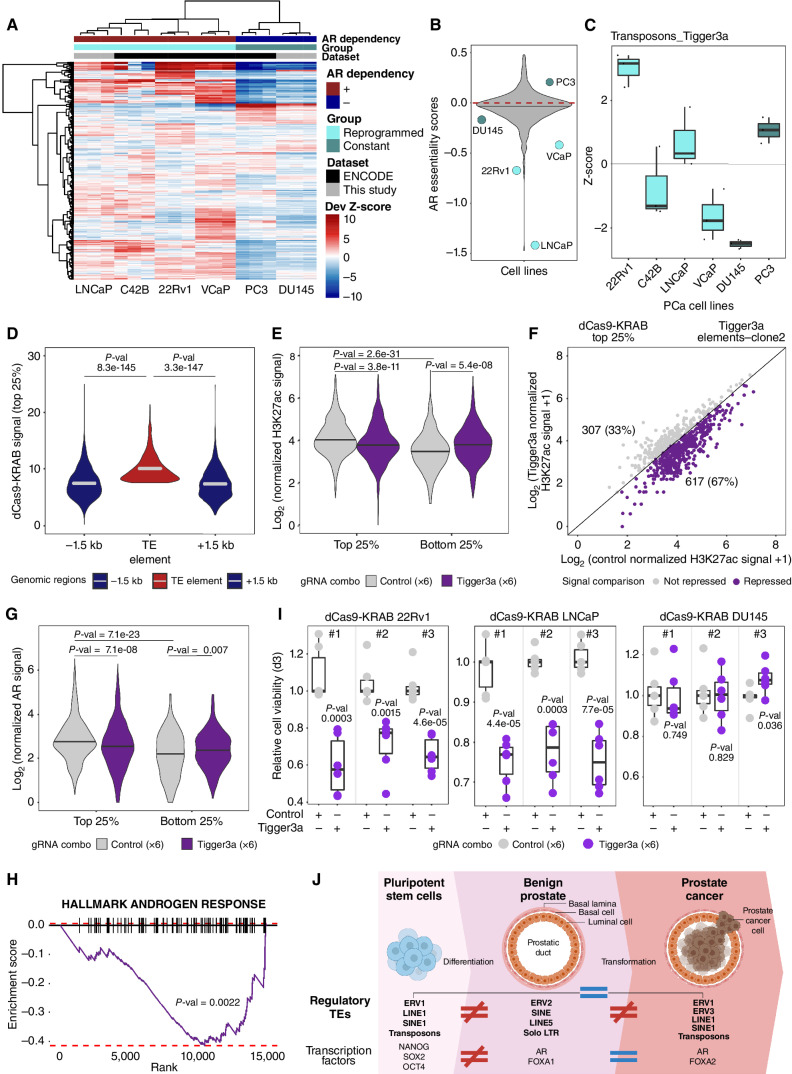
To investigate the role of AR at regulatory TE families, we first determined whether AR dependency for growth was correlated with the reprogrammed subgroup stratification. We used H3K27ac ChIP-seq data generated across a panel of prostate cancer cell lines to quantify the enrichment of TE families at H3K27ac regions (Supplementary Fig. S1A). This assigned LNCaP, C42B, 22Rv1, and VCaP to the reprogrammed subgroup (Fig. 5A). The PC3 and DU145 prostate cancer cell lines were assigned to the constant subgroup (Fig. 5A). Combining these results with AR essentiality scores collected from the DepMAP data (59–61) revealed low essentiality scores corresponding to a high dependency for AR in the reprogrammed subgroup prostate cancer cell lines (Fig. 5A and B; Supplementary Fig. S5A–S5D). In contrast, high scores defining low dependency were observed in the constant subgroup prostate cancer cell lines (Fig. 5A and B; Supplementary Fig. S5A–S5D). To further investigate the process of TE reprogramming in primary prostate cancer, we assessed the enrichment of reprogrammed TEs in chromatin regions marked by the repressive H3K9me3 modification (62) in benign-like and prostate cancer cell lines (Supplementary Fig. S1A). Focusing on the 186 regulatory TE families shared between the CPC-GENE and Porto cohort prostate tumors, we scored the enrichment over H3K9me3-modified chromatin (see Methods). The enrichment of TE families in repressive H3K9me3 chromatin was observed in the “constant” prostate cancer cell line model (DU145) and in benign-like prostate cell lines (Supplementary Fig. S5E). In contrast, limited enrichment was observed for H3K9me3 across TE families for two of three reprogrammed prostate cancer cell lines (LNCaP and VCaP; Supplementary Fig. S5E). Hence, regulatory TE families in reprogrammed prostate cancer cell lines are depleted of H3K9me3 modifications, while the absence of regulatory TE families in constant prostate cancer cells is linked to the enrichment for the repressive H3K9me3 modification (Supplementary Fig. S5E). These results link AR-dependent growth properties in prostate cancer to the chromatin variants over repetitive DNA sequences, defining regulatory TEs specific to the reprogrammed subgroup.
Figure 5.
Tigger3a elements are essential regulatory elements for prostate cancer cell growth. A, Heat map displaying the deviation (Dev) Z-scores of TE families differentially enriched in H3K27ac-positive chromatin between pluripotent stem cells and benign prostate across H3K27ac profiles of prostate cancer cell lines (publicly available or newly generated). B, AR essentiality mediated through RNAi across various cell lines. Each dot indicates a prostate cancer cell line. Prostate cancer lines included in A are labeled with the name of the cell lines and color coded according to similarity to patients with prostate cancer. The red dashed line corresponds to essentiality score = 0 threshold. C, Tigger3a enrichment across H3K27ac profiles of prostate cancer cell lines. Box plots show differential deviation Z-scores in prostate cancer cell lines. D, Violin plot showcasing the dCas9-KRAB signal intensity over top 25% Tigger3a elements compared with matched flanking 1.5-kb regions in clone 2 dCas9-KRAB 22Rv1 (upstream, −1.5 kb; downstream, +1.5 kb; right). P value results of Wilcoxon test are showcased on the violin plot. PCa, prostate cancer. E, Violin plot showcasing the H3K27ac signal distribution in clone 2 dCas9-KRAB 22Rv1 cells nucleofected with control (gray) or Tigger3a (purple) gRNA combinations over Tigger3a elements with high dCas9-KRAB signal (top 25%) or low dCas9-KRAB signal (bottom 25%). P value results of Wilcoxon test are showcased on the violin plot. Note that H3K27ac signal significantly decreases between control and Tigger3a conditions for top 25% Tigger3a elements, while it does not decrease significantly for bottom 25% Tigger3a elements. Interestingly, H3K27ac signal drops significantly also between Control conditions between top 25% and bottom 25% Tigger3a elements. P value results of Wilcoxon test are showcased on the violin plot. F, H3K27ac signal over top 25% dCas9-KRAB bound Tigger3a elements in clone 2 dCas9-KRAB 22Rv1 cells nucleofected with control or Tigger3a gRNA combinations. Every dot corresponds to one Tigger3a element; the x-axis represents the log2 of the normalized H3K27ac signal intensity in two independent nucleofections with control gRNA combination, while the y-axis represents the log2 normalized H3K27ac signal intensity in two independent nucleofections with Tigger3a gRNA combination. G, Violin plot showcasing the AR signal distribution in clone 2 dCas9-KRAB 22Rv1 cells nucleofected with control (gray) or Tigger3a (purple) gRNA combinations over Tigger3a elements with high dCas9-KRAB signal (top 25%). P value results of Wilcoxon tests are showcased on the violin plot. Note that AR signal significantly decreases between control and Tigger3a conditions for top 25% Tigger3a elements. H, Gene set enrichment analysis enrichment plots showcasing significant depletion of androgen response in clone 2 dCas9-KRAB cells nucleofected with Tigger3a gRNA combination compared with control combination. P value results of weighted Kolmogorov–Smirnov test are showcased on the plots. I, Relative cell viability upon dCas9-KRAB mediated chromatin repression at Tigger3a elements (combinations of 6 individual gRNAs targeting Tigger3a elements or scramble) in clonal 22Rv1 and LNCaP (32) and DU145 stably expressing dCas9-KRAB. Every dot represents an independent nucleofection reaction using guides targeting Tigger3a elements or negative control (scramble). Error bars, SD. P values were generated by two-sided t test. J, Graphical representation of the main discoveries of this study showcasing the dynamic of the enrichment of TEs and transcription factors in H3K27ac-positive chromatin in the progression from pluripotent stem cells to benign tissue and to localized primary prostate cancer. Despite the strong similarity between pluripotent stem cells and prostate cancer tissues, TEs act as binding sites for AR in primary prostate cancer. This figure was created with BioRender.com.
We next assessed the requirement for reprogrammed TE families toward prostate cancer cell growth, using the CRISPR/dCas9-KRAB (CRISPRi) chromatin editing technology, to induce chromatin state changes to repress regulatory TEs without affecting their underlying DNA sequence (4, 63, 64). The 22Rv1 and LNCaP cell lines showed the highest enrichment for Tigger3a with H3K27ac ChIP-seq data among all reprogrammed subgroup prostate cancer cell lines, while the constant subgroup DU145 cell lines showed a depletion for Tigger3a elements across its H3K27ac ChIP-seq data (Fig. 5C). Using the H3K9me3 ChIP-seq data, we saw a depletion for the Tigger3a TE family in the repressive chromatin of reprogrammed subgroup prostate cancer cell lines (Supplementary Fig. S5F). This is in contrast to the enrichment of the Tigger3a TE family in H3K9me3 chromatin of constant subgroup prostate cancer cell lines (Supplementary Fig. S5F). Next, we designed a six guide RNA (gRNA) combo against the Tigger3a elements (5,317 individual genomic regions) using the Repguide tool (40, 65) to maximize on-target while limiting off-target effects. Unfortunately, we failed to design gRNAs for other regulatory TE families of interest, such as L1ME4b. As a negative control, we used a combination of six scramble gRNAs (66). From transiently nucleofecting Tigger3a or control gRNA combos in three independent 22Rv1 CRISPRi prostate cancer cell line clones (32) (Supplementary Fig. S5G), we first assessed the recruitment of CRISPRi to Tigger3a elements from CUT&RUN sequencing (CUT&RUN-seq) experiments (67). Comparing the CRISPRi CUT&RUN-seq signal intensity revealed its preferential binding at Tigger3a elements, as opposed to Tigger3b or Charlie7 TEs, with the latter corresponding to off-target controls from the same TE superfamily (Fig. 5D; Supplementary Fig. S6A–S6K). As an additional control, we compared the CRISPRi CUT&RUN-seq signal intensity over Tigger3a elements to the signal over the flanking 1.5 kb. While the top 25% of Tigger3a elements were more significantly bound by CRISPRi than flanking sequences, the opposite was found at the bottom 25% of Tigger3a elements (Fig. 5D; Supplementary Figs. S6D, S6E, S6L–S6Q), suggesting a preferential recruitment of CRISPRi to a subset of the 5,317 Tigger3a TEs. In agreement, CUT&RUN-seq for H3K27ac in 22Rv1 was stronger over the top 25% compared with the bottom 25% of CRISPRi-bound Tigger3a elements (Fig. 5E; Supplementary Fig. S6R and S6S). Furthermore, performing H3K27ac CUT&RUN-seq assays in the 22Rv1 CRISPRi clones revealed a significant loss of H3K27ac signal at the majority of the top 25% Tigger3a elements in cells nucleofected with the target as opposed to control gRNAs (Fig. 5E and F; Supplementary Fig. S6R–S6U). In contrast, no changes in the H3K27ac CUT&RUN-seq signal was observed over the bottom 25% Tigger3a elements under all conditions (Fig. 5E; Supplementary Fig. S6R and S6S). Extending our analysis to Tigger3b and Charlie7 elements in the top 25% for CRISPRi CUT&RUN signal intensity revealed no significant changes in H3K27ac signal except for a weakly significant drop at Tigger3b elements in two of three experiments (Supplementary Fig. S6V–S6X). Collectively, these results argue for the ability to target CRISPRi to Tigger3a preferentially with our gRNA combo and the resulting decrease in H3K27ac signal reflective of active chromatin editing over targeted repetitive DNA sequences. Having established the specificity and efficacy of our chromatin editing strategy, we next investigated the functional impact of targeting Tigger3a on AR binding to the chromatin. CUT&RUN for AR in 22Rv1 CRISPRi clones revealed a significant loss of AR signal over top 25% CRISPRi-bound Tigger3a elements in cells nucleofected with the Tigger3a gRNA combo compared with control gRNA combo (Fig. 5G; Supplementary Fig. S7A and S7B). In parallel, we observed a significant downregulation of genes from the androgen response pathway upon Tigger3a chromatin editing in two of three experiments (Fig. 5H; Supplementary Fig. S7C and S7D; Supplementary Table S24). Genes belonging to the E2F targets and MYC targets V1 pathways (68, 69), which are known to be implicated in cell-cycle progression and cell growth, were also significantly downregulated under the same conditions (Supplementary Fig. S7C–S7E; Supplementary Table S24). In agreement, CRISPRi chromatin editing at Tigger3a elements decreased growth of reprogrammed prostate cancer model cell lines (LNCaP and 22Rv1) by at least 20% compared with control conditions (Fig. 5I). In contrast, growth of constant prostate cancer model cell lines (DU145) was not altered by CRISPRi chromatin editing at Tigger3a elements (Fig. 5I; Supplementary Fig. S5G). Collectively, these results support a direct role for Tigger3a TEs as regulatory elements for AR to control its downstream target genes and in controlling growth of prostate cancer cells.
DISCUSSION
Cell fate commitment relies on transitions in the dependency for pluripotency to lineage-specific transcription factors that forge cell state–specific expression patterns. In this study, we show how repetitive DNA engages across physiologic cell states as regulatory elements. We specifically observed that “regulatory TEs” from chromatin states in pluripotent stem cells are biased toward the ERV superfamilies and define binding sites for pluripotency transcription factors such as NANOG and OCT4, in alignment with previous reports (Fig. 5J; bioRxiv 2021.02.16.431334; refs. 47, 48, 70–75). Beyond LTR7 and ERVH regulatory TE families (44, 45, 47, 76), we identify LTR8 and MER4CL34 families as regulatory elements for the pluripotency factors NANOG, OCT4, and SOX2. In contrast, we found 92 lineage-restricted regulatory TE families across 32 mature cell and tissue states, reinforcing the tissue-specific regulatory potential of TEs. These observations emphasize the distinct role of TE families in pluripotent stem cells and mature tissues. Focusing on benign prostate epithelium, we report the role of regulatory TEs as docking sites for lineage-specific transcription factors, including AR and FOXA1 (Fig. 5J; refs. 52, 53). Collectively, our work supports a model whereby transitions in transcription factor dependencies align with changes in the set of TE families serving as regulatory elements along cell fate commitment. Moreover, the set of TE families differentially enriched between pluripotent stem cells and benign prostate epithelium represents a reference set of regulatory TEs to study when characterizing the role of regulatory TEs in cancer.
Cancer arises through the accumulation of genomic variations that alter the function of DNA sequences, such as chromatin variants found in repetitive DNA (4, 77–79). For instance, some ERV superfamily members are found in repressive chromatin variants in cancer that block their expression to prevent double-stranded RNA production, which would otherwise induce cell growth arrest through the viral mimicry response (20, 35, 36). In contrast, inducing the expression of LINE1 TEs leads to novel insertions across the genome, thereby increasing the load of somatic genetic variants in cancer (80, 81). Similarly, TEs can be co-opted as regulatory elements in cancer to favor oncogene overexpression (39, 40, 82). Focusing on prostate cancer, we show how TEs are hijacked by lineage-specific transcription factors to favor oncogenesis. We specifically report on 186 regulatory TE families in primary prostate cancer, including 164 common with pluripotent stem cells. Despite similarities with pluripotent stem cells, regulatory TE families in primary prostate cancer serve as docking sites for lineage-specific transcription factors, such as AR, instead of pluripotency transcription factors (Fig. 5J). Collectively, our results suggest that cancer arises from the attrition of pluripotent stem cell regulatory TE families, which parallels a reliance on lineage-specific as opposed to pluripotency transcription factors.
Chromatin states can distinguish the plethora of DNA elements that populate the human genome, including transcribed versus silent genes and active versus inactive regulatory elements (5, 6, 8, 9, 11, 15, 83). While prior studies using CRISPR/Cas9 genome editing have established the oncogenic contribution of individual TEs (39, 40), we used CRISPRi to demonstrate that the role of TEs in prostate cancer also relies on their chromatin state. We specifically show how altering the chromatin states at hundreds of Tigger3a TEs disrupts AR binding to the chromatin, interferes with the expression of AR target genes, and blocks the growth of AR-dependent prostate cancer cells. Our results agree with the role of chromatin states in regulating the function of nonrepetitive DNA sequences (63, 84), as well as TEs that control the growth of cancer cells in acute myeloid leukemia (40). Taken together, our results support a direct role for TEs toward oncogenesis and identify options to negate their function using approaches altering their chromatin state.
METHODS
ChIP-seq
H3K27ac and AR in Primary Tissues.
Samples from all CPC-GENE patients were obtained with written informed consent with Research Ethics Board ethical approval (UHN 11–0024). Data from H3K27ac ChIP-seq were added to the previously published data (25, 27) based on patient samples being processed exactly as described in ref. 25 using the same antibody (Abcam, ab4729). Sequencing libraries were prepared using 0.5 to 10 ng of ChIP or input DNA with the Rubicon ThruPLEX FD Kit (Takara) using the manufacturer's recommended protocol. Libraries were then size selected in the range of 240 to 360 bp using a Caliper LabChIP XT DNA 750 Kit (PerkinElmer). Size-selected libraries were sequenced on an Illumina HiSeq 2000 with single-end 50-bp reads or paired-end 100-bp reads. Alignment (human genome - hg38) and peak calling were performed with the same parameters and workflow as described in ref. 85. The same parameters were used for alignment (human genome - hg38) and peak calling for H3K27ac and AR data profiled in the Porto cohort patients (26). The only exception for AR profiling was the peak-calling significance threshold, from q <0.005 for H3K27ac to q <0.01 for AR. Our approach to map peaks of H3K27ac to repetitive sequences relies on single-ended reads, therefore limiting our ability to call peaks restricted to highly repetitive sequences, such asevolutionary young repetitive sequences.
H3K27ac in Prostate Cancer Cell Lines.
Approximately 2 × 106 LNCaP or DU145 cells were used to perform H3K27ac (Abcam, ab4729) ChIP-seq as described in ref. 25. Alignment (human genome - hg38) and peak calling were performed following ENCODE pipeline (https://github.com/ENCODE-DCC/chip-seq-pipeline2) to ensure comparability with ENCODE samples.
H3K9me3 in Prostate Cancer Cell Lines.
Approximately 2 × 106 PWR1E, RWPE1, 22Rv1, LNCaP, VCaP, DU145, and PC3 cells were used to perform H3K9me3 (Abcam, ab8898) ChIP-seq as described in ref. 25. Libraries were generated using the Rubicon ThruPLEX FD Kit (Takara) using the manufacturer's recommended protocol. Libraries were then size-selected in the range of 150 to 360 bp using AMPure XP beads (Beckman Coulter, A63881) and sequenced with paired-end 50-bp reads to reach 45 million total mapped reads per replicate as recommended by ENCODE guidelines. Alignment (human genome - hg38) was performed by using Bowtie2, while peak calling was performed using MACS2 with q ≤0.05 significance threshold.
ATAC Sequencing on PRAD TCGA
Bam files corresponding to PRAD ATAC data were downloaded from the Genomic Data Commons Data Portal, and peaks were called using MACS3 with the following parameters: –shift -75 –extsize 150 –nomodel –call-summits –nolambda –keep-dup all -p 0.01, and a nonoverlapping peak set was produced as described in ref. 55. narrowPeak files were used for TE enrichment as described below.
TE Families and Superfamilies
Bed files of repeats classified as TE families (excluding simple repeats, satellites, sn/rRNA, and repeats of unknown subfamilies) (n = 971) in the hg38 human genome build were downloaded from the UCSC Genome Browser. Families were classified into superfamilies according to Repbase (https://www.girinst.org/repbase/; refs. 86, 87).
TE Enrichment Analysis
Enrichment of TE families was performed using ChromVAR within mappable H3K27ac, ATAC, or H3K9me3 peaks (88) with the modifications described in bioRxiv 2021.02.16.431334. Briefly, we computed the presence/absence of H3K27ac, ATAC, or H3K9me3 peaks, found in at least one sample/patient, in each sample of interest (H3K27ac: ENCODE samples, CPC-GENE or Porto patients’ samples, and ENCODE cancer cell lines; ATAC: TCGA PRAD; H3K9me3: data generated in this study on prostate cell lines) to generate the first binary matrix. The same peaks of interest were used to assess the overlap with each individual TE family, generating the second binary matrix. ChromVAR was then run with default parameters computing a bias-corrected deviation Z-score. We used nonparametric two-sided Wilcoxon signed rank test to compare Z-scores between samples assigned to predefined groups or to groups defined on the basis of TE families differentially enriched between pluripotent stem cells and the benign prostate tissue state (as reported in Figs. 3A and B and 5A; Supplementary Fig. S2D). Heat maps were plotted truncating deviation Z-scores to minimum and maximum values of −10 and 10, respectively, and clustering was based on Euclidean distance. To generate the “reprogramming score,” we combined all TEs commonly enriched in pluripotent stem cells, reprogrammed TEs (enriched in reprogrammed CPC-GENE and Porto patients), or commonly enriched in benign prostate tissue state, reprogrammed CPC-GENE and Porto patients (benign prostate TEs) in separate .bed files and calculated the Z-score over reprogrammed TEs and benign prostate TEs separately. To combine both Z-scores, we used the Stouffer method with equal weights of the reprogrammed Z-score and the inverse benign prostate Z-score (bioRxiv 2021.02.16.431334). To generate the “repressive score,” we followed the same exact steps as the “reprogramming score” but using H3K9me3 chromatin regions, as opposed to H3K17ac or ATAC-based ones.
Clinical Features and Time to Biochemical Recurrence Analysis
Clinical features including time to biochemical recurrence data were provided with all the rest of patients’ clinical data when available. Survival analysis was performed using the Kaplan–Meier estimate method. P values comparing Kaplan–Meier survival curves were calculated using the log-rank (Mantel–Cox) test. Hypoxia score was computed as described in ref. 89 for both CPC-GENE (only for patients included in this study) and Porto patients.
Transcription Factor Cistrome and Motif Enrichment Analysis
The enrichment of transcription factor cistromes or motifs was assessed at the 97 TE families enriched in benign prostate compared to pluripotent stem cells (Fig. 1A; Supplementary Table S5), at the 315 enriched in pluripotent stem cells compared with benign prostate, and at the 164 reprogrammed TE families (Figs. 2A and B and 3F). Each TE family was analyzed individually, restricting the analysis to elements overlapping the consensus set of H3K27ac peaks of interest (e.g., for a TE family enriched in benign prostate, we inquired transcription factor cistromes and motifs enriched at the elements overlapping with the H3K27ac peaks catalog of the benign tissue state). Enrichment of transcription factor cistromes was performed using the Bioconductor package LOLA (https://bioconductor.org/packages/release/bioc/html/LOLA.html; ref. 90), version 1.20.0. A total of 1,135 transcription factor cistromes were obtained from ReMap (remap2020; http://remap.univ-amu.fr/). Enriched transcription factor cistromes at each individual TE family were called using as background all elements belonging to all 971 TE families overlapping with the consensus set of H3K27ac peaks used to select the elements of interest in the previous step (e.g., for each TE family enriched in benign prostate, our background was all the elements belonging to the 971 TE families overlapping with the H3K27ac peaks catalog of the benign tissue state; q-value <0.05, logOR >1.5). The top 5% enriched transcription factor cistromes for benign prostate, pluripotent stem cells, and reprogrammed CPC-GENE or Porto patients TEs are shown in Figs. 1E, 2D and E, and 4D and Supplementary Fig. S1B. For each set of TE families, we performed analysis as negative controls. More precisely, for benign prostate tissue state TEs, we assessed the enrichment of transcription factor cistromes at elements belonging to TE families enriched in benign prostate tissue state overlapping with endoderm H3K27ac peaks (intestine, liver, or lung with matched backgrounds as explained above; Fig. 2D; Supplementary Fig. S1B). For reprogrammed TEs, we assessed the enrichment of transcription factor cistromes at reprogrammed TE family elements overlapping with constant CPC-GENE or Porto patients (Fig. 4A and B). The enrichment of motifs was computed using HOMER motif discovery tool (findMotifsGenome.pl) version 4.7. Enriched motifs within individual TE families were carried out as explained above for transcription factor cistromes (Benjamini q-value <0.05).
GIGGLE scores were calculated using the Web form on the following website: http://dbtoolkit.cistrome.org/. In the section entitled “What factors have a significant binding overlap with your peak set?”, we selected the following parameters: Species - Human hg38, Data type in Cistrome - Transcription factor, chromatin regulator, Peak number of Cistrome sample to use - All peaks in each sample.
Enrichment of Matched AR ChIP-seq
To assess the enrichment of matched AR cistromes over TE families observed to be bound by AR in cancer cell lines (Fig. 4D), we used AR ChIP-seq profiled in Porto patients (for which we have H3K27ac data), described in ref. 26. Alignment (human genome - hg38) and peak calling were performed with the same parameters and workflow described above, with the difference that peaks were called using a q-value <0.01. Enrichment of transcription factor cistromes was performed using the Bioconductor package LOLA (mentioned above). We generated three individual catalogs of peaks by merging peaks called in reprogrammed, constant, or intermediate patients. Enriched reprogrammed, constant, and intermediate AR cistromes at each individual TE family element, overlapping with non–patient-specific reprogrammed Porto H3K27ac regions, were called using as background all elements belonging to all 971 TE families overlapping with non–patient-specific H3K27ac regions found in all Porto patients. logOR enrichment scores for the three groups of patients at individual TE families tested are showcased in Fig. 4G and Supplementary Fig. S4A.
Essentiality Scores
Essentiality scores were obtained from the Broad Institute project Achilles (DepMap; ref. 56). CRISPR (Avana 21Q1) and combined RNAi genetic dependencies data were downloaded from the “Download” page of the website https://depmap.org/portal/achilles/.
Generation of DU145 dCas9-KRAB Clonal Cell Lines
DU145 cells were transduced with lentiviral particles as described in ref. 32. Briefly, lentiviral particles were collected from 293FT (Thermo Fisher Scientific) cotransfected with the pMDG.2 and psPAX2 packaging plasmids (Addgene; #12259 and #12260, a gift from Didier Trono, School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) together with the Lenti-dCas9-KRAB-blast plasmid (Addgene #89567, a gift from Gary Hon, Laboratory of Regulatory Genomics, Cecil H. and Ida Green Center for Reproductive Biology Sciences, Division of Basic Reproductive Biology Research, Department of Obstetrics and Gynecology, University of Texas Southwestern Medical Center, Dallas, TX). After being transduced for 48 hours with equal amount of virus, DU145 cells were exposed to selection using media containing blasticidin (7.5 μg/mL). Upon selection, we perform single-cell seeding into 96-well plates containing selection media. The expression of dCas9-KRAB was then assessed by Western blot analysis, and three clones were selected for validations.
Cell Culture
PWR1E and RWPE1 were cultured in Keratinocyte SFM medium, naive LNCaP and 22Rv1 cells were cultured in RPMI medium, while naive DU145 and PC3 cells were cultured in DMEM, with both RPMI and DMEM media supplemented with 10% FBS and 1% penicillin–streptomycin, at 37°C in a humidified incubator with 5% CO2. These prostate cancer cells originated from ATCC. LNCaP and 22Rv1 dCas9-KRAB clonal cell lines were cultured in RPMI medium (as described above) with the addition of blasticidin (7.5 μg/mL for LNCaP cells, 6 μg/mL for 22Rv1 cells; ref. 32). DU145 dCas9-KRAB clonal cell lines were cultured in DMEM (as described above) with the addition of blasticidin (7.5 μg/mL). All cells are regularly tested for Mycoplasma contamination. The authenticity of these cells was confirmed through short tandem repeat profiling.
Design of gRNAs Targeting TEs
gRNAs targeting Tigger3a TE family were designed using Repguide (https://tanaylab.github.io/repguide/; ref. 65), which ensures high targeting efficiency while minimizing off-targets. To identify gRNAs (addGuides function), we used default parameters and we picked the combination of six gRNAs that grants the maximum number of Tigger3a elements targeted with predicted off-targets on Tigger3b elements and, to a lesser extent, on other Tigger3 TE families. Six negative control gRNAs (scramble) were obtained from ref. 66 and were used as the negative control combination of gRNAs for both the proliferation assay and genomic and transcriptomic profiling. Sequences of gRNAs used in this study can be found in Supplementary Table S25.
Dcas9-KRAB–Mediated Chromatin Repression at TEs
Chromatin repression at TEs was carried out using three of the clonal 22Rv1 and LNCaP dCas9-KRAB cell lines described in ref. 32. gRNA duplexing was performed according to the manufacturer's protocol (Integrated DNA Technologies) by mixing equal amounts of CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) to reach the concentration of 50 μmol/L. For each reaction, 1 or 1.5 μL (for the proliferation assay or for genomic and transcriptomic profiling, respectively) of each of the six Tigger3a or control gRNAs (crRNA–tracrRNA duplexes) were pooled into a single tube. Prior to nucleofection, 1 μL (100 μmol/L) of electroporation enhancer (Integrated DNA Technologies) was added to the mix. The nucleofection reaction was performed into 350,000 cells for the proliferation assay and into 1.5 million cells for genomic and transcriptomic profiling, through SF Solution 4D Nucleofector (Lonza), with the program number EN120. We used 16-well nucleocuvette strips for the proliferation assay and 100 μL nucleocuvette vessels for genomic and transcriptomic profiling (Lonza). Cells were then harvested 72 hours after nucleofection for the proliferation assay or genomic and transcriptomic profiling.
Cell Proliferation Assays
Cells were maintained as described above and seeded in 24-well plates after nucleofection with Tigger3a or control gRNA combinations. Cell viability was assessed using crystal violet as previously described (20). Results presented in Fig. 5G represent the median of six independent nucleofection reactions. P values were obtained performing a two-sided t test.
CUT&RUN
Pulldown for Cas9, AR, and H3K27ac was performed using a previously described CUT&RUN protocol (67) in biological duplicates (two independent nucleofections) of 22Rv1 clonal cell lines stably expressing dCas9-KRAB 3 days after nucleofection with combinations of gRNAs targeting Tigger3a elements (Tigger3a) or negative control (scramble). Cas9 and H3K27ac pulldowns were performed in the same nucleofection reactions in cells nucleofected with combinations of gRNAs targeting Tigger3a elements, while AR pulldown was performed on different nucleofection reactions. Cells nucleofected with the combination of negative control (scramble) gRNAs were used to generate AR or H3K27ac pulldowns. Cells that were not used for Cas9 and/or H3K27ac pulldown were used to extract RNA for RNA sequencing (RNA-seq; see below). This procedure ensures genomic and transcriptomic profiling in the same nucleofection reaction. A total of 250,000 cells per pulldown were collected, resuspended in nuclear binding buffer [20 mmol/L HEPES-KOH (pH 7.9), 10 mmol/L KCl, 1 mmol/L CaCl2, 1 mmol/L MnCl2], and incubated for 10 minutes, rotating at room temperature, with 10 μL of Concanavalin A Beads (Bangs Laboratories, BP531) to promote cell–bead binding. Bead-bound cells were resuspended in antibody buffer [20 mmol/L HEPES (pH 7.5), 150 mmol/L NaCl, 0.5 mmol/L spermidine, 0.01% digitonin, 2 mmol/L EDTA] supplemented with protease inhibitor combination (cOmplete, EDTA-free Protease Inhibitor combination, Roche), with 5 μg of anti-Cas9 antibody (Diagenode, C15200229), 3 μg of anti-AR antibody (EpiCypher, 13–2020), or 3 μg of anti-H3K27ac antibody (Abcam, ab4729) and incubated overnight, rotating at 4°C. The next day, beads were washed once in wash buffer containing digitonin (as antibody buffer without EDTA) and then incubated with pAG-MNase (NEB, 40366S) in digitonin-containing wash buffer for 1 hour, rotating at 4°C. The MNase was activated by adding 2 mmol/L CaCl2 (final concentration), and exposed DNA was digested for 30 minutes at 0°C (ice bath). DNA digestion was inactivated by adding STOP buffer (200 mmol/L NaCl, 20 mmol/L EDTA, 4 mmol/L EGTA, 0.01% digitonin, 50 μg/mL RNaseA, 40 μg/mL glycogen, 2 pg/mL Saccharomyces cerevisiae heterologous DNA). Chromatin fragments released from beads were promoted by incubating beads at 37°C for 10 minutes. Finally, DNA was extracted using the MinElute Kit (Qiagen). Libraries were generated using the Rubicon Thruplex FD Kit (Takara) using the manufacturer's recommended protocol. Libraries were then size selected in the range of 150 to 360 bp using AMPure XP beads (Beckman Coulter, A63881) and sequenced with paired-end 75-bp reads to reach up to 40 million read pairs per sample.
CUT&RUN Analysis
For dCas9-KRAB, H3K27ac, and AR pulldowns, reads were aligned to the human genome (hg38) using the bowtie2 settings described in ref. 67 (q -I 10 -X 700 –local –very-sensitive-local –no-mixed –no-discordant –no-unal –phred33). Spike-in Saccharomyces cerevisiae DNA was aligned with the following parameters -q -I 10 -X 700 –local –very-sensitive-local –no-mixed –no-discordant –no-overlap –no-dovetail –no-unal –phred33. Spike-in calibrated peaks were called using SEACR (v13; ref. 91). For each individual clone, spike-in calibrated dCas9-KRAB signal was quantified over Tigger3a (main target), Tigger3b (secondary target), and Charlie7 (negative control) TE families and 1.5-kb adjacent regions using deeptools (92) as described in the following tutorial for CUT&RUN analysis (Zheng and colleagues; Protocol.io). Tigger3a elements were classified into quartiles based on dCas9-KRAB CUT&RUN signal. The top 25% and bottom 25% quartiles were used for downstream analyses. We further validated the specificity for dCas9-KRAB CUT&RUN signal over Tigger3a elements by comparing the signal against the 1.5-kb adjacent regions. Similar comparisons were done to assess signal differences for the H3K27ac data. To do so, we ran DESeq2 on Tigger3a elements overlapping H3K27ac peaks. We specifically analyzed top and bottom 25% Tigger3a elements based on dCas9-KRAB signal and evaluated global levels of normalized spike-in calibrated H3K27ac signal comparing Tigger3a versus control gRNA combinations. The top 25% Tigger3a elements based on dCas9-KRAB signal showing a loss of H3K27ac signal were defined as “repressed Tigger3a elements.” Similarly, we assessed the potential loss of AR binding over the top 25% Tigger3a elements. We ran DESeq2 on Tigger3a elements overlapping AR peaks, and we evaluated global levels of normalized spike-in calibrated AR signal comparing Tigger3a versus control gRNA combinations.
RNA Extraction, RNA-seq, and Analysis
RNA-seq was performed in a part of the same clonal dCas9-KRAB 22Rv1 nucleofected with combinations of Tigger3a or negative control (scramble) gRNAs (biological duplicates; two independent nucleofections). After isolating cells for CUT&RUN pulldowns (see above), the RNeasy Plus Mini Kit (Qiagen, 74136) was used to collect whole RNA from cells as per manufacturer's instructions. RNA was delivered to the Princess Margaret Genomics Centre (PMGC). The PMGC performed ribosomal RNA depletion using the Ribo-Zero Gold rRNA Removal Kit (Illumina). Libraries were sent for 100-bp paired-end sequencing to reach 60 million read pairs per sample. Reads were first processed using Kallisto to obtain transcript abundances, and then DESeq2 was used to obtain normalized read counts per gene. DESeq2 output was used to perform gene set enrichment analysis (GSEA) on each clone individually. “HALLMARK” pathways were obtained using the package “msigdb” (Bioconductor), and their enrichment was assessed using the “fgsea” pakcage (Bioconductor). The “Androgen response” pathway was always reported together with pathways significantly different in all three dCas9-KRAB 22Rv1 cells (Tigger3a vs. control; Fig. 5H; Supplementary Fig. S7C–S7E).
Protein Extractions and Western Blot Analysis
DU145 dCas9-KRAB cell lines were recovered and lysed in modified RIPA (10 mmol/L Tris-HCl, pH 8.0; 1 mmol/L EDTA; 140 mmol/L NaCl; 1% Triton X-100; 0.1% SDS; 0.1% sodium deoxycholate) containing protease inhibitor combination (cOmplete, EDTA-free Protease Inhibitor combination, Roche). Lysates were sonicated for 5 cycles of 30 seconds on, 30 seconds off using a Diagenode Bioruptor 300. Cell debris was removed by centrifugation at 4°C for 10 minutes at 15,000 rpm, followed by protein quantification using the BCA Protein Assay Kit (Thermo Fisher Scientific). Twenty micrograms of protein per sample were resuspended in 2X Laemmli containing 5% β-mercaptoethanol, boiled for 5 minutes at 95°C, and resolved on precast 5% to 20% gradient gels (Bio-Rad). After transfer, membranes were blocked using 5% skim milk in PBS-Tween 0.05% and incubated overnight with anti-Cas9 antibody (Diagenode, C15200229; 1:2,000 dilution) resuspended in 1% BSA PBS-Tween 0.05%. As a loading control, we incubated the membrane with anti–α-tubulin (Sigma Aldrich, T5168 Sigma-Aldrich, T5168; 1:2,000 dilution) for 1 hour at room temperature. Membranes were then washed and incubated with IRDye 680RD donkey anti-mouse (LI-COR; 1:5,000), resuspended in 1% skim milk in PBS-Tween 0.05%, and incubated for 1 hour at room temperature, covered from light. Membranes were imaged using ODYSSEY CLx (LI-COR), and images were analyzed using Image Studio (LI-COR).
Research Reproducibility and Code Availability
Code for data processing, analysis, and plotting can be found on CodeOcean (https://codeocean.com/capsule/5158405/tree). The analytical pipeline is also available on the CoBE platform (www.pmcobe.ca).
Data Availability
All data generated in this study are deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE224687, including H3K27ac regions profiled in CPC-GENE patients and in six additional benign prostate epithelium samples. Porto patient H3K27ac and AR regions were obtained from Stelloo and colleagues (26) and processed as specified in the Methods section. H3K27ac regions profiled in pluripotent stem cells and mature cell and tissue states were downloaded from ENCODE (https://www.encodeproject.org/) using the access numbers provided in Supplementary Table S1.
Supplementary Material
ENCODE samples accession numbers used in this article
Tissue-state enriched transposable element families vs pluripotent stem cells
Pluripotent stem cells enriched transposable element families versus individual tissue states
pluripotent stem cell- and tissue state-specific transposable element families
Differentially enriched transposable element families between pluripotent stem cells and benign prostate tissue state
Top 5% transcription factor cistromes enriched at pluripotent stem cell transposable elements
Top 5% transcription factor cistromes enriched at benign prostate transposable elements
Top 5% transcription factor cistromes enriched at intestine transposable elements
Top 5% transcription factor cistromes enriched at liver transposable elements
Top 5% transcription factor cistromes enriched at lung transposable elements
AR GIGGLE enrichment scores at AR-bound benign prostate transposable elements
FOXA1 GIGGLE enrichment scores at FOXA1-bound benign prostate transposable elements
Differentially enriched transposable element families between reprogrammed and constant CPC-GENE patients
Differentially enriched transposable element families between reprogrammed and constant Porto patients
Differentially enriched transposable element families between intermediate and constant Porto patients
Onco-exaptated transposable elements in reprogrammed and constant PCa patients
Differentially enriched transposable element families between reprogrammed and constant TCGA PRAD patients (ATAC-seq)
Clinical information CPC-GENE patients
Clinical information Porto patients
Top 5% transcription factor cistromes enriched at reprogrammed transposable element families (CPC-GENE reprogrammed patients)
Top 5% transcription factor cistromes enriched at reprogrammed transposable element families (Porto reprogrammed patients)
Top 5% transcription factor cistromes enriched at reprogrammed transposable element families (CPC-GENE constant patients)
Top 5% transcription factor cistromes enriched at reprogrammed transposable element families (Porto constant patients)
Pathwyas significantly different in all three CRISPRi 22Rv1 clones (Tigger3a vs Control)
List of gRNA sequences used in this article
This figure contains the roadmap for all the different datasets used in this study. It also showcases transcription factor and motif enrichment over transposable element families enriched in different normal tissues states
This figure complements Figure 2 and showcases the enrichment of tranposable element families in TCGA PRAD ATAC data
This figure showcases the entire molecular and clinical characterization of CPC-GENE and Porto patients' groups identified using transposable element families differentially enriched between pluripotent stem cells and benign prostate tissue states
This figure complements Figure 4, showcasing the enrichment of Porto AR cistromes and AR motif over reprogrammed transposable element families (with the exception of Tigger3a and L1MB4 that are showcased in Figure 4)
This figure showcases the enrichment of transposable element families in H3K27ac or H3K9kme3 ChIP-seq data generated in benign-like and prostate cancer cell lines. It also includes dCas9-KRAB expression patterns in all CRISPRi clonal cell lines used in this article
This figure showcases dCas9-KRAB and H3K27ac profiling in CRISPRi 22Rv1 clones #1 and #3 nucleofected with control or Tigger3a gRNA combos (clone #2 is showcased in Figure 5)
This figure complements Figure 5 showcasing AR profiling and transcriptomic analysis results in CRISPRi 22Rv1 clones #1 and #3 nucleofected with control or Tigger3a gRNA combos (clone #2 is showcased in Figure 5)
Acknowledgments
The authors thank the Princess Margaret Genomics Centre and the Princess Margaret Bioinformatics group for providing support and infrastructure for the computational analysis of this work as well as high-throughput sequencing support. We thank members of the M. Lupien lab for their fruitful discussions and feedback. This work is supported by the Dutch Cancer Society KWF/Alpe d'HuZes (10084 and NKI 2014–6711 ALPE; to W. Zwart), the Canadian Institutes of Health Research (FRN-153234 and 168933; to M. Lupien), and the Ontario Institute for Cancer Research Investigator Award through funding provided by the Government of Ontario (to M. Lupien) and the Princess Margaret Cancer Foundation.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T. Keshavarzian reports other support from the Cancer Digital Intelligence Scholarship, the Ontario Graduate Scholarship, the Ontario Student Opportunity Trust Fund, and the University of Toronto Merit Scholarship outside the submitted work. M. Teng reports grants from the Canadian Institutes of Health Research during the conduct of the study. K.J. Kron reports personal fees from Deep Genomics outside the submitted work. J.R. Hawley reports that at the time of publication, he was an employee of Hoffmann-La Roche Limited (Roche Canada). G.V. Raj reports other support from EtiraRx outside the submitted work. W. Zwart reports grants from the Dutch Cancer Society, ZonMW, the Department of Defense, Alpe d'HuZes, and the Prostate Cancer Foundation during the conduct of the study, as well as grants from Astellas Pharma outside the submitted work. M. Lupien reports grants from the Canadian Institutes of Health Research (FRN-153234 and FRN-168933), the Ontario Institute for Cancer Research (Investigator Award), and the Princess Margaret Cancer Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
G. Grillo: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Keshavarzian: Software, formal analysis, writing–review and editing. S. Linder: Resources, data curation, writing–review and editing. C. Arlidge: Formal analysis. L. Mout: Formal analysis, writing–review and editing. A. Nand: Software. M. Teng: Formal analysis. A. Qamra: Software. S. Zhou: Formal analysis, writing–review and editing. K.J. Kron: Data curation. A. Murison: Software. J.R. Hawley: Data curation, software. M. Fraser: Resources, data curation. T.H. van der Kwast: Data curation. G.V. Raj: Resources. H.H. He: Supervision, writing–review and editing. W. Zwart: Resources, data curation, supervision, writing–review and editing. M. Lupien: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Shchuka VM, Malek-Gilani N, Singh G, Langroudi L, Dhaliwal NK, Moorthy SD, et al. Chromatin dynamics in lineage commitment and cellular reprogramming. Genes 2015;6:641–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 2018;173:1535–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takayama N, Murison A, Takayanagi S-I, Arlidge C, Zhou S, Garcia-Prat L, et al. The transition from quiescent to activated states in human hematopoietic stem cells is governed by dynamic 3D genome reorganization. Cell Stem Cell 2021;28:488–501. [DOI] [PubMed] [Google Scholar]
- 4. Grillo G, Lupien M. Cancer-associated chromatin variants uncover the oncogenic role of transposable elements. Curr Opin Genet Dev 2022;74:101911. [DOI] [PubMed] [Google Scholar]
- 5. Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9:473–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008;132:958–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007;39:311–8. [DOI] [PubMed] [Google Scholar]
- 10. Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 2011;21:1273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006;125:315–26. [DOI] [PubMed] [Google Scholar]
- 12. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011;470:279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 2013;154:888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meuleman W, Muratov A, Rynes E, Halow J, Lee K, Bates D, et al. Index and biological spectrum of human DNase I hypersensitive sites. Nature 2020;584:244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Madani Tonekaboni SA, Haibe-Kains B, Lupien M. Large organized chromatin lysine domains help distinguish primitive from differentiated cell populations. Nat Commun 2021;12:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature 2012;488:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roadmap Epigenomics Consortium; Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stütz AM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 2014;506:445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deblois G, Tonekaboni SAM, Grillo G, Martinez C, Kao YI, Tai F, et al. Epigenetic switch-induced viral mimicry evasion in chemotherapy-resistant breast cancer. Cancer Discov 2020;10:1312–29. [DOI] [PubMed] [Google Scholar]
- 21. Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016;529:110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alonso-Curbelo D, Ho YJ, Burdziak C, Maag JLV, Morris JP, Chandwani R, et al. A gene–environment-induced epigenetic program initiates tumorigenesis. Nature 2021;590:642–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 2012;336:736–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patten DK, Corleone G, Győrffy B, Perone Y, Slaven N, Barozzi I, et al. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat Med 2018;24:1469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kron KJ, Murison A, Zhou S, Huang V, Yamaguchi TN, Shiah YJ, et al. TMPRSS2-ERG fusion co-opts master transcription factors and activates NOTCH signaling in primary prostate cancer. Nat Genet 2017;49:1336–45. [DOI] [PubMed] [Google Scholar]
- 26. Stelloo S, Nevedomskaya E, Kim Y, Schuurman K, Valle-Encinas E, Lobo J, et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat Commun 2018;9:4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mazrooei P, Kron KJ, Zhu Y, Zhou S, Grillo G, Mehdi T, et al. Cistrome partitioning reveals convergence of somatic mutations and risk variants on master transcription regulators in primary prostate tumors. Cancer Cell 2019;36:674–89. [DOI] [PubMed] [Google Scholar]
- 28. Pomerantz MM, Qiu X, Zhu Y, Takeda DY, Pan W, Baca SC, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat Genet 2020;52:790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 2015;47:1346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X, Cowper-Sal Lari R, Bailey SD, Moore JH, Lupien M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res 2012;22:1437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 2009;41:882–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou S, Hawley JR, Soares F, Grillo G, Teng M, Madani Tonekaboni SA, et al. Noncoding mutations target cis-regulatory elements of the FOXA1 plexus in prostate cancer. Nat Commun 2020;11:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baca SC, Singler C, Zacharia S, Seo JH, Morova T, Hach F, et al. Genetic determinants of chromatin reveal prostate cancer risk mediated by context-dependent gene regulation. Nat Genet 2022;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hua JT, Ahmed M, Guo H, Zhang Y, Chen S, Soares F, et al. Risk SNP-mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell 2018;174:564–75. [DOI] [PubMed] [Google Scholar]
- 35. Roulois D, Yau HL, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 2015;162:961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015;162:974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alizadeh-Ghodsi M, Owen KL, Townley SL, Zanker D, Rollin SPG, Hanson AR, et al. Potent stimulation of the androgen receptor instigates a viral mimicry response in prostate cancer. Cancer Res Commun 2022;2:706–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Babaian A, Mager DL. Endogenous retroviral promoter exaptation in human cancer. Mob DNA 2016;7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet 2019;51:611–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deniz Ö, Ahmed M, Todd CD, Rio-Machin A, Dawson MA, Branco MR. Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat Commun 2020;11:3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fueyo R, Judd J, Feschotte C, Wysocka J. Roles of transposable elements in the regulation of mammalian transcription. Nat Rev Mol Cell Biol 2022;23:481–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, et al. Ten things you should know about transposable elements. Genome Biol 2018;19:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu X, Sachs F, Ramsay L, Jacques PÉ, Göke J, Bourque G, et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 2014;21:423–5. [DOI] [PubMed] [Google Scholar]
- 45. Wang J, Xie G, Singh M, Ghanbarian AT, Raskó T, Szvetnik A, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 2014;516:405–9. [DOI] [PubMed] [Google Scholar]
- 46. Chèneby J, Ménétrier Z, Mestdagh M, Rosnet T, Douida A, Rhalloussi W, et al. ReMap 2020: a database of regulatory regions from an integrative analysis of Human and Arabidopsis DNA-binding sequencing experiments. Nucleic Acids Res 2020;48:D180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan Y-S, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 2010;42:631–4. [DOI] [PubMed] [Google Scholar]
- 48. Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 2015;522:221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem 2010;285:16135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature 2005;435:944–7. [DOI] [PubMed] [Google Scholar]
- 51. Layer RM, Pedersen BS, DiSera T, Marth GT, Gertz J, Quinlan AR. GIGGLE: a search engine for large-scale integrated genome analysis. Nat Methods 2018;15:123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci 2006;63:2317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heinlein CA, Chang C. Androgen receptor (AR) coregulators: an overview. Endocr Rev 2002;23:175–200. [DOI] [PubMed] [Google Scholar]
- 54. Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017;541:359–64. [DOI] [PubMed] [Google Scholar]
- 55. Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, et al. The chromatin accessibility landscape of primary human cancers. Science 2018;362:eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell 2017;170:564–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roche PJ, Hoare SA, Parker MG. A consensus DNA-binding site for the androgen receptor. Mol Endocrinol 1992;6:2229–35. [DOI] [PubMed] [Google Scholar]
- 58. Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, et al. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep 2007;8:871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate 2003;57:205–25. [DOI] [PubMed] [Google Scholar]
- 60. Decker KF, Zheng D, He Y, Bowman T, Edwards JR, Jia L. Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions. Nucleic Acids Res 2012;40:10765–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith R, Liu M, Liby T, Bayani N, Bucher E, Chiotti K, et al. Enzalutamide response in a panel of prostate cancer cell lines reveals a role for glucocorticoid receptor in enzalutamide resistant disease. Sci Rep 2020;10:21750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010;463:237–40. [DOI] [PubMed] [Google Scholar]
- 63. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013;154:442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 2015;12:1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fuentes DR, Swigut T, Wysocka J. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. Elife 2018;7:e35989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 2018;13:1006–19. [DOI] [PubMed] [Google Scholar]
- 68. Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 1999;19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G2–M checkpoints. Genes Dev 2002;16:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jacques PÉ, Jeyakani J, Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet 2013;9:e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 2014;24:1963–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016;351:1083–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C, et al. Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell 2018;23:276–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gomez NC, Hepperla AJ, Dumitru R, Simon JM, Fang F, Davis IJ. Widespread chromatin accessibility at repetitive elements links stem cells with human cancer. Cell Rep 2016;17:1607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cao Y, Chen G, Wu G, Zhang X, McDermott J, Chen X, et al. Widespread roles of enhancer-like transposable elements in cell identity and long-range genomic interactions. Genome Res 2019;29:40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci U S A 2014;111:12426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ishak CA, Classon M, De Carvalho DD. Deregulation of retroelements as an emerging therapeutic opportunity in cancer. Trends Cancer Res 2018;4:583–97. [DOI] [PubMed] [Google Scholar]
- 78. Grundy EE, Diab N, Chiappinelli KB. Transposable element regulation and expression in cancer. FEBS J 2021;289:1160–79. [DOI] [PubMed] [Google Scholar]
- 79. Burns KH. Repetitive DNA in disease. Science 2022;376:353–4. [DOI] [PubMed] [Google Scholar]
- 80. Rodriguez-Martin B, Alvarez EG, Baez-Ortega A, Zamora J, Supek F, Demeulemeester J, et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet 2020;52:306–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Payer LM, Steranka JP, Kryatova MS, Grillo G, Lupien M, Rocha PP, et al. Alu insertion variants alter gene transcript levels. Genome Res 2021;31:2236–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Babaian A, Romanish MT, Gagnier L, Kuo LY, Karimi MM, Steidl C, et al. Onco-exaptation of an endogenous retroviral LTR drives IRF5 expression in Hodgkin lymphoma. Oncogene 2016;35:2542–6. [DOI] [PubMed] [Google Scholar]
- 83. Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009;459:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nuñez JK, Chen J, Pommier GC, Zachery Cogan J, Replogle JM, Adriaens C, et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 2021;184:2503–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hawley JR, Zhou S, Arlidge C, Grillo G, Kron KJ, Hugh-White R, et al. Reorganization of the 3D genome pinpoints noncoding drivers of primary prostate tumors. Cancer Res 2021;81:5833–48. [DOI] [PubMed] [Google Scholar]
- 86. Jurka J. Repeats in genomic DNA: mining and meaning. Curr Opin Struct Biol 1998;8:333–7. [DOI] [PubMed] [Google Scholar]
- 87. Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 2015;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 2017;14:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bhandari V, Hoey C, Liu LY, Lalonde E, Ray J, Livingstone J, et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet 2019;51:308–18. [DOI] [PubMed] [Google Scholar]
- 90. Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics 2016;32:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Meers MP, Tenenbaum D, Henikoff S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 2019;12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ENCODE samples accession numbers used in this article
Tissue-state enriched transposable element families vs pluripotent stem cells
Pluripotent stem cells enriched transposable element families versus individual tissue states
pluripotent stem cell- and tissue state-specific transposable element families
Differentially enriched transposable element families between pluripotent stem cells and benign prostate tissue state
Top 5% transcription factor cistromes enriched at pluripotent stem cell transposable elements
Top 5% transcription factor cistromes enriched at benign prostate transposable elements
Top 5% transcription factor cistromes enriched at intestine transposable elements
Top 5% transcription factor cistromes enriched at liver transposable elements
Top 5% transcription factor cistromes enriched at lung transposable elements
AR GIGGLE enrichment scores at AR-bound benign prostate transposable elements
FOXA1 GIGGLE enrichment scores at FOXA1-bound benign prostate transposable elements
Differentially enriched transposable element families between reprogrammed and constant CPC-GENE patients
Differentially enriched transposable element families between reprogrammed and constant Porto patients
Differentially enriched transposable element families between intermediate and constant Porto patients
Onco-exaptated transposable elements in reprogrammed and constant PCa patients
Differentially enriched transposable element families between reprogrammed and constant TCGA PRAD patients (ATAC-seq)
Clinical information CPC-GENE patients
Clinical information Porto patients
Top 5% transcription factor cistromes enriched at reprogrammed transposable element families (CPC-GENE reprogrammed patients)
Top 5% transcription factor cistromes enriched at reprogrammed transposable element families (Porto reprogrammed patients)
Top 5% transcription factor cistromes enriched at reprogrammed transposable element families (CPC-GENE constant patients)
Top 5% transcription factor cistromes enriched at reprogrammed transposable element families (Porto constant patients)
Pathwyas significantly different in all three CRISPRi 22Rv1 clones (Tigger3a vs Control)
List of gRNA sequences used in this article
This figure contains the roadmap for all the different datasets used in this study. It also showcases transcription factor and motif enrichment over transposable element families enriched in different normal tissues states
This figure complements Figure 2 and showcases the enrichment of tranposable element families in TCGA PRAD ATAC data
This figure showcases the entire molecular and clinical characterization of CPC-GENE and Porto patients' groups identified using transposable element families differentially enriched between pluripotent stem cells and benign prostate tissue states
This figure complements Figure 4, showcasing the enrichment of Porto AR cistromes and AR motif over reprogrammed transposable element families (with the exception of Tigger3a and L1MB4 that are showcased in Figure 4)
This figure showcases the enrichment of transposable element families in H3K27ac or H3K9kme3 ChIP-seq data generated in benign-like and prostate cancer cell lines. It also includes dCas9-KRAB expression patterns in all CRISPRi clonal cell lines used in this article
This figure showcases dCas9-KRAB and H3K27ac profiling in CRISPRi 22Rv1 clones #1 and #3 nucleofected with control or Tigger3a gRNA combos (clone #2 is showcased in Figure 5)
This figure complements Figure 5 showcasing AR profiling and transcriptomic analysis results in CRISPRi 22Rv1 clones #1 and #3 nucleofected with control or Tigger3a gRNA combos (clone #2 is showcased in Figure 5)
Data Availability Statement
All data generated in this study are deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE224687, including H3K27ac regions profiled in CPC-GENE patients and in six additional benign prostate epithelium samples. Porto patient H3K27ac and AR regions were obtained from Stelloo and colleagues (26) and processed as specified in the Methods section. H3K27ac regions profiled in pluripotent stem cells and mature cell and tissue states were downloaded from ENCODE (https://www.encodeproject.org/) using the access numbers provided in Supplementary Table S1.



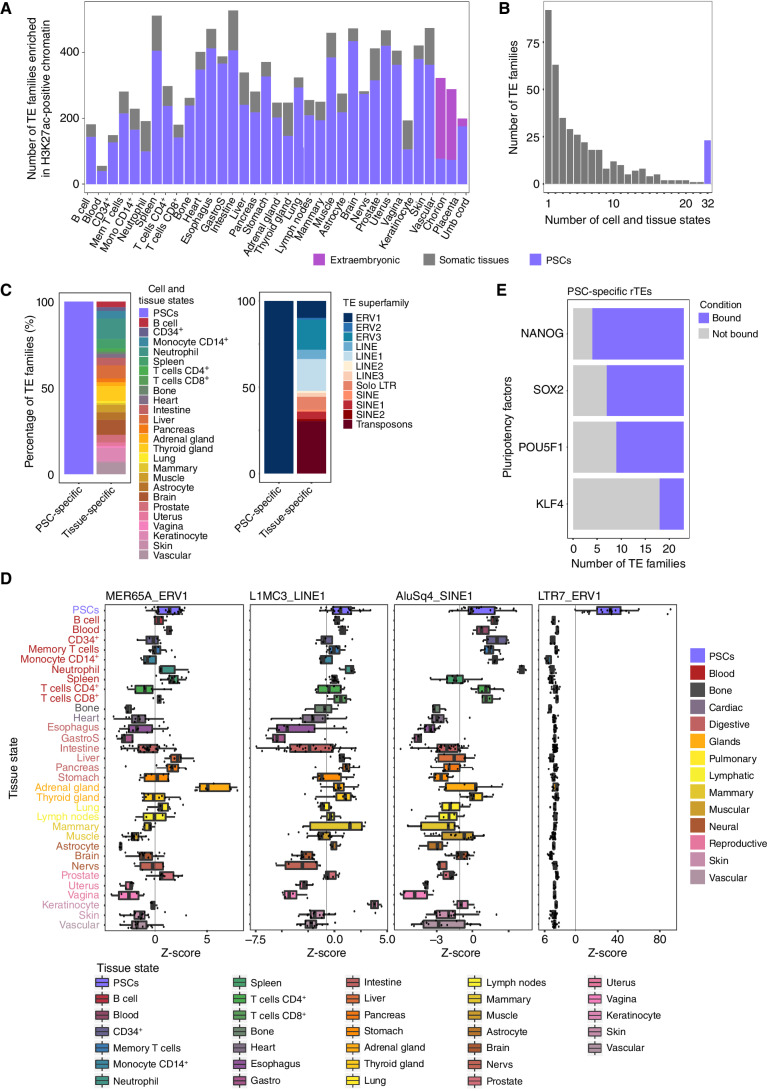
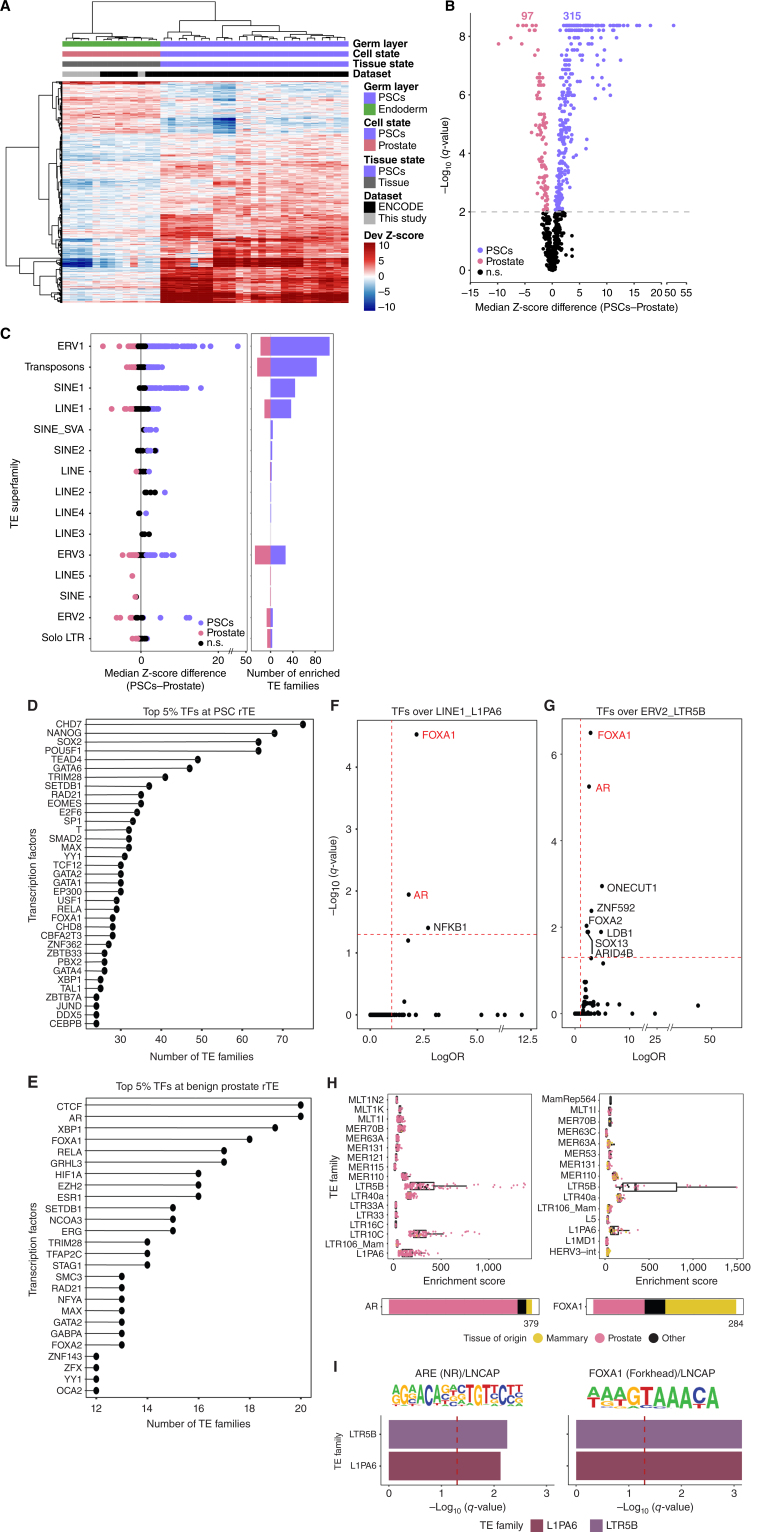
![Figure 3. TE families are reprogrammed in a subset of patients with prostate cancer. A and B, Heat maps displaying the deviation (Dev) Z-scores of TE families differentially enriched in H3K27ac-positive chromatin between pluripotent stem cells and benign prostate tissue state across H3K27ac profiles of CPC-GENE patients (A; n = 48) or Porto patients (B; n = 92). BCR, biochemical recurrence; NE score, neuroendocrine score; Neg, negative; Pos, positive; REP score, reprogramming score. C and D, Volcano plots showing median difference in deviation Z-scores for each TE family enriched in H3K27ac-positive chromatin between reprogrammed (REP) and constant (CONST) patients (CPC-GENE patients in C, Porto patients in D) vs. the −log10 q-value for that difference. The gray dashed line corresponds to −log10(q-value) = 2 (q-value = 0.01). The number of TE families enriched in reprogrammed or constant patients is reported at the top. n.s., not significant. E, UpSet plot showing the intersection of TE families enriched in reprogrammed CPC-GENE and Porto patients. Note that the vast majority of TE families are commonly enriched. F, UpSet plot showing the intersection of TE families enriched in pluripotent stem cells (PSC), benign prostate, and reprogrammed prostate cancer patients. Note that 164 TE families are commonly enriched in patients with reprogrammed prostate cancer and PSCs, corresponding to reprogrammed TE families, while 22 TE families are shared between patients with prostate cancer and benign prostate. Red font highlights the set of TE families described in the corresponding results section (more precisely, 164 TE families common to prostate cancer patients and pluripotent stem cells and 22 TE families common to prostate cancer patients and benign prostate epithelium). G–I, Box plot displaying the reprogramming score in reprogrammed and constant CPC-GENE (G) and Porto patients [with intermediate (INT) patients; H], and in pluripotent stem cells and benign prostate (I). P value results of Wilcoxon test are showcased on the box plot. J, Breakdown of common prostate cancer genetic aberrations called from whole-genome sequencing data in each of the 48 CPC-GENE samples separated by TE-based clustering (left). Comparison of the frequency of genetic aberrations using Fisher exact test (right). The red dashed line corresponds to −log10(P) = 1.3 (P = 0.05) threshold. Note that no genetic aberration tested was found significantly different between reprogrammed and constant patients. NA, not available.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/a716/10618745/c84471259228/2470fig3.jpg)