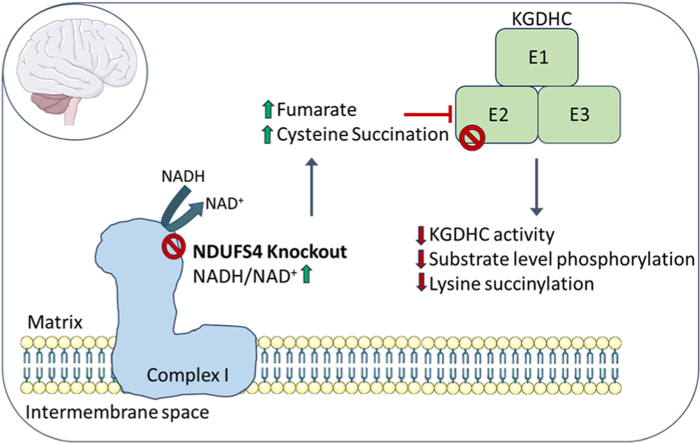
Abstract
The NDUFS4 knockout (KO) mouse phenotype resembles the human Complex I deficiency Leigh Syndrome. The irreversible succination of protein thiols by fumarate is increased in select regions of the NDUFS4 KO brain affected by neurodegeneration. We report that dihydrolipoyllysine-residue succinyltransferase (DLST), a component of the α-ketoglutarate dehydrogenase complex (KGDHC) of the tricarboxylic acid (TCA) cycle, is succinated in the affected regions of the NDUFS4 KO brain. Succination of DLST reduced KGDHC activity in the brainstem (BS) and olfactory bulb (OB) of KO mice. The defective production of KGDHC derived succinyl-CoA resulted in decreased mitochondrial substrate level phosphorylation (SLP), further aggravating the existing oxidative phosphorylation (OXPHOS) ATP deficit. Protein succinylation, an acylation modification that requires succinyl-CoA, was reduced in the KO mice. Modeling succination of a cysteine in the spatial vicinity of the DLST active site or introduction of succinomimetic mutations recapitulates these metabolic deficits. Our data demonstrate that the biochemical deficit extends beyond impaired Complex I assembly and OXPHOS deficiency, functionally impairing select components of the TCA cycle to drive metabolic perturbations in affected neurons.
Keywords: Fumarate, Protein succination, Leigh syndrome, Complex I, Alpha-ketoglutarate dehydrogenase, Substrate level phosphorylation
Graphical abstract
1. Introduction
Leigh syndrome is a mitochondrial disease predominantly caused by single gene defects in the oxidative phosphorylation (OXPHOS) machinery or the pyruvate dehydrogenase complex. It is characterized by bilateral necrotizing lesions of the basal ganglia and brainstem (BS), lactic acidosis, ataxia, seizures, and respiratory failure [1]. Leigh syndrome is associated with mutations in at least 23 genes that lead to mitochondrial Complex I deficiency [1], with mutations in NDUFS4, a gene that encodes the small assembly protein NADH dehydrogenase (ubiquinone) iron-sulfur protein 4 (Ndufs4) [2,3], among the most frequent.
The homozygous NDUFS4 knockout mouse (Ndufs4 KO) recapitulates many biochemical and clinical aspects of Leigh syndrome, including lactic acidosis, BS degeneration, motor retardation and fatal respiratory failure at 8 weeks after birth; the neuropathology also extends to the olfactory bulb (OB) and some cerebellar nuclei [4,5]. Selective deletion of Ndufs4 in glutamatergic neurons leads to BS inflammation, motor and respiratory deficits and early death, whereas ablation of Ndufs4 in GABAergic neurons causes basal ganglia inflammation, hypothermia, and severe epileptic seizures. This highlights the combined contributions of different neuronal populations to the complex pathology in this model [6].
While the genetic defects underlying Leigh syndrome are well documented, the biochemical mechanisms linking the bioenergetic deficit to the loss of select neuronal populations remains ill defined. Increased oxidative stress has been implicated in the pathogenesis of the Ndufs4 KO [5,7,8], attributed in part to elevated local hyperoxia and increased free iron [8,9], however, measures of oxidative stress markers have yielded conflicted results. In contrast, reductive stress is a well-established component of impaired OXPHOS during mitochondrial disease [10,11]. The ratio of NADH/NAD+ is elevated in the Ndufs4 KO model [12], and we have described that loss of Ndufs4 contributes to alterations in the NAD+-dependent tricarboxylic acid (TCA) cycle. Specifically, we have detected elevated levels of fumarate-derived protein succination in the pathologically lesioned regions of the Ndufs4 KO brain, namely brainstem and olfactory bulb [13]. Protein succination is a covalent chemical modification of protein cysteine thiols following non-enzymatic modification by fumarate, generating S-2-succinocysteine (2SC) (Fig. S1A, [[14], [15], [16]]). The formation of 2SC from endogenously increased fumarate leads to reduced function of select proteins [[17], [18], [19]]. We have previously confirmed increased succination of select proteins in lesioned Ndufs4 KO brain regions such as the vestibular nucleus, e.g., mitochondrial voltage-dependent anion channels (VDAC) 1 and 2 [13]. Thiol reactivity with protonated fumarate is enhanced in an acidic microenvironment [20], and the Ndufs4 KO is a well-characterized model of metabolic acidosis. Therefore, the metabolically demanding conditions in select neurons under bioenergetic stress may favor the formation of this irreversible modification.
In the current study, we demonstrate that mitochondrial dihydrolipoyllysine-residue succinyltransferase (DLST), a component of the α-ketoglutarate dehydrogenase complex (KGDHC), is succinated in the Ndufs4 KO brain. KGDHC is a significant rate limiting step of the TCA cycle in the brain [21], suggesting that impaired activity of this complex would have pronounced effects when OXPHOS is defective, indeed homozygous loss of DLST is embryonically lethal [22]. In humans, mutation of the gene for oxoglutarate dehydrogenase (OGDH) in two siblings resulted in a ∼50 % decrease in OGDH activity and manifested as a neurological mitochondrial disease [23]. Two recent studies profiling regional brain differences by global metabolomic profiling reinforce the central importance of the KGDHC axis in the Ndufs4 KO mouse model. Johnson et al. have documented defective α-ketoglutarate metabolism to glutamate and glutamine. Moreover, they demonstrate that the beneficial effect of rapamycin treatment in this model was via the augmentation of α-ketoglutarate [24]. Terburgh et al. also describe in detail defective α-ketoglutarate-derived succinate and glutamate production and increased branched chain amino acids in parallel with decreased BCAA-CoAs, which would yield even less entry of anaplerotic succinyl CoA to the defective TCA cycle [25]. Here, we report the striking functional impact of DLST succination and defective KGDHC activity on distinct mitochondrial metabolic processes, providing a mechanistic link between the documented metabolomic defects in this model and the region-specific accumulation of select succination. These results biochemically define how mitochondrial ATP deficiency is exacerbated beyond the existing genetic Complex I bioenergetic defect.
2. Results
2.1. Dihydrolipoyllysine-residue succinyltransferase is succinated in the Ndufs4 KO mouse brain
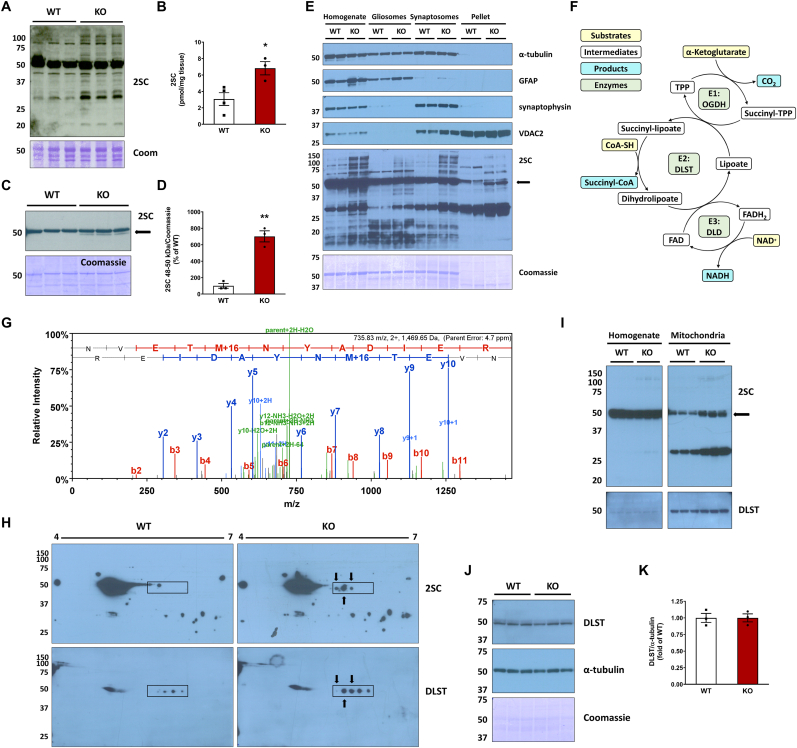
Protein succination is selectively increased in the brainstem of the Ndufs4 KO mouse versus WT, as detected by anti-2SC antibody detecting the succinate moiety following fumarate reactivity with cysteine (Fig. 1A). Quantification of total protein-bound 2SC levels by LC-MS/MS demonstrates an increase from 3.02 pmol/mg tissue in WT to 6.82 pmol/mg tissue in the KO mouse brainstem (P < 0.05, Fig. 1B). We have previously described the abundant and consistent succination of tubulin isoforms in the brainstem (BS) and the olfactory bulb (OB) of both WT and Ndufs4 KO mice [13]. Using immunoblotting with an anti-2SC antibody the pronounced succinated tubulin band is resolved at ∼50 kDa in BS protein homogenates, appearing slightly larger and expanding to a lower MW in the KO mice vs. WT (Fig. 1A). To determine if additional proteins were succinated at this molecular weight and masked by the intensity of tubulin succination, we performed a tubulin polymerization assay that we had previously employed [18] in which the initial high speed centrifugation pellet is substantially depleted of tubulin. Immunoblotting with anti-2SC following this tubulin depletion approach allowed the resolution of any residual succinated tubulin from a new succinated protein that was only present in the KO mouse brainstem (Fig. 1C, arrow in 2SC panel). Densitometric quantification of this 48–50 kDa succinated band demonstrated a significant increase in KO vs. WT brainstem (Fig. 1D). A longer exposure shows enrichment of this ∼48–50 kDa succinated band following depletion of cytosolic tubulin by polymerization (Fig. S1B, arrow in 2SC panel). In a separate experiment to determine where succinated proteins might be localized, we isolated gliosomes and synaptosomes from WT and Ndufs4 KO mouse BS to determine where succinated proteins might be localized. Glial fibrillary acidic protein (GFAP), an astrocytic marker, was enriched in the gliosomes, whereas the presence of the neuronal presynaptic marker synaptophysin was prominent in the synaptosomes (Fig. 1E, GFAP and synaptophysin panels). A final pellet from these enrichments contains other organelles including the mitochondria, and we noted the outer mitochondrial membrane marker VDAC2 was present in the original tissue homogenate and the synaptosomes but was most abundant in this remaining pellet (Fig. 1E, VDAC2 panel). Overall, the pellets were rich in mitochondria but devoid of cytosolic, synaptic and astroglial markers. After probing with anti-2SC antibody, the succinated band at ∼48–50 kDa was strikingly enriched in the pellet fraction of Ndufs4 KO but not in WT mice (Fig. 1E, arrow in 2SC panel). Cytosolic tubulin depletion facilitated the detection of the succinated band at ∼48–50 kDa (Fig. 1E, α-tubulin panel), as the abundant presence of succinated tubulin both in WT and KO tissue homogenates, gliosomes and synaptosomes overwhelms the detection of this ∼48–50 kDa band (Fig. 1, Fig. 2SC panel). Similar results were obtained when we performed the same enrichment with the cerebellum (CB), another affected brain region in Ndufs4 KO mice (Figs. S1C and D).
Fig. 1.
Identification of dihydrolipoyllysine-residue succinyltransferase (DLST) succination in the brainstem (BS) of the Ndufs4 KO mouse.A, Total levels of protein succination (2SC) in brainstem (BS) homogenates from WT and Ndufs4 KO mice, note the abundance of succinated tubulin in WT and KO at ∼50 kDa. Ndufs4 KO BS show enrichment of several additional succinated proteins vs. WT mice (representative blot, n = 3/group). B, LC-MS/MS quantification of total 2SC levels in WT and Ndufs4 KO brainstem (n = 3–4/group). C, Detection and D, densitometric quantification of a distinct succinated band at ∼48–50 kDa (arrow) in Ndufs4 KO BS following cytosolic tubulin depletion (n = 3). E, Characterization of the purity of total tissue protein homogenates, gliosomes, synaptosomes, and a mitochondria-rich pellet fraction obtained from BS by analysis of α-tubulin, GFAP, synaptophysin and VDAC2 expression. In the 2SC panel, a ∼48–50 kDa succinated band is present in the pellet fractions (2SC panel, KO lanes, arrow). F, Structure and function of the α-ketoglutarate dehydrogenase complex (KGDHC). For each subunit the substrates are highlighted in yellow, intermediates/cofactors in white and products in light blue and the enzymatic components of the complex in light green. G, MS/MS spectrum of the peptide NVETMoxNYADIER corresponding to DLST in a ∼48–50 kDa band isolated from the mitochondria-rich pellets used in C. H, Co-localization (arrows) of succinated spots with DLST-immunoreactive spots in the region of ∼48–50 kDa and pH 5.5–6.3 (rectangular area) in Ndufs4 KO BS. I, Analysis of BS tissue homogenates, where tubulin precludes DLST detection, alongside purified mitochondrial fractions showing a succinated band at ∼48–50 kDa in KO lanes (2SC panel, arrow), which overlapped with DLST detection. J, Total DLST and α-tubulin content in BS homogenates of WT and Ndufs4 KO mice. K, Quantification of the DLST band intensity in J (n = 3/group). See also Fig. S1.In A, C, and E, H, I, J molecular weight markers are shown on the left side. Coomassie staining was used to verify even load of the lanes. In B, D, K, results expressed as mean ± SEM were compared by unpaired t-test (*P < 0.05, **P < 0.01). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
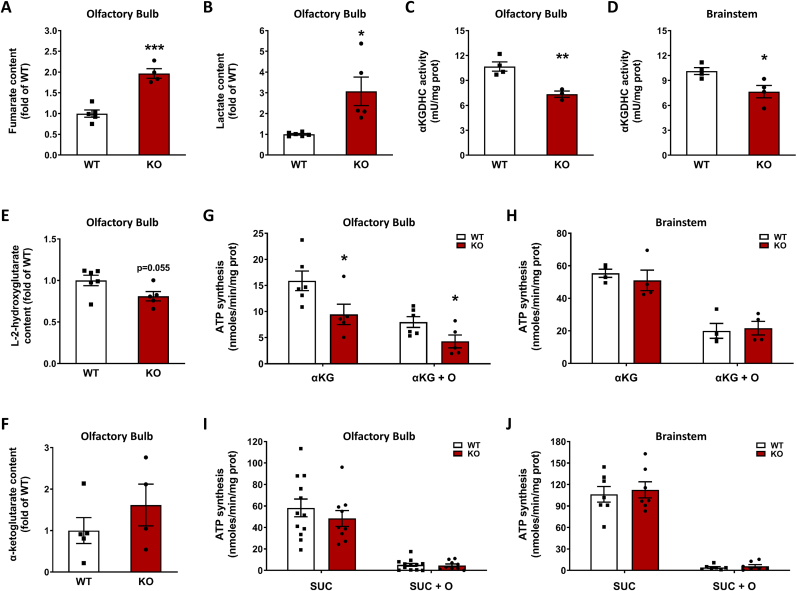
Fig. 2.
α-ketoglutarate dehydrogenase complex (KGDHC) activity is reduced as a consequence of fumarate derived protein succination.A-B, Quantification of fumarate (A, n = 4–5) and lactate (B, n = 5–6) content in the olfactory bulb of Ndufs4 KO and WT mice. C-D, KGDHC activity in the olfactory bulb (C, n = 4) and brainstem (D, (n = 3–4) of Ndufs4 KO and WT mice. E-F, Quantification of L-2-hydroxyglutarate (E, n = 5–6) and α-ketoglutarate (F, n = 4–5) content in the olfactory bulb of Ndufs4 KO and WT mice. G-H, Mitochondrial total ATP synthesis (no oligomycin) and substrate level phosphorylation (SLP, +8 μM oligomycin = O) in the olfactory bulb (G, n = 5–6) and the brainstem (H, n = 4) in the presence of 5 mM α-ketoglutarate (αKG) as respiratory substrate. I-J, Same as in G-H but using 5 mM succinate (SUC) as respiratory substrate (n = 7–12).In panels A-J, results expressed as mean ± SEM, and comparisons performed by unpaired t-test (*p < 0.05, **p < 0.01 and ***p < 0.001 vs. WT).
The mitochondria-rich pellet fractions were further separated by SDS-PAGE, with parallel lanes used for anti-2SC immunoblotting or Coomassie staining, prior to band excision ∼48–50 kDa region and LC-MS/MS mass spectrometry. Several abundant mitochondrial proteins with significant XCorr scores were identified from the in-gel trypsin digests, but a specific succination site could not be assigned as the detection of endogenous levels of negatively charged succinated peptides is difficult in positive ion mode mass spectrometry. For each of these proteins we manually calculated the isoelectric point (pI) of the mature mitochondrial form (minus the signal sequence) to determine if any aligned with a distinct series of succinated spots that we repeatedly resolved following 2D-electrophoresis and immunoblotting (Fig. 1H, boxed region in upper 2SC panels). Several succinated spots were present in the KO this area, whereas in the WT only one spot was present at a slightly higher molecular weight (Fig. 1, Fig. 2SC panels, WT vs. KO boxed regions). We have previously noted that this succinated spot series appeared around a pI ∼6 when resolved away from tubulin isoforms whose pI is 4.7–5.4 (Fig. 1, Fig. 2SC panels, [13,18]). The protein dihydrolipoyllysine-residue succinyltransferase (DLST), a component of the α-ketoglutarate dehydrogenase complex (KGDHC), had a computed isoelectric point (pI) ∼5.98 for the mature mitochondrial form (removing transit peptide 1–68). The KGDHC comprises several copies of three subunits (Fig. 1F): 2-oxoglutarate dehydrogenase (OGDH, E1), DLST (E2), and dihydrolipoamide dehydrogenase (DLD, E3) [26]. The substrates, intermediates and products in the reactions occurring at E1, E2 and E3 are summarized in Fig. 1F schematic. A representative DLST-specific peptide NVETMOXNYADIER ([M+2H]2+: 735.8322, XCorr (+2) = 3.94) identified by LC-MS/MS is shown in Fig. 1G, other DLST peptides confirmed by mass spectrometry are summarized in Fig. S1E. Applying a DLST-specific antibody to the 2-D immunoblots we detected precise co-localization with three succinated spots in the KO (Fig. 1H, overlap marked by arrows in KO 2SC and DLST panels). In contrast, the four spots detected by anti-DLST do not overlap with any succinated spot in the WT (Fig. 1H, WT DLST and 2SC panel). We further analyzed both BS tissue homogenates and purified mitochondrial fractions from WT and Ndufs4 KO BS to examine if the succinated protein paralleled DLST localization. Fig. 1I shows a short exposure of BS tissue homogenates from KO and WT mice without detectable changes in succination (2SC panel, homogenate lanes) due to the abundance of the succinated tubulin band [13,18]. In contrast, mitochondrial purification allowed for better resolution of the distinctly succinated ∼48–50 kDa band in the KO BS (Fig. 1, Fig. 2SC panel, mitochondria lanes, black arrow). The additional succinated band at ∼30–32 kDa had been previously identified as mitochondrial VDAC1 and 2 [13]. After stripping, a specific anti-DLST antibody showed similar levels of this protein in both WT and Ndufs4 KO mitochondria, overlapping with the succinated bands (DLST and 2SC panels, mitochondria lanes). We confirmed that total DLST protein levels do not change in the BS between genotypes, as shown in Fig. 1J and K. In addition, we did not detect any changes in OGDH levels (E1 subunit) or other TCA cycle enzymes, indicating TCA cycle enzyme levels are not reduced in the KO model (Fig. S1F). In summary, DLST is succinated by fumarate in the BS of Ndufs4 KO mice compared to WT mice.
2.2. Succination reduces the activity of the α-ketoglutarate dehydrogenase complex, decreasing substrate level phosphorylation
We hypothesized that DLST succination driven by increased fumarate might affect the functionality of the KGDHC, since components of this complex are susceptible to oxidative modification [[27], [28], [29], [30]]. We confirmed increased fumarate concentration in the olfactory bulbs (OB) of the Ndufs4 KO mouse (96.7 % greater than in WT OB, p < 0.001, Fig. 2A), which accumulated in parallel with increased lactate (Fig. 2B). Pyruvate and malate levels were also significantly increased in the Ndufs4 KO OB (Figs. S2A and D). We then measured KGDHC activity in both the OB and BS of WT and Ndufs4 KO mice and found that in both brain regions it was significantly lower in the KO mice (31.2 % and 24.2 % decrease in OB and BS, p < 0.01 and p < 0.05 respectively, Fig. 2C and D). These reductions were specific for KGDHC activity in OB and BS of the Ndufs4 KO mouse, as the activity of citrate synthase, a TCA cycle enzyme commonly used to normalize the activity of other mitochondrial enzymes, was unaffected in the same brain regions (Figs. S2E and F for OB and BS, respectively). A recent report has shown increases, and not a decrease, in citrate synthase activity in the affected regions of the Ndufs4 KO brain [25], further demonstrating that the KGDHC is selectively impacted. Since the Ndufs4 KO mice have a metabolic acidosis, it could be expected that this would favor the conversion of any accumulated α-ketoglutarate towards L-2-hydroxyglutarate [31]. Quantification of L-2-hydroxyglutarate showed a trend (P = 0.055) to decrease in the OB of the Ndufs4 KO mouse (Fig. 2E), whereas α-ketoglutarate concentration was unchanged (Fig. 2F). Decreased 2-hydroxyglutarate has recently been documented in the Ndufs4 KO mouse [32] and suggests that any α-ketoglutarate conversion to this metabolite may rapidly be redirected to supply the mitochondrial Q-pool via the activity of L-2 hydroxyglutarate dehydrogenase.
Since decreased KGDHC activity would lead to reduced formation of succinyl CoA, this in turn would likely decrease the conversion of succinyl-CoA into succinate in the TCA cycle. This step is catalyzed by the enzyme succinyl-CoA ligase (SUCLA), which is responsible for the synthesis of GTP or ATP by substrate level phosphorylation (SLP). In rodents and human brain, succinyl-CoA ligase preferentially produces ATP, whereas in anabolic tissues the main product is GTP ([33]; Ostergaard, 2008). We measured total and SLP-linked ATP synthesis in mitochondria isolated from both OB and BS of WT and Ndufs4 KO mice. Under the conditions used, total ATP synthesis represents the sum of ATP production by OXPHOS and SLP, and the residual ATP synthesis in the presence of the ATP synthase inhibitor oligomycin represents SLP [34]. Using α-ketoglutarate as a substrate, total ATP synthesis was decreased by 42.5 % (p < 0.05), and SLP was decreased by 48.3 % (p < 0.05) in the OB of the KO mice (Fig. 2G). These data demonstrate that, in addition to the Complex I defect in OXPHOS derived ATP production, the generation of TCA cycle derived intramitochondrial ATP is also compromised. In contrast, we observed no significant differences in total ATP synthesis and SLP in the BS of Ndufs4 KO vs. WT mice (Fig. 2H). This may reflect the heterogeneity of the total mitochondrial protein used fraction, in the BS select nuclei are more affected by pathology, e.g., the vestibular nucleus. When succinate was used as a respiratory substrate, no differences in total ATP synthesis were observed for OB (Fig. 2I) and BS (Fig. 2J). As expected, succinate did not support SLP irrespective of the genotype and brain region (Fig. 2I and J) [34]. Analysis of the succination profile of the isolated mitochondria used for these SLP analyses showed a succinated band at ∼48–50 kDa that was only present Ndufs4 KO mice (Fig. S2H), underscoring the selectivity of succination.
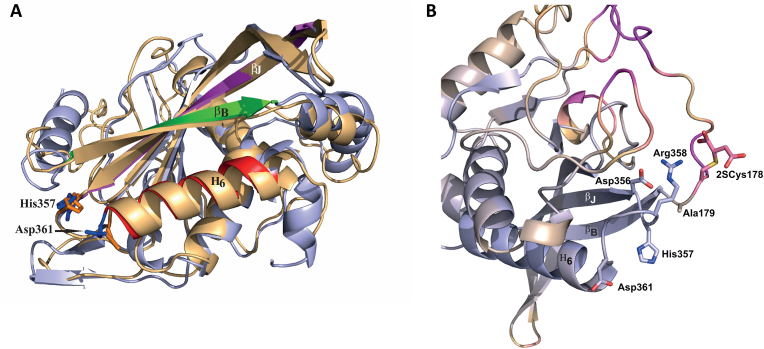
To better understand the impact of DLST succination on the KGDHC functionality, we performed molecular dynamics simulation of human DLST (hDLST) based on the recently characterized 3D structure [35], and preliminary 3D assessment of this recent model demonstrated that Cys178 was located in the spatial vicinity of the active site. The mature forms of both murine and human DLST contain 2 cysteines and we assumed that the striking homology of human and mouse DLST would allow us to draw conclusions applicable to our mouse model (Fig. S3A). We focused on the effects of Cys178 succination (2SCys178) by modeling the covalent succination modification at this site. The possible role of a second cysteine succination (i.e., Cys37) on the functionality of hDLST could not be evaluated due to the very high flexibility of the region before Asn161 that leads to unstable simulation. Molecular dynamics simulation of hDLST showed that the interaction pattern was altered in select regions of the protein when Cys178 was succinated. For instance, the Arg358-Asp356 salt bridge was perturbed due to the immediate spatial vicinity of the succinated Cys178 residue, leading to a new ionic interaction of this residue with Arg358 (distance of 2.71 Å, Fig. S3C). In addition, in the unmodified hDLST, Ala179 and Asp356 (distance of 2.92 Å, Fig. S3C) formed a H-bond that stabilized the relative positions of His357 (and the loop to which His357 belongs), the βB and βJ strands; this is important for proper channeling of the Coenzyme A (CoA) and the succinyl-dihydrolipoamide moiety (SLAM) substrates ([35], see structure in Fig. 3A and B). Conversely, succination of Cys178 resulted in the disruption of this H-bond, as the average distance between Ala179 and Asp356 increased to 4.81 Å (Fig. S3C); this change affects the three chains in the hDLST trimer in a similar manner (Fig. S3D). Rearrangement of the local interaction pattern modified the positions of the aforementioned beta strands and increased the distance between channel-forming side chains, which might lead to less efficient coordination of the CoA and SLAM substrates, resulting potentially in increased KM values. The position/orientation of His357, part of the DLST active site, was also changed in the succinated hDLST (Fig. 3A) and may alter this residue's capacity to either deprotonate the CoA substrate or stabilize the tetrahedral oxyanionic catalytic intermediate [35]. Unfortunately, we could not draw reliable conclusions regarding the interactions between the monomers, since the dynamic simulations were found to be too unstable; this might be a result of omitting residues 150–160, which participate in the stabilization of the trimeric structure. Overall, the observed structural changes were mostly consistent for the three protein chains, showing the reliability of the simulation. The hDLST regions most affected by succination encompass the first 12 and last 8 residues studied, which in the unmodified structure are stable helical structures (Fig. S3E). Although the overall active site region showed shifts lower than 0.5 Å on average in the succinated structure, select residues involved in substrate coordination showed much greater shifts (Fig. 3B). This simulation work strongly supports that succination of Cys178 impacts both catalytically important active site residues and the substrate-binding channels.
Fig. 3.
A, Superimposition of the succinated Cys178 (2SCys178) form of human DLST (hDLST) in orange and the unmodified hDLST in blue. Red, green, and purple colors in the unmodified form indicate the secondary structural elements that form the two substrate channels. B, The catalytic center of hDLST and its immediate spatial vicinity in the succinated (2SCys178) form, showing the degree of displacement relative to the unmodified structure. Blue color represents displacement <0.5 Å and purple indicates displacement >6.5 Å. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
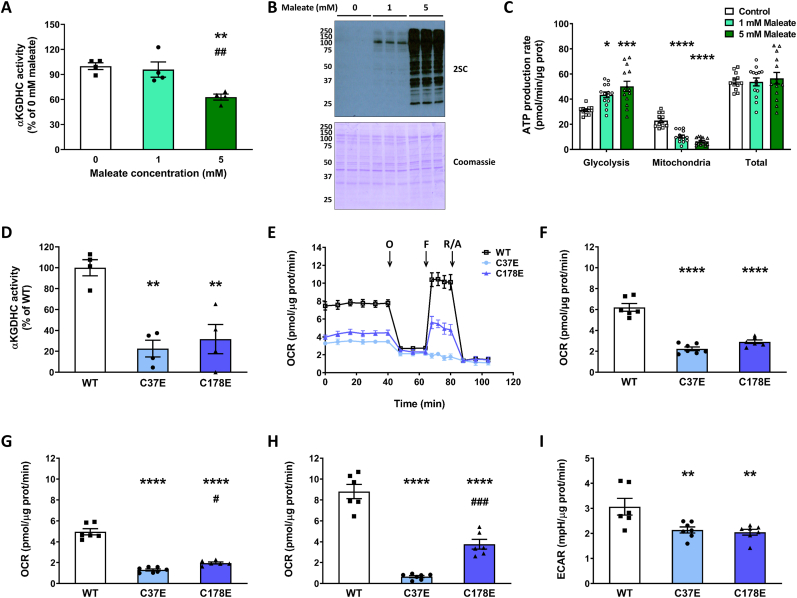
2.3. The fumarate isomer maleate and DLST succinomimetics reduce the activity of the KGDH complex
We used maleate, the cis isomer of fumarate that has increased reactivity at physiological pH and is non-toxic compared to fumarate ester treatment [36] to increase intracellular succination and determine if this impacted KGDHC activity. Treatment of cells with 5 mM maleate for 16 h led to a significant reduction in KGDHC activity (37.11 % vs. untreated controls, p < 0.01, Fig. 4A), whereas 1 mM maleate did not decrease KGDHC activity. Reduced KGDHC activity with 5 mM maleate was paralleled by a marked elevation in protein succination (Fig. 2, Fig. 4SC panel), whereas less succination was detected with 1 mM maleate, but was still increased above controls. This exogenous addition of maleate differs from the endogenous production of fumarate observed in the Ndufs4 KO mice and could be expected to impact cytosolic proteins more than mitochondrial proteins. We examined the sources of ATP production in maleate treated cells and found that maleate caused a shift towards increased ATP synthesis linked to glycolysis (p < 0.05 for 1 mM maleate vs. 0 mM maleate, and p < 0.001 for 5 mM maleate vs. 0 mM maleate), and reduced ATP synthesis linked to mitochondrial function (p < 0.0001 for both comparisons), but no changes in the total amount of ATP produced (Fig. 4C) or cell survival. Total DLST levels were not affected by maleate treatment (Fig. S4A, DLST panel), similar to our findings in the BS of the Ndufs4 KO mouse (Fig. 1K). These data demonstrate specific reductions in KGDHC function in parallel with decreased oxidative phosphorylation, and a compensatory shift towards glycolysis during acute maleate induced succination. Separately, we also included the use of dimethyl fumarate (DMF), a reactive fumarate ester that results in protein succination [37]. Incubation of a KGDHC standard with DMF caused a dose-dependent decrease in the activity of the complex (Fig. S4F). Furthermore, we confirmed decreased KGDHC activity in N1E-115 neurons incubated with DMF (Fig. S4G). When we replaced DMF with dimethyl succinate, a DMF analog that does not modify cysteine residues, there was no loss of KGDHC activity (Fig. S4G). Increased succination of a band at ∼48–50 kDa was observed in N1E-115 neurons after incubation with 50 and 100 μM DMF (Fig. S4H, arrow in 2SC panel), and the band co-localized with DLST immunolabeling (Fig. S4H, DLST panel). While fumarate esters are more reactive and are not comparable to physiological fumarate increases, they do demonstrate that elevated fumarate-derived succination contributes to reduced KGDHC activity.
Fig. 4.
Maleate treatment and succinomimetic mutation of DLST reduce the activity of the KGDHC.A-C, KGDHC activity (A, n = 4), protein succination (B), and ATP production rate linked to glycolysis and mitochondrial function (C) in DLST KO H838 cells expressing WT hDLST, after treatment for 16 h with 0–5 mM maleate (n = 12). D, KGDHC activity in DLST KO H838 cells expressing WT hDLST or hDLST forms where Cys37 or Cys178 were mutated to Glu to mimic succination (n = 4). E, Respiratory profile of the cells described in (D). The oxygen consumption rate (OCR) was measured using a Seahorse XF analyzer, in basal conditions and after sequential injection of oligomycin (O), FCCP (F), and rotenone/antimycin A as described in Methods. F–H, Basal respiration (F), oxygen consumption coupled to ATP production (G), and maximal respiration (H), in the experiment described in (E). I, Extracellular acidification rate (ECAR) after oligomycin injection in the experiment described in (E), n = 6/group.In panels A, C, D, and F–I, data expressed as mean ± SEM and comparisons by one-way ANOVA and post hoc Student-Neuman-Keuls test (A and C, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 for 1 mM and 5 mM maleate vs. 0 mM maleate, and ##p < 0.01 for 5 mM maleate vs. 1 mM maleate; D, and F–I, **p < 0.01, and ****p < 0.0001 for mutants vs. WT, and #p < 0.05 and ###p < 0.001 for C178E vs. C37E). In panel B, the molecular weight markers are shown on the left side.
We employed site-specific mutants to further study the impact of DLST succination on KGDHC functionality. H838 DLST-KO cells were previously generated by CRISPR-Cas9 technology [[38], [39]] and were transfected with a construct to express either wildtype (WT) hDLST, or with constructs where Cys37 or Cys178 were mutated to Glu to introduce a negative charge that mimics succination (the succinomimetics C37E and C178E). Glu is a larger amino acid than Asp and better mimics succination of cysteines [20]. We examined KGDHC activity in the cells expressing either WT hDLST, or the succinomimetic mutants C37E hDLST or C178E hDLST. As shown in Fig. 4D, KGDHC activity was reduced in the mutant lines to 22.65 (C37E, p < 0.01) and 30.58 % (C178E, p < 0.01) of the values obtained for the WT construct. In addition, the respiratory profile of the mutants was greatly affected (Fig. 4E), with significant decreases in basal respiration (Fig. 4F), respiration associated with ATP synthesis (Fig. 4G), maximal respiration (Fig. 4H), and the spare respiratory capacity (Fig. S4B). No changes in respiration associated with proton leak or non-mitochondrial respiration (Figs. S4C and D) were observed in the mutants. The extracellular acidification rate (ECAR) in basal conditions was not affected in the mutants (Fig. S4E), but it was decreased after the addition of oligomycin (Fig. 4I). This data confirms that the succinomimetic mutation of either cysteine in DLST profoundly impacts activity and the metabolic respiratory status of the cells.
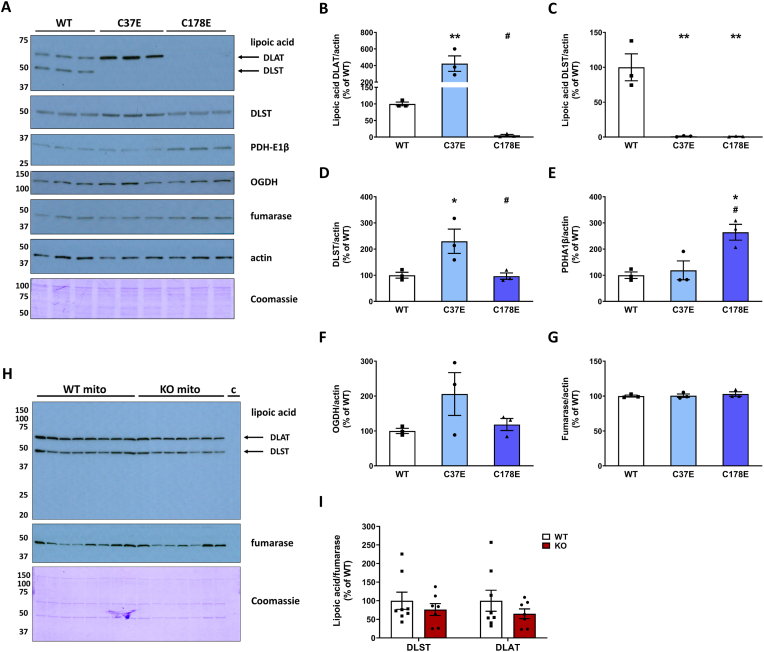
Although we were unable to perform simulated molecular dynamics succination of Cys37, we observed that the C37E mutant was as effective in reducing KGDHC activity as the C178E mutant. We noted that Cys37 is only 6 residues away from the lipoylated lysine (Lys43lipoyl) in both mouse and human DLST [CEIETDKlipoyl]. We reasoned that succination of Cys37 in the Ndufs4 KO might interfere with DLST lipoylation, or that the reduced form of lipoate itself might be susceptible to succination. The production of succinyl-CoA by KGDHC employs the formation of a DLST-mediated succinyl-dihydrolipoate intermediate (Fig. 1F), therefore we used an anti-lipoic acid antibody to investigate the lipoylation state of DLST. We first examined lipoylation in the cells expressing WT hDLST or the C37E and C178E succinomimetics as these would be expected to show a maximal effect. The antibody also detects another prominent lipoylated protein, dihydrolipoyllysine-residue acetyltransferase (DLAT), a lipoyl-containing subunit of the pyruvate dehydrogenase complex (PDH). The C178E mutation led to an almost complete loss of lipoic acid immunoreactivity in both DLST and DLAT (Fig. 5A–C, lipoic acid panel), whereas the lipoylation state increased for DLAT and decreased for DLST for the C37E mutant (Fig. 5A–C, lipoic acid panel). Total DLST levels were not affected in the C178E mutant and appeared to be slightly increased in the C37E mutant (Fig. 5A and D, DLST panel). The levels of the PDH-E1β subunit increased slightly in the C178E mutant, perhaps in a compensatory manner, whereas levels of the OGDH subunit of KGDHC and fumarase did not change markedly (Fig. 5A, 5E-G). It is unclear why DLAT lipoylation was differentially impacted in the DLST-specific mutants, perhaps the loss of DLST activity promotes a separate oxidative lipoate modification precluding detection [40,41]. The pronounced reductions in DLST lipoylation do parallel the decreased KGDHC activity measured in these striking mutants. We next investigated if lipoylation was altered in the brain of the Ndufs4 KO mouse, however, we found that the lipoylated DLST and DLAT signal intensities were not reduced in the KO vs. WT BS (Fig. 5H and I). Extensive attempts to detect peptides containing succinated Cys37 or succinated Lys43lipoyl were also unsuccessful, although the difficulty in detecting lipoylated DLST modifications has previously been noted in ABHD11 deficient cells [40]. The results of our simulation (Fig. 3A and B) and the preserved lipoylation profile of DLST in the Ndufs4 KO brain further suggest that Cys178, and not Cys 37 or Lys43lipoyl, is the likely succination site in the Ndufs4 KO mouse.
Fig. 5.
A, Detection of lipoic acid and mitochondrial markers (DLST, PDH-E1β, OGDH and fumarase) in the DLST KO H838 cells expressing WT hDLST, DLST Cys37Glu or DLST Cys178Glu (n = 3/group). Actin and Coomassie staining were used to verify even loading of the gel. B-G, Densitometric quantification of the mitochondrial markers shown in A relative to cytosolic protein levels (actin). H, Detection of lipoic acid moieties associated with DLST and DLAT (dihydrolipoyllysine-residue acetyltransferase, a subunit of the pyruvate dehydrogenase complex) in the BS of WT and Ndufs4 KO mice (n = 7–8). In addition to the mitochondrial fractions (“mito”), a lane with a cytosolic fraction (“c”) was included as a negative control. I, Normalization of the signal intensity in H, relative to fumarase to account for mitochondrial protein levels. Normalization to Coomassie did not alter the lipoylation status (not shown). In panel A and H, molecular weight markers are shown on the left side.
2.4. Defective DLST function and reduced KGDHC activity contribute to protein hyposuccinylation
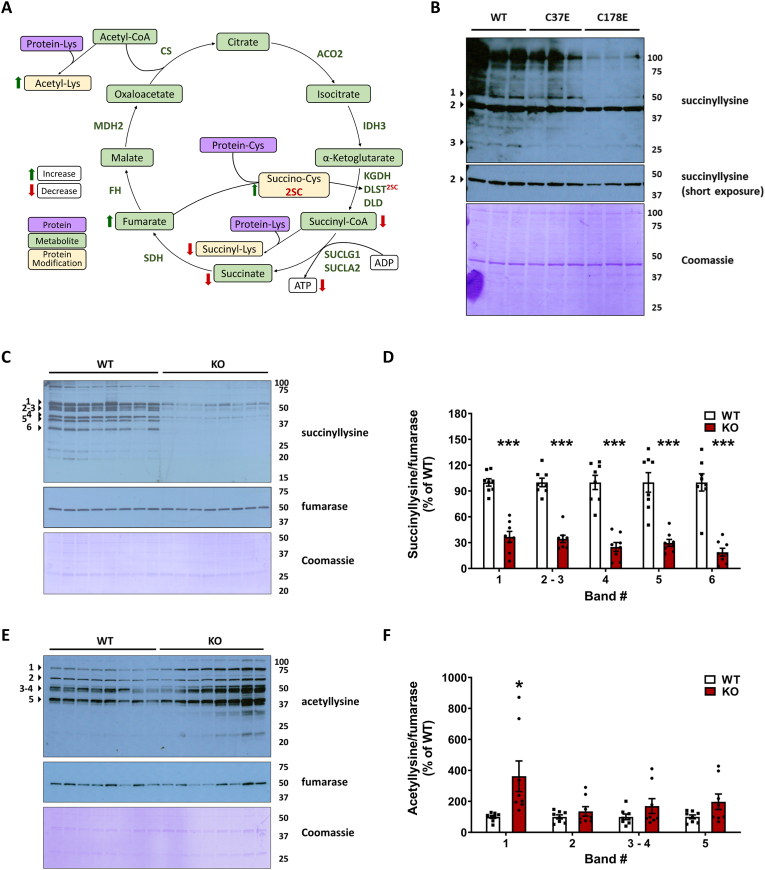
We predicted that decreased synthesis of succinyl-CoA would contribute to reduced protein succinylation, an acyl CoA derived lysine modification that is distinct from cysteine succination [42]. Protein succinylation has been implicated in further modulating the activity of mitochondrial enzymes [43,44]. DLST can act as a protein succinyl transferase [43]; therefore, the succination of DLST could contribute directly to decreased mitochondrial protein succinylation. This is outlined in Fig. 6A, where the impact of increased fumarate-mediated generation of 2-succinocysteine (2SC) on DLST (DLST2SC) contributed to defective KGDHC activity, resulting in lower ATP via reduced SUCLA activity, decreased succinate levels and decreased lysine succinylation (Fig. 6A). We investigated the levels of mitochondrial protein succinylation in DLST KO H838 cells expressing WT hDLST or the C37E and C178E mutants, since the mutants showed a striking reduction in KGDHC activity. The succinylation profile of mitochondrial proteins was notably decreased for the C178 mutant vs. WT across several select bands (Fig. 6B), with a smaller reduction in succinylation in the C37E mutant vs. WT (Fig. 6B). This suggests the C178E mutant resulted in a more significant deficit in succinyl-CoA levels. Cytosolic proteins were also hyposuccinylated in the C178E mutant (Fig. S5C). We also examined the succinylation profile of mitochondrial proteins in DLST KO H838 cells expressing WT hDLST after 16 h treatment with 0–5 mM maleate. No significant change in succinylated proteins was detected (Fig. S5A), similar results were obtained with N1E-115 neurons treated for 16 h with 0–100 μM DMF (Fig. S5B), indicating that despite the reduction in KGDHC activity, this short treatment duration was not sufficient to impact the turnover of succinylated proteins.
Fig. 6.
DLST succination and mutations that mimic DLST succination alter the levels of protein acylation in vivo and in vitro.A, Schematic illustrating TCA cycle deficits in the Ndufs4 KO mouse. Red arrows indicate a measured decrease and green arrows indicate a measured increase. The metabolic impact of a 2-fold increase in fumarate (P < 0.001) contributes to a 2-fold increase in protein succination (P < 0.05). The specific succination of DLST (DLST2SC) contributes to a 25–30 % decrease in KGDHC activity (P < 0.001), and results in lower ATP via a 48 % reduction in SUCLA activity (P < 0.05), decreased lysine succinylation (hyposuccinylation), and a 50 % reduction in succinate levels (P < 0.001). Lysine acetylation of select proteins increases as a consequence of diminished TCA cycle function. These metabolic perturbations impacting the TCA cycle alter the chemical modification of proteins, extending the biochemical deficit beyond the loss of the Complex I component Ndufs4. Relationships between metabolites (green boxes), protein lysine or cysteine residues (purple boxes) and protein modifications (yellow boxes) are indicated. B, Protein lysine succinylation in mitochondrial fractions obtained from DLST KO H838 cells expressing WT hDLST, C37E or C178E hDLST succinomimetic mutants. C, Protein lysine succinylation in mitochondrial fractions obtained from the brainstem of WT and Ndufs4 KO mice. D, Quantification of prominent bands in C (indicated by arrowheads). E, Protein lysine acetylation in mitochondrial fractions obtained from brainstem of WT and Ndufs4 KO mice. F, Quantification of prominent bands in E (indicated by arrowheads).In B, Coomassie Brilliant Blue staining (prominent band at 42 kDa) was used for the normalization of the succinyllysine signal. In C and E, re-probing to detect fumarase was used for mitochondrial protein normalization, followed by Coomassie Brilliant Blue staining. Molecular weight markers are shown on the right side. See also Fig. S4. In D and F, results expressed as mean ± SEM were compared by unpaired t-test (*p < 0.05 and ***p < 0.001 vs. WT). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We next investigated the mitochondrial protein succinylation profile in the brain of WT and Ndufs4 KO mice. Fig. 6C demonstrates a remarkable global hyposuccinylation in the BS of the KO mice. Densitometric quantification of several prominent bands demonstrated >50 % reduction in succinylation (Fig. 6D). The Ndufs4 KO also showed decreased succinylation of cytosolic proteins in the BS, although less succinylation was detected in cytosolic vs. mitochondrial fractions (Figs. S5D and S5E). Lysine succinylation changes were less evident in the nuclear protein fraction (Figs. S5F and S5G), emphasizing that the greatest effect on succinylation status is within the mitochondria where fumarate is endogenously elevated in parallel with defective DLST function. Overall, reduced BS succinylation due to lower succinyl-CoA levels is further represented by decreased levels of succinate in the OB of the Ndufs4 KO (Fig. S5H). Given that metabolite flux through KGDHC is impaired, we expected that acetyl CoA-derived mitochondrial lysine acetylation would be increased (Fig. 6A), this is shown for select proteins in the Ndufs4 KO versus the WT (Fig. 6E and F) and is aligned with the hyperacetylation described in a glutamatergic neuron specific Ndufs4 KO [45]. In summary, cysteine succination of DLST (KGDHC E2) impairs the activity of the KGDHC, further compromising ATP generation via reduced substrate level phosphorylation and decreasing global mitochondrial lysine succinylation.
3. Discussion
The results of the current study demonstrate that compromised DLST function due to cysteine succination by fumarate is sufficient to pronouncedly decrease KGDHC activity in the BS and OB of the Ndufs4 KO mouse, the two brain regions with the most profound pathological changes in this model of Leigh syndrome [5]. These important results link an OXPHOS genetic defect to impaired TCA function and defective succinyl-CoA production, a TCA intermediate necessary for SUCLA-mediated SLP [22,34] and succinylation reactions (see schematic in Fig. 6A, [46]). Our findings align with recent metabolomic efforts that suggest that defective glutamate/α-ketoglutarate metabolism distinguishes pathologically affected brain regions from unaffected regions [24,25]. While redox changes modulate TCA cycle activity depending on the energy status of the cell, protein succination represents a static event that contributes to functional insufficiency of the affected proteins, elucidating an unrecognized pathological contributor in Complex I deficiency. The increase in fumarate levels despite significantly reduced KGDHC activity and succinate production likely reflect metabolic rewiring of the TCA cycle under conditions of reductive stress and metabolic acidosis, which limits the oxidative decarboxylation of malate [31] and allows both malate and fumarate accumulate (Fig. 2A and S2D). In contrast, the oxidation of succinate is favored in the Ndufs4 KO to supply electrons for OXPHOS via FADH2 generation, supporting our observations of decreased succinate and increased fumarate and malate.
Previous reports on ATP synthesis levels in the Ndufs4 KO mouse vary depending on the tissue analyzed, with no differences in total cellular ATP content in immortalized fibroblasts (Valsecchi et al., 2012) and skeletal muscle [4,47,48]. However, neither fibroblasts nor skeletal muscle show overt pathology, nor did we find evidence of increased succination in the skeletal muscle of the Ndufs4 KO compared to WT mice [13]. Maximal mitochondrial ATP production in the whole brain is slightly decreased in Ndufs4 KO versus WT mice [49], and CI-linked ATP synthesis was also slightly decreased in isolated permeabilized neurons and astrocytes from mice with a spontaneous mutation leading to disruption of NDUFS4 [50]. We report that both total and SLP-linked ATP synthesis in mitochondria isolated from the OB of the Ndufs4 KO mouse were decreased when α-ketoglutarate, a preferential substrate for SLP, was used. A previous study using brain mitochondria from DLD ± and DLST ± mice showed diminished mitochondrial ATP efflux with fuel combinations that support SLP compared to WT mitochondria, and these mice also had a 20–48 % decrease in KGDHC activity, which is consistent with our findings [22]. Our data provides novel mechanistic insight on the metabolic complexity of CI deficiency, elucidating how the mitochondria suffer from not only defective OXPHOS, but also have inadequate compensatory ATP generation via SLP. Reduced ATP production in the most susceptible neurons would accelerate their functional decline.
The role of KGDHC deficiency in neurodegeneration has been studied in the preclinical setting, including the genetic modulation of DLD and DLST; in both cases the KO mice were not viable [51,52]. DLST ± and DLD ± do not show overt symptoms but are susceptible to mitochondrial toxins used to replicate neurodegenerative diseases [52,53]. Pharmacological inhibition of KGDHC activity in vitro alters mitochondrial morphology and increases fission and mitophagy [54]. Interestingly, the degree of KGDHC inhibition in the brains of Ndufs4 KO mice is aligned with that reported using pharmacological inhibitors of the complex to impair mitochondrial function [54]. Similar to human OGDH mutation [23], human patients with mutations in the LIPT1/2 genes that encode a lipoyltransferase necessary for DLST lipoylation develop Leigh Syndrome-like encephalopathies [55,56], supporting the important role for KGDHC deficiency in driving neuropathology. Brain regions from patients with Alzheimer's disease (AD) show ∼57 % reduced KGDHC activity [57,58]. Of note, the expression of DLST in the human brain appears to be restricted exclusively to neurons [59], supporting the notion that neuronal dysfunction is the triggering event for pathological lesion formation. Interestingly, DLST (E2) also serves as the E2 component of the 2-oxoadipate dehydrogenase complex [60], involved in lysine catabolism, and Ndufs4 KO metabolomics studies have shown evidence of lysine accumulation [25], although dysregulated redox dynamics in the Ndufs4 KO brain may also contribute to impairments in this pathway.
Our current studies elucidate that mitochondrial stress driven by a genetic Complex I deficiency contributes to fumarate-driven reductions in KGDHC activity. Remarkably, neither of the two cysteines in mature DLST are part of the active site, but a recently determined structure demonstrated that Cys178 is in the spatial vicinity of the active site [35], and Cys37 is proximal to the known lipoylation site. We employed both mathematical modeling and site-directed mutagenesis to further interrogate the impact of cysteine conversion to the larger, negatively charged succinocysteine. Modeling Cys178 succination demonstrated the formation of a new ionic interaction between the succinated cysteine and Arg358, displacing the normal Arg358-Asp356 salt bridge. Cys178 succination also altered the orientation of active site His357 and disrupted the H-bond that stabilizes His357 and the loop in which it resides and is predicted to result in less efficient coordination of the CoA and SLAM substrates and increased KM values. These modeling observations strongly support the decreased succinyl CoA mediated succinylation profile and further succinate reductions observed in the Ndufs4 KO brainstem. While our studies focused on the brain regions affected by neuropathology, defective succinyl CoA production could also be anticipated to reduce global heme production, potentially contributing further to the iron dyshomeostasis recently described the Ndufs4 KO mouse [8]. We further employed a succinomimetic mutagenesis strategy to determine if switching cysteine to a larger, negatively charged amino acid (Glu) would alter KGDHC function. Remarkably, mutation of either cysteine had pronounced effects on KGDHC activity, mitochondrial respiratory capacity and DLST lipoylation status. Although the C37E mutation had the greatest impact on respiration, we noted that the striking hyposuccinylation profile observed in the C178E mutant was aligned with the hyposuccinylation observed in the Ndufs4 KO BS, and our modeling data suggested that altered DLST Cys178 succination might impair the ability of the active site to yield succinyl CoA. Since we were unable to define by mass spectrometry which DLST cysteine is succinated, these data provide confidence that loss of either cysteine is sufficient to reduce the function of this central TCA cycle metabolic junction, and succination of Cys178 parallels the metabolic defects observed in the Ndufs4 KO brainstem.
The profound mitochondrial hyposuccinylation profile detected in the brainstem (Fig. 6C) has not been described in mitochondrial encephalopathies, and the additive functional impact of this decrease remains unknown in a model where the Complex I deficit already results in a lower ATP yield. KGDHC can succinylate proteins in vitro more efficiently than free succinyl-CoA, likely due to the succinyl-transferase activity of the E2 component [43]. In agreement with our observations in the Ndufs4 KO, KGDHC inhibitors reduce succinylation of cytosolic and mitochondrial proteins in primary neurons [43]. Mellid et al. have recently described hyposuccinylation due to DLST mutations in heritable human pheochromocytomas and paragangliomas (PPGL), noting marked decreases in succinylation of TCA cycle and glycolytic enzymes that may contribute to the tumorigenic capacity of these cells [61]. Yang et al. have described pronounced hyposuccinylation in the mitochondrial fraction of human Alzheimer's Disease brains, with the maximal decrease in succinylated peptides found on ATP synthase subunits, ATP-synthase subunit d (ATP5H) and ATP-synthase subunit b (ATP5F1) [62], however, the impact on ATP synthase has not yet been studied. Interestingly, some of the largest decreases in succinylation detected by Mellid et al. in the heritable DLST PPGL cells were also in ATP synthase components (ATP5H, ATP5B, ATP5O). A defect in the function of the ATP synthase might further exacerbate the Complex I deficiency, negatively impacting oxidative metabolism.
In contrast to the hyposuccinylation we report, hypersuccinylation has been documented in SUCLA2 deficiency, a mtDNA depletion syndrome [63]. This would be expected in a model where succinyl CoA is the accumulating metabolite versus the deficiency that we detect with reduced KGDHC function. It will be important to define which target proteins are differentially affected in these models as this may contribute to the heterogeneity of the molecular and clinical phenotype of these encephalopathies. While the SUCLA2 insufficiency has some clinical similarity with NDUFS4 deficiency, other features such as sensorineural deafness and mtDNA depletion differ from the CI defect. In contrast to cysteine succination, succinylation is reversible by specific sirtuin (SIRT) enzymes [46]. Overexpression of SIRT5 increased survival and improved mitochondrial oxygen consumption in a zebrafish model of SUCLA2 deficiency, but did not increase mtDNA content, reinforcing that modulation of succinylation has functional effects on OXPHOS parameters [63].
Hyperacetylation of select proteins was also detected in the mitochondria from the Ndufs4 KO brainstem (Fig. 6E). Gella et al. have previously shown increased acetylation in the Ndufs4 KO glutamatergic neurons, with increased acetylated peptides from the ATP synthase complex and pyruvate dehydrogenase (PDH) subunit hyperacetylation. The increase in acetylation corresponded to a significant decrease in PDH activity. Therefore, despite the bidirectional changes in succinylation and acetylation status detected in the Ndufs4 KO brainstem, the net effect suggests an additional insult to oxidative respiration in already compromised neurons.
Hypoxia has been shown to prolong lifespan and reduce brain lesions in the Ndufs4 KO mouse [64,65]. Reduction in the oxygen content in the air normalizes the hyperoxia present in the brain of the Ndufs4 KO mouse to levels found in WT littermates breathing normal air [9]. Interestingly, in vitro studies with N2a neural cells showed that hypoxia leads to increased mitochondrial protein succinylation [66], therefore it will be of interest to determine if hypoxia treatment can alleviate the decreased lysine succinylation observed in the Ndufs4 KO mouse. In addition, dimethyl α-ketoglutarate (DMKG), a cell permeable form of α-ketoglutarate, has been shown to delay neurological symptoms and increase the median lifespan of Ndufs4 KO by ∼40 days [12], and rapamycin treatment also pronouncedly increases α-ketoglutarate levels [24]. Overall increases in α-ketoglutarate would be expected to increase succinyl CoA and confer benefit by providing electrons to support Complex-II dependent respiration in neurons where ∼70 % residual activity is present. Future targeted studies to reduce fumarate accumulation are expected to prevent the sustained damage caused by covalent protein succination during reductive stress, and improve the response to supplementary therapies, such as DMKG, to benefit neuronal survival during Leigh Syndrome.
4. Conclusions
The biochemical mechanisms driving neuropathology in mitochondrial oxidative phosphorylation (OXPHOS) disorders, such as Leigh syndrome, are not well understood beyond the known ATP deficit. Therapeutic approaches to treat mitochondrial diseases employ vitamins and antioxidants, yet these do not significantly slow the rate of neurological decline. In contrast, reductive stress is associated with metabolic acidosis and altered tricarboxylic acid (TCA) cycle metabolism. We demonstrate that fumarate levels are increased in brain regions of the NDUFS4 knockout mouse model of mitochondrial Complex I deficiency. Increased fumarate reacts covalently with protein thiols to generate the chemical modification 2-succinocysteine (protein succination). We report that the specific succination of dihydrolipoyllysine-residue succinyltransferase (DLST) irreversibly reduces the activity of the α-ketoglutarate dehydrogenase complex (KGDHC), confirmed by modeling cysteine succination and succinomimetic DLST mutagenesis. This persistent deficit worsens the mitochondrial OXPHOS derived ATP deficit by limiting compensatory substrate level phosphorylation (SLP) via succinyl CoA ligase. Defective KGDHC activity also reduces succinyl CoA mediated lysine succinylation, a distinct acyl modification, in the affected brain regions of the Ndufs4 knockout mouse. The data mechanistically demonstrates how altered TCA cycle metabolism drives the chemical modification of proteins to disrupt metabolic homeostasis; and provides a novel biochemical basis for neuronal metabolic decline during Complex I OXPHOS deficiency.
Author contributions
G.G.P and N.F. designed the research. G.G.P., A.M.M., R.S.M., H.H.S., W.E.C, M.D.W., O.O., and S.M. performed experiments. G.G.P., R.S.M., A.M.M., H.H.S., W.E.C, M.D.W., O.O., A.A. and N.F. analyzed data. A.C. provided new reagents. G.G.P., A.M.M., and N.F. wrote the paper.
Declaration of competing interest
None.
Acknowledgements
This work was supported by the National Institutes of Health (R01NS126851, R56NS116174, R01NS092938, to N.F), F31DK108559 (to A.M.M)), P20GM109091-S2 and National Science Foundation (1828059) and a University of South Carolina Research Foundation ASPIRE-I awards (to N.F and G.G.P). This study was supported by the Hungarian Scientific Research Fund (OTKA grant 143627, to A.A.) and the Ministry of Innovation and Technology of Hungary (TKP2021-EGA-25 grant, to A.A. (project no. TKP2021-EGA-25 has been implemented with the support provided by the Ministry of Innovation and Technology of Hungary from the National Research, Development, and Innovation Fund, financed under the TKP2021-EGA funding scheme)). Additional support from project PI22/01490 (to A.C) from the Instituto de Salud Carlos III (ISCIII) through the “Acción Estratégica en Salud” (AES), cofounded by the European Regional Development Fund (ERDF).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102932.
MATERIALS AND METHODS
Materials
Unless otherwise stated all reagents were purchased as analytical grade from Millipore Sigma or Fisher Scientific. The preparation of the polyclonal anti-2SC antibody (Ab) and 13C,15N–2SC has been described previously [67]. DLST rabbit Ab was from Bethyl Laboratories (Cat#A304-309A-T). Acetylated-lysine rabbit Ab (Cat#9441), Fumarase rabbit mAb (Cat#4567), GAPDH rabbit mAb (Cat#5174), GFAP mouse mAb (Cat#3670), NeuN rabbit mAb (Cat#24307), Synaptophysin mouse mAb (Cat#12270), SDHA (Cat#11998), α-tubulin mouse mAb antibody (Cat#3873), and βIII-tubulin rabbit mAb (Cat#5666) were obtained from Cell Signaling Technology. Lipoic acid rabbit Ab (Cat#437695) was obtained from Millipore Sigma. VDAC1 mouse mAb was obtained from UC Davis/NIH NeuroMab Facility (Cat#75–204). Actin goat Ab (Cat#sc-1616), MBP mouse mAb (Cat#sc-376995), MOG mouse mAb (Cat#sc-376138) and PDH-E1β (Cat#sc-65243) were obtained from Santa Cruz Biotechnology. OGDH rabbit Ab was obtained from Proteintech (Cat#15212-1-AP). Pan succinyllysine rabbit Ab was obtained from PTM BIO (Cat#PTM-401). Succinic acid-13C4 (CLM1571) and l-lactic acid-13C3, (CLM1579) were purchased from Cambridge Isotope Laboratories.
Mice
Animal care and use procedures were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by The University of South Carolina Animal Care and Use Committee. Ndufs4+/− mice (129/Sv x C57BL/6 mixed background), originally obtained from Dr. Richard Palmiter (University of Washington, Seattle, WA) [4,5], were mated in trios (one male and two females), switched to a breeder's rodent diet (LabDiet 5058) and weaned at 21 days of age. Weaned male and female mice were housed in a pathogen-free animal facility under a 12h light/dark cycle (lights on from 7AM to 7PM) and received Teklad 8904 rodent diet and water ad libitum. Genotyping was performed as previously described [13], and both male and female mice were used. Ndufs4+/+ and Ndufs4−/− mice were sacrificed by decapitation under deep isoflurane anesthesia at 8 weeks of age. The brain was removed from the skull, and the brainstem (BS), olfactory bulbs (OB) and cerebellum were dissected. Other tissues including liver were also collected. For some protocols fresh brain regions were used immediately; on other occasions the tissues were snap frozen in liquid nitrogen and stored at −80 °C until further use.
N1E-115 Cells
N1E-115 neuroblastoma cells (subclone N1E-115-1, Cat#08062511, Sigma Aldrich) were expanded in non-differentiation medium containing 90 % DMEM and 10 % FBS, as previously described [13,37]. At 80 % confluence, the cells were differentiated into neurons in the presence of 2 % FBS and 1.25 % dimethyl sulfoxide in DMEM for 5 days.
H838 Cells
DLST-KO was generated in H838 human lung epithelial adenocarcinoma cells (Cat#CRL-5844, ATCC) by CRISPR-Cas9 technology [38]. The vectors used for the expression of DLST mutants C37E and C178E were obtained through mutagenesis of a previously generated DLST cDNA construct [39] using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) and the following primers Human DLST C37E mutant Fw CAGTTGCAGAAGATGAAGTGGTTGAGGAGATTGAAACTGACAAGACATC, Human DLST C37E mutant Rv GATGTCTTGTCAGTTTCAATCTCCTCAACCACTTCATCTTCTGCAACTG, Human DLST C178E Fw CGTCTGAAGGAGGCCCAGAATACAGAGGCAATGCTGACAACTTTTAATGAG, Human DLST C178E Rv CTCATTAAAAGTTGTCAGCATTGCCTCTGTATTCTGGGCCTCCTTCAGACG. Lentivirus-based constructs (for both the Cys mutants and the WT hDLST) were made according to the standard protocol of the RNAi Consortium from the Broad Institute (http://portals.broadinstitue.org/gpp/public/resources/protocols). Briefly, HEK293T cells were transfected with a lentiviral vector mix including the corresponding DLST cDNA construct and Lipofectamine 2000 (ratio DNA:Lipofectamine was 1:2.5). Media was replaced 16 h after transfection. The virus-containing supernatant was collected 24 h later and added to the H838 DLST-KO cells together with 8 mg/mL of Polybrene. This step was repeated the following day for a second infection. Drug selection started 12 h later, with RPMI-1640 medium containing 10 % FBS and 400 μg/ml G418. Selection proceeded until all uninfected control cells were dead. The cells were then switched to RPMI-1640 medium containing 10 % FBS and 200 μg/ml G418.
Mouse brain homogenates for immunoblotting
Cleared homogenates were prepared from frozen brain regions in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl pH 7.4, with the addition of 150 mM NaCl, 1 mM EDTA, 0.1 % Triton X-100, 0.1 % SDS, 0.5 % sodium deoxycholate, 2 mM diethylenetriaminepentaacetic acid and a protease inhibitor cocktail). Homogenization was performed by pulse sonication at 2 W using a Model 100 sonic dismembrator (Fisher Scientific, Fair Lawn, NJ) for 30 s prior to resting on ice for 30 min. The homogenate was clarified from nuclei and unbroken cells by centrifugation at 900×g for 10 min at 4 °C. Protein in the supernatants was precipitated with 9 vol of cold acetone for 10 min on ice. After centrifugation at 3,000×g for 10 min and removal of the acetone, the protein pellet was resuspended in RIPA buffer. These cleared homogenate fractions were used for detection of succinated proteins (Fig. 1A and G and S1D).
Considering the abundance of tubulin in total brain homogenates, the high level of tubulin succination, and the location of the ∼48–50 kDa succinated band that we describe in this manuscript immediately below tubulin, we followed three different protocols to enrich the ∼48–50 kDa band and at the same time deplete them of tubulin.
First, we subjected mouse BS samples to an in vitro tubulin polymerization protocol that we previously described [18]. Briefly, frozen mouse BS samples were reduced to a powder with a pestle in a mortar containing liquid nitrogen. The pulverized tissue was then resuspended in cold Mes/glutamate buffer (0.1 M Mes, pH 6.8, containing 0.5 mM MgCl2, 1 mM EGTA, 1 M glutamate, 1 mM DTT and a protease inhibitor cocktail), in a volume ratio of 1:1.5 (powder:buffer). The suspension was pulse sonicated for 5 intervals of 10 s. The total protein homogenate was then centrifuged at 30,000×g at 4 °C for 15 min to generate a pellet (P1) that was analyzed by immunoblotting (Fig. 1B), and the supernatant (S1) was subjected to microtubule polymerization by addition of 20 μM taxol and 1 mM GTP (Cytoskeleton Cat#BST06), followed by incubation for 30 min at 37 °C. Following further centrifugation at 30,000×g for 30 min at 37 °C, the supernatant of microtubules (SM) was removed, desalted through Zeba Spin Columns (MW cut-off 7 kDa, Thermo Scientific, 89882S) and analyzed by immunoblotting (Fig. S1B); the microtubular pellet (MP) composed of >95 % tubulin was not used.
Second, we prepared gliosomes and synaptosomes from BS and cerebellum samples according to Carney et al. [68], with minor modifications. Briefly, fresh BS and cerebellum samples were homogenized in 0.32 M sucrose, 1 mM EDTA, 10 mM HEPES pH 7.4 using a glass-teflon homogenizer; the resulting crude homogenates were centrifuged at 1,000×g at 4 °C for 5 min. The supernatants (cleared homogenates) were then loaded on top of a gradient composed of layers containing 3, 7, 10 and 20 % Percoll in the homogenization buffer; a fraction of the cleared homogenate was saved for immunoblotting comparative purposes. The gradients were centrifuged at 33,500×g at 4 °C for 6 min without brakes, and the interfaces between 3 and 7 % Percoll (gliosomes), 10 and 20 % (synaptosomes) and the loose pellet at the bottom of the tubes were collected and washed with the homogenization buffer to remove the Percoll. All the collected fractions and the cleared homogenates were analyzed by immunoblotting (Fig. 1C and S1C).
Third, we prepared purified mitochondrial fractions according to Kayser et al. [69], with minor changes. Briefly, fresh BS samples were homogenized in 225 mM mannitol, 75 mM sucrose, 1 mM EGTA, 5 mM HEPES pH 7.2 containing 1 mg/ml fatty acid-free BSA and a protease inhibitor cocktail using a glass-teflon homogenizer; the resulting crude homogenates were centrifuged at 1,100×g at 4 °C for 2 min. The supernatants (cleared homogenates) were added to Percoll to a 5 % concentration, and then layered on top of 15 % Percoll; a fraction of the cleared homogenate was saved for immunoblotting comparative purposes. The gradients were centrifuged at 18,500×g at 4 °C for 10 min; the top layer, the interface and the lower layer were removed and the loose pellets at the bottom of the tubes were collected and washed with 250 mM sucrose, 0.1 mM EGTA, 5 mM HEPES pH 7.2 to remove the Percoll. The mitochondrial pellets and the cleared homogenate were analyzed by immunoblotting for succinated proteins (Fig. 1F).
Cell Treatments to Increase Protein Succination
N1E-115 cells were treated during the final 16 h of differentiation with 0–100 μM dimethyl fumarate (DMF) prepared in Dulbecco's PBS (DPBS) as a 100X stock and passed through a 0.22 μm pore nitrocellulose filter for sterility; some of the wells received dimethyl succinate (DMS) in the same range of concentrations as a negative control. H838 DLST-KO cells receiving a construct to restore WT hDLST were treated during 16 h with 0–5 mM maleate prepared in DPBS and filtered as described above. The cell collection and subcellular fractionation is described in detail in the section “Measurement of the KGDHC activity in purified bovine heart complex and cells”.
Subcellular Fractionation of Cells for Immunobloting
The subcellular fractionation was performed according to Manuel et al. [70], with minor modifications. H838-DLST KO cells expressing the hDLST WT and cysteine mutants were rinsed twice with PBS and then with homogenization buffer (10 mM Tris-HCl, pH 7.4, containing 0.32 M sucrose, 1 mM EDTA, 1 mM dithiothreitol and a protease inhibitor cocktail). Homogenization buffer was added and cells were gently scraped and homogenized in glass-glass homogenizers, followed by six syringe passes through a 25 G needle. This initial homogenate was centrifuged at 600×g for 10 min at 4 °C in a microcentrifuge, and the supernatant was further centrifuged at 18,000×g for 20 min in a microcentrifuge to isolate mitochondrial enriched pellets; the supernatant was saved as cytosol. These fractions were used for immunoblotting (Fig. 3, Fig. 4A, and S4C)
One-dimensional PAGE and Western Blotting
Western blotting was performed as described previously [13,18,37]. Ten to fifty μg of proteins were run on 12 % gels (Criterion 567–1043, BioRad) and transferred to PVDF membranes. Immunoblotting was performed with antibodies listed in Materials. In most cases, membranes were stripped with 62.5 mM Tris, pH 6.8, containing 2 % SDS and 0.7 % 2-mercaptoethanol for 15 min at 65 °C prior to reprobing. Chemiluminescent signals were captured on photographic film (HyBlot CL).
Two-dimensional Gel Electrophoresis and Western Blotting
Isoelectric focusing on pI 4–7, 11 cm strips and two-dimensional (2D) gel electrophoresis was performed in an Ettan IPGphor II device (Amersham Biosciences) as described previously [13,18]. The 2D gels were transferred onto PVDF membranes to detect protein succination by western blotting; the blots were then stripped and subjected to DLST detection.
Measurement of the α-ketoglutarate dehydrogenase complex activity in brain regions
The activity of the α-ketoglutarate dehydrogenase complex (KGDHC) was measured by following the formation of NADH at 340 nm according to Yang et al. [52], with minor modifications. BS and OB frozen samples were homogenized in 50 mM Tris-HCl pH 7.2 containing 1 mM dithiothreitol, 0.2 mM EGTA, 0.4 % Triton X-100 and a protease inhibitor cocktail using a glass-teflon homogenizer. The crude homogenates were centrifuged at 2,000×g for 4 min at 4 °C; the supernatants were used to determine KGDHC activity. One hundred μl of a reaction mix containing 50 mM Tris-HCl pH 7.8, 1 mM MgCl2, 1 mM CaCl2, 0.5 mM EDTA, 0.3 mM thiamine pyrophosphate, 0.1 % Triton X-100 and 1 mM dithiothreitol were added to the wells of a 96-well plate, followed by 50 μl of an assay mix containing 50 mM Tris-HCl pH 7.8, 3 mM NAD+ and 0.75 mM coenzyme A, and then 25 μl of the 2,000×g supernatants (see above) containing 5–10 μg protein (or homogenization buffer for the blanks). A baseline reading every min for 10 min with the temperature set at 30 °C was taken at 340 nm in a plate reader (Tecan Safire2, TECAN Systems Inc.). After the addition of 25 μl 3 mM α-ketoglutarate in 50 mM Tris-HCl pH 7.8 (or 50 mM Tris-HCl pH 7.8 for “no substrate” controls), further readings were taken for an additional period of 60 min. The readings in the absence of α-ketoglutarate were subtracted from those in the presence of the substrate, and these corrected values were plotted as a function of time to calculate the Vmax from the slopes. Each sample was run in triplicates that were averaged; the n size was 3–4 for each experimental group. The results were converted to mU/mg of protein considering ENAD(340) = 6.22 cm−1mM−1 and a path length of 0.4 cm.
Measurement of the KGDHC activity in purified bovine heart complex and cells
In vitro succination effects on the activity of a commercially available bovine heart KGDHC standard was studied after incubation for 16 h at room temperature with a range of 0–2 mM DMF following the protocol described above for brain regions; dithiothreitol was replaced with 300 μM tris(2-carboxyethyl) phosphine (TCEP) to avoid the possible reaction of DMF with dithiothreitol. The results of the KGDHC activity were expressed as percentage of the control group (no DMF added). The KGDHC activity was also measured in N1E-115 cells treated with DMF or DMS and H838 cells treated with maleate (see “Cell Treatments” above) with minor modifications. Briefly, the medium was removed; the cells were rinsed 3 times with DPBS and gently scraped with 50 mM Tris-HCl pH 7.2 containing 1 mM dithiothreitol, 0.2 mM EGTA, 0.4 % Triton X-100 and a protease inhibitor cocktail. The extracts were homogenized in a glass-glass homogenizer, the resulting homogenates were centrifuged at 2,000×g for 4 min at 4 °C, and the supernatants were used to determine KGDHC activity following the protocol described above for brain regions. The results of the KGDHC activity were expressed as percentage of the control group (no DMF, DMS or maleate added).
Measurement of citrate synthase activity in brain regions
The activity of citrate synthase was measured according to Leek et al. (2001), with minor modifications. Briefly, frozen brain regions were homogenized in 100 mM monobasic/dibasic potassium phosphate pH 7.4, containing 5 mM EDTA, 5 mM EGTA and a protease inhibitor cocktail with a glass-teflon homogenizer (ratio buffer to tissue was 20:1). The initial homogenate was sonicated twice for 20 s to disrupt the mitochondrial membrane, and then centrifuged at 2,000×g for 4 min at 4 °C. The resulting supernatants were then frozen, thawed, and subjected to a new cycle of sonication, freezing and thawing, with a final dilution 1:20 in the homogenization buffer before proceeding to the assay. The measurement was performed by mixing 180 μl of 100 mM Tris pH 8.1, containing 0.1 mM 5.5′-dithio-bis(2-nitrobenzoic acid) (DTNB), 0.2 mM acetyl-coenzyme A and 0.5 mM oxaloacetic acid with 20 μl of the samples (3–6 μg protein); blank wells did not contain oxaloacetic acid. The side reaction between coenzyme A and DTNB, with formation of a mercaptide ion, was followed at 30 °C in a plate reader at 412 nm at 20 s intervals for 20 cycles. The readings in the absence of oxaloacetic acid were subtracted from those in the presence of the substrate, and these corrected values were plotted as a function of time to calculate the Vmax from the slopes. Each sample was run in triplicates that were averaged; the n size was 4 for each experimental group. The results were converted to mU/mg of protein considering Emercaptide ion(412) = 13.6 cm−1mM−1 and a path length of 0.4 cm.
Molecular simulation of Cys178 succination effects on DLST functionality
Molecular dynamics simulations were performed as described previously [35,71], with adaptations as described below. Molecular graphics software Visual Molecular Dynamics (VMD; [72]) and UCSF Chimera [73] were used to model the structure of both the unmodified human DLST (hDLST) and the succinated Cys178 variant; the 6H05 PDB cryo-EM structure was used as a starting point. hDLST trimer coordinate and protein structure files were then generated according to the symmetry operators as well as the atom types and connectivity definitions of the CHARMM36 m force field [74]. The effects of succination on Cys178 were characterized via running an ab initio calculation using the Gaussian 03W software package [75] to carry out a density functional theory modeling at the B3LYP/6-31G++(d,p) level of theory. The obtained results together with the CHARMM36 m force field data of cysteine and glutamic acid were used to set the parameters − force constants, equilibrium bond lengths and angles, partial charges, van der Waals parameters − of the succinated cysteine residue.
Preliminary results indicated that the region before Asn161 has very high flexibility that led to unstable dynamics simulation. This prompted us to model only residues 161–386. The trimeric protein was embedded in an ionized (K+ and Cl−, at 150 mM concentration) water sphere, applying the TIP3P water model. The system was set up in a way that the ionization states of the amino acid side chains mimicked the pH∼7 condition, with the δ-N of the histidine imidazole ring being protonated. The program Nanoscale Molecular Dynamics (NAMD; [76]) was used to run a 50 ns long simulation of the unmodified and succinated hDLST. Energy minimization of the constructed protein was carried out using the conjugate gradient method in 200,000 cycles. The subsequent molecular dynamic simulation run was carried out under Linux using the CUDA-enabled 2.14 version of the NAMD software package. The simulation parameters were set up as follows: (i) in terms of time steps, 1 fs intervals were applied, (ii) calculation of electrostatic interactions (in this spatially limited system) was performed by the particle mesh Ewald method [77] applying a real-space cut-off distance of 12 Å and grid width of 1 Å, (iii) switching distance for non-bonded electrostatics and van der Waals interactions was 10 Å and the scaled 1–4 exclusion scheme was applied, and (iv) final temperature of the system was set to 310.15 K. During the pre-equilibration run for gaining a microcanonical ensemble, the temperature was augmented from 10.15 K to the final temperature in 10 K increments; 2,000 simulation steps were performed at each temperature, except for the final one, where an additional 190,000 steps were also carried out. The main run was controlled via Langevin dynamics under isobaric-isothermal condition; the temperature and pressure were maintained at 310.15 K and 101,325 Pa (1 atm; Langevin piston Nose-Hoover method included in the NAMD package; [76,78]), respectively. The trajectory was sampled between 35.7 and 36.7 ns (stable region highlighted in Fig. S2J) and the average coordinates, calculated by using the VMD smooth trajectory script, were energy minimized in 20,000 cycles. The final succinated structure was analyzed via superimposing to the unmodified monomeric/dimeric/trimeric structure. Interatomic distances and root-mean-square deviations (RMSD) were computed by UCSF Chimera. Structure visualization was performed by the program Pymol.
Substrate level phosphorylation measurements
Mitochondrial isolation and measurement of ATP synthesis was performed according to Komlódi and Tretter [34], with minor modifications. Freshly obtained BS and OB from Ndufs4 KO mice and WT littermates were immediately homogenized in 5 mM Tris-HCl pH 7.4 containing 225 mM mannitol, 75 mM sucrose and 1 mM EGTA using a glass-teflon homogenizer. The initial homogenates were centrifuged at 1,300×g for 3 min, and the pellets were resuspended, washed twice in the homogenization buffer and saved as a nuclear fraction (see below). The supernatants of the 1,300×g centrifugation were further centrifuged at 20,000×g for 10 min. The resultant supernatant was saved as cytosol (see below), and the pellet was resuspended in 15 % Percoll and then layered on top of a discontinuous Percoll gradient consisting of 23 % and 40 % layers. The gradients were then centrifuged at 30,700×g for 8 min using no brake at the end. Immunoblotting experiments determined that the interface between the 23 % and 40 % Percoll and the loose pellet in the 40 % Percoll layer were enriched in mitochondrial markers and devoid of myelin (Fig. S2G). Consequently, these two fractions were pooled together, topped with homogenization buffer and centrifuged for 10 min at 16,600×g; and the pellet was re-suspended in homogenization buffer and subsequently centrifuged again at 6,300×g for 10 min. The resulting purified mitochondrial pellet was re-suspended in 5 mM Tris-HCl pH 7.4 containing 225 mM mannitol and 75 mM sucrose, and immediately used for mitochondrial ATP synthesis measurements. The assay medium consisted of 20 mM HEPES pH 7.0 containing 0.1 mM EGTA, 125 mM KCl, 2 mM K2HPO4, 1 MgCl2, and 0.025 % fatty acid-free bovine serum albumin, with the addition of 3 mM NADP+, 5 mM glucose, 300 μM AP5 (P1,P5-Di(adenosine-5’) pentaphosphate; an inhibitor of adenylate kinase), 0.75 U/ml hexokinase, and 0.25 U/ml glucose-6-phosphate dehydrogenase, in a final volume of 200 μl. ATP formation was detected through the phosphorylation of glucose by added hexokinase, coupled to the oxidation of the resulting glucose 6-phosphate by added glucose 6-phosphate dehydrogenase, with formation of NADPH. NADPH synthesis was followed at 340 nm in a plate reader as described for the measurement of α-ketoglutarate dehydrogenase complex activity (see above). Basal measurements started upon addition of the mitochondrial samples (5 μg/well) and ADP (2 mM) for 10 min at 37 °C, followed by the addition of α-ketoglutarate or succinate (both at 5 mM); some wells also received oligomycin (8 μM) to determine total ATP synthesis (oxidative phosphorylation + substrate level phosphorylation) in the absence of oligomycin, and substrate level phosphorylation in the presence of oligomycin. Corrections in the absence of substrate were applied, and these corrected values were plotted as a function of time to calculate the ATP synthesis from the slopes. Each sample was run in triplicates that were averaged; the n size was 7–12 for each experimental group. The results were converted to nmoles ATP/min/mg of protein using an ATP curve (range: 0–20 nmoles). The purified mitochondrial fractions were also used for immunoblotting experiments (succinylation in Fig. 4C, acetylation in Fig. 4E, succination in Fig. S2H, and detection of protein lipoylation in Fig. S3F). The nuclear fractions obtained after the centrifugation at 1,300×g were used for immunoblotting experiments (succinylation in Fig. S4F), whereas the cytosols obtained after the centrifugation at 20,000×g were used for immunoblotting experiments (succinylation in Fig. S4D).
Metabolic Measurements with the Seahorse Extracellular Flux Analyzer
H838 cells were seeded on V7 Seahorse cell culture microplates at a density of 30,000 cells/well. After 2 days in culture, two different experiments were performed with these cells.
Experiment 1 was a mitochondrial stress assay designed to test the effect of DLST cysteine mutations on mitochondrial functionality. The cells were switched to XF Basal Medium supplemented with 10 mM glucose and 2 mM glutamine (same concentrations as in the RPMI medium used to grow the cells), and the oxygen consumption rate (OCR) was measured with a Seahorse XFe24 Extracellular Flux Analyzer in basal conditions (no drugs injected), and after injections of oligomycin (2 μM), carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, 0.25 μM), and rotenone/antimycin A (3 μM/4 μM) to determine ATP production, proton leak, maximal respiration, spare respiratory capacity and non-mitochondrial respiration, as previously described [37].
Experiment 2 was an ATP production rate assay designed to test the contribution of glycolysis and mitochondria to the total ATP synthesis in cells expressing WT hDLST when treated for 16 h with 0–5 mM maleate to increase protein succination. The cells were switched to XF DMEM supplemented with 10 mM glucose, 2 mM glutamine and 1 mM sodium pyruvate. The extracellular acidification rate (ECAR) and the OCR were then measured in basal conditions and after sequential injections of oligomycin (1.5 μM), and rotenone/antimycin A (0.5 μM for both drugs) to determine the relative contribution of glycolysis and mitochondrial metabolism to ATP synthesis, and the total amount of ATP produced.
For both experiments, the wells washed 3 times with cold PBS after completion of the assay and the plates were stored at −70 °C prior to the measurement of the total protein content to normalize individual measurements.
Determination of Protein Content
The protein content in all the brain and cell homogenates, including samples for immunoblotting and those for the determinations of enzymatic activity was determined by the method of Lowry et al. [79].
Metabolite Quantification
TCA cycle metabolite quantification was performed in an adaptation of previous methods [70,80]. Metabolites were extracted by adding 1 ml ice-cold 80 % methanol to snap-frozen olfactory bulbs. The tissue was dissociated in pre-cooled glass-teflon homogenizers and transferred to 1.5 ml Eppendorf tubes. Homogenized samples were briefly pulse-sonicated and frozen for 15 min at −70 °C. After thawing, the samples were centrifuged at 15,000×g for 10 min at 4 °C. The supernatants were transferred to new Eppendorf tubes and the extracts were dried by centrifugal evaporation. The protein pellet was resuspended in RIPA buffer for quantification by the Lowry method.
The quantification of L-2-hydroxyglutarate by LC-MS was performed following chiral derivatization with diacetyl-L-tartaric anhydride (DATAN). 50 μM L-2-hydroxyglutarate-13C5 internal standard was added to tissues prior to extraction, and to all racemic standards. Dried tissue metabolite extracts and standards were resuspended in 50 μl DATAN prepared in acetonitrile:acetic acid (4:1, v/v) to 50 mg/ml and heated to 70 °C for 2 h. The samples were cooled to room temperature and centrifuged at 1000×g for 5 min. The derivatized samples were further diluted to 100 μl with acetonitrile. LC separation was performed on Thermo Vanquish Flex liquid chromatograph using a PEEK coated SeQuant ZIC HILIC column (Merck Millipore). The column was 2.1 mm × 150 mm with 3.5 μm particles and was preceded by an equivalent 20 mm long guard column. One μl of sample was separated by isocratic elution with 15 % mobile phase A (10 % 200 mM formic acid:90 % LC-MS grade water) and 85 % mobile phase B (10 % 200 mM formic acid:90 % acetonitrile) at a total flow rate of 200 μl/min. The formic acid was titrated to pH 3.25 with ammonium hydroxide. Negative ion electrospray mass spectra were acquired on a Thermo Q-Exactive HF-X quadrupole-Orbitrap in MS/MS mode with the Orbitrap resolution set at 7,500. Precursor ions were isolated by the quadrupoles and fragmented in the HCD cell at 10 ev. The product ions were mass analyzed in the Orbitrap. XCalibur 4.3 software was used to construct chromatograms of the transitions 363 > 147 (L-2-HG) and 368 > 152 (L-2-HG-13C5). The peak areas were normalized to the areas of the 13C internal standard and the analyte mass was normalized to the tissue weights. The analysis of brainstem protein-bound 2SC was performed following protein hydrolysis in 6 M HCl at 110 °C for 18hrs, 10 pmol 13C3,15N–2SC was added to all samples as an internal standard. After drying, solid phase extraction on 1 mL C18 columns (Waters) was performed [67] and the extracted amino acids were dried in vacuo before derivatization. Ethanol/acetyl chloride (100:20 v/v) was mixed on ice and 25 μL was added to each tube. The tubes were vortexed thoroughly and incubated at 55 °C for 2 h before being dried in vacuo, in an adaptation of the method described by Ref. [81]. The reaction products were resuspended in acetonitrile/water (20:80 v/v) before separation by reverse-phase HPLC and positive ion mode LC-MS/MS analysis. The transitions 322 → 205 (2SC), 326 → 205 (13C3,15N–2SC) were monitored and the peak areas were normalized to the areas of the 13C internal standard, and the analyte mass was calculated from a standard curve and normalized to the tissue weights.
The quantification of TCA cycle intermediates was performed by GC-MS at the David H. Murdock Research Institute (DHMRI, Kannapolis, NC). Prior to derivatization the extracts were resuspended in ethyl acetate and transferred to GC-MS vials. The samples were dried with N2, and an internal standard (100 μM succinate-13C4) was added to each of the samples and standards. The samples and standards were derivatized with 200 μl of methylamine (20 mg/ml in pyridine) at 30 °C for 90 min, followed by drying under N2. This was followed by the addition of 120 μl of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) with 1 % trimethylchlorosilane (TMCS); the mixture was incubated at 70 °C for 60 min. The derivatized product was stored in a −20 °C freezer for 1 h prior to GC/MS analysis. An Agilent 7890A GC system, coupled to an Agilent 5975C electron ionization (EI) mass selective detector (MSD) was used to analyze the TMS-derivatized samples. Select ion monitoring (SIM) was performed and the peak areas obtained were normalized to the internal standard. Absolute quantitation for all metabolites was performed based on standard curves obtained from the normalized reference standards, and the final metabolite concentrations were normalized to tissue weights or total protein content.
Protein identification by liquid chromatography-tandem mass spectrometry (LC-MS/MS)
The in-gel protein digestion method used was previously described [13,37]. Briefly, samples of the pellets obtained from the synaptosomal-enriched preparations (20 μg of protein) were resolved by SDS-PAGE in 7.5 % gels; the gels were stained with Coomassie Brilliant Blue and the bands of interest in the ∼48–50 kDa region were excised. The gel pieces were destained, washed with 50 mM ammonium bicarbonate in 50 % acetonitrile, and dehydrated with 100 % acetonitrile; proteins were then reduced with 10 mM dithiothreitol and alkylated with 170 mM 4-vinylpyridine. Protein digestion was carried out overnight at 37 °C in the presence of 500 ng sequencing grade modified trypsin in 50 mM ammonium bicarbonate. After gel extraction with 5 % formic acid in 50 % acetonitrile, the samples were analyzed in a blinded manner on a Dionex Ultimate 3000-LC system (Thermo Scientific) coupled to a Velos Pro Orbitrap mass spectrometer (Thermo Scientific). The LC solvents were 2 % acetonitrile/0.1 % formic acid (Solvent A) and 80 % acetonitrile/0.1 % formic acid (Solvent B); the water used for these solvents was LC-MS grade. Peptides were first trapped on a 2 cm Acclaim PepMap-100 column (Thermo Scientific) with Solvent A at 3 μl/min. At 4 min the trap column was placed in line with the analytical column, a 75 μm C18 stationary-phase LC PicoChip Nanospray column (New Objective). The peptides were eluted with a gradient from 98%A:2%B to 40%A:60%B over 30 min, followed by a 5 min ramp to 10%A:90%B that was held for 10 min. The Orbitrap was operated in data-dependent acquisition MS/MS analysis mode and excluded all ions below 200 counts. Following a survey scan (MS1), up to 8 precursor ions were selected for MS/MS analysis. All spectra were obtained in the Orbitrap at 7500 resolution. The DDA data were analyzed using Proteome Discover 1.4 software with SEQUEST algorithm against the uniprot_ref_mouse database (2014-10-03 version, 52,474 proteins) with XCorr validation >2 (+2) or >2.5 (+3). An allowance was made for 2 missed cleavages following trypsin digestion. No fixed modifications were considered. The variable modifications of methionine oxidation (MOX), proline hydroxylation (POX), cysteine pyridylethylation (CPE, 105.058) or cysteine succination by fumarate (C2SC, 116.011) were considered with a mass tolerance of 15 ppm for precursor ions and a mass tolerance of 10 ppm for fragment ions. The results were filtered with a false discovery rate of 0.01. For all DLST peptides identified the spectra were manually inspected to confirm the abundance of product ions. The software Scaffold 4D was used to generate the spectrum in Fig. 1E.
Quantification and Statistical Analysis
Results were expressed as means ± standard error of the mean (SEM). For the enzymatic activities, a description of the calculations is presented in the experimental method details. Statistical comparisons between two experimental groups were performed using unpaired t tests; when three or more groups were compared, one-way ANOVA followed by Student-Neuman-Keuls’ test was applied. Differences were considered statistically significant when a P value < 0.05 was achieved. The n size for each experiment is shown in the Figures and represents the number of individual biological replicates, typically n = 5/group. Variations in group sizes are due to the variable KO mouse numbers in breeding cohorts, and fresh brain tissue regions were required for enzymatic and mitochondrial assays. All data points are shown, no outliers were considered or removed. For Fig. 2C, n = 4 mice were used, but a genotyping error was discovered for 1 KO mouse after it was used for the assay, this data point was removed leading to n = 3 for the KO group. The software Image J (NIH) was used for the quantification of band intensities by densitometry [82]. Uncropped blots of total succination profiles were not quantified in this manner due to the abundance of succinated tubulin and other intensely modified bands that saturate the pixels and obscure discrete changes when quantified. Relevant proteins at a lower intensity were quantified, such as DLST, succinated DLST, protein lipolylation of DLST and DLAT, mitochondrial markers and select succinylated bands. Blots were cropped for single protein blots, uncropped blots are shown for total succination and succinylation profiles. The software GraphPad Prism V9.1.1 was used for statistical analysis and graphs.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Lake N.J., Compton A.G., Rahman S., Thorburn D.R. Leigh syndrome: one disorder, more than 75 monogenic causes. Ann. Neurol. 2016;79:190–203. doi: 10.1002/ana.24551. [DOI] [PubMed] [Google Scholar]
- 2.Calvaruso M.A., Willems P., van den Brand M., Valsecchi F., Kruse S., Palmiter R., Smeitink J., Nijtmans L. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet. 2011;21:115–120. doi: 10.1093/hmg/ddr446. [DOI] [PubMed] [Google Scholar]
- 3.Ortigoza-Escobar J.D., Oyarzabal A., Montero R., Artuch R., Jou C., Jiménez C., Gort L., Briones P., Muchart J., López-Gallardo E., et al. Ndufs4 related Leigh syndrome: a case report and review of the literature. Mitochondrion. 2016;28:73–78. doi: 10.1016/j.mito.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Kruse S.E., Watt W.C., Marcinek D.J., Kapur R.P., Schenkman K.A., Palmiter R.D. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metabol. 2008;7:312–320. doi: 10.1016/j.cmet.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quintana A., Kruse S.E., Kapur R.P., Sanz E., Palmiter R.D. Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10996–11001. doi: 10.1073/pnas.1006214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolea I., Gella A., Sanz E., Prada-Dacasa P., Menardy F., Bard A.M., Machuca-Márquez P., Eraso-Pichot A., Mòdol-Caballero G., Navarro X., et al. Defined neuronal populations drive fatal phenotype in a mouse model of Leigh syndrome. Elife. 2019;8 doi: 10.7554/eLife.47163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haas R., Das D., Garanto A., Renkema H.G., Greupink R., van den Broek P., Pertijs J., Collin R.W.J., Willems P., Beyrath J., et al. Therapeutic effects of the mitochondrial ROS-redox modulator KH176 in a mammalian model of Leigh Disease. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-09417-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly C.J., Couch R.K., Ha V.T., Bodart C.M., Wu J., Huff S., Herrel N.T., Kim H.D., Zimmermann A.O., Shattuck J., Pan Y.C., Kaeberlein M., Grillo A.S. Iron status influences mitochondrial disease progression in Complex I-deficient mice. Elife. 2023;12 doi: 10.7554/eLife.75825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain I.H., Zazzeron L., Goldberger O., Marutani E., Wojtkiewicz G.R., Ast T., Wang H., Schleifer G., Stepanova A., Brepoels K., et al. Leigh Syndrome mouse model can be rescued by interventions that normalize brain hyperoxia, but not HIF activation. Cell Metabol. 2019;30:824–832. doi: 10.1016/j.cmet.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Titov D.V., Cracan V., Goodman R.P., Peng J., Grabarek Z., Mootha V.K. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science. 2016;352:231–235. doi: 10.1126/science.aad4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma R., Reinstadler B., Engelstad K., Skinner O.S., Stackowitz E., Haller R.G., Clish C.B., Pierce K., Walker M.A., Fryer R., Oglesbee D., Mao X., Shungu D.C., Khatri A., Hirano M., De Vivo D.C., Mootha V.K. Circulating markers of NADH-reductive stress correlate with mitochondrial disease severity. J. Clin. Invest. 2021 doi: 10.1172/JCI136055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C.F., Caudal A., Abell L., Nagana Gowda G.A., Tian R. Targeting NAD+ metabolism as interventions for mitochondrial disease. Sci. Rep. 2019;9:3073. doi: 10.1038/s41598-019-39419-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piroli G.G., Manuel A.M., Clapper A.C., Walla M.D., Baatz J.E., Palmiter R.D., Quintana A., Frizzell N. Succination is increased on select proteins in the brainstem of the NADH dehydrogenase (ubiquinone) Fe-S protein 4 (Ndufs4) knockout mouse, a model of Leigh Syndrome. Mol. Cell. Proteomics. 2016;15:445–461. doi: 10.1074/mcp.M115.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alderson N.L., Wang Y., Blatnik M., Frizzell N., Walla M.D., Lyons T.J., Alt N., Carson J.A., Nagai R., Thorpe S.R., et al. S-(2-Succinyl)cysteine: a novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch. Biochem. Biophys. 2006;450:1–8. doi: 10.1016/j.abb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Frizzell N., Rajesh M., Jepson M.J., Nagai R., Carson J.A., Thorpe S.R., Baynes J.W. Succination of thiol groups in adipose tissue proteins in diabetes: succination inhibits polymerization and secretion of adiponectin. J. Biol. Chem. 2009;284:25772–25781. doi: 10.1074/jbc.M109.019257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merkley E.D., Metz T.O., Smith R.D., Baynes J.W., Frizzell N. The succinated proteome. Mass Spectrom. Rev. 2014;33:98–109. doi: 10.1002/mas.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manuel A.M., Walla M.D., Faccenda A., Martin S.L., Tanis R.M., Piroli G.G., Adam J., Kantor B., Mutus B., Townsend D.M., et al. Succination of protein disulfide isomerase links mitochondrial stress and endoplasmic reticulum stress in the adipocyte during diabetes. Antioxidants Redox Signal. 2017;27:1281–1296. doi: 10.1089/ars.2016.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piroli G.G., Jepson M.J., Manuel A.M., Walla M.D., Brock J.W.C., Rajesh M.P., Tanis R.M., Cotham W.E., Frizzell N. Identification of protein succination as a novel modification of tubulin. Biochem. J. 2014;462:231–245. doi: 10.1042/BJ20131581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ternette N., Yang M., Laroyia M., Kitagawa M., O'Flaherty L., Wolhulter K., Igarashi K., Saito K., Kato K., Fischer R., et al. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep. 2013;3:689–700. doi: 10.1016/j.celrep.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkarni R.A., Bak D.W., Wei D., Bergholtz S.E., Briney C.A., Shrimp J.H., Alpsoy A., Thorpe A.L., Bavari A.E., Crooks D.R., et al. A chemoproteomic portrait of the oncometabolite fumarate. Nat. Chem. Biol. 2019;15:391–400. doi: 10.1038/s41589-018-0217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheu K.F.R., Blass J.P. The α-ketoglutarate dehydrogenase complex. Ann. N. Y. Acad. Sci. 1999;893:61–78. doi: 10.1111/j.1749-6632.1999.tb07818.x. [DOI] [PubMed] [Google Scholar]
- 22.Kiss G., Konrad C., Doczi J., Starkov A.A., Kawamata H., Manfredi G., Zhang S.F., Gibson G.E., Beal M.F., Adam-Vizi V., et al. The negative impact of α-ketoglutarate dehydrogenase complex deficiency on matrix substrate-level phosphorylation. Faseb. J. 2013;27:2392–2406. doi: 10.1096/fj.12-220202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap Z.Y., Strucinska K., Matsuzaki S., Lee S., Si Y., Humphries K., Tarnopolsky M.A., Yoon W.H. A biallelic pathogenic variant in the OGDH gene results in a neurological disorder with features of a mitochondrial disease. J. Inherit. Metab. Dis. 2021;44:388–400. doi: 10.1002/jimd.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson S.C., Kayser E.B., Bornstein R., Stokes J., Bitto A., Park K.Y., Pan A., Sun G., Raftery D., Kaeberlein M., Sedensky M.M., Morgan P.G. Regional metabolic signatures in the Ndufs4(KO) mouse brain implicate defective glutamate/α-ketoglutarate metabolism in mitochondrial disease. Mol. Genet. Metabol. 2020;130:118–132. doi: 10.1016/j.ymgme.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terburgh K., Coetzer J., Lindeque Z., van der Westhuizen F.H., Louw R. Aberrant BCAA and glutamate metabolism linked to regional neurodegeneration in a mouse model of Leigh syndrome. Biochim. Biophys. Acta, Mol. Basis Dis. 2021;1867 doi: 10.1016/j.bbadis.2021.166082. [DOI] [PubMed] [Google Scholar]
- 26.Reed L.J., Oliver R.M. Structure-function relationships in pyruvate and α-ketoglutarate dehydrogenase complexes. Adv. Exp. Med. Biol. 1982;148:231–241. doi: 10.1007/978-1-4615-9281-5_19. [DOI] [PubMed] [Google Scholar]
- 27.Ambrus A., Tretter L., Adam-Vizi V. Inhibition of the alpha-ketoglutarate dehydrogenase-mediated reactive oxygen species generation by lipoic acid. J. Neurochem. 2009;109:222–229. doi: 10.1111/j.1471-4159.2009.05942.x. [DOI] [PubMed] [Google Scholar]
- 28.Chinopoulos C., Tretter L., Adam-Vizi V. Depolarization of in situ mitochondria due to hydrogen peroxide-induced oxidative stress in nerve terminals: inhibition of alpha-ketoglutarate dehydrogenase. J. Neurochem. 1999;73:220–228. doi: 10.1046/j.1471-4159.1999.0730220.x. [DOI] [PubMed] [Google Scholar]
- 29.Humphries K.M., Szweda L.I. Selective inactivation of α-ketoglutarate dehydrogenase and pyruvate dehydrogenase: reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry. 1998;37:15835–15841. doi: 10.1021/bi981512h. [DOI] [PubMed] [Google Scholar]
- 30.Starkov A.A., Fiskum G., Chinopoulos C., Lorenzo B.J., Browne S.E., Patel M.S., Beal M.F. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J. Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadtochiy S.M., Schafer X., Fu D., Nehrke K., Munger J., Brookes P.S. Acidic pH is a metabolic switch for 2-hydroxyglutarate generation and signaling. J. Biol. Chem. 2016;291:20188–20197. doi: 10.1074/jbc.M116.738799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terburgh K., Lindeque Z., van der Westhuizen F.H., Louw R. Cross-comparison of systemic and tissue-specific metabolomes in a mouse model of Leigh syndrome. Metabolomics. 2021;17:101. doi: 10.1007/s11306-021-01854-8. [DOI] [PubMed] [Google Scholar]
- 33.Lambeth D.O., Tews K.N., Adkins S., Frohlich D., Milavetz B.I. Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues. J. Biol. Chem. 2004;279:36621–36624. doi: 10.1074/jbc.M406884200. [DOI] [PubMed] [Google Scholar]
- 34.Komlódi T., Tretter L. Methylene blue stimulates substrate-level phosphorylation catalyzed by succinyl-CoA ligase in the citric acid cycle. Neuropharmacology. 2017;123:287–298. doi: 10.1016/j.neuropharm.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Nagy B., Polak M., Ozohanics O., Zambo Z., Szabo E., Hubert A., Jordan F., Novaček J., Adam-Vizi V., Ambrus A. Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: implications for the overall hKGDHc structure. Biochim. Biophys. Acta Gen. Subj. 2021;1865 doi: 10.1016/j.bbagen.2021.129889. [DOI] [PubMed] [Google Scholar]
- 36.Bergholtz S.E., Briney C.A., Najera S.S., Perez M., Linehan W.M., Meier J.L. An Oncometabolite Isomer Rapidly Induces a Pathophysiological Protein Modification. ACS Chem Biol. 2020;15:856–861. doi: 10.1021/acschembio.0c00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piroli G.G., Manuel A.M., Patel T., Walla M.D., Shi L., Lanci S.A., Wang J., Galloway A., Ortinski P.I., Smith D.S., et al. Identification of novel protein targets of dimethyl fumarate modification in neurons and astrocytes reveals actions independent of Nrf2 stabilization. Mol. Cell. Proteomics. 2019;18:504–519. doi: 10.1074/mcp.RA118.000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen E.L., Ulanet D.B., Pirman D., Mahoney C.E., Coco J., Si Y., Chen Y., Huang L., Ren J., Choe S., et al. Differential aspartate usage identifies a subset of cancer cells particularly dependent on OGDH. Cell Rep. 2016;17:876–890. doi: 10.1016/j.celrep.2016.09.052. [DOI] [PubMed] [Google Scholar]
- 39.Remacha L., Pirman D., Mahoney C.E., Coloma J., Calsina B., Currás-Freixes M., Letón R., Torres-Pérez R., Richter S., Pita G., et al. Recurrent germline DLST mutations in individuals with multiple pheochromocytomas and paragangliomas. Am. J. Hum. Genet. 2019;104:1008–1010. doi: 10.1016/j.ajhg.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey P.S.J., Ortmann B.M., Martinelli A.W., Houghton J.W., Costa A.S.H., Burr S.P., Antrobus R., Frezza C., Nathan J.A. ABHD11 maintains 2-oxoglutarate metabolism by preserving functional lipoylation of the 2-oxoglutarate dehydrogenase complex. Nat. Commun. 2020;11:4046. doi: 10.1038/s41467-020-17862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaubel R.A., Rustin P., Isaya G. Mutations in the dimer interface of dihydrolipoamide dehydrogenase promote site-specific oxidative damages in yeast and human cells. J. Biol. Chem. 2011;286:40232–40245. doi: 10.1074/jbc.M111.274415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson G.E., Xu H., Chen H.L., Chen W., Denton T.T., Zhang S. Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. J. Neurochem. 2015;134:86–96. doi: 10.1111/jnc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J., Chen Y., Tishkoff D.X., Peng C., Tan M., Dai L., Xie Z., Zhang Y., Zwaans B.M.M., Skinner M.E., Lombard D.B., Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gella A., Prada-Dacasa P., Carrascal M., Urpi A., González-Torres M., Abian J., Sanz E., Quintana A. Mitochondrial proteome of affected glutamatergic neurons in a mouse model of Leigh syndrome. Front. Cell Dev. Biol. 2020;8:660. doi: 10.3389/fcell.2020.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y., Gibson G.E. Succinylation links metabolism to protein functions. Neurochem. Res. 2019;44:2346–2359. doi: 10.1007/s11064-019-02780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alam M.T., Manjeri G.R., Rodenburg R.J., Smeitink J.A., Notebaart R.A., Huynen M., Willems P.H., Koopman W.J. Skeletal muscle mitochondria of NDUFS4-/- mice display normal maximal pyruvate oxidation and ATP production. Biochim. Biophys. Acta. 2015;1847:526–533. doi: 10.1016/j.bbabio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Terburgh K., Lindeque Z., Mason S., van der Westhuizen F., Louw R. Metabolomics of Ndufs4-/- skeletal muscle: adaptive mechanisms converge at the ubiquinone-cycle. Biochim. Biophys. Acta, Mol. Basis Dis. 2019;1865:98–106. doi: 10.1016/j.bbadis.2018.10.034. [DOI] [PubMed] [Google Scholar]
- 49.Manjeri G.R., Rodenburg R.J., Blanchet L., Roelofs S., Nijtmans L.G., Smeitink J.A., Driessen J.J., Koopman W.J., Willems P.H. Increased mitochondrial ATP production capacity in brain of healthy mice and a mouse model of isolated complex I deficiency after isoflurane anesthesia. J. Inherit. Metab. Dis. 2016;39:59–65. doi: 10.1007/s10545-015-9885-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bird M.J., Wijeyeratne X.W., Komen J.C., Laskowski A., Ryan M.T., Thorburn D.R., Frazier A.E. Neuronal and astrocyte dysfunction diverges from embryonic fibroblasts in the Ndufs4fky/fky mouse. Biosci. Rep. 2014;34 doi: 10.1042/BSR20140151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson M.T., Yang H.S., Magnuson T., Patel M.S. Targeted disruption of the murine dihydrolipoamide dehydrogenase gene (Dld) results in perigastrulation lethality. Proc. Natl. Acad. Sci. U.S.A. 1997;94:14512–14517. doi: 10.1073/pnas.94.26.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L., Shi Q., Ho D.J., Starkov A.A., Wille E.J., Xu H., Chen H.L., Zhang S., Stack C.M., Calingasan N.Y., et al. Mice deficient in dihydrolipoyl succinyl transferase show increased vulnerability to mitochondrial toxins. Neurobiol. Dis. 2009;36:320–330. doi: 10.1016/j.nbd.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klivenyi P., Starkov A.A., Calingasan N.Y., Gardian G., Browne S.E., Yang L., Bubber P., Gibson G.E., Patel M.S., Beal M.F. Mice deficient in dihydrolipoamide dehydrogenase show increased vulnerability to MPTP, malonate and 3-nitropropionic acid neurotoxicity. J. Neurochem. 2004;88:1352–1360. doi: 10.1046/j.1471-4159.2003.02263.x. [DOI] [PubMed] [Google Scholar]
- 54.Banerjee K., Munshi S., Xu H., Frank D.E., Chen H.L., Chu C.T., Yang J., Cho S., Kagan V.E., Denton T.T., et al. Mild mitochondrial metabolic deficits by α-ketoglutarate dehydrogenase inhibition cause prominent changes in intracellular autophagic signaling: potential role in the pathobiology of Alzheimer's disease. Neurochem. Int. 2016;96:32–45. doi: 10.1016/j.neuint.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Habarou F., Hamel Y., Haack T.B., Feichtinger R.G., Lebigot E., Marquardt I., Busiah K., Laroche C., Madrange M., Grisel C., et al. Biallelic mutations in LIPT2 cause a mitochondrial lipoylation defect associated with severe neonatal encephalopathy. Am. J. Hum. Genet. 2017;101:283–290. doi: 10.1016/j.ajhg.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stowe R.C., Sun Q., Elsea S.H., Scaglia F. LIPT1 deficiency presenting as early infantile epileptic encephalopathy, Leigh disease, and secondary pyruvate dehydrogenase complex deficiency. Am. J. Med. Genet. A. 2018;176:1184–1189. doi: 10.1002/ajmg.a.38654. [DOI] [PubMed] [Google Scholar]
- 57.Butterworth R.F., Besnard A.M. Thiamine-dependent enzyme changes in temporal cortex of patients with Alzheimer's disease. Metab. Brain Dis. 1990;5:179–184. doi: 10.1007/BF00997071. [DOI] [PubMed] [Google Scholar]
- 58.Gibson G.E., Sheu K.F.R., Blass J.P., Baker A., Carlson K.C., Harding B., Perrino P. Reduced activities of thiamine-dependent enzymes in the brains and peripheral tissues of patients with Alzheimer's disease. Arch. Neurol. 1988;45:836–840. doi: 10.1001/archneur.1988.00520320022009. [DOI] [PubMed] [Google Scholar]
- 59.Dobolyi A., Bago A., Palkovits M., Nemeria N.S., Jordan F., Doczi J., Ambrus A., Adam-Vizi V., Chinopoulos C. Exclusive neuronal detection of KGDHC-specific subunits in the adult human brain cortex despite pancellular protein lysine succinylation. Brain Struct. Funct. 2020;225:639–667. doi: 10.1007/s00429-020-02026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nemeria N.S., Gerfen G., Nareddy P.R., Yang L., Zhang X., Szostak M., Jordan F. The mitochondrial 2-oxoadipate and 2-oxoglutarate dehydrogenase complexes share their E2 and E3 components for their function and both generate reactive oxygen species. Free Radic. Biol. Med. 2018;115:136–145. doi: 10.1016/j.freeradbiomed.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 61.Mellid S., García F., Leandro-García L.J., Díaz-Talavera A., Martínez-Montes Á.M., Gil E., Calsina B., Monteagudo M., Letón R., Roldán-Romero J.M., Santos M., Lanillos J., Valdivia C., Martínez-Puente N., de Nicolás-Hernández J., Jiménez S., Pérez-Martínez M., Honrado E., Coloma J., Cerezo A., Santiveri C.M., Esteller M., Campos-Olivas R., Caleiras E., Montero-Conde C., Rodríguez-Antona C., Muñoz J., Robledo M., Cascón A. DLST mutations in pheochromocytoma and paraganglioma cause proteome hyposuccinylation and metabolic remodeling. Cancer Commun (Lond) May 4. 2023 doi: 10.1002/cac2.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y., Tapias V., Acosta D., Xu H., Chen H., Bhawal R., Anderson E.T., Ivanova E., Lin H., Sagdullaev B.T., Chen J., Klein W.L., Viola K.L., Gandy S., Haroutunian V., Beal M.F., Eliezer D., Zhang S., Gibson G.E. Altered succinylation of mitochondrial proteins, APP and tau in Alzheimer's disease. Nat. Commun. 2022;13:159. doi: 10.1038/s41467-021-27572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gut P., Matilainen S., Meyer J.G., Pällijeff P., Richard J., Carroll C.J., Euro L., Jackson C.B., Isohanni P., Minassian B.A., et al. SUCLA2 mutations cause global protein succinylation contributing to the pathomechanism of a hereditary mitochondrial disease. Nat. Commun. 2020;11:5927. doi: 10.1038/s41467-020-19743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jain I.H., Zazzeron L., Goli R., Alexa K., Schatzman-Bone S., Dhillon H., Goldberger O., Peng J., Shalem O., Sanjana N.E., et al. Hypoxia as a therapy for mitochondrial disease. Science. 2016;352:54–61. doi: 10.1126/science.aad9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferrari M., Jain I.H., Goldberger O., Rezoagli E., Thoonen R., Cheng K.H., Sosnovik D.E., Scherrer-Crosbie M., Mootha V.K., Zapol W.M. Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E4241–E4250. doi: 10.1073/pnas.1621511114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen H., Xu H., Potash S., Starkov A., Belousov V.V., Bilan D.S., Denton T.T., Gibson G.E. Mild metabolic perturbations alter succinylation of mitochondrial proteins. J. Neurosci. Res. 2017;95:2244–2252. doi: 10.1002/jnr.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagai R., Brock J.W., Blatnik M., Baatz J.E., Bethard J., Walla M.D., Thorpe S.R., Baynes J.W., Frizzell N. Succination of protein thiols during adipocyte maturation: a biomarker of mitochondrial stress. J. Biol. Chem. 2007;282:34219–34228. doi: 10.1074/jbc.M703551200. [DOI] [PubMed] [Google Scholar]
- 68.Carney K.E., Milanese M., van Nierop P., Li K.W., Oliet S.H., Smit A.B., Bonanno G., Verheijen M.H. Proteomic analysis of gliosomes from mouse brain: identification and investigation of glial membrane proteins. J. Proteome Res. 2014;13:5918–5927. doi: 10.1021/pr500829z. [DOI] [PubMed] [Google Scholar]
- 69.Kayser E.B., Sedensky M.M., Morgan P.G. Region-specific defects of respiratory capacities in the Ndufs4(KO) mouse brain. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manuel A.M., Walla M.D., Dorn M.T., Tanis R.M., Piroli G.G., Frizzell N. Fumarate and oxidative stress synergize to promote stability of C/EBP homologous protein in the adipocyte. Free Radic. Biol. Med. 2020;14:70–82. doi: 10.1016/j.freeradbiomed.2019.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ambrus A., Mizsei R., Adam-Vizi V. Structural alterations by five disease-causing mutations in the low-pH conformation of human dihydrolipoamide dehydrogenase (hLADH) analyzed by molecular dynamics - implications in functional loss and modulation of reactive oxygen species generation by pathogenic hLADH forms. Biochem. Biophys. Rep. 2015;2:50–56. doi: 10.1016/j.bbrep.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 73.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 74.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmuller H., MacKerell A.D. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frisch M., Trucks G., Schlegel H., Scuseria G., Robb M., Cheeseman J., Montgomery J., Jr., Vreven T., Kudin K., Burant J.J.I. Wallingford, CT; 2004. Gaussian 03, Revision C. 02. [Google Scholar]
- 76.Phillips J.C., Hardy D.J., Maia J.D.C., Stone J.E., Ribeiro J.V., Bernardi R.C., Buch R., Fiorin G., Henin J., Jiang W., et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020;153 doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herce H.D., Garcia A.E., Darden T. The electrostatic surface term: (I) Periodic systems. J. Chem. Phys. 2007;126 doi: 10.1063/1.2714527. [DOI] [PubMed] [Google Scholar]
- 78.Rudin A., Choi P. In: The Elements of Polymer Science & Engineering. third ed. Rudin A., Choi P., editors. Academic Press; Boston: 2013. Chapter 6 - Diffusion in polymers; pp. 275–304. [Google Scholar]
- 79.Lowry O.H., Rosenbrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 80.Oldham W.M., Loscalzo J. Quantification of 2-hydroxyglutarate enantiomers by liquid chromatography-mass spectrometry. Bio. Protoc. 2016;6 doi: 10.21769/BioProtoc.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y., Chen Y.N., Cheng J., Yan J.X., Xue C.Y., Pan H.Y., Shen X.Y., Zhou J., Jiang P., Zhou Y.L., Zhang X.X. Ultrasensitive HPLC-MS quantification of S-(2-Succino) cysteine based on ethanol/acetyl chloride derivatization in fumarate accumulation cells. Anal. Chem. 2023;95:1817–1822. doi: 10.1021/acs.analchem.2c03573. 2023. [DOI] [PubMed] [Google Scholar]
- 82.Tanis R.M., Piroli G.G., Day S.D., Frizzell N. The effect of glucose concentration and sodium phenylbutyrate treatment on mitochondrial bioenergetics and ER stress in 3T3-L1 adipocytes. Biochim. Biophys. Acta. 2015;1853:213–221. doi: 10.1016/j.bbamcr.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.