Abstract
Purpose
To develop and evaluate a deep learning algorithm for Meibomian gland characteristics calculation.
Design
Evaluation of diagnostic technology.
Subjects
A total of 1616 meibography images of both the upper (697) and lower (919) eyelids from a total of 282 individuals.
Methods
Images were collected using the LipiView II device. All the provided data were split into 3 sets: the training, validation, and test sets. Data partitions used proportions of 70/10/20% and included data from 2 optometry settings. Each set was separately partitioned with these proportions, resulting in a balanced distribution of data from both settings. The images were divided based on patient identifiers, such that all images collected for one participant could end up only in one set. The labeled images were used to train a deep learning model, which was subsequently used for Meibomian gland segmentation. The model was then applied to calculate individual Meibomian gland metrics. Interreader agreement and agreement between manual and automated methods for Meibomian gland segmentation were also carried out to assess the accuracy of the automated approach.
Main Outcome Measures
Meibomian gland metrics, including length ratio, area, tortuosity, intensity, and width, were measured. Additionally, the performance of the automated algorithms was evaluated using the aggregated Jaccard index.
Results
The proposed semantic segmentation–based approach achieved average aggregated Jaccard index of mean 0.4718 (95% confidence interval [CI], 0.4680–0.4771) for the ‘gland’ class and a mean of 0.8470 (95% CI, 0.8432–0.8508) for the ‘eyelid’ class. The result for object detection–based approach was a mean of 0.4476 (95% CI, 0.4426–0.4533). Both artificial intelligence–based algorithms underestimated area, length ratio, tortuosity, widthmean, widthmedian, width10th, and width90th. Meibomian gland intensity was overestimated by both algorithms compared with the manual approach. The object detection–based algorithm seems to be as reliable as the manual approach only for Meibomian gland width10th calculation.
Conclusions
The proposed approach can successfully segment Meibomian glands; however, to overcome problems with gland overlap and lack of image sharpness, the proposed method requires further development. The study presents another approach to utilizing automated, artificial intelligence–based methods in Meibomian gland health assessment that may assist clinicians in the diagnosis, treatment, and management of Meibomian gland dysfunction.
Financial Disclosure(s)
The authors have no proprietary or commercial interest in any materials discussed in this article.
Key Words: Artificial intelligence, Deep learning, Image processing, Meibography, Meibomian gland imaging, Meibomian glands, Meibomian gland structure
Application of artificial intelligence (AI) is one of the most promising areas of health innovation, particularly in medical imaging. Artificial intelligence–based techniques are advancing rapidly and can improve medical imaging for disease screening, progression, and management.1, 2, 3, 4 A considerable amount of literature has been published on utility of AI in meibography.5,6,7, 8, 9, 10, 11, 12, 13, 14, 15 Deep learning models can be trained using large data sets of labeled meibography images to automatically identify and segment the Meibomian glands. This enables the calculation of various Meibomian gland metrics such as length, area, tortuosity, intensity, or width, providing more quantitative and objective measures for diagnosis and monitoring of Meibomian gland dysfunction (MGD). In addition, deep learning models can also be used to predict the severity of MGD and the likelihood of developing related ocular surface diseases, based on the extracted Meibomian gland features. This can help clinicians make more informed decisions about treatment and management strategies for their patients. The aforementioned studies have proposed various approaches to quantify visible Meibomian gland structure, which have successfully replaced other conventional approaches such as thresholding, edge detection, region growing, and clustering.16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26 These approaches rely on predefined rules and heuristics to identify and segment different regions of an image. In contrast, deep learning methods use neural networks to learn representations of features directly from the image data, without the need for manual feature engineering. This allows for more accurate and robust segmentation performance, especially when dealing with complex and variable image features. The analysis of morphometric parameters of the Meibomian glands can improve the diagnosis of MGD in several ways. First, quantitative measurements of Meibomian gland metrics such as length, area, tortuosity, intensity, and width, obtained through image analysis techniques like deep learning, provide more objective and accurate measures of the gland structure and function. This can help clinicians identify and track changes in Meibomian gland morphology over time and better distinguish between normal and abnormal gland structures. Second, the analysis of Meibomian gland morphometric parameters can aid in the grading and classification of MGD severity, helping clinicians to differentiate between mild, moderate, and severe cases of the disease. This is particularly important because MGD is a multifactorial condition that can present with a range of symptoms and signs, making diagnosis challenging. Finally, the analysis of Meibomian gland metrics can also be used to monitor the efficacy of MGD treatments over time. By tracking changes in Meibomian gland morphology before and after treatment, clinicians can determine the effectiveness of different therapies and adjust treatment plans accordingly. Overall, the analysis of morphometric parameters of the Meibomian glands provides valuable quantitative information that can improve the accuracy, objectivity, and efficiency of MGD diagnosis, grading, and treatment monitoring. Many objective methods and metrics have been proposed to quantify Meibomian gland morphology15,18, 19, 20, 21, 22,24, 25, 26, 27; specifically, metrics such as length, width, tortuosity, area of loss, contrast, and density have been proposed to describe individual Meibomian gland characteristics.9,20,22,24,28,29 Dai et al14 have recently found that AI-based calculations of Meibomian gland length, width, and area ratio were significantly correlated with subjective symptoms, tear break up time, lid margin abnormalities, and Meibomian gland expressibility. Prabhu et al6 demonstrated that healthy patients have higher numbers and longer and fewer tortuous Meibomian gland structures than those with disease when metrics are calculated using a deep learning approach. Furthermore, Wang et al9 showed that low Meibomian gland contrast was the primary indicator for AI-based ghost Meibomian gland (glands that have a faint appearance in meibography) identification.
Proper assessment of Meibomian glands can help to diagnose MGD and other conditions that affect the health of the eyes. This can involve various techniques, such as meibography, which is a noninvasive imaging technique that allows clinicians to visualize the structure and function of the glands. By accurately diagnosing MGD and other conditions that affect the Meibomian glands, clinicians can develop targeted treatment plans that address the underlying causes of the condition. This may involve the use of medications, warm compresses, lid hygiene, or other interventions designed to improve gland function and reduce symptoms.
The LipiView II system (Johnson & Johnson Vision), formally termed the LipiView II Ocular Surface Interferometer with Dynamic Meibomian Imaging, can be used to perform noncontact meibography, measure lipid layer thickness, analyze blink dynamics, and take a photograph of the external ocular surface. The device works under 3 different imaging modes: dynamic illumination (Fig 1A), adaptive transillumination (Fig 1B), and dual-mode Dynamic Meibomian imaging (Fig 1C), which combines dynamic illumination and adaptive transillumination.30 The dynamic illumination offers a more accurate estimation of gland morphology as it reduces the amount of glare and backscatter from the Meibomian glands by emitting light from various locations. The adaptive transillumination imaging automatically compensates for lid thickness variations, thanks to 3 independently controlled light sources for each part of the eyelid. The dual mode captures 2 separate images, 1/30 second apart, and then combines them into a single image to increase the amount of visible detail and image contrast. It has been shown that glands that seemed truncated or absent with standard noncontact infrared meibography were more defined and visible with dual mode.31
Figure 1.
Different imaging techniques showing Meibomian glands captured with LipiView II. Images captured with dynamic illumination, adaptive transillumination, and dual mode, respectively. (A) Direct illumination. (B) Transillumination. (C) Dual mode.
In this study, 2 approaches for objective Meibomian gland structure analysis are compared: a semantic segmentation–based approach and an object detection–based approach. In addition, the performance of both approaches was compared against manual annotations of Meibomian glands by an independent reader. The interreader agreement was also evaluated. In the long term, the proposed methods could be a useful tool in assisting clinicians with MGD diagnosis, treatment, and management.
Methods
Data set and Preprocessing
A total of 1616 meibography images of both upper (697) and lower eyelids (919) were collected using the LipiView II Ocular Surface Interferometer at 2 optometry settings in the United Kingdom (Eurolens Research at the University of Manchester and BBR Optometry Ltd) between 2017 and 2022. For this study, only images acquired using dynamic illumination mode were included. This mode corresponds to the direct illumination method of Meibomian gland imaging and is one of the imaging modes available in LipiView, which also offers transillumination. In dynamic illumination mode, surface lighting is generated from multiple light sources to minimize reflection. This study was approved by the University Research Ethics Committee of The University of Manchester before data extraction. All procedures adhered to the tenets of the Declaration of Helsinki, and all participants provided written informed consent before data extraction. Consent was obtained retrospectively from all participants for their anonymized images to be analyzed for the purpose of deep learning–model development. Multiple images from 282 individuals for both eyes were used in the study. The images were sourced from a diverse range of patients, including both healthy individuals and those with varying degrees of Meibomian gland dropout, who were predominantly from standard optometry practice and spanned a wide age range. The images were received and processed as JPG files. The first preprocessing step involved center-cropping to 640 × 900 pixels to focus on the relevant area. For the semantic segmentation–based approach, the following image augmentation methods were applied: random left/right reflection and random X/Y translation of ±10 pixels.
Data Annotations
All images were manually annotated using an interactive tool, the Image Labeler app, provided by MATLAB R2022a (The MathWorks, Inc). Deeplabv3+ model in the study was trained on a labeled data set of meibography images captured with direct illumination using LipiView II, with 3 distinct classes: ‘gland,’ ‘eyelid,’ and ‘background.’ In contrast, the Mask R-CNN model was trained only on the gland class. The ‘gland’ class refers to the long, hyperreflective structures that correspond to the Meibomian glands. These structures are typically located within the tarsal plate of the eyelid and have a distinct appearance compared with other structures within the image. The ‘eyelid’ class refers to regions of the image that correspond to the skin and soft tissue surrounding the eye but do not contain the Meibomian gland structures. The ‘background’ class refers to any other region of the image that is not part of the gland or eyelid structures. This can include areas of the image that contain artifacts, noise, or other structures that are not relevant to the analysis of the Meibomian glands. The Smart Polygon tool was used to estimate the shape of an object of interest within a drawn polygon. The Smart Polygon tool identifies an object of interest using regional graph-based segmentation (‘GrabCut’).32 Estimated regions of interest were further corrected with the Brush tool to make sure that gland labels were as precise as possible. A single label was assigned to each individual Meibomian gland in every image to provide ground truth data for instance-wise level network evaluation (see Video 1 [available at www.ophthalmologyscience.org]).
Data Partitioning
All the provided data were partitioned into 3 sets: the training set, which was used to create a model; the validation set, which was used for hyperparameter optimization; and the test set, which was used to obtain an unbiased assessment of model performance. The proportions of these partitions were 70/10/20%, with each set comprising data from 2 optometry settings. In other words, the 70/10/20% proportions were applied to each set separately, resulting in a balanced distribution of data from both optometry settings. This resulted in 1162 images being used for training, 153 being used for validation, and 301 being used for testing. The images were split based on patient identifier, as such all images collected from one participant could be included in one set only.
Architectural Details
Image segmentation is the process of grouping image pixels based on their content to process it for tasks such as image classification and object detection. Recent image segmentation methods can be classified into semantic and instance segmentation. In this work 2 state-of-the-art deep learning convolutional neural network (CNN) architectures were used: Deeplabv3+33 for semantic segmentation and Mask R-CNN34 for instance segmentation. Transfer learning was used to overcome the issue with relatively small data sets. Pretrained Inception-ResNet-v2 was selected as a base network for Deeplabv3+. It is a CNN that is trained on more than a million images from the ImageNet database35; as such, it reduces training time and, more importantly, the number of required images. Inception-ResNet-v2 was selected as the backbone for Deeplabv3+ due to limited choices available in the software package and also because it has shown strong performance in various computer vision tasks including medical image segmentation, as reported in the literature.36,37, 38, 39, 40 Additionally, the architecture has also been used successfully in other ocular image segmentation tasks, such as the segmentation of the optic disc and cup in glaucoma diagnosis,41,42 retinal detachment detection and diabetic retinopathy.43,44 A pretrained Mask R-CNN object detector was trained on the Common Objects in Context data set with a ResNet-50 network as the feature extractor. ResNet-101 was also considered, but it did not improve the model performance.
Evaluation Metrics
There are various ways to evaluate machine learning model performance. It is particularly important to evaluate how well a machine learning model generalizes to new, previously unseen data. As such, the model was evaluated on a test data set (20% of all images) on both pixel and object level. To quantify model performance, the following metrics were calculated: the Dice score, Intersection-Over-Union, Precision, Recall, and aggregated Jaccard index.45 Aggregated Jaccard Index was the primary metric of interest as it penalizes false positive and false negative gland detections. A receiver operating characteristic curve and a precision-recall curve were also plotted to give a more informative picture of the algorithm’s performance.
Meibomian Gland Metrics Calculation
The following Meibomian gland metrics were introduced to quantify changes over time: Meibomian gland length, length ratio, area, intensity, tortuosity, and width. Meibomian gland length was defined as the pixel-wise length of gland topological skeleton.15 The length ratio was the length of the gland with respect to eyelid height at the position of the gland, measured along the orientation of the gland (see Fig 2). Length ratio was introduced to overcome problems with inconsistency of eyelid eversion and head position.20 The individual Meibomian gland area was defined as the number of pixels within each region of interest. The mean gray level of each labeled gland (the brightness of all pixels within a single gland, range 0–255) was defined as Meibomian gland intensity. Tortuosity was defined as the ratio of the arc-length of the gland (length of the curve or length of gland topological skeleton) to the distance between the end points.15 Meibomian gland width was calculated as the average of all widths of the gland along all points on the gland central line, similar to the definition of Xiao et al.21 The gland width was measured as the distance between the gland mask edges along the line perpendicular to the topological skeleton to compensate for gland curvature so that the width was calculated as the actual distance between gland contour. The perpendicular direction was calculated for each point of the gland’s topological skeleton using principal component analysis of 8 neighboring points (see Video 2 [available at www.ophthalmologyscience.org]). Meibomian gland width was further divided into mean (widthmean), median (widthmedian), and 10th (width10th) and 90th (width90th) percentile of all widths. Median was proposed to deal with potential outliers, and the 10th and 90th percentiles were introduced to detect a potential change in the thinnest and thickest region, respectively. The 10th percentile of Meibomian gland width refers to the width value below which 10% of all measured widths fall. This provides insight into the thinnest regions of the gland, which may be particularly susceptible to dysfunction or damage. Similarly, the 90th percentile of Meibomian gland width refers to the width value above which 90% of all measured widths fall. This provides information about the thickest regions of the gland, which may be important for understanding gland structure and function. By including both the 10th and 90th percentile widths, the analysis is able to provide a more comprehensive picture of the Meibomian gland structure, which can aid in diagnosis and monitoring of MGD.
Figure 2.
Meibomian gland ratio calculation. The blue color represents the eyelid, orange the height of the eyelid at the position of the gland, and yellow corresponds to the gland of interest. This image represents a rotated object after performing principal component analysis (PCA) on the data. Principal component 1 (PC1) and PC2 are the 2 most important axes of the rotated object, with PC1 representing the direction of greatest variability in the data and PC2 representing the direction of second-greatest variability. The values along PC1 and PC2 can be used to describe the object’s shape and orientation.
Semantic segmentation–based approach
Score thresholding was performed on predictions from a semantic segmentation network to filter out pixels that are less likely to be part of the desired class. This helps to reduce false positives and improve the accuracy of the segmentation results. The score threshold, selected on the validation set, was set to 0.75. The selection of the score was based on an analysis of the model’s performance on the independent validation set. We chose the threshold that resulted in the best average aggregated Jaccard index (aAJI) (Fig S3, available at www.ophthalmologyscience.org). Thresholded binary masks of Meibomian glands predicted by Deeplabv3+ were further processed to separate merged glands. The fragmentation algorithm has been developed based on work described by Xiao et al21 and Llorens-Quintana et al.20 Figure 4 shows the difference before and after application of fragmentation algorithm on selected Meibomian glands.
Figure 4.
The fragmentation algorithm. The input to the algorithm is a binary mask of a glandular structure. The algorithm begins by filling any holes in the input mask and labeling each connected region of pixels as a separate gland. For each gland, the algorithm applies a rotation to align the major axis of the gland with the horizontal axis. The algorithm then computes the number of segments in each row of the rotated gland, and, if this number exceeds a specified threshold, the gland is fragmented into smaller subglands. To fragment the gland, the algorithm first erodes the gland until it consists of multiple disconnected regions. It then applies an external gradient to each of these regions, followed by filling any holes in the gradient, and, finally, subtracts an internal gradient from the filled gradient to obtain a fragmented subgland. If the number of segments in a row of the rotated gland does not exceed the threshold, the gland is not fragmented and is returned as is. The output of the algorithm is a binary mask of the fragmented gland structure. (A) Original image with a few glands close to each other. (B) Semantic segmentation before application of fragmentation algorithm. (C) Semantic segmentation after application of fragmentation algorithm.
Object detection–based approach
The detection threshold was set to 0.75. To further reduce false positive detections by Mask R-CNN the area of interest was limited by the area of the eyelid detected by the Deeplabv3+ network. Mask R-CNN was also trained to detect the eyelid; however, the results were not satisfactory; thus, Deeplabv3+ was used instead. We found that the performance of Mask R-CNN in detecting the eyelid region was not satisfactory, despite our best efforts to fine-tune the model and adjust the training parameters. It is possible that Mask R-CNN’s instance segmentation approach, which is designed to detect multiple objects in an image, may have been better suited for detecting the gland structures, which are typically more complex and have a greater variety of shapes and sizes compared with the eyelid region. On the other hand, the eyelid region is a relatively simple structure that can be accurately detected using a semantic segmentation approach, which is what Deeplabv3+ uses. Multiple detections for one eyelid were common with Mask R-CNN. Additionally, it is possible that the training data for the eyelid region was not diverse enough or did not contain enough examples to allow Mask R-CNN to effectively learn to detect this specific structure.
Inter-reader Agreement
To measure the level of agreement between 2 annotators (K.S. and M.L.R.), who manually labeled 20 random images from the test set aAJI,45,46 Dice similarity coefficient,47 Cohen’s Kappa coefficient,48 and interclass correlation coefficient49 were calculated for each model. A similar approach has been proposed by Setu et al7 for test–retest reliability. In addition, the agreement between each annotator and each algorithm was assessed using the same metrics.
Statistical Analysis
Bland–Altman50 analysis was used to assess agreement between manual and automated methods for Meibomian gland metrics calculation. The same analysis was carried out for Meibomian gland metrics calculated on manual annotations performed by 2 readers.
The 95% confidence interval (CI) of the means for each interreader agreement metric and metric for model performance assessment were calculated by performing bootstrap resampling.51 Bias corrected and accelerated percentile methods were used to correct for bias and skewness in the distribution of bootstrap estimates. Statistical analysis was performed using MATLAB R2022a with Statistics and Machine Learning Toolbox (The MathWorks, Inc).
Results
Network Training Details
Both models were trained on a High Performance Computing cluster utilizing 1 GPU (NVIDIA A100 80GB) for 300 (Deeplabv3+) and 100 (Mask R-CNN) epochs. The training process of Deeplabv3+ network lasted 44 hours 52 minutes, whereas Mask R-CNN lasted 34 hours and 56 minutes. Stochastic gradient descent with momentum optimizer was used to optimize both models. Momentum was set to 0.5 and 0.9 for Deeplabv3+ and Mask R-CNN, respectively. The exploratory analysis on the validation set found that results were not sensitive to the number of epochs for dropping the learning rate and the dropping factor; as such, they were set to default values. The learning rate was updated every 10 (Deeplabv3+) and 50 (Mask R-CNN) epochs by multiplying by a factor of 0.3. Mini-batch size was set to 8 for both models. The training data were shuffled before each training epoch.
Network Performance
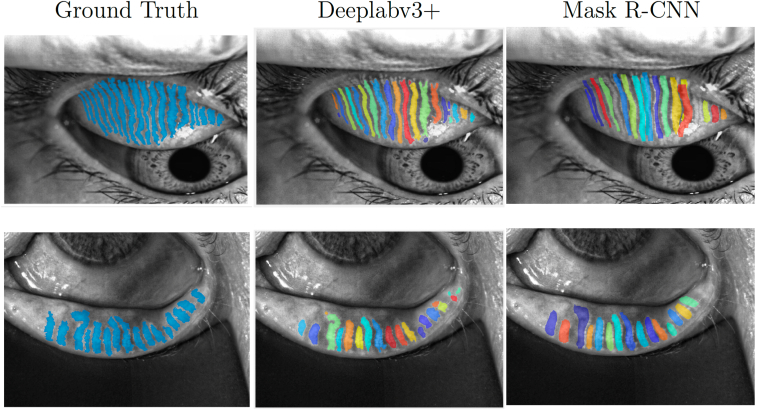
The aAJI on the test set was a mean of 0.4718 (95% CI, 0.4680–0.4771) for the ‘gland’ class and a mean of 0.8470 (95% CI, 0.8432–0.8508) for the ‘eyelid’ class for semantic segmentation–based approach. The aAJI for object detection-based approach was a mean of 0.4476 (95% CI, 0.4426–0.4533). Figure 5 shows example images with aAJI results for each model. Figure 6 demonstrates examples of software failures. In general, both models performed better on the lower eyelid images.
Figure 5.
Examples of model performance on images from the test set. The average aggregated Jaccard indexes (aAJIs) for the model based on Deeplabv3+ were 0.6925 and 0.6493 for the upper and lower eyelid, respectively. The Mask R-CNN-based approach achieved aAJIs of 0.5027 and 0.6743 for the upper and lower eyelid, respectively.
Figure 6.
Examples of model failures. The first row shows examples of failures with the semantic segmentation–based approach, whereas the second row shows examples of failures with the object detection–based approach. (A) Merged glands. (B) Misdetection. (C) Oversegmentation. (D) Overlapping glands. (E) Merged and missing glands (arrows). (F) Poor performance on an image out of focus.
Meibomian Gland Metrics Calculation
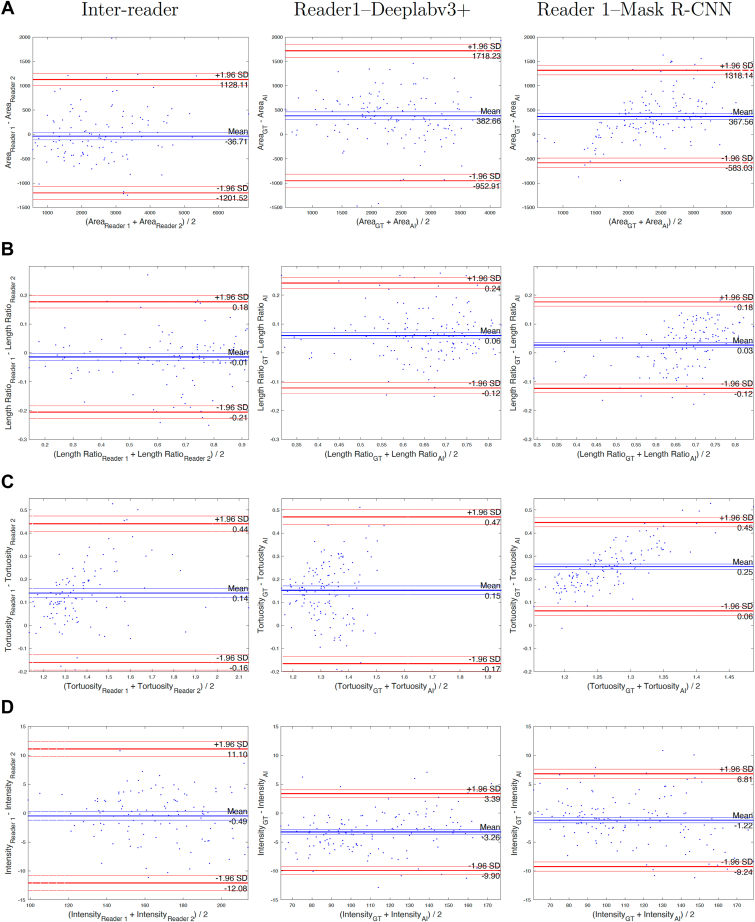
To quantify Meibomian gland structure, Meibomian gland area, length ratio, tortuosity, intensity, and width were calculated. The results were compared with the manual algorithm in which glands were annotated by a single masked reader. Columns 2 and 3 of Figure 7 show the Bland–Altman analysis for all metrics. Both AI-based algorithms underestimated area, length ratio, tortuosity, widthmean, widthmedian, width10th, and width90th. Meibomian gland intensity was overestimated by both algorithms compared with the manual approach. The object detection–based algorithm seems to be as reliable as the manual approach only for the Meibomian gland width10th calculation.
Figure 7.
Bland–Altman analysis of Meibomian gland metrics between 2 readers (interreader) between ground truth (GT) and object detection-based approach (artificial intelligence [AI]) (Reader1-Deeplabv3+) and between GT and semantic segmentation–based approach (Reader1-Mack R-CNN). Analysis was performed on 20 images of the lower eyelids.
Interreader Agreement
Column 1 of Figure 7 shows the Bland–Altman analysis for all metrics when Meibomian glands were annotated by 2 independent and masked readers. The results show that there is no bias between 2 readers for most metrics except for Meibomian gland tortuosity. Table 1 and 2 show interreader reliability. Both tables show the same values for interreader reliability to facilitate easy comparison between reader and algorithm agreement for each approach.
Table 1.
Interreader Agreement of Manual Annotations and Reader–Algorithm Agreement for Semantic Segmentation–Based Algorithm.
| Metric | Interreader |
Reader 1: Algorithm |
Reader 2: Algorithm |
|||
|---|---|---|---|---|---|---|
| ‘gland’ | ‘eyelid’ | ‘gland’ | ‘eyelid’ | ‘gland’ | ‘eyelid’ | |
| aAJI | 0.5487 (0.5310,0.5620) | 0.8774 (0.8688,0.8834) | 0.4503 (0.4368,0.4663) | 0.8691 (0.8606,0.8764) | 0.4435 (0.4265,0.4572) | 0.8546 (0.8465,0.8631) |
| DSC | 0.6230 (0.6033,0.6404) | 0.9340 (0.9302,0.9384) | 0.5612 (0.5478,0.5732) | 0.9291 (0.9225,0.9321) | 0.5323 (0.5154,0.5469) | 0.9207 (0.9161,0.9248) |
| Kappa | 0.6224 (0.6033,0.6404) | 0.9228 (0.9182,0.9286) | 0.5604 (0.5449,0.5724) | 0.9108 (0.9060,0.9166) | 0.5315 (0.5139,0.5453) | 0.8994 (0.8934,0.9050) |
| ICC | 0.7264 (0.7123,0.7343) | 0.9240 (0.9195,0.9291) | 0.5863 (0.5731,0.5978) | 0.9129 (0.9076,0.9176) | 0.5608 (0.5463,0.5750) | 0.9010 (0.8951,0.9067) |
Each value represents 2-sided mean with 95% confidence interval.
aAJI = average aggregated Jaccard index; DSC = Dice similarity coefficient; ICC = interclass correlation coefficient; Kappa = Cohen’s Kappa coefficient.
Table 2.
Interreader Agreement of Manual Annotations and Reader–Algorithm Agreement for Object Detection-Based Algorithm.
| Metric | Interreader |
Reader 1: Algorithm |
Reader 2: Algorithm |
|||
|---|---|---|---|---|---|---|
| ‘gland’ | ‘eyelid’ | ‘gland’ | ‘eyelid’ | ‘gland’ | ‘eyelid’ | |
| aAJI | 0.5487 (0.5310,0.5620) | 0.8774 (0.8688,0.8834) | 0.4294 (0.4132,0.4545) | — | 0.4102 (0.3912,0.4372) | — |
| DSC | 0.6230 (0.6033,0.6404) | 0.9340 (0.9302,0.9384) | 0.5860 (0.5694,0.6041) | — | 0.5516 (0.5268,0.5687) | — |
| Kappa | 0.6224 (0.6033,0.6404) | 0.9228 (0.9182,0.9286) | 0.5848 (0.5662,0.5998) | — | 0.5503 (0.5309,0.5716) | — |
| ICC | 0.7264 (0.7123,0.7343) | 0.9240 (0.9195,0.9291) | 0.5943 (0.5793,0.6154) | — | 0.5613 (0.5368,0.5787) | — |
Each value represents 2-sided mean with 95% confidence interval.
aAJI = average aggregated Jaccard index; DSC = Dice similarity coefficient; ICC = interclass correlation coefficient; Kappa = Cohen’s Kappa coefficient.
Discussion
This study compared 2 popular deep learning neural network architectures for object segmentation. Both semantic segmentation and object detection–based approaches provided biased results when compared with a manual approach. Both approaches underestimated most gland metrics except for intensity, which is overestimated by AI-based approaches. In this work, we evaluated whether AI-based algorithms can give a more consistent performance than manual assessment. The discrepancy between manual and fully automated approaches are rather small for most metrics. In particular, Meibomian gland width has been a metric for which the bias does not seem to be clinically significant. In this study, the bias between manual and AI-based measurements of Meibomian gland width was found to be in the range of 0.17 to 4.75 pixels, which is relatively small. Although a bias of this magnitude may be noticeable when analyzing subtle changes in gland structure over time or in response to treatment, it is unlikely to have a clinically significant impact when assessing Meibomian glands en masse. This is because the variations in gland width that are observed in healthy individuals and those with MGD are typically larger than the range of bias observed in this study, as reported in previous research.15,17,21 Therefore, even if there is a slight underestimation or overestimation of Meibomian gland width by AI-based approaches, it is unlikely to significantly affect the overall assessment of Meibomian gland structure and function in a clinical setting. It should be noted that after incorporating a calibration factor, a change of 1 pixel in the image corresponds to a change of 0.028 mm in the gland width. Therefore, the observed bias of 0.17 to 4.75 pixels translates to a bias of 0.005 to 0.134 mm, which is still relatively small. The calibration factor was only used to provide a more intuitive interpretation of the measurements in a physical context. The limits of agreement for this data set ranged between −11.15 and 16.12 in pixels, which is a relatively small range of variation compared with the observed variations in gland width in healthy individuals and those with dry eye disease.15,17,21 These findings suggest that the deep learning model had good performance in calculating the width of Meibomian glands. However, it should be noted that some studies have reported variations in gland width depending on the stage of MGD. For instance, in the initial stages of atrophy, the width of glands may start to increase as the glands fade out, leading to more variation in early-stage cases.15 Additionally, another study reported that gland thickness increases in intermediate MGD but decreases in severe MGD.21 Finally, a study found that the diameter deformation index, which is the standard deviation of the local gland widths within a single gland, is more common in early-stage MGD.17 These variations should be taken into account when interpreting the results of the deep learning model.
Of the many recently published works on the utility of AI in meibography, only a few focused on quantifying individual Meibomian gland structure.7,9,14,15 The recurring Meibomian gland metrics are length, width, and tortuosity. In addition, some other individual gland parameters were proposed: the number of glands,7,15 local contrast,9 and the number of ghost glands.9 In this study, we also proposed a single Meibomian gland area, intensity, and detailed width calculation. All of the aforementioned studies trained their models on images captured with Keratograph 5M (OCULUS). In contrast, this study analyzed images captured with the LipiView II device. So far, only one other study trained a deep learning model on a similar type of images.10 However, they did not focus on individual Meibomian gland characteristics, rather focusing on overall gland area, which was then used for automated meiboscore evaluation.10 Comparison of model performance described in this study with previously published works is not simple due to the lack of a universal metric agreement when it comes to deep learning–model performance evaluation. In this study, aAJI was used to evaluate the segmentation performance to prioritize accurate instance segmentation, as opposed to mere semantic segmentation. The results of our models indicate that interreader agreement is not much different from algorithm versus ground truth, indicating that automated Meibomian gland segmentation may be highly dependent on the subjective ground truth data and, furthermore, AI-based algorithms are as reliable as the manual approach.
Both models performed better on the lower eyelid images. One possible reason is that the lower eyelid may have had more clear and distinguishable gland structures compared with the upper eyelid, allowing the deep learning model to better detect and classify the gland regions. Additionally, the image features and characteristics of the lower eyelid may have been more consistent and easier to learn for the deep learning model, leading to improved performance. Finally, it is possible that the lower eyelid simply had more training examples in the data set compared with the upper eyelid, which could have improved the model’s ability to generalize and make accurate predictions.
With regard to the research methods, some limitations need to be acknowledged. Although both models performed well on good quality images, they failed to successfully segment Meibomian glands on poor quality images, particularly those out of focus, as can be seen in Figure 6. However, many clinical images are far from ideal, which could justify the discrepancy between manual and automatic approaches. In this study, real, imperfect, clinical images were used for model training. To overcome this limitation, either more images could have been used for training or improved training and quality control procedures to ensure that the optometrist was performing meibography in the best possible way. As previously discussed,52 there are many ways to optimize image capture, for example, performing appropriate eyelid eversion or using a better tool for eyelid eversion, appropriate eye gaze, and head position.52
In many recently published studies on deep learning–based Meibomian gland segmentation, good quality images were preselected, meaning that it is likely that the developed algorithm performance would suffer when used in a standard optometric practice. The proposed approaches in this paper are not good for detection of fine details. Both semantic segmentation and object detection–based approaches fail to segment every individual gland seen in the image. This can happen when glands are too close to each other or if they appear in the peripheral region that is usually out of focus (Fig 6). However, the current analysis of Meibomian gland visible structure is limited to either the overall area of the gland region or to individual parameters of central glands, so the mentioned algorithm failures may not play a significant role in the current gland assessment. Perhaps future models will focus on all the glands’ individual characteristics. Another limitation of this study is that only images captured with the LipiView II were included to train the model, meaning that the algorithm is likely to need further optimization to process images from another device, such as the Keratograph 5M.
The relatively small size of the data set may be another reason why both models failed to detect all Meibomian glands in the image. Meibography is not typically part of a standard eye examination, so it is difficult to build a large database of images required for deep learning tasks, and no databases of meibography images are publicly available to our knowledge. Further data collection is required to determine exactly how data set size affects model performance. The strengths of the study include the in-depth analysis of interreader agreement. Previous studies of the application of deep learning in meibography suffer from methodologic limitations.8,9,11, 12, 13, 14, 15,53 Most of these studies have not investigated the interreader and intrareader reliability. Only some looked closer at these relationships and confirmed the poor interreader5,10 and intrareader reliability.7 Another strength of this study is the data partitioning that was based on the patient identifier. Most previous studies performed a random split of the data into training and evaluation sets without clearly stating if they have included multiple images from a single patient, which could lead to potential bias in the stated model performance.5,7,9,11, 12, 13, 14 In this study, all images for each patient could end up only in one set; as such, there was no risk that model performance would be biased. A key strength of the present study was the in-depth calculation of Meibomian gland width that considers all widths along the gland; as such, comprehensive analysis of possible changes in the thickest and thinnest part of the gland could be possible (e.g., when analyzing longitudinal changes that could happen as a response to treatment).
Despite its limitations, the study certainly contributes to the development of AI-based approaches in meibography. The use of deep learning models for the analysis of meibography images has the potential to improve the accuracy and efficiency of Meibomian gland assessment in clinical practice. There is still the need for a proper and universal evaluation method that would allow comparison between different approaches. Currently, different studies use different evaluation metrics, making it difficult to compare the performance of different AI-based approaches. Moreover, the use of different instruments for image capture can also affect the accuracy and reliability of the measurements. Further research should be undertaken to explore how clinicians could benefit from individual gland analysis and how to overcome the issues with the current approaches that still face some difficulties with distinguishing single glands seen in each image.
In conclusion, this study demonstrates another approach for automated, AI-based Meibomian gland analysis focusing on individual Meibomian gland characteristics. Given its accuracy and efficiency, the proposed methods could be successfully utilized in clinical settings for aggregate Meibomian gland assessment. Further development is required to solve the issues with overlapping and out of focus glands.
Acknowledgments
The authors acknowledge the assistance given by Research IT and the use of the Computational Shared Facility at The University of Manchester. The authors acknowledge the assistance of the clinical, logistical, and administrative colleagues at Eurolens Research in the acquisition of data for this study and Emily Willerton of BBR Optometry Ltd for her work in seeking participant consent for use of images for the project.
Manuscript no. XOPS-D-23-00072R1.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosures:
All authors have completed and submitted the ICMJE disclosures form.
The authors made the following disclosures: C.A.B.: Employment – Johnson & Johnson Vision Inc. C.MC.: Research grants/support – Alcon, CooperVision Inc, Johnson & Johnson Vision Care Inc. Menicon, Visco Vision.
P.B.M.: Research grants/support – Alcon, CooperVision Inc, Menicon, Visco Vision; Honoraria – Alcon, CooperVision Inc, Johnson & Johnson Vision.
M.L.R.: Research grants/support – Alcon Inc, CooperVision Inc, Johnson & Johnson Vision Inc, Menicon Co Ltd, Visco Vision Inc; Travel expenses – CooperVision Inc; Patents – Compositions and uses and methods relating thereto US11253452B2.
M.F.: Research grants/support – InnovateUK, NIHR; Consultant – Marion Surgical Ltd; Honoraria – UCB; Board membership – Spotlight Pathology Ltd, Sentira XR (VR-Evo Ltd); Shares – Spotlight Pathology Ltd, Sentira XR (VREvo Ltd), Fixtuur (Shortbite Ltd).
Supported by Johnson & Johnson Vision, Inc.
HUMAN SUBJECTS: Human subjects were included in this study. This study was approved by the University Research Ethics Committee of The University of Manchester before data extraction. All procedures adhered to the tenets of the Declaration of Helsinki, and all participants provided written informed consent before data extraction. Consent was obtained retrospectively from all participants for their anonymized images to be analyzed for the purpose of deep learning model development.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Swiderska, Blackie, Maldonado-Codina, Morgan, Read, Fergie
Data collection: Swiderska
Analysis and interpretation: Swiderska, Blackie, Maldonado-Codina, Morgan, Read, Fergie
Obtained funding: Blackie, Maldonado-Codina, Morgan, Read
Overall responsibility: Morgan
Supplementary Data
The relationship between score threshold and average aggregated Jaccard index (aAJI) on the validation set. The highest aAJI was achieved at a score threshold of 0.75.
An example of manual annotation process and single Meibomian gland label assignment.
Meibomian gland width calculation using principal component analysis (PCA). The perpendicular direction to the gland’s topological skeleton is calculated for each point using PCA of 8 neighboring points. The Meibomian gland width is then divided into mean, median, 10th percentile, and 90th percentile widths to provide comprehensive information about gland structure and function.
References
- 1.Becker A.S., Marcon M., Ghafoor S., et al. Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Invest Radiol. 2017;52:434–440. doi: 10.1097/RLI.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 2.Kim H.E., Kim H.H., Han B.K., et al. Changes in cancer detection and false-positive recall in mammography using artificial intelligence: a retrospective, multireader study. Lancet Digit Health. 2020;2:e138–e148. doi: 10.1016/S2589-7500(20)30003-0. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt-Erfurth U., Sadeghipour A., Gerendas B.S., et al. Artificial intelligence in retina. Prog Retin Eye Res. 2018;67:1–29. doi: 10.1016/j.preteyeres.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Sharif M.S., Qahwaji R., Ipson S., Brahma A. Medical image classification based on artificial intelligence approaches: a practical study on normal and abnormal confocal corneal images. Appl Soft Comput. 2015;36:269–282. [Google Scholar]
- 5.Wang J., Yeh T.N., Chakraborty R., et al. A deep learning approach for Meibomian gland atrophy evaluation in meibography images. Transl Vis Sci Technol. 2019;8:37. doi: 10.1167/tvst.8.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prabhu S.M., Chakiat A., S S., Vunnava K.P., Shetty R. Deep learning segmentation and quantification of Meibomian glands. Biomed Signal Process Control. 2020;57 [Google Scholar]
- 7.Setu M.A.K., Horstmann J., Schmidt S., et al. Deep learning-based automatic meibomian gland segmentation and morphology assessment in infrared meibography. Sci Rep. 2021;11:7649. doi: 10.1038/s41598-021-87314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan Z.K., Umar A.I., Shirazi S.H., Rasheed A., Qadir A., Gul S. Image based analysis of meibomian gland dysfunction using conditional generative adversarial neural network. BMJ Open Ophthalmol. 2021;6 doi: 10.1136/bmjophth-2020-000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Li S., Yeh T.N., et al. Quantifying Meibomian gland morphology using artificial intelligence. Optom Vis Sci. 2021;98(9):1094–1103. doi: 10.1097/OPX.0000000000001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha R.K., Chowdhury A.M.M., Na K.S., et al. Automated quantification of meibomian gland dropout in infrared meibography using deep learning. Ocul Surf. 2022;26:283–294. doi: 10.1016/j.jtos.2022.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Yu Y., Zhou Y., Tian M., et al. Automatic identification of meibomian gland dysfunction with meibography images using deep learning. Int Ophthalmol. 2022;42:3275–3284. doi: 10.1007/s10792-022-02262-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z., Lin X., Yu X., et al. Meibomian gland density: an effective evaluation index of Meibomian gland dysfunction based on deep learning and transfer learning. J Clin Med. 2022;11:2396. doi: 10.3390/jcm11092396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Graham A.D., Yu S.X., Lin M.C. Predicting demographics from meibography using deep learning. Sci Rep. 2022;12 doi: 10.1038/s41598-022-18933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai Q., Liu X., Lin X., et al. A novel Meibomian gland morphology analytic system based on a convolutional neural network. IEEE Access. 2021;9:23083–23094. [Google Scholar]
- 15.Setu MAK, Horstmann J, Stern ME, Steven P. Automated analysis of meibography images: comparison between intensity, region growing and deep learning-based methods [abstract] Ophthalmologe. 2019;116:25–218. [Google Scholar]
- 16.Cieżar K., Pochylski M. 2nd short-time Fourier transform for local morphological analysis of meibomian gland images. PLOS ONE. 2022;17 doi: 10.1371/journal.pone.0270473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Y., Wang Q., Luo Z., et al. Quantitative analysis of morphological and functional features in meibography for Meibomian gland dysfunction: diagnosis and grading. EClinicalmedicine. 2021;40 doi: 10.1016/j.eclinm.2021.101132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cieżar K., Pochylski M. 2D fourier transform for global analysis and classification of meibomian gland images. Ocular Surf. 2020;18:865–870. doi: 10.1016/j.jtos.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Llorens-Quintana C., Rico-Del-Viejo L., Syga P., et al. Meibomian gland morphology: the influence of structural variations on gland function and ocular surface parameters. Cornea. 2019;38:1506–1512. doi: 10.1097/ICO.0000000000002141. [DOI] [PubMed] [Google Scholar]
- 20.Llorens-Quintana C., Rico Del Viejo L., Syga P., et al. A novel automated approach for infrared-based assessment of meibomian gland morphology. Transl Vis Sci Technol. 2019;8:17. doi: 10.1167/tvst.8.4.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao P., Luo Z., Deng Y., et al. An automated and multiparametric algorithm for objective analysis of meibography images. Quant Imaging Med Surg. 2021;11:1586–1599. doi: 10.21037/qims-20-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X., Fu Y., Li L., et al. A novel quantitative index of Meibomian gland dysfunction, the Meibomian gland tortuosity. Transl Vis Sci Technol. 2020;9:34. doi: 10.1167/tvst.9.9.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koprowski R., Wilczyński S., Olczyk P., et al. A quantitative method for assessing the quality of meibomian glands. Comput Biol Med. 2016;75:130–138. doi: 10.1016/j.compbiomed.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Arita R., Suehiro J., Haraguchi T., et al. Objective image analysis of the meibomian gland area. Br J Ophthalmol. 2014;98(6):746–755. doi: 10.1136/bjophthalmol-2012-303014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koh Y.W., Celik T., Lee H.K., et al. Detection of meibomian glands and classification of meibography images. J Biomed Opt. 2012;17(8) doi: 10.1117/1.JBO.17.8.086008. [DOI] [PubMed] [Google Scholar]
- 26.Celik T., Lee H.K., Petznick A., Tong L. BioImage informatics approach to automated meibomian gland analysis in infrared images of meibography. J Optom. 2013;6:194–204. [Google Scholar]
- 27.Daniel E., Maguire M.G., Pistilli M., et al. Grading and baseline characteristics of meibomian glands in meibography images and their clinical associations in the Dry Eye Assessment and Management (DREAM) study. Ocul Surf. 2019;17:491–501. doi: 10.1016/j.jtos.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh T.N., Lin M.C. Repeatability of Meibomian gland contrast, a potential indicator of Meibomian gland function. Cornea. 2019;38:256–261. doi: 10.1097/ICO.0000000000001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García-Marqués J.V., García-Lázaro S., Martínez-Albert N., Cerviño A. Meibomian glands visibility assessment through a new quantitative method. Graefes Arch Clin Exp Ophthalmol. 2021;259:1323–1331. doi: 10.1007/s00417-020-05034-7. [DOI] [PubMed] [Google Scholar]
- 30.Grenon SM, Korb DR, Grenon J, et al. Eyelid illumination systems and methods for imaging meibomian glands for meibomian gland analysis. 2014. TearScience Inc. Patent No. US 2014/0330129 A1, Filed May 5, 2014, Issued November 6, 2014.
- 31.Grenon S., Liddle S., Grenon J., et al. A novel meibographer with dual mode standard noncontact surface infrared illumination and infrared transillumination. Invest Ophthalmol Vis Sci. 2014;55:26. [Google Scholar]
- 32.Rother C., Kolmogorov V., Blake A. “Grabcut”: interactive foreground extraction using iterated graph cuts. ACM Trans Graph. 2004;23:309–314. [Google Scholar]
- 33.Chen LC, Zhu Y, Papandreou G, et al. In: Vision – ECCV. Ferrari V, Hebert M, Sminchisescu C, Weiss Y, editors. Springer; 2018. Encoder-decoder with atrous separable convolution for semantic image segmentation; pp. 833–851. [Google Scholar]
- 34.He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. IEEE International Conference on Computer Vision [ICCV], 2017:2980–2988.
- 35.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception-v4, inceptionresnet and the impact of residual connections on learning. arXiv:1602.07261.
- 36.Al-Masni M.A., Kim D.H., Kim T.S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput Methods Programs Biomed. 2020;190 doi: 10.1016/j.cmpb.2020.105351. [DOI] [PubMed] [Google Scholar]
- 37.Siciarz P., McCurdy B. U-net architecture with embedded Inception-ResNet-v2 image encoding modules for automatic segmentation of organs-at-risk in head and neck cancer radiation therapy based on computed tomography scans. Phys Med Biol. 2022;67 doi: 10.1088/1361-6560/ac530e. [DOI] [PubMed] [Google Scholar]
- 38.Bose S.R., Kumar V.S. Efficient inception v2 based deep convolutional neural network for real-time hand action recognition. IET Image Process. 2020;14:688–696. [Google Scholar]
- 39.Alruwaili M., Shehab A., Abd El-Ghany S. COVID-19 diagnosis using an enhanced Inception-ResNetV2 deep learning model in CXR images. J Healthc Eng. 2021;2021 doi: 10.1155/2021/6658058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Kang X, Ren F. Employing Inception-ResNet-v2 and Bi-LSTM for medical domain visual question answering. Cappellato L, Ferro N, Nie JY, Soulier L, editors. Conference and Labs of the Evaluation Forum. 2018;2125(107) [Google Scholar]
- 41.Singh H., Saini S.S., Lakshminarayanan V. Rapid classification of glaucomatous fundus images. J Opt Soc Am A Opt Image Sci Vis. 2021;38:765–774. doi: 10.1364/JOSAA.415395. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen TD, Jung K, Bui PN, et al. Retinal disease early detection using deep learning on ultra-wide-field fundus images. medRxiv. 2023.03.09.23287058.
- 43.Zhou W.D., Dong L., Zhang K., et al. Deep learning for automatic detection of recurrent retinal detachment after surgery using ultra-widefield fundus images: a single-center study. Adv Intell Syst. 2022;4 [Google Scholar]
- 44.Jiang H, Yang K, Gao M, et al. An interpretable ensemble deep learning model for diabetic retinopathy disease classification. In: 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society [EMBC]. 2019:2045–2048. [DOI] [PubMed]
- 45.Kumar N., Verma R., Sharma S., et al. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging. 2017;36(7):1550–1560. doi: 10.1109/TMI.2017.2677499. [DOI] [PubMed] [Google Scholar]
- 46.Kumar N., Verma R., Anand D., et al. A multi-organ nucleus segmentation challenge. IEEE Trans Med Imaging. 2020;39:1380–1391. doi: 10.1109/TMI.2019.2947628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou K.H., Warfield S.K., Bharatha A., et al. Statistical validation of image segmentation quality based on a spatial overlap index1. Acad Radiol. 2004;11:178–189. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McHugh M.L. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- 49.Koo T.K., Li M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;327:307–310. [PubMed] [Google Scholar]
- 51.DiCiccio T.J., Efron B. Bootstrap confidence intervals. Statist Sci. 1996;11:189–228. [Google Scholar]
- 52.Swiderska K., Read M.L., Blackie C.A., Maldonado-Codina C., Morgan P.B. Latest developments in meibography: a review. Ocular Surf. 2022;25:119–128. doi: 10.1016/j.jtos.2022.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Yeh C.H., Yu S.X., Lin M.C. Meibography phenotyping and classification from unsupervised discriminative feature learning. Transl Vis Sci Technol. 2021;10:4. doi: 10.1167/tvst.10.2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The relationship between score threshold and average aggregated Jaccard index (aAJI) on the validation set. The highest aAJI was achieved at a score threshold of 0.75.
An example of manual annotation process and single Meibomian gland label assignment.
Meibomian gland width calculation using principal component analysis (PCA). The perpendicular direction to the gland’s topological skeleton is calculated for each point using PCA of 8 neighboring points. The Meibomian gland width is then divided into mean, median, 10th percentile, and 90th percentile widths to provide comprehensive information about gland structure and function.