Abstract
Aging is associated with gradual changes in liver structure, altered metabolites and other physiological/pathological functions in hepatic cells. However, its characterized phenotypes based on altered metabolites and the underlying biological mechanism are unclear. Advancements in high-throughput omics technology provide new opportunities to understand the pathological process of aging. Here, in our present study, both metabolomics and phosphoproteomics were applied to identify the altered metabolites and phosphorylated proteins in liver of young (the WTY group) and naturally aged (the WTA group) mice, to find novel biomarkers and pathways, and uncover the biological mechanism. Analysis showed that the body weights, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) increased in the WTA group. The grips decreased with age, while the triglyceride (TG) and cholesterol (TC) did not change significantly. The increase of fibrosis, accumulation of inflammatory cells, hepatocytes degeneration, the deposition of lipid droplets and glycogen, the damaged mitochondria, and deduction of endoplasmic reticulum were observed in the aging liver under optical and electron microscopes. In addition, a network of metabolites and phosphorylated proteomes of the aging liver was established. Metabolomics detected 970 metabolites in the positive ion mode and 778 metabolites in the negative ion mode. A total of 150 pathways were pooled. Phosphoproteomics identified 2618 proteins which contained 16621 phosphosites. A total of 164 pathways were detected. 65 common pathways were detected in two omics. Phosphorylated protein heat shock protein HSP 90-alpha (HSP90A) and v-raf murine viral oncogene homolog B1(BRAF), related to cancer pathway, were significantly upregulated in aged mice liver. Western blot verified that protein expression of MEK and ERK, downstream of BRAF pathway were elevated in the liver of aging mice. However, the protein expression of BRAF was not a significant difference. Overall, these findings revealed a close link between aging and cancer and contributed to our understanding of the multi-omics changes in natural aging.
Keywords: aging, Mouse, liver, metabolomics, Phosphoproteomics
Graphical abstract
Highlights
-
•
General phenotype changed in naturally aging mice.
-
•
Aging metabolites and phosphorylated proteins were identified.
-
•
65 common pathways of metabolomics and phosphoproteomics were detected.
-
•
Cancer-related pathways are closely related to liver aging.
-
•
BRAF and the MAPK pathway deserve further attention.
1. Introduction
Aging is a complex, multifactorial, and progressive physiological process in which organisms gradually decline over time and experience various aging-related diseases [1]. Thus, understanding the mechanism is crucial to preventing and delaying the aging process. Being a complex metabolic organ, the liver is also predisposed to changes in structure and function during the process of aging [2]. Some liver-related metabolic syndromes, such as nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), and even hepatocellular carcinoma (HCC) increase with age [3]. Therefore, starting from the liver may be important in exploring the relevant mechanisms of aging and developing therapeutic targets to treat aging-related diseases.
In recent years, various omics techniques such as genomics, transcriptomics, metabolomics, and proteomics have been used to unravel aging mechanisms [4]. Metabolites are cellular intermediates of metabolic reactions composed of various structural elements and biochemical pathways [5]. Targeted and untargeted metabolome studies on aging have been carried out recently [[6], [7], [8], [9], [10]]. As we know, succinylation, phosphorylation, ubiquitination, and other protein post-translational modifications (PTMs) can affect aging and lifespan. For instance, advanced glycation endproducts (AGEs) and the accumulation of redox modifications are characteristic of aging and metabolic diseases [11], and the progression of Alzheimer's disease is connected to hyperphosphorylation of tau [12]. Some metabolic changes that occur during aging can affect protein function, such as the changes in some histone and non-histone modifications, and thy interact to initiate the vicious cycle of aging. According to a focused metabolome investigation of Drosophila, changes in the amounts of metabolites linked to methionine metabolism were strongly associated with age. The down-regulation of two enzymes that regulate methionine metabolism, dAhcyL1/L2, could significantly extend the life span and inhibit the trimethylation modification (H3K4me3) level of histone [13]. The proteomics method was employed to investigate the alterations in liver proteins in age-related and diet-restricted rats in a study, concluding that these proteins were mostly attached to glucose and fatty acid metabolism [14]. Proteins catalyze various physiological reactions in the body, and their phosphorylation modifications are the most extensive, participating in almost all biological processes. Currently, phosphatase has been considered a target for the treatment of age-related diseases, such as Parkinson's disease [12,15]. However, there are few studies on the changes in the natural aging liver phosphorylation. And the metabolome, on the other hand, is influenced by a variety of genetic and environmental factors and thus cannot accurately represent the complex aging process. Therefore, the combination of metabolism and phosphorylation may provide new insights into natural liver aging.
Thus, this study aimed to use these combinatorial techniques to establish a phosphoprotein metabolism regulatory network generating phosphoproteomic and metabolic maps and elucidate a comprehensive mechanism of the natural aging liver to identify potential targets for liver-related aging diseases.
2. Materials and methods
2.1. Animals
Tengli Trade Co. Ltd (Jinan, China) provided the 3-month-old male C57BL/6J mice that were placed in a pathogen-free environment and fed a regular mouse chow diet and tap water. The 3- and 22-month-old mice were attributed to wild-type young (WTY, n = 10) and wild-type aging (WTA, n = 10) groups. All experimental animal protocols were approved by the Laboratory Animal Ethical and Welfare Committee of Shandong University Cheeloo College of Medicine (Jinan, China).
2.2. Intervention methods
Before the animals were sacrificed, the grip tester (Jiangsu Cyonce Biotechnology Co., Ltd, Nanjing, China) was used to measure the grip [16], and the body weight was measured using JM-A10002 electronic balance (Beijing Jinkelida Electronic Technology Co., Ltd, Beijing, China). After mice were anesthetized with 10 % chloral hydrate, blood samples were collected from the inferior vena, centrifuged at 3000 rpm for 10 min to obtain serum, and stored at −80 °C. The weights of the livers were recorded, and the livers were quickly removed to be washed in phosphate-buffered saline (PBS). A part of tissues was used for preparing optical and electron microscopy sections. The other tissues were frozen in liquid nitrogen after dissection for further omics analyses.
2.3. Liver function indicators assay
The chemray 800 automatic chemistry analyzer (Rayto Life and Analytical Sciences Co., Ltd, Shenzhen, China) was used to measure serum levels of ALT, AST, TC, and TG. The corresponding commercial enzymatic Kits also came from Rayto Life and Analytical Sciences Co., Ltd, Shenzhen, China.
2.4. Histopathological observation
After Masson [17] and hematoxylin-eosin (HE) staining [18], the stained slides were examined under an optical microscope (Olympus DX45, Tokyo, Japan). According to the conventional transmission electron microscopy (TEM) sample preparation method [19], the sample slices were observed by TEM (JEOL-1200E, Beijing, China).
2.5. Metabolomics method
2.5.1. Sample preparation
An equivalent of 80 mg of liver tissues frozen in liquid nitrogen was cut on dry ice and homogenized with 200 μL H2O and five ceramic beads using the MP homogenizer (24*2, 6.0 M/S, 60s, twice). 1 mL of cold extraction solvent methanol/acetonitrile/H2O (2:2:1, v/v/v) was added to the sample for precipitating protein. The metabolites were extracted from tissues by adding 800 μL methanol/acetonitrile (1:1, v/v) sonicated at 4 °C (30min/once, twice) followed by centrifugation at 14000×g at 4 °C for 15 min. The samples were resolubilized in 100 μL of acetonitrile/water (1:1, v/v) solvent for LC-MS analysis after the supernatant was dried in 4 °C vacuum centrifuge.
2.5.2. LC-MS/MS analysis
A quadrupole time-of-flight mass spectrometer (AB Sciex TripleTOF 6600) coupled to an Ultra-high Performance Liquid Chromatography system (UHPLC) (1290 Infinity LC, Agilent Technologies) was used to analyze the extracts. Samples were analyzed on a 2.1 mm × 100 mm ACQUIY UPLC (1290 Infinity LC, Agilent Technologies, Beijing, China) BEH 1.7 μm column (Waters, Ireland) for hydrophilic interaction chromatography (HILIC) separation. The mobile phase contained A = 25 mM ammonium acetate and 25 mM ammonium hydroxide in water and B = acetonitrile. 85 % B for 1 min, and linearly decreased to 65 % in 11 min, then reduced to 40 % in 0.1 min, retained for 4 min, then increased to 85 % in 0.1 min, with a 5 min re-equilibration interval. The injection volume was 2 L, the column temperature was 25 °C, the automatic sampler temperature was 5 °C, and the flow rate was 0.4 mL/min. The quadrupole time-of-flight mass spectrometer was operated in both positive and negative ion modes. The following were the settings for the electrospray ionization (ESI) source: Source temperature of 600 °C, IonSpray Voltage Floating (ISVF) of 5500 V, Ion Source Gases 1 and 2 (Gas1 and Gas2) of 60 each, and Curtain Gas (CUR) of 30. The accumulation time for the TOF MS scan was set at 0.20 s/spectra, and the MS acquisition was configured to cover the m/z range of 60–1000 Da. The apparatus was set up for auto MS/MS acquisition with the m/z range of 25–1000 Da and the accumulation time for product ion scan at 0.05 s/spectra. The information-dependent acquisition (IDA) parameters were set as follows: the collision energy (CE) was fixed at 35 V with ±15 eV; declustering potential (DP), 60 V (+) and −60 V (−); exclude isotopes within 4 Da, candidate ions to monitor per cycle: 10.
2.5.3. Data processing and analysis
ProteoWizard MSConvert was used to convert the raw MS data to MzXML files, which were then imported into XCMS software. Only the variables with >50 % of the non-zero measurement values in at least one group were preserved among the extracted ion features. Accurate m/z value (<10 ppm) and MS/MS spectra compared to an in-house database established with authentic standards were utilized to identify the compound identification of the metabolites. The 7-fold cross-validation and response permutation testing were used to evaluate the robustness of the model. The processed data were analyzed using univariate statistical analysis, multivariate data analysis (including pareto-scaled principal component analysis (PCA), orthogonal partial least-squares discriminant analysis (OPLS-DA)), differential metabolite correlation analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis after sum-normalization. The differences between groups were determined by variable importance (VIP) > 1 (OPLS-DA) and p < 0.05 (Student's t-test). The correlation between the two variables was determined by Pearson's correlation analysis. P-value <0.05 was considered statistically significant.
2.6. Phosphoproteomics method
2.6.1. Protein extraction and peptide enzymatic hydrolysis
After lysing the liver tissue samples in UA lysis buffer (8 m urea, 150 m MTrismuryl HCl pH8.0), the protein concentration was determined using the BCA Protein Assay Kit (Bio-Rad, USA). Then 20 μg proteins with 5x loading buffer of each sample were boiled for 15 min. The proteins were separated on 4%–20 % SDS-PAGE gel (constant voltage 180V, 35 min). Proteins were evaluated by Coomassie Blue R-250 staining was used to visualize protein bands. Equal aliquots of each sample were pooled into one sample (called Pool sample) for DDA library generation and quality control. 400ug proteins digestion by trypsin were performed according to filter-aided sample preparation (FASP) procedure After dithiothreitol (DTT) adding to change the sample's final concentration to 20 mM, the sample was cooled to room temperature by gently shaking at 37 °C for 60 min. Then cysteine and iodoacetamide (IAA) were added to the sample until the concentration is 50 mM, and incubated for 30min in the dark. The concentration of UA in protein samples was diluted to less than 1.5 M by adding NH4HCO3 buffer (50 mM). Lys-C was added to the sample according to the ratio of 1: 100, digested at 37 °C for 4 h, then added trypsin at the ratio of 1: 50, and digested at 37 °C for 16 h to obtain the lysates. The peptide concentration was measured by OD280.
2.6.2. Phosphopeptides enrichment
All samples including the Pool sample were enriched phosphopeptides with a High-SelectTM Fe-NTA Phosphopeptides Enrichment Kit (Thermo Scientific A32992). After lyophilized, the phosphopeptides were resuspended in 0.1 % formic acid (FA) LC-MS Water.
2.6.3. Spectral library
The enriched phosphorylated peptides from pool samples were divided into 10 fractions using high-pH reversed-phase chromatography (HPRP). All fractions were collected, desalted, and freeze-dried. Then, 2 μg peptide was taken from each fraction, mixed with an appropriate amount of iRT (BiognosysKi-3002-2) standard peptides, and detected by DDA mass spectrometry on a Q-Exactive HF mass spectrometer (Thermo Scientific). Thermo Scientific Q Exactive HF mass spectrometer connected to an Easy-nLC 1200 chromatography system (Thermo Scientific) was used to build the DDA library. The operation process was as follows: A was 0.1 % formic acid, B was 84 % ACN, and 0.1 % formic acid. Peptides were equilibrated with buffer A and separated by buffer B in EASY-SprayTM C18 Trap Column (Thermo Scientific, P/N 164946, 3 μm, 75 μm*2 cm) and EASY-SprayTM C18 reverse phase column (Thermo Scientific, ES803, 2 μm, 75 μm*50 cm). The gradient progressed from 8 % to 30 % in 60 min, then to 100 % B over 20 min, and then to 100 % B for 40 min at a flow rate of 300 nL/min. The operating mode was positive ion mode. The range of m/z was 300–1800 full scan, 60,000 resolution (@200 m/z), normalized AGC (automatic gain control) target was 3e6, and maximum ion implantation time was 50 ms. Peptides were chosen for MS/MS interrogation after each full MS scan using higher energy collisional dissociation (HCD, normalized collision energy:27), with 20 MS/MS scans per cycle. MS/MS scans were performed with an isolation width of 1.3 m/z, an AGC target of 3e6, a resolving power of 30,000 (@m/z 200), and a maximum ion injection time of 120 ms. The raw mass spectrometry data from each DDA dataset were used to build libraries for DIA searches using SpectronautTM 14.4.200727.47784 by searching against a Uniprot database. Search parameters were set as follows: enzyme: trypsin, max miss cleavage site: 1, fixed modification: Carbimidomethyl (C), dynamic modification: Oxidation (M), phosphate (STY) and Acetyl (Protein N-term), FDR (false discovery rate): <1 %, and PTM localization probability cutoff: >0.75.
2.6.4. DIA data acquisition
2 μg enriched phosphorylated peptides from each sample were mixed with iRT (Biognosys Ki-3002-2) standard peptides and analyzed individually detected by DIA. Each sample for DIA analysis was injected into a Thermo Scientific Q-Exactive HF mass spectrometer connected to an Easy-nLC 1200 chromatography system (Thermo Scientific). The chromatographic separation conditions were as described above. The chromatographic samples were analyzed by DIA mass spectrometry with a Q-Exactive HFspectrometer. The parameters were as follows: detection mode: positive ions, the scan range of primary mass spectrometry: 350–1650 m/z, Ms resolution: 60,000 (@200 m/z), AGC (automatic gain control) target: 3e6, maximum ion injection time: 50 ms. MS2 adopted DIA data collection mode and set 58 DIA collection windows, Ms resolution was 60,000 (@200 m/z), AGC (automatic gain control) target was 3e6, maximum ion injection time: 50 ms, maximum ion injection time was auto, MS2 Activation Type was HCD (normalized collision energy:30).
Raw data from each DIA dataset were used for DIA searches using SpectronautTM 14.4.200727.47784 by searching against the DDA library. Search parameters were set as follows: retention time prediction type: dynamic iRT, interference on MS2 level correction: enabled, cross run normalization: enabled, Q Value cutoff: 0.01.
2.6.5. Bioinformatic analysis
The expression factor (Fold Change, FC) > 1.5 (up-regulated >1.5 or down-regulated <0.67) and P value < 0.05 (T-test) were used as the criteria to screen the significant differentially modified peptides. The quantitative information of the target protein set was normalized (−1, 1). The complex heatmap R packet (RVersion3.4) was used to generate a hierarchical clustering heatmap. CELLO (http://cello.life.nctu.edu.tw/) method was used to predict the subcellular localization. Pfam database was used for protein domain analysis. Blast2GO was used for Gene Ontology (GO) annotation. KEGG Automatic Annotation Server (KAAS) software was applied to expound the KEGG pathway of the connected proteins. Fisher's exact test was used to enrich the annotation. Only functional categories and pathways with p-values less than 0.05 were deemed significant. The direct and indirect interactions of proteins were analyzed using the Cytoscape software (version number: 3.2.1) and the STRING database (http://string-db.org/).
2.7. Combination analysis of two omics
The online KEGG (http://www.kegg.jp/) was used to query and map all differentially expressed proteins and metabolites. The two omics' KEGG annotation and enrichment results were combined with R software (version 3.5.1). Bar plot and Venn diagram were created. Log2 normalization was performed for the different abundant proteins and metabolites to concatenate into one matrix. Then correlation coefficient was calculated with the Pearson algorithm to construct the matrix heat map. The Pearson correlation coefficient among differentially expressed proteins and metabolites was loaded into the CytoScape (version 3.5.1) and a correlation network was calculated.
2.8. Western blot
The steps were described in our previous study [20]. The primary antibodies against BRAF (proteintech), MEK (Abcam), ERK (proteintech), GAPDH (proteintech), and the mouse IgG (H + L) secondary antibody (Pierce, Rockford, USA), rabbit IgG (H + L) secondary antibody (Pierce, Rockford, USA) were used.
2.9. Statistical analysis
SPSS version 25.0 (IBM, New York, USA) was performed to statistical analysis. Origin 2021 (Origin Software, OriginLab, USA) generated statistical diagrams. The results were expressed by mean ± standard error of the mean. Student's t-test was used to analyzed the comparisons between groups. P values < 0.05 were considered significant.
3. Results
3.1. General phenotype characteristics
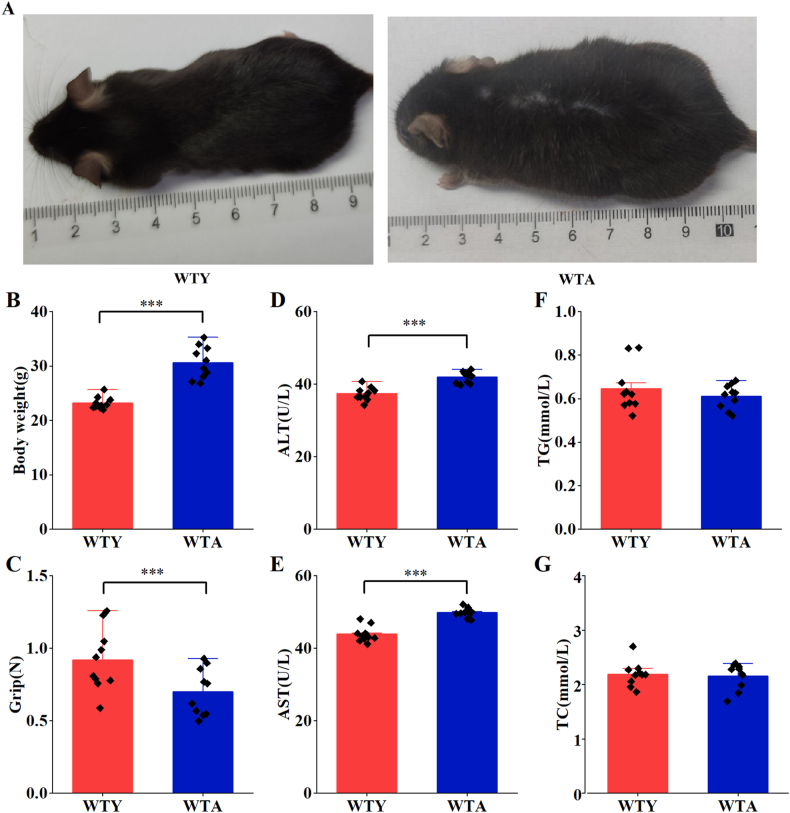
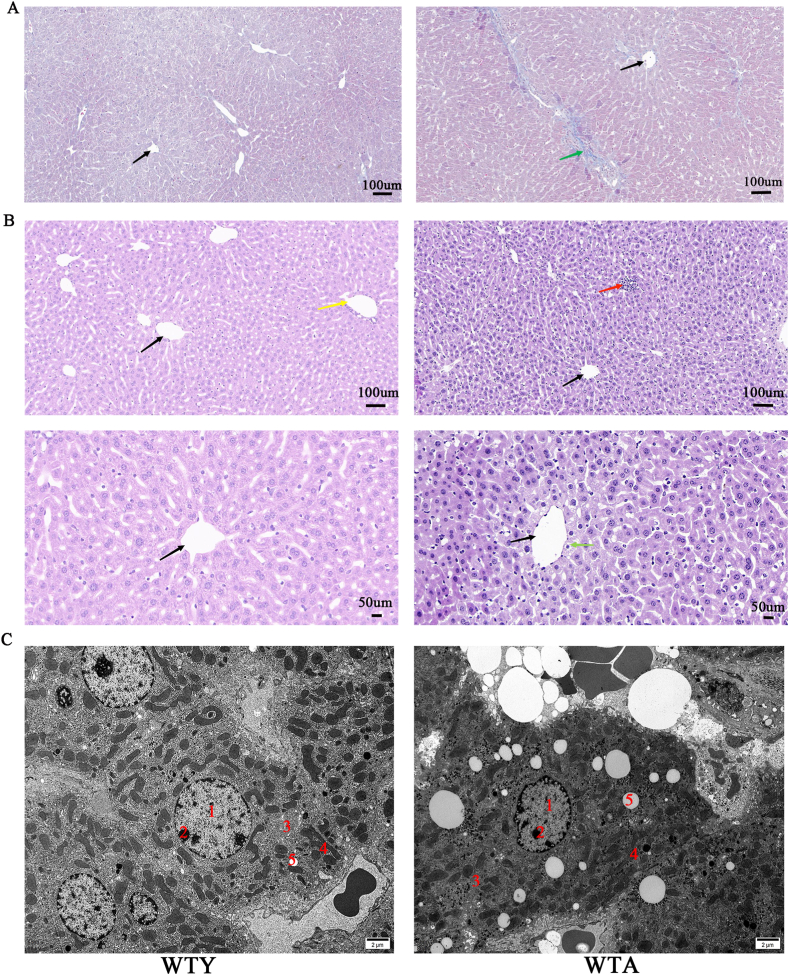
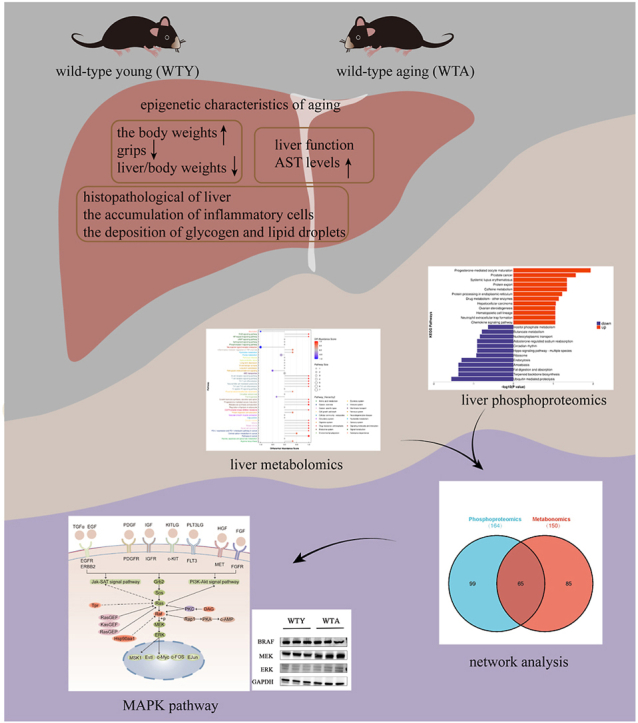
To explore the phenotypic characteristics of aging mice, some indicators were measured. The appearance of the aged mice changed markedly. The hair of old mice became white and sparse (Fig. 1A). The body weights increased significantly with age (Fig. 1B), but the grips of the WTA group decreased compared with the WTY group (Fig. 1C). ALT (Fig. 1D) and AST (Fig. 1E) increased with age, but TG (Fig. 1F) and TC (Fig. 1G) were not affected by aging. The optical microscope results revealed that adult mice had normal cytoarchitecture of the liver parenchyma with a polygonal morphology and distinct cellular boundaries, while the aging group showed a significantly collagen fiber aggregation (Fig. 2A) and focal inflammatory cell infiltration (Fig. 2B). Hepatocytes degeneration, swelling, loose cytoplasm and light staining were observed around the central vein in the aging group (Fig. 2B). Hepatocytes from adult animals had “normal” nucleus, mitochondria, nucleolus, endoplasmic reticulum, lysosomes, and other ultrastructures. The arrangement of mitochondria and endoplasmic reticulum was more regular. In contrast, dramatically increased lipid droplets and glycogen were observed in elderly mice hepatocytes. In the WTA group, the mitochondrial membrane was damaged, the mitochondrial crest decreased and the number of endoplasmic reticulum decreased (Fig. 2C).
Fig. 1.
General epigenetic characteristics changes in the two groups. (A) Changes in the appearance of mice between the two groups. The hair of aging mice became white and sparse. (B) The body weight increased but grip (C) decreased in the WTA group. The serum levels of ALT (D) and AST (E) increased with age, but the TG (F) and TC (G) levels have no significant changes in the two groups. WTA: wild-type aging, WTY: wild-type young. (***) P < 0.05, n = 10 per group.
Fig. 2.
Histopathological alterations in aging mice. (A) Liver interstitial collagen evaluated in the aged mice (Masson staining, scale bar = 100 μm). Black arrow: central veins of hepatic lobules, green arrow: deposition of increased collagen fibers. (B) Degeneration of hepatocytes and inflammatory cell infiltration were observed in the aged mice (H-E staining, scale bar = 100 μm/50 μm). Black arrow: central veins of hepatic lobules, yellow arrow: conduit area, red arrow: a focal infiltration of inflammatory cells between the hepatic cord, green arrow: hepatocytes around the central vein watery degeneration, swelling, loose cytoplasm and light staining. (C) The WTA group showed deposited lipid droplets and glycogen, damaged mitochondria and reduced endoplasmic reticulum (electron microscope, scale bar = 2 μm/500 nm).1—nucleus, 2—nucleolus, 3—endoplasmic reticulum, 4—mitochondria, 5—lipid droplets, 6—aggregated glycogen, 7—lysosome, red arrow—damaged mitochondria with loss of membrane and reduction of crest. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Metabolomics analysis
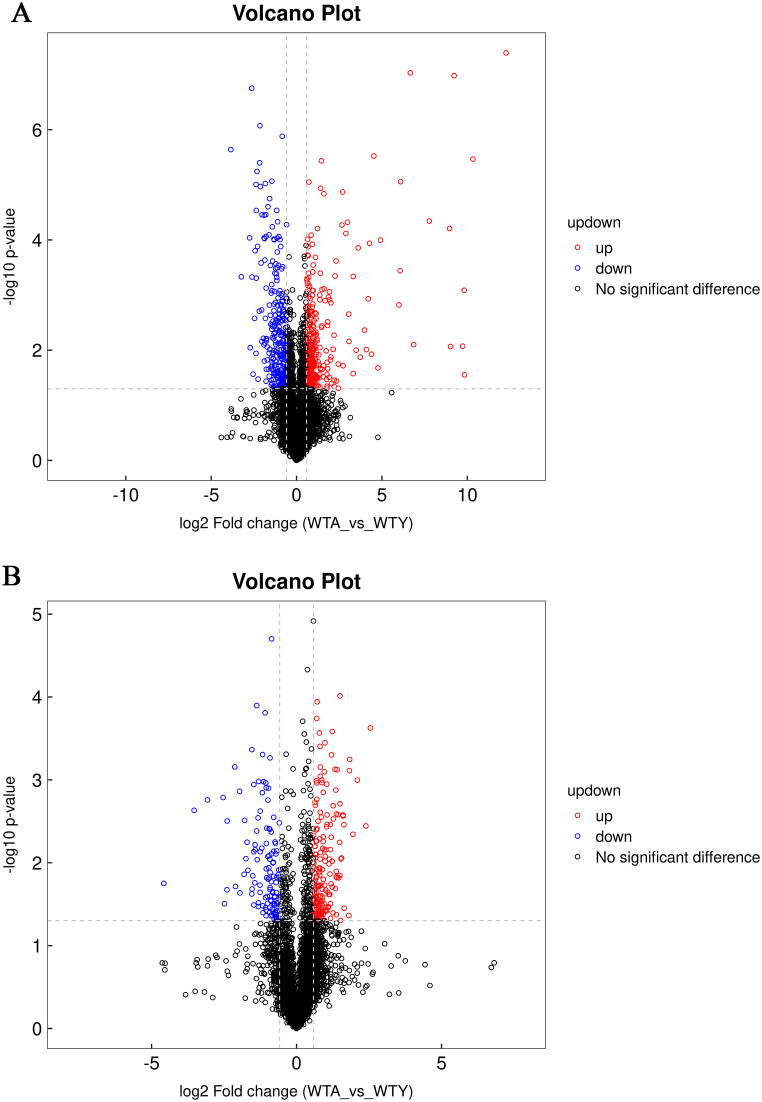
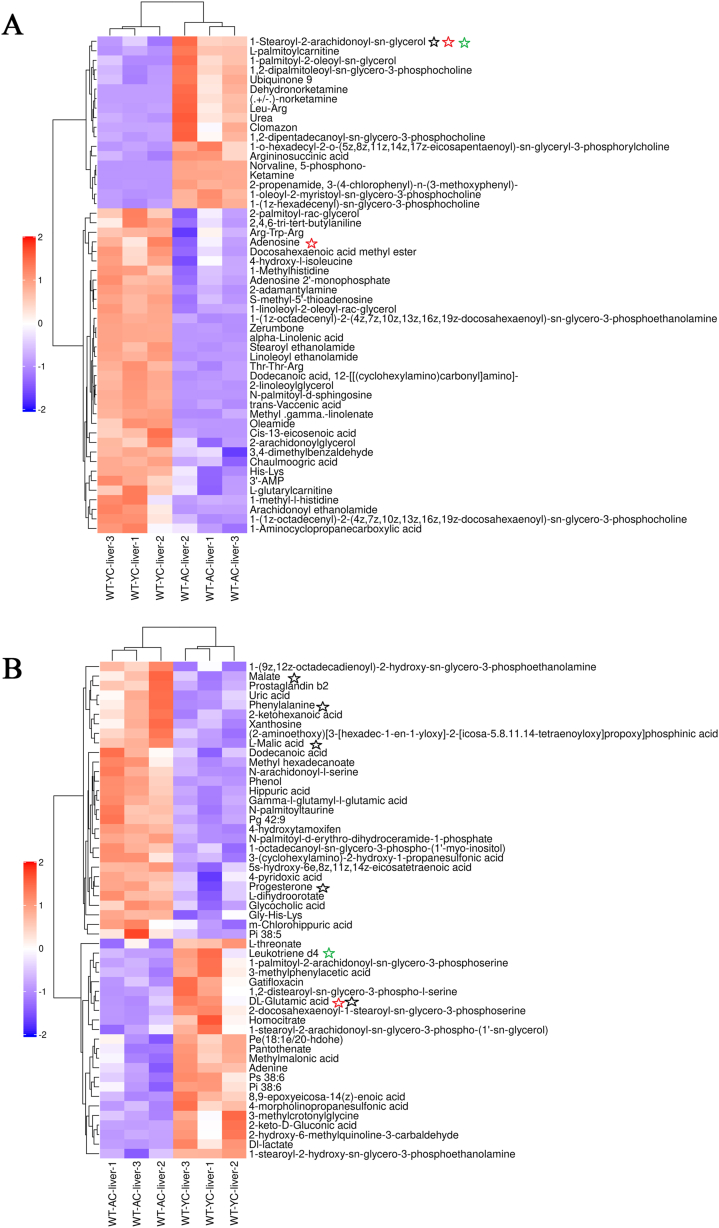
A total of 970 metabolites were identified in the positive ion mode and 778 metabolites in the negative ion mode. Supplemental data summarized the relevant information of the metabolites. These metabolites mainly included lipids and lipid-like molecules, organic acids and derivatives, organoheterocyclic compounds, benzenoids, and so on. The visual representation of the differential metabolites of FC > 1.5 or FC < 0.67 and p < 0.05 was in the form of a volcano chart (positive ion mode, Fig. 3A; negative ion mode, Fig. 3B). To identify potential metabolites associated with aging, OPLS-DA VIP >1 and p < 0.05 were set as the screening criteria to select various significantly changed metabolites. The heat map provided a visual representation of significantly different metabolites in the positive (Fig. 4A) and negative (Fig. 4B) ion modes.
Fig. 3.
The Volcano plot of different metabolites. Volcano plot in (A) positive and (B) negative ion mode (red: FC > 1.5 and P < 0.05, blue: FC < 0.67 and P < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
Hierarchical clustering heatmaps of metabolites with a significant difference. (A) positive and (B) negative ion mode (red: upregulated, blue: downregulated) of hierarchical clustering heatmaps. Red marks indicated metabolites involved in signal transduction pathways. Black marks related to cancer. Green labeling participated in immune system-related pathways. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
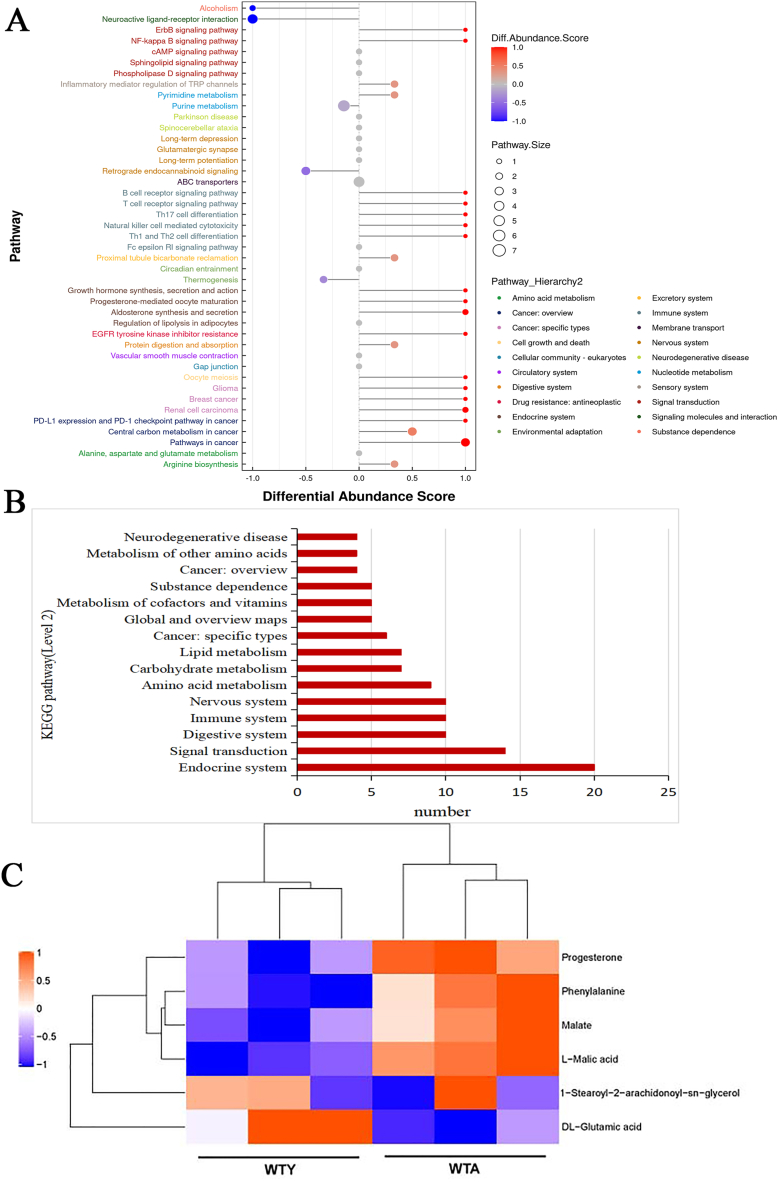
Combining the differential metabolites screened by the positive and negative ion modes, a total of 150 metabolic pathways were pooled and 43 processes were enriched, which participated in signal transduction, immune system, amino acid metabolism, carbohydrate metabolism, lipid metabolism, cancer, cell growth and death et al. (Fig. 5A). Fig. 5B showed the different pathways according to the level 2 classification. There are 14 pathways in the 150 pathways were involved in signal transduction, including phospholipase D, cAMP, sphingolipid, FoxO, MAPK, ErbB, Ras, Rap1, Calcium, NF-kappa B, HIF-1, VEGF, Apelin, and cGMP-PKG signaling pathway. Our findings identified 10 immune system-related pathways, including Fc epsilon RI signaling pathway, chemokine signaling pathway, platelet activation, C-type lectin receptor signaling pathway, natural killer cell mediated cytotoxicity, Th1 and Th2 cell differentiation, Th17 cell differentiation, T cell receptor signaling pathway, B cell receptor signaling pathway, Fc gamma R-mediated phagocytosis. Most of these pathways were closely related to the occurrence of inflammation. Surprisingly, 1-stearoyl-2-arachidonoyl-sn-glycerol (DAG), a diradylglycerols/lineolic acids and derivatives were up-regulated in most of these pathways. Notably, some of these processes, such as pathways in cancer, central carbon metabolism in cancer, renal cell carcinoma, breast cancer, glioma, choline metabolism in cancer, PD-L1 expression and PD-1 checkpoint pathway in cancer, are related to the occurrence and development of tumor. Further analysis of the differential metabolites of these cancer-related metabolic pathways showed significant changes in the differential metabolites of l-malic acid, progesterone, malate, 1-stearoyl-2-arachidonoyl-sn-glycerol, phenylalanine, and dl-glutamic acid. The hierarchical clustering heatmap of these metabolites is shown in Fig. 5C. Thus, it can be deduced that the occurrence of tumors is homologous to the process of aging.
Fig. 5.
Display metabolic pathways and metabolites related to cancer. (A) Differential abundance score maps of all differential metabolic pathways. (B) The pathways in the level 2 classification. The classification names for more than three pathways are displayed in figure. (C) Metabolic heatmaps of differential metabolites related to cancer (red: upregulated, blue: downregulated).
3.3. Phosphoproteomics analysis
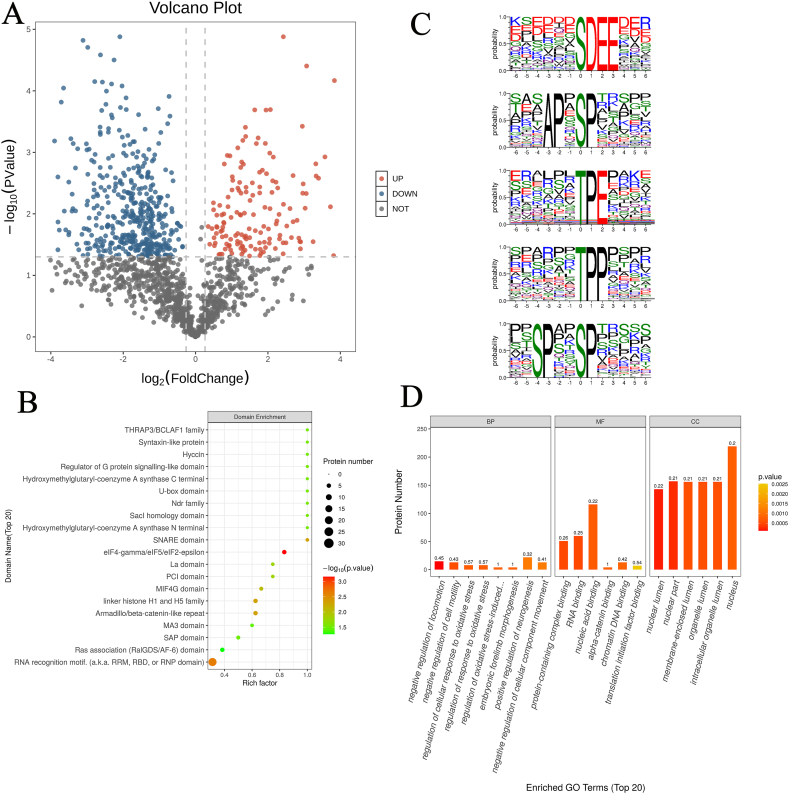
Phosphoproteomics was utilized to further investigate the mechanism of liver aging. A total of 16621 phosphosites were identified from 2618 proteins. Supplemental data summarized the relevant information of the proteins. Among these, 94.88 % of the proteins have two or more phosphorylation modification sites. The Volcano plot showed that 139 phosphopeptides were upregulated (FC > 1.5 and P < 0.05), and 437 phosphopeptides were downregulated (FC < 0.67 and P < 0.05) (Fig. 6A). The WTA group comprised 169 unique differentially modified peptides (≥half of the samples in the WTA group are not null), and the WTY group consisted of 747 differentially modified peptides (≥half of the samples in the WTY group are not null). Based on the domain enrichment analysis, the differentially expressed modified peptides were mainly enriched in eIF4-gamma/eIF5/eIF2-epsilon, RNA recognition motif, SNARE domain, Armadillo/beta-catenin-like repeat, and linker histone H1 and H5 family (Fig. 6B). To understand the sequence characteristics around the phosphosites, conservative motif analysis was carried out using the MEME software. The analysis of all the identified phosphorylation sites showed that most of the phosphorylation occurred in the region around the serine (S) and threonine (T) residues (Fig. 6C). S and T were in the 0 positions.
Fig. 6.
Phosphoproteomics show related changes in the liver of aging mice. (A) Volcano plot of significant differences in phosphorylated peptides (red: upregulated, blue: downregulated). (B) Domain enrichment map of differentially expressed proteins. The closer the bubble color is to red, the higher the significance level of the enrichment. (C) Statistics of motif analysis results of phosphorylated modified peptides. The figure shows the top five enriched motifs. (D) GO analysis of identified phosphoproteins, including BP, MF, and CC. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
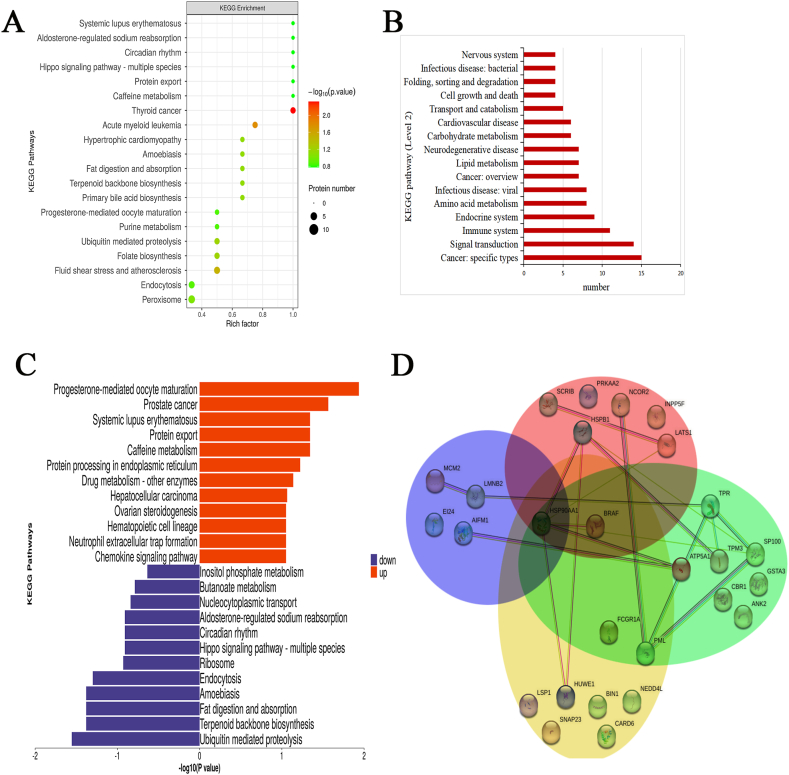
The characteristics of the function/pathway level of the modified protein were reflected by the GO and KEGG enrichment analyses. The biological process (BP) analysis showed that the majority of the altered phosphoproteins regulated the processes, such as locomotion, cell motility, oxidative stress et al. The phosphorylation sites of some proteins in inflammation-related biological processes have also been changed, such as high affinity immunoglobulin gamma Fc receptor I (FCGR1), glutathione peroxidase (GPX1), 60 kDa chaperonin (A0A6P5Q938), heterogeneous nuclear ribonucleoprotein K (A0A6I9LDE6), argininosuccinate synthase (ASSY). Specific sites of these proteins were shown in Table 1. Molecular function (MF) analysis revealed that these proteins were associated with protein-containing complex binding, RNA binding, nucleic acid binding, alpha-catenin binding, chromatin DNA binding, and translation initiation factor binding. Cellular component (CC) analysis indicated that these were mainly annotated as intracellular proteins (Fig. 6D). KEGG enrichment analysis suggested that a total of 164 pathways were detected, of which thyroid cancer, acute myeloid leukemia, fluid shear stress and atherosclerosis were significantly enriched (Fig. 7A). The various pathways were depicted in the level 2 classification (Fig. 7B). In the 164 pathways, 14 pathways participated signal transduction (MAPK, Foxo, mTOR, PI3K-AKt, Hippo, ErbB, Rap, cAMP, phosphatidylinositol, AMPK, notch, VEGF, Apelin, Hippo). 11 pathways mediated immune system, including NOD-like receptor signaling pathway, Fc gamma R-mediated phagocytosis, chemokine signaling pathway, platelet activation, antigen processing and presentation, neutrophil extracellular trap formation, C-type lectin receptor signaling pathway, hematopoietic cell lineage, natural killer cell mediated cytotoxicity, IL-17 signaling pathway, Th17 cell differentiation. 4 pathways involved in cell growth and death, including apoptosis, necroptosis, cell cycle, p53 signaling pathway. Fc gamma R-mediated phagocytosis, neutrophil extracellular trap formation, antigen processing and presentation, IL-17 signaling pathway, Th17 cell differentiation, apoptosis, necroptosis,p53 and FoxO signaling pathways were involved in inflammation. The upregulated and downregulated proteins were assessed using KEGG enrichment analysis (Fig. 7C). The proteins with upregulated phosphorylation levels on their sites were involved in thyroid cancer, acute myeloid leukemia, pathway in cancer, prostate cancer, whereas the proteins with downregulated phosphorylation levels are involved in ubiquitin-mediated proteolysis, terpenoid backbone biosynthesis, fat digestion and absorption, amoebiasis. We discovered that these pathways, similar to the results of metabolomics, played a role in the emergence and progression of cancer, such as transcriptional misregulation in cancer, proteoglycans in cancer. Moreover, changes in the pathway of hepatocellular carcinoma were also found. Further analysis of the phosphorylated proteins related to cancer pathways revealed that BRAF, HS90A, TPR, A0A068EW80, and ATPA had changed. The following list depicts a significant change in the phosphorylated expression of these cancer-related proteins (Table 2). The PPI diagram demonstrated the clear relationships between phosphorylated proteins relevant to the immune system, signal transduction, cell growth and death, and cancer pathways (Fig. 7D). HSP90A and BRAF, the center of network, were involved in many pathways of immune system, signal transduction, cell growth and death, and cancer.
Table 1.
List of significantly regulated proteins in inflammation-related biological processes of old mice liver samples using DIA-based phosphoproteomics analysis.
| Accessions | GeneName | ProteinNames | Modifications | PTMLocations | Fold Change (WTA/WTY) | t-test p value |
|---|---|---|---|---|---|---|
| P26151 | Fcgr1 | FCGR1_MOUSE | _ANS[Phospho (STY)]FQQVR_ | (S347) | 2.630904789 | 0.00369479 |
| A0A6P5QC65 | Gpx1 | GPX1_MOUSE | _PLTGGEPVSLGS[Phospho (STY)]LR_ | (S32) | 4.129211966 | 0.025512239 |
| A0A6P5QC65 | Gpx1 | GPX1_MOUSE | _NALPTPSDDPTALMT[Phospho (STY)]DPK_ | (T143) | 1.668292473 | 0.023672885 |
| A0A6P5QC65 | Gpx1 | GPX1_MOUSE | _LS[Phospho (STY)]AAAQSTVYAFSAR_ | (S7) | 1.655447157 | 0.010419741 |
| A2AKH4 | Snap23 | A2AKH4_MOUSE | _ATWGDGGDNS[Phospho (STY)]PSNVVSK_ | (S110); (S121) | 11003.3450520833 | 0.363564601389563 |
| A0A6P5Q938 | Hspd1 | A0A6P5Q938_MUSCR | _NAGVEGS[Phospho (STY)]LIVEK_ | (S488) | 0.298876544065367 | 0.0271400442638472 |
| A0A6I9LDE6 | Hnrnpk | HNRPK_MOUSE | _DYDDMS[Phospho (STY)]PR_ | (S283); (S259); (S260); (S284); | 0.202655506175861 | 0.000274312680619401 |
| P16460 | Ass1 | ASSY_MOUSE | _QHGIPIPVT[Phospho (STY)]PK_ | (T174) | 1.58933648245381 | 0.007293343641764 |
| P16460 | Ass1 | ASSY_MOUSE | _Q[Gln- > pyro-Glu]HGIPIPVT[Phospho (STY)]PK_ | (T174) | 0.527255982404853 | 0.0143278981421657 |
| P16460 | Ass1 | ASSY_MOUSE | _APNS[Phospho (STY)]PDVLEIEFK_ | (S219) | 0.409737640889429 | 0.000894758857725513 |
Fig. 7.
Display pathways and PPI diagrams between phosphorylated proteins. (A) KEGG pathway enrichment bubble map of differentially expressed phosphoproteins. (B) The pathways in the level 2 classification. The classification names for more than three pathways are displayed in figure. (C) KEGG pathway enrichment map of upregulated and downregulated proteins. (D) Protein-Protein Interaction (PPI) network between immune system, signal transduction, cell growth and death, and cancer pathways. Yellow: immune system, red: signal transduction, blue: cell growth and death, green: cancer. HSP90A and BRAF listed in the center of network.
Table 2.
List of significantly regulated proteins in cancer-related pathways of old mice liver samples using DIA-based phosphoproteomics analysis.
| Accessions | GeneName | ProteinNames | Modifications | PTMLocations | Fold Change (WTA/WTY) | t-test p value |
|---|---|---|---|---|---|---|
| P28028 | Braf | BRAF_MOUSE | _S[Phospho (STY)]PQKPIVR_ | (S135) | 8.161065261 | 0.008104432 |
| P07901 | Hsp90aa1 | HS90A_MOUSE | _ESDDKPEIEDVGS[Phospho (STY)]DEEEEEKK_ | (S263) | 7.788584848 | 0.004622558 |
| B2RXY7 | Cbr1 | B2RXY7_MOUSE | _KFS[Phospho (STY)]GDVVLAAR_ | (S30) | 6.99000184 | 0.012767077 |
| P07901 | Hsp90aa1 | HS90A_MOUSE | _DKEVS[Phospho (STY)]DDEAEEK_ | (S231) | 3.406402242 | 0.029635354 |
| P30115 | Gsta3 | GSTA3_MOUSE | _FLQPGS[Phospho (STY)]QR_ | (S202) | 2.811665714 | 0.048385791 |
| P26151 | Fcgr1 | FCGR1_MOUSE | _ANS[Phospho (STY)]FQQVR_ | (S347) | 2.630904789 | 0.00369479 |
| Q80U72 | Scrib | SCRIB_MOUSE | _NS[Phospho (STY)]LESISSIDR_ | (S1206) | 2.232036288 | 0.027956347 |
| Q58E70 | Tpm3 | Q58E70_MOUSE | _EQAEAEVAS[Phospho (STY)]LNR_ | (S51) | 2.00485194 | 0.04392409 |
| Q03265 | Atp5f1a | ATPA_MOUSE | _TGTAEMSS[Phospho (STY)]ILEER_ | (S53) | 0.644444502 | 0.049941145 |
| Q8C8R3 | Ank2 | ANK2_MOUSE | _S[Phospho (STY)]PQGLELPLPNR_ | (S2364) | 0.584217795 | 0.046960981 |
| A0A068EW80 | Pml | A0A068EW80_MOUSE | _ATS[Phospho (STY)]PPHLDGTSNPESTVPEKK_ | (S528) | 0.420165598 | 0.040340587 |
| F6ZDS4 | Tpr | TPR_MOUSE | _Q[Gln- > pyro-Glu]TPQAPQS[Phospho (STY)]PR_ | (S2141) | 0.28922622 | 0.005587672 |
| E9Q4Y0 | Sp100 | E9Q4Y0_MOUSE | _GGDTS[Phospho (STY)]DTESSIIIR_ | (S314) | 0.162136819 | 0.025229997 |
| Q03265 | Atp5f1a | ATPA_MOUSE | _AVDS[Phospho (STY)]LVPIGR_ | (S198) | 0.10018786 | 0.001793458 |
| Q3UW87 | Cyp3a13 | Q3UW87_MOUSE | _VVSRDETVS[Phospho (STY)]DE_ | (S501) | 0.079717205 | 9.01803E-05 |
3.4. Network analysis of metabolomic and phosphoproteomics
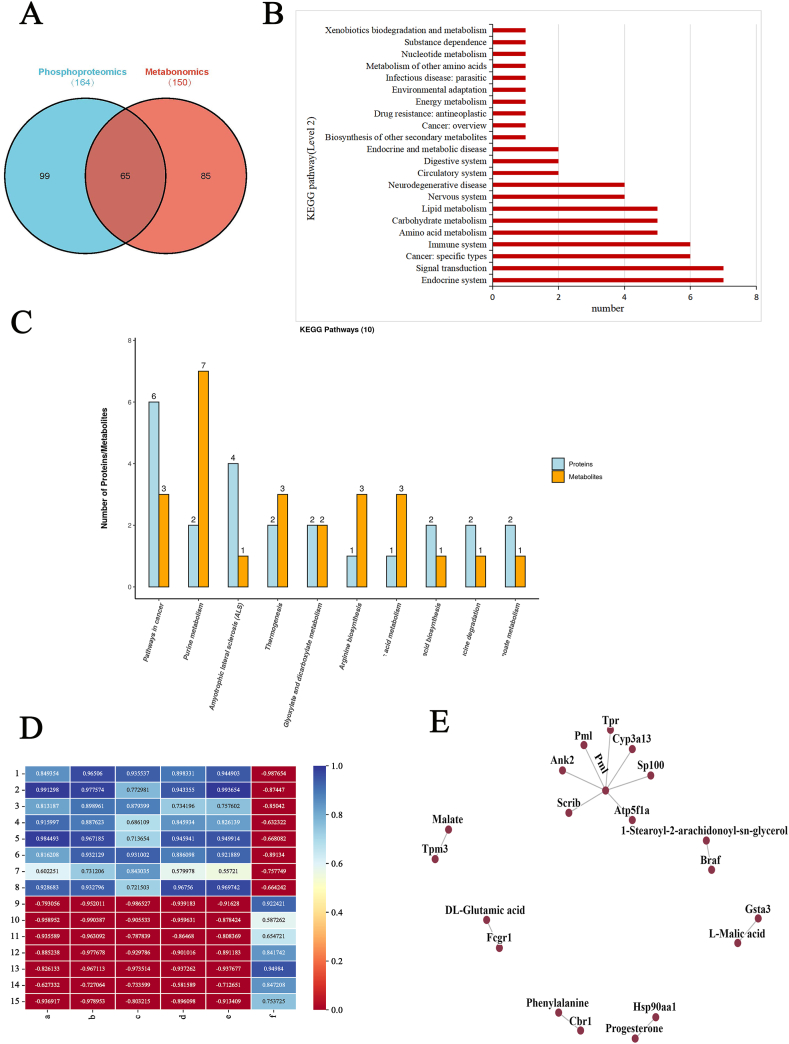
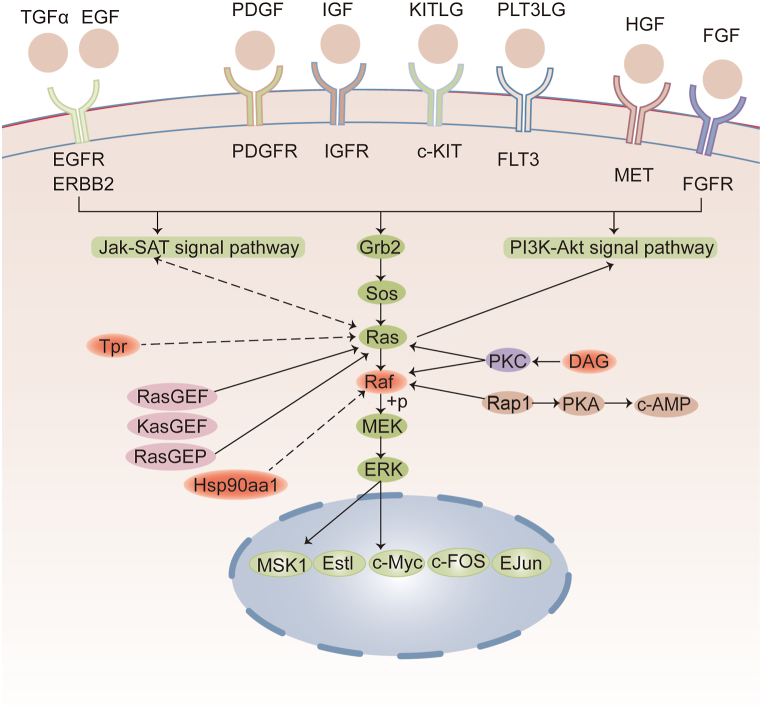
To further understand the mechanisms underlying aging, we integrated phosphoproteomics and metabolomics data and extracted the pathways that contain the two omics. Subsequently, 65 pathways were detected (Fig. 8A), including in the endocrine system, signal transduction, cancer, immune system, amino acid metabolism, lipid metabolism et al. (Fig. 8B). Signal transduction is engaged in eight of the 65 pathways, including MAPK, FoxO, cAMP, ErbB, Rap1, VEGF and Apelin signaling pathway. Immune system mediation involved 6 different pathways, including Fc gamma R-mediated phagocytosis, chemokine signaling pathway, platelet activation, C-type lectin receptor signaling pathway, natural killer cell mediated cytotoxicity and Th17 cell differentiation. At the same time, there are six pathways related to cancer, including pathways in cancer, prostate cancer, hepatocellular carcinoma, renal cell carcinoma, glioma, non-small cell lung cancer and breast cancer. The specific pathways were shown in the Supplemental data. The top 10 KEGG pathways that were annotated to the most modifiers and metabolites at the same time were visually displayed in the form of a bar chart (Fig. 8C). It can be seen that the pathways in cancer contain more differential proteins and metabolites. The analysis of the metabolites of these common pathways revealed an upregulated expression of these metabolites in aging mice compared to young mice, and most were also differential metabolites of the cancer-related pathways, such as l-malic acid, progesterone, and dl-glutamic acid. The analysis of these common pathways revealed that most phosphorylated proteins were differential proteins of cancer-related pathways. The correlation coefficient matrix heat map (Fig. 8D) and interaction network diagram (Fig. 8E) explored the regulatory relationship between cancer-related proteins and metabolites. These metabolites and phosphorylated proteins are closely related to each other. The pathways in cancer were significantly enriched in metabolomics-related and phosphorylation-upregulated pathways. The pathways in cancer is the overview of cancer. Thus, it is confirmed that aging is related to tumors. Surprisingly, the majority of these immune system, cancer and signal transduction pathways exhibit phosphorylation modifications in Serine/threonine-protein kinase B-raf protein (BRAF) at S135, and Heat shock protein HSP 90-alpha protein (HSP90A) at S263 and S231, and DAG was up-regulated in the WTA group. BRAF phosphorylated proteins are mainly involved in the MAPK signaling pathway (Fig. 9), which was also detected in the two omics. Together, these findings suggested that the MAPK pathway plays a major role in natural aging.
Fig. 8.
Network analysis of two omics. (A) Venn diagram of the KEGG pathways in two omics. (B) Level 2 of common pathways in two omics. (C) The histograms of the first ten KEGG pathways with the most proteins and metabolites annotated concurrently in the common pathway. (D) Correlation coefficient matrix heat map with cancer-related proteins and metabolites (rectangle's number: the correlation value, blue: positive correlation, red: negative correlation; the darker the color, the stronger the correlation). (1: P28028_S[Phospho(STY)]PQKPIVR_2, 2: P30115_FLQPGS[Phospho(STY)]QR_2, 3: P07901_ESDDKPEIEDVGS[Phospho(STY)]DEEEEEKK_3, 4: P07901_DKEVS[Phospho(STY)]DDEAEEK_2, 5: B2RXY7_KFS[Phospho(STY)]GDVVLAAR_2, 6: P26151_ANS[Phospho(STY)]FQQVR_2, 7: Q58E70_EQAEAEVAS[Phospho(STY)]LNR_2, 8: Q80U72_NS[Phospho(STY)]LESISSIDR_2, 9: Q8C8R3_S[Phospho(STY)]PQGLELPLPNR_2, 10: A0A068EW80_ATS[Phospho(STY)]PPHLDGTSNPESTVPEKK_3, 11: F6ZDS4_Q[Gln- > pyroGlu]TPQAPQS[Phospho(STY)]PR_2, 12: Q3UW87_VVSRDETVS[Phospho(STY)]DE_2, 13: E9Q4Y0_GGDTS[Phospho(STY)]DTESSIIIR_2, 14: Q03265_TGTAEMSS[Phospho(STY)]ILEER_2, 15: Q03265_AVDS[Phospho(STY)]LVPIGR_2, a: 1-Stearoyl-2-arachidonoyl-sn-glycerol, b: l-Malic acid, c: Progesterone, d: Malate, e: Phenylalanine, f: dl-Glutamic acid) (E) Network interaction diagram between cancer-related proteins and metabolites. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 9.
Schematic of the MAPK pathway. The pink markers are the molecules detected in the study. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Changes of proteins in the MAPK pathway
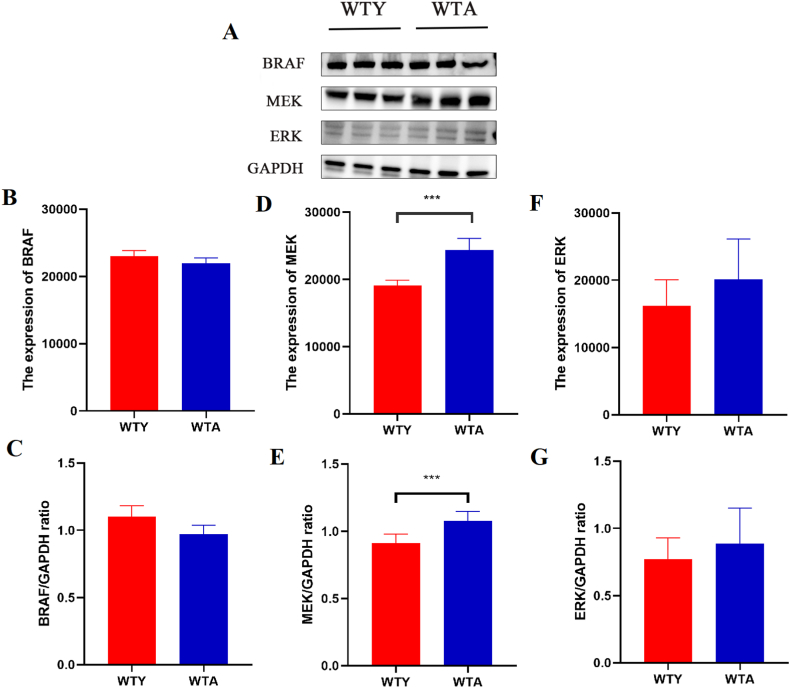
To further verify the changes of BRAF phosphorylation in the liver of aging mice, the protein expressions of BRAF and its downstream MEK and ERK were determined using Western blot (Fig. 10A). There was no significant difference in the BRAF protein between the two groups (Fig. 10B and C). It was surprising to discover that MEK protein expression was significantly upregulated in the WTA group (Fig. 10D and E), indicating that BRAF phosphorylation caused the change in MEK. Similarly, we also found that the expression of ERK in aging mice tended to be up-regulated (P>0.05) (Fig. 10F and G). It has also been demonstrated that the MAPK pathway is important and that the aging liver is closely related to cancer-related pathways.
Fig. 10.
Changes of proteins in the MAPK pathway. (A) Western blot (WB) detected the expression levels of BRAF, MEK, and ERK. The original WB images were available in the supplementary file. (B, C) BRAF protein expression in young and old mice. (D, E) MEK protein expression in young and old mice. (F, G) ERK protein expression in young and old mice. (***) P < 0.05.
4. Discussion
The subject of aging has gained increasing academic interest over the years. Some organs (such as liver [21,22], kidney [23], and brain [24]), some biological fluids [25] (such as serum and urine), and some aging-related diseases (such as Alzheimer's disease [26]) were used to explore the changes of aging. However, an in-depth and comprehensive understanding of the mechanism is yet lacking. Recently in a review article, Wiley and Campisi pointed out that senescence changes cellular and body metabolic states, disrupts metabolic homeostasis, and metabolic dysfunction in turn drives senescence, thus a feedback loop is developed [27]. These metabolic balances, which certainly involve the participation of numerous proteins and metabolites, can help identify potential causal mechanisms during aging. Several studies have been conducted on metabolism during liver aging [[28], [29], [30], [31], [32]]. Proteome [22,33,34] and post-translational modification of proteins, such as glycation [35], succinate of protein thiols [36], the acetylation/acylation of lysine side chains [37] have also been studied in aging. Modifications in phosphorylation, the most prevalent post-translational modification of proteins, have also been investigated in aging as well. For example, researchers have examined phosphoproteomic alterations in aged skeletal muscle [38,39] and Alzheimer's disease [40,41]. However, few researches have been conducted on phosphorylation alterations in the aging liver. Research has shown that the hallmarks of aging in the liver include epigenetic alterations, loss of proteostasis, deregulated nutrient sensing caused metabolic disorder, mitochondrial dysfunction, and so on [42,43]. However, most of the previous studies only focus on a specific aspect of the mechanism of liver aging.The metabolic disorder and loss of proteostasis are important mechanisms of liver aging, which can be revealed by metabolomic and phosphoproteomic, few studies have conducted a joint of the two aspects. Herein, we characterized the phenotype, metabolomic, and phosphoproteomic alterations in young vs. old mice, thereby exploring the mechanisms of aging liver.
Several studies have proved that with the aggravation of aging, general phenotypes also change significantly. Shoji et al. analyzed the body weight of male C57BL/6J mice, found that the body weight of middle-aged mice (10–11 and 16–17 months) and old‐aged mice (22 months) were higher than young mice (1–2 months), and old‐aged mice were lighter than middle‐aged mice [44]. In our results, the body weight of 22-month-old mice was also higher than that of 3-month-old mice, which was consistent with the above results. But due to the lack of an intermediate age control group, the clear relationship between aging and body weight needs to be further confirmed. Grip strength reflects the overall strength of the body, upper limb function, and nutritional status. The decline in grip strength has been regarded as a sign of aging [45]. Grip strength has become a tool to evaluate the healthy life of aging mice [46]. Aging mice had significantly smaller grip strength than young mice, which is consistent with previous studies [16,44]. The changes in liver function reflect the level of liver injury, which are different in different researches [28,[47], [48], [49]]. Consistent with the findings of Luo's study, we discovered that ALT and AST levels in the serum increased with age [28]. However, serum TC and TG levels didn't show a downward or upward trend with age. These results were consistent with the previous report [48]. The changes in the normal tissue structure of the aging liver were detected by optical and electron microscopy, such as loose swelling of hepatocytes and infiltration of inflammatory cells, which were consistent with the former studies [28,48]. Hepatic inflammation can lead to liver fibrosis, and fibrosis increases with the age of the mouse liver, which was consistent with our results [50]. Similarly, the deposition of lipid droplets in the aging liver was also consistent with the study published in 2019 [49]. As we have observed, liver mitochondria also change with age. Mitochondrial membrane damage and mitochondrial crest rupture were consistent with previous studies [42,48].
Omics techniques have become a favorite tool for aging research, which has great potential in evaluating multifactor aging. Some other studies have pointed out that the aging process involves metabolic changes related to glucose, amino acids, fat, and redox reactions [5,51]. According to our metabolomics results, metabolites and their related pathways, including lipids, organic acids, and organic oxygen compounds, were also detected. The changes in the genome and epigenome will lead to dysregulation of mitochondrial function and imbalance of the nutrition perception pathway, resulting in cell senescence and low-grade inflammation, leading to liver senescence [42]. Studies have shown that chronic inflammation and fibrosis of the liver are closely related to liver aging [42,52]. Macrophages also play an important role in aging liver, autophagy and phagocytosis are generally changed in senescent hepatocytes, which can induce the impaired of signal transduction [53]. In the metabolomic results, a variety of inflammation-related pathways have been detected, such as FoxO, ErbB, NF-kappa B, etc. In our phosphoproteomics results, the sites in S273, S288, S304 and S319 of myc box-dependent-interacting protein 1(A0A3Q4EBK4) and S347 of high affinity immunoglobulin gamma Fc receptor I (FCGR1) were up-regulated in the autophagy-related Fc gamma R-mediated phagocytosis pathway. The S263 and S231 phosphorylation sites of HSP90A were up-regulated in antigen processing and presentation, IL-17 signaling pathway and Th17 cell differentiation. Necroptosis can lead to chronic inflammation and fibrosis of the liver [50]. There are four pathways related to cell senescence and death, apoptosis, necroptosis, cell cycle, p53 signaling pathway. Among them, the phosphorylation levels of the S519 and S523 sites of apoptosis-inducing factor 1 (AIFM1), the S427 site of Lamin-B2 (LMNB2) and the S27 site of DNA replication licensing factor (MCM2) were down-regulated, the S338 and S 320 sites of Etoposide-induced protein 2.4 (EI24) were up-regulated. When liver injury and inflammation continue to worsen, the liver does not recover normally, leading to fibrosis. IL-17, p53, and NF-kappaB signal pathways are also crucial in hepatic fibrosis [54], and all of these pathways were discovered in our findings. The expression of senescence markers in the liver is beneficial to the clonal expansion of normal hepatocytes, creating a microenvironment that promotes cancer development, increasing the overall risk for cancer development associated with aging [55]. Moreover, we found cancer-related metabolic pathways. According to Aging Atlas, which is a multi-omics database on aging [56], we observed that the metabolites allied to cancer-related metabolic pathways, such as phenylalanine and malate, were associated with aging. In addition, alterations were detected in 1-stearoyl-2-arachidonoyl-sn-glycerol (DAG), l-malic acid, progesterone, and dl-glutamic acid, suggesting that these metabolites might become biomarkers of aging. In particular, DAG is involved in multiple common pathways of two omics and may be worth further exploration. Ando et al. analyzed the metabolomes and lipidomes of the liver in young and aged mice, DAG levels were elevated during aging [57]. Ishizuka et al. fund that the concentrations of DAG were markedly increased following high-fat diet in elder mice [58]. According to our findings, DAG participates in many signaling pathways, but further research is needed to determine how exactly it works. Some studies reported aging-related pathways (such as the AMPK, FoxO, p53, mTOR, and MAPK), and aging-related proteins (such as SIRT, CREB, and APOE) [[59], [60], [61]], were also detected in our results. Consistent with the results of metabolomics, some genes related to cancer pathways (such as Pml, Hsp90aa1, Cbr1, Fcgr1, Tpm3, Sp100, Atp5f1a, Scrib, and Ank2) were also detected in the Aging Atlas database. Some pathways in phosphoproteomics were also associated with cancer, prompting us to explore the correlation between aging and cancer.
Next, we conducted a cross-study of the pathways of two omics. Consequently, 65 common metabolic pathways were detected. Upregulation of HSP90A at S263 and S231 and BRAF at S135 sites was found in various signal pathways. Notably, the pathways in cancer were significantly enriched. A few studies have shown that aging and cancer are inseparable [[62], [63], [64]], suggesting that oncogene activation and inactivation induce senescence. Based on the Uniport database [65], HSP90A promotes the maturation, structural preservation, and regulation of specific target proteins. Heat shock proteins (HSPs) participate in the process of protein folding. Protein folding disorder in the liver leads to the accumulation of misfolded proteins and lipotoxicity, amplification of ROS and inflammation, leading to the occurrence of aging-related diseases [42]. Quantitative proteomics of rat livers have shown that the increase of HSPs levels represent the occurrence of chronic stress, which lead to premature senility and shorter life expectancy [66]. HSP90 has become a therapeutic target for cardiac aging [67]. Our previous study has shown that exosomes down-regulated the level of HSP90A at S263 in aging liver, indicating that HSP90 is also expected to be a target for the treatment of liver aging and age-related diseases [68]. Interestingly, BRAF participates in multiple signal pathways. BRAF is a critical kinase in the Ras/Raf/MEK/ERK cascade, phosphorylates MAP2K1, and activates the MAPK signal transduction pathway, involving the transduction of mitogenic signals from the cell membrane to the nucleus [69]. Previous studies have shown that the suppression of the Ras/Raf/MEK/ERK pathway may prevent the induction of aging [60,61]. In human plasma, human skeletal muscle, mouse hippocampi, and white adipose tissue, the overrepresented RAF was observed with increased age [61]. Many BRAF inhibitors can exert anti-tumor and anti-aging effects. In normal cells, oncogene-induced senescence can be triggered by BRAF-V600E, ouabain and chloroquine, as selectively toxic compounds, can inhibit autophagy, and eliminate BRAF‐senescent cells [70]. Studies have shown that trametinib inhibited the MEK kinase, disrupts the related RAF-MEK complex, and prolongs the Drosophila lifespan through the Ras-Erk-ETS pathway, which affects the insulin/IGF-1 (IIS) signaling pathway [71]. Jae et al. have shown that BRAF inhibition, SB590885, induces the recovery of mitochondrial function and metabolic reprogramming, thus improving senescence [72]. Blagosklonny et al. reported that cell senescence was caused by the overactivation of growth-promoting pathways, such as MEK/MAPK and PI-3K/mTOR, which consists of hyper-activated Ras, Raf, and MEK induces cellular senescence [73]. In our results, the S135 phosphorylation site of BRAF is significantly up-regulated, suggesting that this site may be a target site for some anti-aging drugs. At the same time, the results of WB showed that the expression of MEK (the downstream protein of MEK) was up-regulated in aging mice, but not the expression of BRAF. Phosphorylation was the most common protein modification type of BRAF protein, which confirms that the phosphorylation of BRAF protein may be changed. Thus, it can be concluded that the aging mechanism is linked to the occurrence of tumors, and RAF and its related MAPK family may be the specific mechanism underlying the correlation between aging and cancer. Our findings suggested that BRAF and HSP90A have a high association and participate in various signal pathways, but the specific interaction between them and whether they share a role in aging requires additional investigation.
Nevertheless, the present study has some limitations. Firstly, phosphoproteomics can not determine whether the change in phosphorylated protein contents is caused by the overall protein abundance or the percentage of phosphorylation at the specific site. Further validation of total protein abundance is needed to support our results. we did not describe total proteome analysis, and only performed WB verification on BRAF protein and its downstream sites. Second, we only performed WB verification on BRAF protein and its downstream sites. The changes in phosphorylation sites haven't been verified, because it's challenging to locate phosphorylated antibodies at particular protein locations. In the next step, the related protein sites may be verified by the parallel reaction monitoring method. In addition, the correlation between HSP90A and BRAF has not been further verified. It would be of great interest to further explore the molecular mechanisms for the relationship between HSP90A and BRAF.
5. Conclusions
In summary, the general phenotype of aging mice changed significantly. The combined results of metabolomics and phosphorylated proteomics revealed the changes in metabolites and phosphorylated proteins in natural liver aging, which provided genomic resources for further study of the mechanism of aging. We also concluded that aging might be related to tumor development, BRAF, HSP90A, and the MAPK pathway in the occurrence of aging.
Funding
This research was funded by the Key Technology Research and Development Program of Shandong [grant number 2017GSF218101]; the National Natural Science Foundation of China [grant number 81700725]; the Natural Science Foundation of Shandong Province [grant number ZR2017BH003]; the Natural Science Foundation of Shandong Province [grant number ZR2021MH008]; the Key Technology Research and Development Program of Shandong [grant number 2022CXGC010510] and the Natural Science Foundation of Shandong Province [grant number ZR20222M193].
Data availability statement
The data associated with our study have been deposited into publicly available repositories. Metabolomics data have been deposited to the EMBL-EBI463 MetaboLights database with the identifier MTBLS5001. The complete dataset can be accessed here: https://www.ebi.ac.uk/metabolights/MTBLS5001.The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD034392.
Ethics statement
This study was reviewed and approved by the Laboratory Animal Ethical and Welfare Committee of Shandong University Cheeloo College of Medicine, with the approval number: 20157.
CRediT authorship contribution statement
Cong-min Tang: Formal analysis, Investigation, Methodology, Writing – original draft, Conceptualization, Data curation. Zhen Zhang: Conceptualization, Formal analysis, Funding acquisition, Investigation, Writing – review & editing, Data curation. Yan Sun: Formal analysis, Investigation, Methodology. Wen-jing Ding: Methodology. Xue-chun Yang: Methodology. Yi-ping Song: Formal analysis, Investigation. Ming-ying Ling: Funding acquisition. Xue-hui Li: Formal analysis, Investigation. Rong Yan: Methodology. Yu-jing Zheng: Formal analysis, Investigation. Na Yu: Resources. Wen-hua Zhang: Resources. Yong Wang: Funding acquisition, Resources. Shao-peng Wang: Resources. Hai-qing Gao: Resources. Chuan-li Zhao: Supervision. Yan-qiu Xing: Conceptualization, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Bohua Zhao and Zheng Li for the permission to use the microscope for the experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21011.
Contributor Information
Chuan-li Zhao, Email: chuanlizhao@email.sdu.edu.cn.
Yan-qiu Xing, Email: xingyanqiu@sina.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Wu L., Xie X., Liang T., et al. Integrated multi-omics for novel aging biomarkers and antiaging targets. Biomolecules. 2021;12(1) doi: 10.3390/biom12010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azman Khairunnuur Fairuz, Safdar Afifa, Zakaria Rahimah. D-galactose-induced liver aging model: its underlying mechanisms and potential therapeutic interventions. Exp. Gerontol. 2021:150. doi: 10.1016/j.exger.2021.111372. [DOI] [PubMed] [Google Scholar]
- 3.Sheedfar F., Di Biase S., Koonen D., et al. Liver diseases and aging: friends or foes? Aging Cell. 2013;12(6):950–954. doi: 10.1111/acel.12128. [DOI] [PubMed] [Google Scholar]
- 4.Kudryashova K.S., Burka K., Kulaga A.Y., et al. Aging biomarkers: from functional tests to multi-omics approaches. Proteomics. 2020;20(5–6) doi: 10.1002/pmic.201900408. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava S. Emerging insights into the metabolic alterations in aging using metabolomics. Metabolites. 2019;9(12) doi: 10.3390/metabo9120301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Z., Zhai G., Singmann P., et al. Human serum metabolic profiles are age dependent. Aging Cell. 2012;11(6):960–967. doi: 10.1111/j.1474-9726.2012.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menni C., Kastenmüller G., Petersen A.K., et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 2013;42(4):1111–1119. doi: 10.1093/ije/dyt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman J.M., Soltow Q.A., Li S., et al. Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, Drosophila melanogaster. Aging Cell. 2014;13(4):596–604. doi: 10.1111/acel.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni Lochlainn M., BowyerC R.C.E., Steves J. Dietary protein and muscle in aging people: the potential role of the gut microbiome. Nutrients. 2018;10(7) doi: 10.3390/nu10070929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nik Mohd Fakhruddin N.N.I., Shahar S., Ismail I.S., et al. Urine untargeted metabolomic profiling is associated with the dietary pattern of successful aging among Malaysian elderly. Nutrients. 2020;12(10) doi: 10.3390/nu12102900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanhooren V., Navarrete Santos A., Voutetakis K., et al. Protein modification and maintenance systems as biomarkers of ageing. Mech. Ageing Dev. 2015;151:71–84. doi: 10.1016/j.mad.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 12.SantosA A.L., Lindner B. Protein posttranslational modifications: roles in aging and age-related disease. Oxid. Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/5716409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkhitko A.A., Binari R., Zhang N., et al. Tissue-specific down-regulation of S-adenosyl-homocysteine via suppression of dAhcyL1/dAhcyL2 extends health span and life span in Drosophila. Genes Dev. 2016;30(12):1409–1422. doi: 10.1101/gad.282277.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y., Kwon O.K., Chae S., et al. Quantitative proteomic analysis of changes related to age and calorie restriction in rat liver tissue. Proteomics. 2018;18(5–6) doi: 10.1002/pmic.201700240. [DOI] [PubMed] [Google Scholar]
- 15.Braithwaite S.P., Voronkov M., Stock J.B., et al. Targeting phosphatases as the next generation of disease modifying therapeutics for Parkinson's disease. Neurochem. Int. 2012;61(6):899–906. doi: 10.1016/j.neuint.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 16.Takeshita H., Yamamoto K., Nozato S., et al. Modified forelimb grip strength test detects aging-associated physiological decline in skeletal muscle function in male mice. Sci. Rep. 2017;7 doi: 10.1038/srep42323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J., Xian L., Zheng R., et al. Canthaxanthin shows anti-liver aging and anti-liver fibrosis effects by down-regulating inflammation and oxidative stress in vivo and in vitro. Int. Immunopharm. 2022;110 doi: 10.1016/j.intimp.2022.108942. [DOI] [PubMed] [Google Scholar]
- 18.Mohamed Slaoui, Bauchet Anne-Laure, Fiette Laurence. Drug Safety Evaluation: Methods and Protocols, Jean-Charles Gautier. Springer New York; New York, NY: 2017. Tissue sampling and processing for histopathology evaluation; pp. 101–114. [DOI] [PubMed] [Google Scholar]
- 19.Tizro Parastou, Choi Cecilia, Khanlou Negar. In: Biobanking: Methods and Protocols. Yong William H., editor. Springer New York; New York, NY: 2019. Sample preparation for transmission electron microscopy; pp. 417–424. [DOI] [PubMed] [Google Scholar]
- 20.Ding W.J., Li X.H., Tang C.M., et al. Quantification and proteomic characterization of β-hydroxybutyrylation modification in the hearts of AMPKα2 knockout mice. Mol. Cell. Proteomics. 2023;22(2) doi: 10.1016/j.mcpro.2023.100494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rui L. Energy metabolism in the liver. Compr. Physiol. 2014;4(1):177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin B., FordR K.I., Robinson A.S. Quantitative proteomics to study aging in rabbit liver. Mech. Ageing Dev. 2020;187 doi: 10.1016/j.mad.2020.111227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takemon Y., Chick J.M., Gerdes Gyuricza I., et al. Proteomic and transcriptomic profiling reveal different aspects of aging in the kidney. Elife. 2021;10 doi: 10.7554/eLife.62585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaev N.K., StelmashookE E.V., Genrikhs E. Neurogenesis and brain aging. Rev. Neurosci. 2019;30(6):573–580. doi: 10.1515/revneuro-2018-0084. [DOI] [PubMed] [Google Scholar]
- 25.Adav S.S., Wang Y. Metabolomics signatures of aging: recent advances. Aging Dis. 2021;12(2):646–661. doi: 10.14336/AD.2020.0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adav S.S., ParkS J.E., Sze K. Quantitative profiling brain proteomes revealed mitochondrial dysfunction in Alzheimer's disease. Mol. Brain. 2019;12(1):8. doi: 10.1186/s13041-019-0430-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campisi C.D. WileyJ. The metabolic roots of senescence: mechanisms and opportunities for intervention. Nat. Metab. 2021;3(10):1290–1301. doi: 10.1038/s42255-021-00483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo D., Li J., Chen K., et al. Study on metabolic trajectory of liver aging and the effect of fufang zhenzhu tiaozhi on aging mice. Front. Pharmacol. 2019;10:926. doi: 10.3389/fphar.2019.00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Son N., Hur H.J., Sung M.J., et al. Liquid chromatography-mass spectrometry-based metabolomic analysis of livers from aged rats. J. Proteome Res. 2012;11(4):2551–2558. doi: 10.1021/pr201263q. [DOI] [PubMed] [Google Scholar]
- 30.Song J., Zhou Y.Z., Pang Y.Y., et al. The anti-aging effect of Scutellaria baicalensis Georgi flowers extract by regulating the glutamine-glutamate metabolic pathway in d-galactose induced aging rats. Exp. Gerontol. 2020;134 doi: 10.1016/j.exger.2020.110843. [DOI] [PubMed] [Google Scholar]
- 31.Zhao F., Chang Y., Gao L., et al. Protective effects of Scutellaria baicalensis Georgi extract on D-galactose induced aging rats. Metab. Brain Dis. 2018;33(5):1401–1412. doi: 10.1007/s11011-018-0229-z. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F., Kerbl-Knapp J., Akhmetshina A., et al. Tissue-specific landscape of metabolic dysregulation during ageing. Biomolecules. 2021;11(2) doi: 10.3390/biom11020235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Couteur D.G., Solon-Biet S.M., Parker B.L., et al. Nutritional reprogramming of mouse liver proteome is dampened by metformin, resveratrol, and rapamycin. Cell Metab. 2021;33(12):2367–2379.e4. doi: 10.1016/j.cmet.2021.10.016. [DOI] [PubMed] [Google Scholar]
- 34.Williams E.G., Pfister N., Roy S., et al. Multiomic profiling of the liver across diets and age in a diverse mouse population. Cell Syst. 2022;13(1):43–57.e6. doi: 10.1016/j.cels.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.FanV X., Monnier M. Protein posttranslational modification (PTM) by glycation: role in lens aging and age-related cataractogenesis. Exp. Eye Res. 2021;210 doi: 10.1016/j.exer.2021.108705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jové M., Pradas I., Mota-Martorell N., et al. Succination of protein thiols in human brain aging. Front. Aging Neurosci. 2020;12:52. doi: 10.3389/fnagi.2020.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blasl A.T., Schulze S., Qin C., et al. Post-translational lysine ac(et)ylation in health, ageing and disease. Biol. Chem. 2022;403(2):151–194. doi: 10.1515/hsz-2021-0139. [DOI] [PubMed] [Google Scholar]
- 38.Bareja A., Draper J.A., Katz L.H., et al. Chronic caloric restriction maintains a youthful phosphoproteome in aged skeletal muscle. Mech. Ageing Dev. 2021;195 doi: 10.1016/j.mad.2021.111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell M.D., Martín-Pérez M., Egertson J.D., et al. Elamipretide effects on the skeletal muscle phosphoproteome in aged female mice. Geroscience. 2022;44(6):2913–2924. doi: 10.1007/s11357-022-00679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ping L., Kundinger S.R., Duong D.M., et al. Global quantitative analysis of the human brain proteome and phosphoproteome in Alzheimer's disease. Sci. Data. 2020;7(1):315. doi: 10.1038/s41597-020-00650-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D., Jo Y.S., Jo H.S., et al. Comparative phosphoproteomics of neuro-2a cells under insulin resistance reveals new molecular signatures of Alzheimer's disease. Int. J. Mol. Sci. 2022;23(2) doi: 10.3390/ijms23021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt N.J., Kang S.W.S., Lockwood G.P., et al. Hallmarks of aging in the liver. Comput. Struct. Biotechnol. J. 2019;17:1151–1161. doi: 10.1016/j.csbj.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biomarker Consortium Aging, Bao H., Cao J., et al. Biomarkers of aging. Sci. China Life Sci. 2023;66(5):893–1066. doi: 10.1007/s11427-023-2305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyakawa H. ShojiT. Age-related behavioral changes from young to old age in male mice of a C57BL/6J strain maintained under a genetic stability program. Neuropsychopharmacol Rep. 2019;39(2):100–118. doi: 10.1002/npr2.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bohannon R.W. Grip strength: an indispensable biomarker for older adults. Clin. Interv. Aging. 2019;14:1681–1691. doi: 10.2147/CIA.S194543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellantuono I., De Cabo R., Ehninger D., et al. A toolbox for the longitudinal assessment of healthspan in aging mice. Nat. Protoc. 2020;15(2):540–574. doi: 10.1038/s41596-019-0256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preuss H.G., Kaats G.R., Mrvichin N., et al. Circulating ALT levels in healthy volunteers over life-span: assessing aging paradox and nutritional implications. J. Am. Coll. Nutr. 2019;38(8):661–669. doi: 10.1080/07315724.2019.1580169. [DOI] [PubMed] [Google Scholar]
- 48.Lysek-Gladysinska M., Wieczorek A., Jozwik A., et al. Aging-related changes in the ultrastructure of hepatocytes and cardiomyocytes of elderly mice are enhanced in ApoE-deficient animals. Cells. 2021;10(3) doi: 10.3390/cells10030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seo E., Kang H., Choi H., et al. Reactive oxygen species-induced changes in glucose and lipid metabolism contribute to the accumulation of cholesterol in the liver during aging. Aging Cell. 2019;18(2) doi: 10.1111/acel.12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohammed S., Thadathil N., Selvarani R., et al. Necroptosis contributes to chronic inflammation and fibrosis in aging liver. Aging Cell. 2021;20(12) doi: 10.1111/acel.13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houtkooper R.H., Argmann C., Houten S.M., et al. The metabolic footprint of aging in mice. Sci. Rep. 2011;1:134. doi: 10.1038/srep00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim I.H., Xu J., Liu X., et al. Aging increases the susceptibility of hepatic inflammation, liver fibrosis and aging in response to high-fat diet in mice. Age (Dordr). 2016;38(4):291–302. doi: 10.1007/s11357-016-9938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stahl E.C., Haschak M.J., Popovic B., et al. Macrophages in the aging liver and age-related liver disease. Front. Immunol. 2018;9:2795. doi: 10.3389/fimmu.2018.02795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.SekiD E., Brenner A. Recent advancement of molecular mechanisms of liver fibrosis. J Hepatobiliary Pancreat Sci. 2015;22(7):512–518. doi: 10.1002/jhbp.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasciu D., Montisci S., Greco M., et al. Aging is associated with increased clonogenic potential in rat liver in vivo. Aging Cell. 2006;5(5):373–377. doi: 10.1111/j.1474-9726.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- 56.Consortium Aging Atlas Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 2021;49(D1):D825–D830. doi: 10.1093/nar/gkaa894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ando A., Satomi M. OkaY. Deoxysphingolipids and ether-linked diacylglycerols accumulate in the tissues of aged mice. Cell Biosci. 2019;9:61. doi: 10.1186/s13578-019-0324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishizuka K., Kon K., Lee-Okada H.C., et al. Aging exacerbates high-fat diet-induced steatohepatitis through alteration in hepatic lipid metabolism in mice. J. Gastroenterol. Hepatol. 2020;35(8):1437–1448. doi: 10.1111/jgh.15006. [DOI] [PubMed] [Google Scholar]
- 59.Christensen K., JohnsonJ T.E., Vaupel W. The quest for genetic determinants of human longevity: challenges and insights. Nat. Rev. Genet. 2006;7(6):436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tabibzadeh Siamak. vol. 26. 2021. pp. 50–96. (Signaling Pathways and Effectors of Aging). 1. [DOI] [PubMed] [Google Scholar]
- 61.Moaddel R., Ubaida-Mohien C., Tanaka T., et al. Proteomics in aging research: a roadmap to clinical, translational research. Aging Cell. 2021;20(4) doi: 10.1111/acel.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatsirisupachai K., Lesluyes T., Paraoan L., et al. An integrative analysis of the age-associated multi-omic landscape across cancers. Nat. Commun. 2021;12(1):2345. doi: 10.1038/s41467-021-22560-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calcinotto A., Kohli J., Zagato E., et al. Cellular senescence: aging, cancer, and injury. Physiol. Rev. 2019;99(2):1047–1078. doi: 10.1152/physrev.00020.2018. [DOI] [PubMed] [Google Scholar]
- 64.Serrano M. Unraveling the links between cancer and aging. Carcinogenesis. 2016;37(2):107. doi: 10.1093/carcin/bgv100. [DOI] [PubMed] [Google Scholar]
- 65.Consortium Uniprot UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gat-Yablonski G., Finka A., Pinto G., et al. Quantitative proteomics of rat livers shows that unrestricted feeding is stressful for proteostasis with implications on life span. Aging (Albany NY) 2016;8(8):1735–1758. doi: 10.18632/aging.101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dabravolski S.A., Sukhorukov V.N., Kalmykov V.A., et al. Heat shock protein 90 as therapeutic target for CVDs and heart ageing. Int. J. Mol. Sci. 2022;23(2) doi: 10.3390/ijms23020649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ling M., Tang C., Yang X., et al. Integrated metabolomics and phosphoproteomics reveal the protective role of exosomes from human umbilical cord mesenchymal stem cells in naturally aging mouse livers. Exp. Cell Res. 2023;427(1) doi: 10.1016/j.yexcr.2023.113566. [DOI] [PubMed] [Google Scholar]
- 69.Zaman A., WuT W., Bivona G. Targeting oncogenic BRAF: past, present, and future. Cancers. 2019;11(8) doi: 10.3390/cancers11081197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.L'hote V., Courbeyrette R., Pinna G., et al. Ouabain and chloroquine trigger senolysis of BRAF-V600E-induced senescent cells by targeting autophagy. Aging Cell. 2021;20(9) doi: 10.1111/acel.13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Slack C., Alic N., Foley A., et al. The ras-erk-ETS-signaling pathway is a drug target for longevity. Cell. 2015;162(1):72–83. doi: 10.1016/j.cell.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J.W., Kuk M.U., Choy H.E., et al. Mitochondrial metabolic reprograming via BRAF inhibition ameliorates senescence. Exp. Gerontol. 2019;126 doi: 10.1016/j.exger.2019.110691. [DOI] [PubMed] [Google Scholar]
- 73.Blagosklonny M.V. Aging-suppressants: cellular senescence (hyperactivation) and its pharmacologic deceleration. Cell Cycle. 2009;8(12):1883–1887. doi: 10.4161/cc.8.12.8815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with our study have been deposited into publicly available repositories. Metabolomics data have been deposited to the EMBL-EBI463 MetaboLights database with the identifier MTBLS5001. The complete dataset can be accessed here: https://www.ebi.ac.uk/metabolights/MTBLS5001.The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD034392.