Abstract
The peroxisome serves a significant role in the occurrence and development of cancers. Specifically, the peroxisomal biogenesis factor 13 (PEX13) is crucial to the occurrence of peroxisomes. However, the biological function of PEX13 in cancers remains unclear. To address this, various portals and databases such as The Cancer Genome Atlas Program, The Genotype-Tissue Expression project, the Gene Expression Profiling Interactive Analysis 2, cBioPortal, the Genomic Identification of Significant Targets In Cancer 2.0, Tumor Immune Estimation Resource 2, SangerBox, LinkedOmics, DAVID and STRING were applied to extract and analyze PEX13 data in tumors. The correlations between PEX13 and prognosis, genetic alterations, PEX13-related gene enrichment analysis, weighted gene co-expression network analysis (WGCNA), protein interaction, long non-coding (lnc)RNA/circular (circ)RNA-micro (mi)RNA network and tumor immunity were explored in various tumors. The lncRNA-miRNA-PEX13 and circRNA-miRNA-PEX13 regulatory networks were identified via miRabel, miRDB, TargetScan and ENCORI portals and Cytoscape tool. In vitro assays were applied to verify the biological functions of PEX13 in pancreatic adenocarcinoma (PAAD) cells. The findings revealed that PEX13 is upregulated in various tumors and high PEX13 mRNA expression is associated with poor prognosis in patients with multiple cancers. Genetic alterations in PEX13 such as amplification, mutation and deep deletion have been found in multiple cancers. PEX13-related genes were associated with T cell receptor, signaling pathway and hippo signaling pathway through ‘biological process’ subontology of Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses. Through WGCNA analysis, it was discovered that PEX13 hub genes were mainly enriched in the Rap1, ErbB and AMPK signaling pathways in PAAD. Immune analysis showed that PEX13 was significantly related to tumor infiltration immune cells, immune checkpoint genes, microsatellite instability, TMB and tumor purity in a variety of tumors. Cell Counting Kit-8, wound healing, Transwell and colony formation assays displayed that PEX13 knockdown could suppress PAAD cell proliferation, migration, invasion, and colony formation in vitro, respectively. Overall, PEX13 is a potential predictor of immunotherapeutic and prognostic biomarkers in various malignant tumors, including ACC, KICH, LGG, LIHC and PAAD.
Keywords: PEX13, prognosis, immune, PAAD, ncRNA-miRNA- PEX13 network
Introduction
At present, 14 human peroxisomal biogenesis factor (PEX) genes that encode peroxin proteins that serve a role in various stages of peroxisome biogenesis have been identified, including peroxisome matrix protein input, membrane formation and peroxisome proliferation (1,2). Peroxisome membrane proteins are also thought to function as signaling platforms in reactive oxygen species (ROS)-induced autophagy and antiviral immunity (3,4). Peroxisome is a metabolic organelle involved in cellular redox balance and lipid metabolism (5,6). The function of peroxisome is important for ether phospholipid synthesis, fatty acid oxidation, bile acid synthesis and ROS homeostasis (7). Although peroxisomes are involved in cell metabolism, their functional effects in cancer remain unclear compared with those of other metabolic organelles such as mitochondria (8,9).
In the last decade, peroxisome has also been recognized as a central regulator of immunity (9). Lipid metabolites of peroxisomes, such as polyunsaturated fatty acids (PUFAs), are precursors of important immune mediators, including leukotrienes (LTs), and suppressors (10–12). Peroxisome redox metabolism regulates cellular immune signaling, such as activation of B-cell activated NF-κB (13). In addition, the development and activation of innate and adaptive immune cells are regulated by peroxisome β-oxidation and ether lipid synthesis (12,14). These findings open up avenues for targeting peroxisome interventions for immune disorders, inflammation and cancer. In addition, Lee et al (2) found that PEX13 is required for selective autophagy (virus autophagy) of Sindbis virus and damaged mitochondria (mitochondrial autophagy), and PEX13 mutant I326T and W313G associated with the disease are defective in mitochondria autophagy. The mitophagy function of PEX13 is shared with another peroxin family member, PEX3, but not with the other two peroxins required for general autophagy, namely PEX14 and PEX19 (15). Since PEX13 gene has a significant role in various stages of peroxisome biogenesis and autophagy, and peroxisome or autophagy has a crucial role in tumor occurrence and development, the present study aimed to investigate the biological functions that PEX13 may play in multiple cancers, especially in pancreatic adenocarcinoma (PAAD) (16,17).
In the present study, systematic bioinformatics analysis was conducted on patient data retrieved from The Cancer Genome Atlas (TCGA) database to verify the biological function and prognostic significance of PEX13 in various tumors. The present study comprehensively explored the functional significance of alterations in PEX13 expression level in various tumors in terms of PEX13 expression level, prognosis, genetic alterations, PEX13-related gene enrichment analysis, weighted correlation network analysis (WGCNA), protein interaction, long non-coding (lnc)RNA/circular (circ)RNA-micro (mi)RNA network and tumor immunity. As a general surgery team, the present authors are particularly interested in PAAD and focused. Finally, the relationships between the expression of PEX13 and the biological functions of PAAD were verified through in vitro experiments.
Materials and methods
PEX13 mRNA and protein expression and prognostic analyses
The PEX13 mRNA expression data for 33 tumors retrieved from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) database were analyzed using the SangerBox portal (http://SangerBox.com/Tool) (18). These tumors included adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large b-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck cancer (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma (MESO), ovarian cancer (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), uterine carcinosarcoma (UCS) and uveal melanoma (UVM). PEX13 mRNA expression distributions were visualized in violin plots. Subsequently, the Gene Expression Profiling Interactive Analysis 2 (GEPIA2) portal (genepattern.org) was applied to further explore the PEX13 mRNA expression in 33 tumors (19). The immunohistochemical (IHC) staining data for the analysis of PEX13 expression in various tumor tissues and corresponding normal tissues were obtained from The Human Protein Atlas (THPA) portal (https://www.proteinatlas.org/) (20). The analyzed tumors included glioma and breast, colorectal, liver, lung, pancreatic, skin, stomach, thyroid and renal cancer. Survival prognosis analysis was performed using GEPIA2 and SangerBox. Through GEPIA2, the overall survival (OS) map data for PEX13 in multiple tumors of TCGA database was obtained. The low- and high-expression cohorts of PEX13 were obtained using the expression thresholds of cutoff-high (>50%) and cutoff-low (<50%) values. Cox regression analysis was used to analyze the OS, disease-free interval (DFI), progression-free interval (PFI) and disease-specific survival (DSS) data in various cancers using SangerBox portal.
Genetic alteration analysis
The characteristics of PEX13 genetic alterations were explored in multiple tumors using the cBioPortal database (https://www.cbioportal.org/), including mutation, amplification and deep deletion (21). The structural variant data, mutation data and copy number alteration (CNA) data were obtained from the cBioPortal database. Data of somatic CNAs and somatic mutations of PAAD were obtained from TCGA datasets (gdc.cancer.gov/). The CNAs were related to the PEX13 expression and the threshold CNA peaks were analyzed through GISTIC 2.0 (https://cloud.genepattern.org/) (22). According to the PEX13 expression level, the patients with PAAD were divided into the first 25% PEX13low (n=46) and the last 25% PEX13high (n=46) groups. To download and visualize the somatic mutations of patients with PAAD, the ‘maftools’ package (bioconductor.org/packages/release/bioc/vignettes/maftools/inst/doc/maftools.html#7_Visualization) was applied in R software (4.2.1) (https://www.r-project.org/) (23).
PEX13-related genes enrichment analysis
Through the GEPIA2 portal, the top 200 PEX13-related genes were explored in TCGA database. Subsequently, the TIMER2 (http://timer.cistrome.org/) portal was used to generate a heatmap of the top 10 PEX13-related genes in various cancers, including KCMF1, KIAA1841, MPHOSPH10, MRPL19, PPP1CB, PUS10, RAB1A, SLC30A6, SMEK2 and YIPF4 (24). The correlation coefficient (R) and P-value were calculated. The Gene Ontology (GO)-biological process (BP) subontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of 200 PEX13-correlated genes in pan-cancers were explored via DAVID (https://david.ncifcrf.gov/home.jsp) and SangerBox portal. Moreover, the top 50 PEX13 negatively related genes and top 50 PEX13 positively related genes were explored and heatmaps were generated in PAAD using LinkedOmics (www.linkedomics.org/login.php) (25). The GO-BP and KEGG enrichment analysis of the top 50 PEX13 negatively related genes and top 50 PEX13 positively related genes in PAAD were explored via DAVID and SangerBox portal. Linkedomics (linkedomics.org) portal was used to analyze the enrichment pathways of PEX13 positively and negatively related genes in PAAD (25).
WGCNA
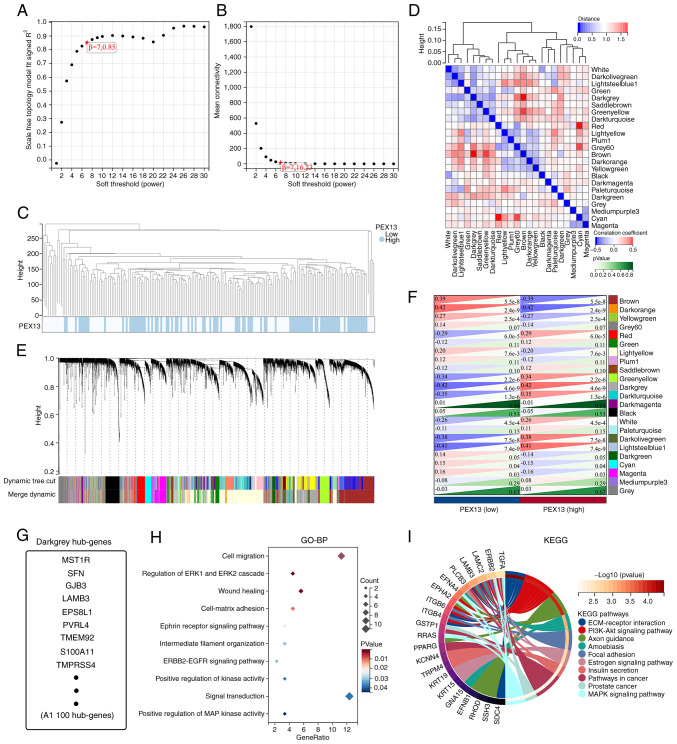
The hub genes affected by PEX13 expression level were obtained through WGCNA using SangerBox portal. First, the PAAD gene expression profile in TCGA database was used to calculate the median absolute deviation (MAD) of each gene. The outlying genes and samples were removed using the GoodSamplesGenes method of the ‘WGCNA’ R software package (genetics.ucla.edu/labs/horvath/CoexpressionNetwork/Rpackages/WGCNA) (26). WGCNA was then applied to build a scale-free co-expression network. As a soft-thresholding parameter, β could emphasize strong gene-gene associations and penalize weak correlations. To ensure a scale-free network, the power of β=7 (0.85) was selected as the soft-thresholding parameter and the power of β=7 (16.23) was selected as the soft-thresholding parameter in mean connectivity. Moreover, the PAAD samples were clustered using Pearson's correlation and average linkage method. The average linkage hierarchical clustering identified 23 modules and the differences among these were analyzed according to the expression level of PEX13, among which darkgrey module had the strongest association with PEX13 expression. Consequently, the darkgrey module was chosen as the important module for extracting hub genes.
Protein-protein interaction and ncRNA-miRNA-PEX13 regulatory networks analyses
Through the STRING database (https://string-db.org/), 50 PEX13 interacting proteins were identified in pan-cancers. Using Cytoscape (3.10.1; cytoscape.org/download.html), a PEX13 interacting protein network map was generated. The key protein-protein interaction network of PEX13 was extracted in this protein-protein interaction network using Cytoscape. Molecular docking technique was applied to predict the binding sites between PEX13 and PEX2, PEX12, PEX14 and ABCD3. By using the HDOCK server (http://hdock.phys.hust.edu.cn/), the binding sites between PEX13 and PEX2, PEX12, PEX14 and ABCD3 were analyzed. Using the PyMOL tool (2.5.5; pymol.org/2/), the binding sites between PEX13 and PEX2, PEX12, PEX14 and ABCD3 were visualized (27).
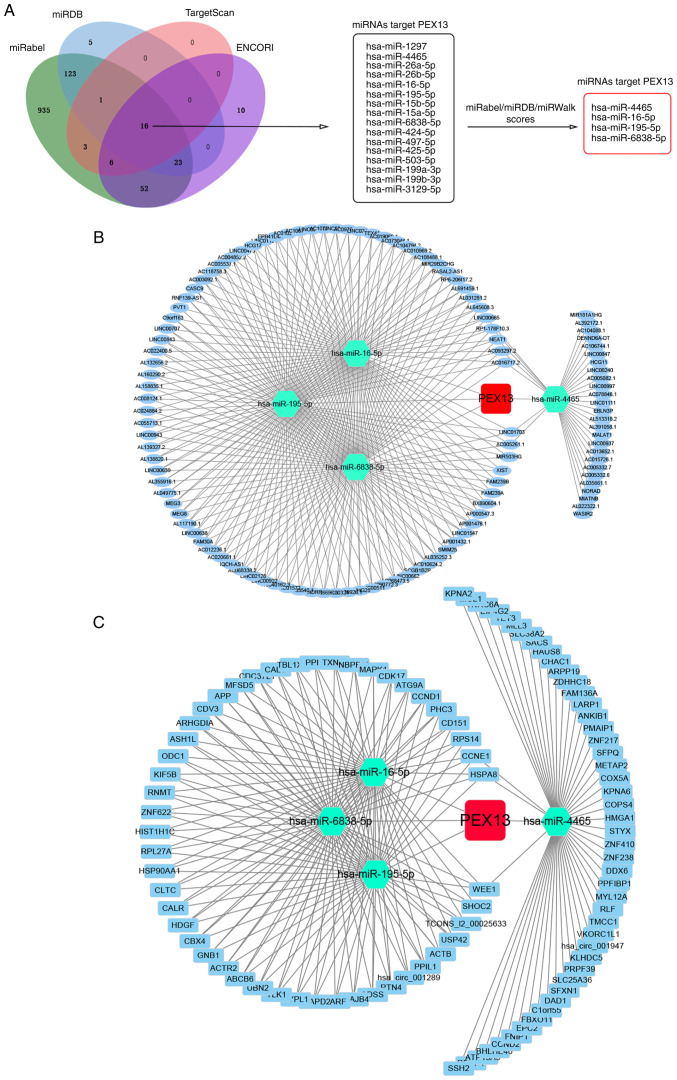
In addition, lncRNA/circRNA-miRNA-PEX13 networks were investigated. Through the niRabel (http://bioinfo.univ-rouen.fr/mirabel/index.php), miRDB (http://mirdb.org/), TargetScan (http://www.targetscan.org/vert_72/) and ENCORI (https://starbase.sysu.edu.cn/) databases, 16 miRNAs that might target PEX13 mRNA were identified. Subsequently, four miRNAs were further screened and selected through miRabel, miRDB and miRWalk scores, including hsa-miR-4465, hsa-miR-16-5p, hsa-miR-195-5p and hsa-miR-6838-5p. Using the ENCORI online tool, 105 lncRNAs and 96 circRNAs that targeted hsa-miR-4465, hsa-miR-16-5p, hsa-miR-195-5p and hsa-miR-6838-5p were predicted, and the lncRNA-miRNA-PEX13 and circRNA-miRNA-PEX13 regulatory networks were visualized via Cytoscape tool.
Immune-related analysis
The SangerBox portal was used to investigate the relationships between PEX13 mRNA expression and tumor infiltration immune cells (TIICs) in various cancers, including T_cells_CD8, T_cells_CD4_memory_resting, B_cell_memory, Tregs, NK_cells_activated, macrophages and dendritic_cells_activated. Subsequently, the correlations between PEX13 mRNA expression and immune checkpoint (ICP) genes, microsatellite instability (MSI), tumor mutation burden (TMB) and tumor purity in multiple TCGA tumors we investigated using SangerBox. Furthermore, TIMER2 portal was applied to analyze the associations between PEX13 and the infiltration levels of B cell and macrophage in pan-cancers, especially in PAAD via different algorithms, such as XCELL, CIBERSORT and EPIC. Moreover, the relationships between PEX13 and stromal score, immune score and ESTIMATE score in multiple cancers were explored using SangerBox, especially in lung adenocarcinoma (LUAD), stomach and esophageal carcinoma (STES), sarcoma (SARC) and lung squamous cell carcinoma (LUSC).
Cell culture
The human PAAD cell lines of PANC1 and BxPC3 were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Comprehensive cell line authentication was performed and cells were periodically checked for mycoplasma contamination. The DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (FBS; Satorius AG) was used to culture the PAAD cell lines. The PAAD cells were incubated in a cell incubator at 37°C with 5% CO2.
Small-interfering (si)RNAs delivery, RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
siRNAs used to knock down the expression of PEX13 were obtained from Shanghai GenePharma Co., Ltd. A total of 2×105 PAAD cells were seeded in 6-well plates, 4.5 µl Lipofectamine™ RNAiMAX reagent (Thermo Fisher Scientific, Inc.) and 40 pmol siRNA were mixed for 10 min in each well at room temperature. Subsequently, the mixture was added to the cells cultured with FBS-free medium and the cells were replaced with full medium at 24 h after transfection. The PAAD cells were collected after 48 h with TRIzol® reagent (Takara Bio, Inc.). The GoScript reverse transcription system (Promega Corporation) was applied to generate PEX13 cDNA and the GoTaq® qPCR Master Mix (Promega Corporation) was used to detect the knockdown efficiency of PEX13 siRNAs through qPCR using the ABI QuantStudio 3 system. The siRNA sequences were as follows: si-PEX13-1 sense, 5′-ACUUGAUUCCACCGUUUUCCUTT-3′ and antisense, 5′-GAAAACGGUGGAAUCAAGUAATT-3′; si-PEX13-2 sense, 5′-UUAUUCUAGUCCUGAAUUCCUTT-3′ and antisense, 5′-GAAUUCAGGACUAGAAUAAAGTT-3′; and siRNA negative control (si-NC) sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. The thermocycling conditions used for qPCR were: Initial denaturation at 95°C for 10 min; 40 cycles of amplification for 15 sec at 95°C and 45 sec at 60°C, and an extension step at 72°C for 2 min. The relative expression levels of the PEX13 mRNA were calculated using the 2−ΔΔCq method and normalized to the internal control GAPDH (28). The primer sequences were as follows: GAPDH forward, 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and reverse, 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ reverse; PEX13 forward, 5′-GCCCCACTTTCCAATCTGCT-3′ and reverse, 5′-AGAATAGGTGGGGGCACTCT-3′.
Cell Counting Kit-8 (CCK-8) assay
A total of 2×103 PANC1 or BxPC3 cells were plated into 96-well plates. PEX13 expression was knocked down using the PEX13 siRNAs. Subsequently, the optical density (OD) at 450 wavelengths was measured for 5 consecutive days (at days 0, 1, 2, 3, 4 and 5) after incubation for 1 h at 37°C with the CCK-8 reagent (ApexBio Technology) using a multiplate reader.
Wound healing and Transwell assays
A total of 2.5×105 PANC1 or BxPC3 cells were plated in 6-well plates. A 200 µl pipette tip was used to scratch the cell monolayer. After 0, 12 and 24 h incubation in an FBS-free medium, the images of wound closure were captured and further explored via an inverted microscope (Olympus Corporation) and Image J software (1.8.0; National Institutes of Health). For the Transwell assay, 8-µm pore filters coated with Matrigel matrix glue (Corning, Inc.) were used. Matrigel was added into the upper chamber for 40 min at 37°C. A total of 5×104 PANC1 and BxPC3 cells in 300 µl serum-free DMEM were seeded into the upper chamber and 600 µl DMEM with 10% FBS medium was added to the lower chamber. After 24 h, cells were fixed with 4% paraformaldehyde at room temperature and stained with 1% crystal violet at room temperature. The images were captured using a microscope (BX53; Olympus Corporation).
Colony formation assay
In 6-well plates, 1×103 PANC1 and BxPC3 cells were seeded in each well with 2.5 ml DMEM containing 10% FBS. The cells were incubated for 2 weeks at 37°C in a humidified chamber with 5% CO2. The colonies were fixed with 4% paraformaldehyde for 30 min at room temperature before being stained with crystal violet for 10 min at room temperature. A cell mass of >50 cells was considered a colony, and Image J (1.8.0) was used for counting colonies.
Statistical analysis
All of the experimental data were statistically analyzed using GraphPad Prism 8.0 (Dotmatics). Comparisons between two groups were made using an unpaired t-test and the statistical method used for survival analysis was Kaplan-Meier. Cox regression analysis was performed using logrank test to obtain prognostic significance. The correlation analyses in the manuscript were applied using Pearson's correlation analysis. One-way ANOVA with Bonferroni's post-hoc test was used to compare the differences between multiple groups. Two-way ANOVA with Bonferroni's post-hoc test was used to compare the differences between CCK-8 groups (si-NC and si-PEX13-1/si-PEX13-2). The data are presented as mean ± standard deviation from three individual experiments and each experiment was repeated at least three times. P<0.05 was considered to indicate a statistically significant difference.
Results
PEX13 expression patterns in various cancers
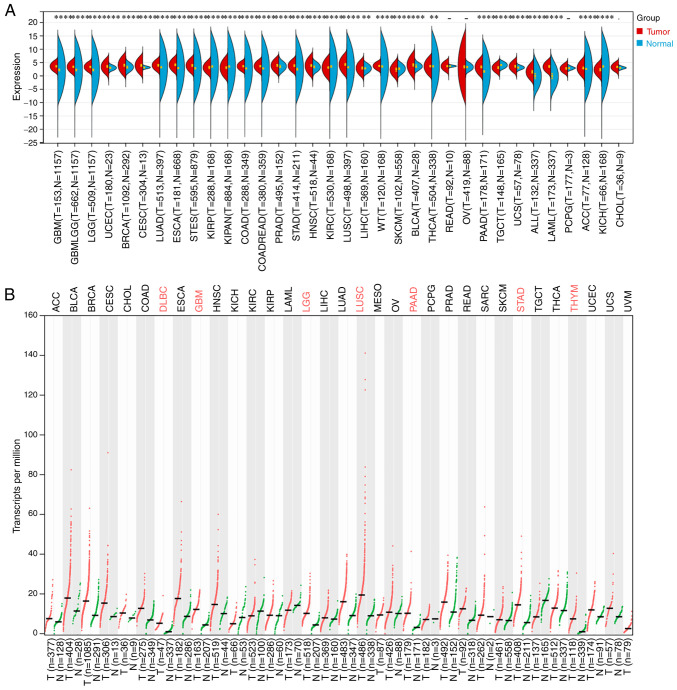
Through TCGA and GTEx databases analysis, PEX13 was revealed to be highly expressed in multiple tumors, including GBM, LGG, GBMLGG, BRCA, UCEC, LUAD, CESC, ESCA, COAD, STES, READ, STAD, PRAD, HNSC, LUSC, LIHC, high-risk wilms tumor (WT), BLCA, THCA, PAAD, UCS, LAML, acute lymphoblastic leukemia (ALL) and ACC. In addition, the expression level of PEX13 was downregulated in KIRP, pan-kidney cohort (KIPAN), KIRC, SKCM, TGCT and KICH compared with the corresponding normal tissues (Fig. 1A). In GEPIA2 portal, it was discovered that PEX13 was remarkable upregulated in DLBC, GBM, LGG, LUSC, PAAD, STAD and THYM (Fig. 1B). In addition, the results of IHC showed that the expression of PEX13 protein in breast cancer, colorectal cancer, glioma, liver cancer, lung cancer, pancreatic cancer, skin cancer, stomach cancer, thyroid cancer and renal cancer was higher than that in normal tissues (Fig. S1). The present findings indicated that PEX13 mRNA and protein expression levels are increased in a variety of tumors, suggesting that PEX13 may function as an oncogene in various cancers, including PAAD.
Figure 1.
PEX13 expression levels in normal tissue and tumors. (A) Comparison of PEX13 expression level among 33 tumor tissues and normal tissues via The Cancer Genome Atlas and Genotype-Tissue Expression databases. (B) The differences of PEX13 expression levels in tumor and normal samples by GEPIA2. **P<0.01, ***P<0.001 and ****P<0.0001. PEX, peroxisomal biogenesis factor.
PEX13 expression and prognosis across multiple types of cancer
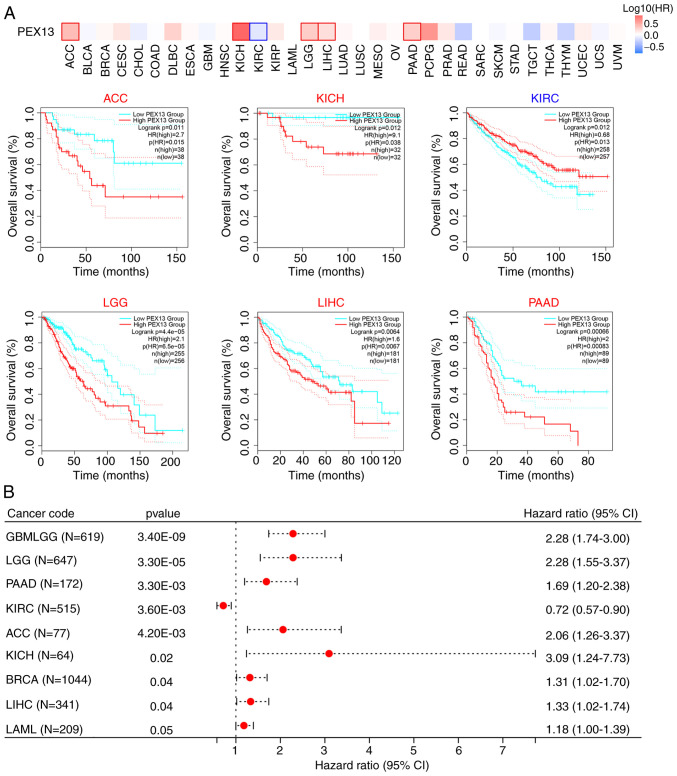
The prognostic value of PEX13 in various tumors was analyzed by comparing the OS of patients with PEX13 expression. Analysis of the pan-cancer cohort using the GEPIA2 database demonstrated that high PEX13 mRNA expression levels was associated with a significantly lower OS in patients with ACC, KICH, LGG, LIHC and PAAD, and high PEX13 mRNA expression levels were associated with a significantly higher OS in patients with KIRC (Fig. 2A). Cox regression analysis in SangerBox database suggested that PEX13 mRNA expression level was related to OS, DFI, DSS and PFI in patients with various tumors. Regarding the OS, the findings revealed that high PEX13 mRNA expression was linked to a shorter OS in GBMLGG, LGG, PAAD, ACC, KICH, BRCA, LIHC and LAML (Fig. 2B). Regarding the DFI, high PEX13 mRNA expression was linked to a shorter DFI in PAAD, KIPAN, KIRP and ACC (Fig. S2A). In PFI, the high PEX13 expression was related to shorter PFI in GNMLGG, ACC, LGG, PAAD, LIHC, KICH and UVM (Fig. S2B). Furthermore, a high PEX13 mRNA expression was correlated with a shorter DSS in GBMLGG, LGG, PAAD, ACC, KICH and LIHC (Fig. S2C). It was also observed that high PEX13 mRNA expression was significantly correlated with shorter OS, DFI, PFI, and DSS in PAAD and ACC. The present results revealed that the level of PEX13 mRNA expression was related to the prognosis of various tumors. Furthermore, high PEX13 mRNA expression was linked to a worse prognosis in patients with PAAD.
Figure 2.
Prognosis analysis of PEX13 in pan-cancers. (A) Survival maps of overall survival according to PEX13 expression in various cancers via Gene Expression Profiling Interactive Analysis 2. (B) Cox regression analysis according to PEX13 expression in multiple tumors. PEX, peroxisomal biogenesis factor.
PEX13 genetic alterations in various cancers
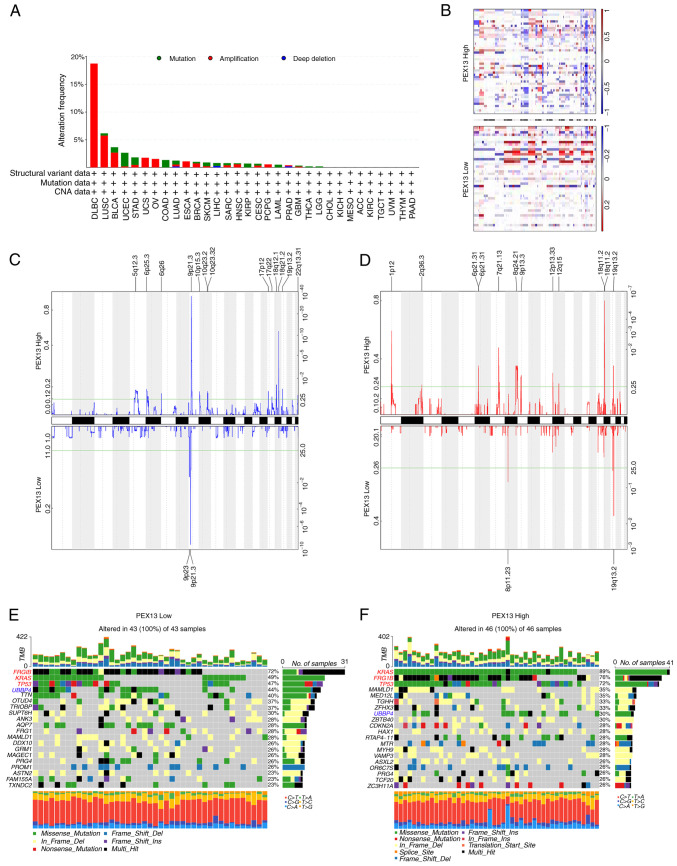
Mutations, deletions or amplification of oncogenes or tumor suppressor genes have been linked to the growth and progression of several tumors (29). Hence, the present study analyzed diverse types of genetic alterations in the PEX13 gene using the cBioPortal portal, including amplification, mutation and deep deletion. The most common genetic alteration of PEX13 gene in DLBC (18.75%), LUSC (5.75%), BLCA (2.68%), UCS (1.75%), ovarian serous cystadenocarcinoma (1.54%) and ESCA (1.1%) was amplification. In UCEC (2.46%), STAD (1.36%), COADREAD (1.35%) and KIRP (0.71%), the most common genetic alteration of PEX13 gene was mutation (Fig. 3A). Subsequently, the TCGA-PAAD dataset was interrogated to investigate the correlation between PEX13 and specific genomic features such as copy number variations (CNVs) and somatic mutations. Fig. 3B shows the comparison of the CNV profiles in the PEX13high (n=46) and PEX13low (n=43) groups. In the PEX13high group, deletion peaks were found in the 5p12.3, 6p25.3, 9p21.3, 10p15.3, 10q23.2, 18q12.1, 18q21.2 and 19p13.2 chromosomal locations, while amplification peaks were found in the 1p12, 6p21.31, 7q21.13, 8q24.21, 9p13.3, 12p13.33, 18q11.2 and 19q13.2 chromosomal regions (Fig. 3C-D). In the PEX13low group, deletion peaks were found in the 9p23 and 9p21.3 chromosomal locations, while frequent amplification peaks were found in the 8p11.23 and 19q13.2 chromosomal regions (Fig. 3C and D). These findings suggested that high PEX13 expression led to more deletions and amplifications of multiple sites in the genome of patients with PAAD. The PEX13low group indicated that high frequency of somatic mutations in the FRG1B (72%), KRAS (49%), TP53 (47%), UBBP4 (44%) and TTN (40%) genes and the PEX13high group suggested that high frequency of mutations in the KRAS (89%), FRG1B (76%), TP53 (72%), MAMLD1 (35%), MED12L (35%) and UBBP4 (30%) genes (Fig. 3E and F). These findings revealed that high PEX13 expression may lead to the increased mutation probability of KRAS, FRG1B, TP53 and other genes, and to the decreased mutation probability of UBBP4 gene in PAAD, which may be a potential mechanism leading to the occurrence and development of tumors.
Figure 3.
Distinct genomic profiles associated with PEX13 expression in pan-cancers, including PAAD. (A) Genetic alterations of PEX13 in different cancers in TCGA database were discovered using the cBioPortal tool, including mutation, structural variant, amplification and deep deletion. (B) The copy number alteration profile analysis between 25% PEX13low and 25% PEX13high groups in a PAAD cohort was preformed using GISTIC2.0. Frequencies of (C) deletion and (D) amplification of PAAD associated with PEX13 expression (blue, deletion; red, amplification). Detection of somatic mutations in PAAD, including (E) 25% PEX13low and (F) 25% PEX13high group. PEX, peroxisomal biogenesis factor. TCGA, The Cancer Genome Atlas; PAAD, pancreatic adenocarcinoma.
These findings suggested the presence of PEX13 gene mutations, amplification and deletion in multiple tumors. The PAAD tissues revealed different somatic mutations and CNVs depending on the level of PEX13 expression. This suggested that alterations in the PEX13 gene could influence the occurrence and progression of multiple cancers, particularly PAAD.
Pathway enrichment analysis of PEX13-associated genes
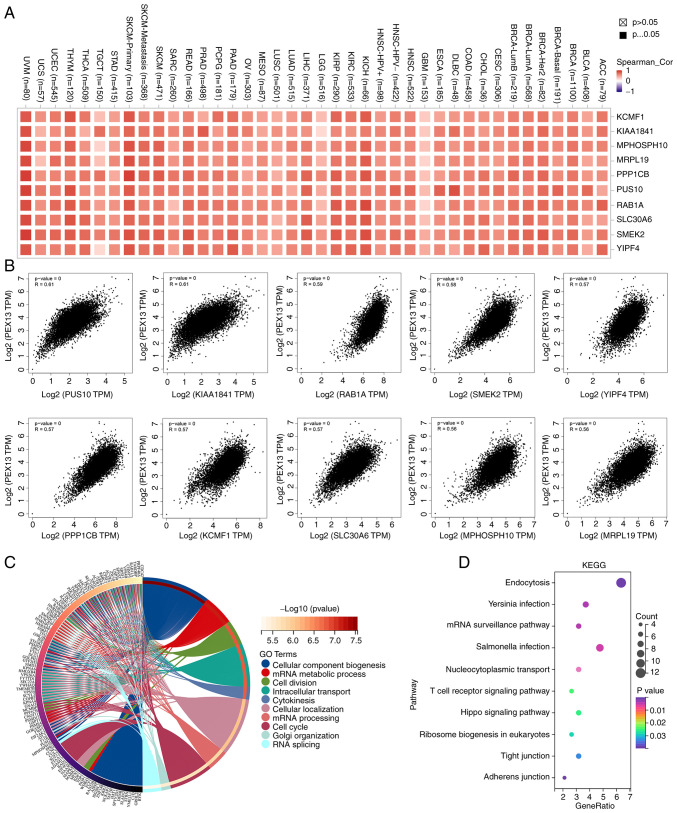
The PEX13-related genes were analyzed in 33 cancers through GEPIA2 database and 200 PEX13-related genes were obtained. The top 10 genes with the highest correlation among 200 PEX13-related genes were generated correlation heat maps in 33 cancers via TIMER2, including KCMF1, KIAA1841, MPHOSPH10, MRPL19, PPP1CB, PUS10, RAB1A, SLC30A6, SMEK2 and YIPF4 (Fig. 4A). Fig. 4B shows that PEX13 expression was positively correlated with PUS10 (R=0.61), KIAA1841 (R=0.61), RAB1A (R=0.59), SMEK2 (R=0.58), YIPF4 (R=0.57), PPP1CB (R=0.57), KCMF1 (R=0.57), SLC30A6 (R=0.57), MPHOSPH10 (R=0.56) and MRPL19 (R=0.56) in GEPIA2. The GO-BP analysis of 200 PEX13-related genes in 33 tumors showed that PEX13-related genes were mainly enriched in mRNA metabolic process, cellular component biogenesis, cell division, intracellular transport, cytokinesis, mRNA processing, cell cycle and RNA splicing (Fig. 4C). PEX13-related genes were mainly enriched in Endocytosis, mRNA surveillance pathway, nucleocytoplasmic transport, T cell receptor, signaling pathway and hippo signaling pathway based on the results of the KEGG enrichment analysis (Fig. 4D).
Figure 4.
The enrichment analysis of PEX13-related genes in pan-cancers. (A) Correlation heat map displayed the top 10 PEX13-related genes in various tumors via the TIMER2.0 portal. (B) Correlations between PEX13 expression and top 10 PEX13-related genes in pan-cancers via GEPIA2. The (C) Gene Ontology-Biological Process and (D) KEGG enrichment analyses of 200 PEX13-related genes in various cancers. PEX, peroxisomal biogenesis factor; TPM, transcripts per million; KEGG, Kyoto Encyclopedia of Genes and Genomes; UVM, uveal melanoma; UCS, uterine carcinosarcoma; UCEC, uterine corpus endometrial carcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; STAD, stomach adenocarcinoma; SKCM, skin cutaneous melanoma; SARC, sarcoma; READ, rectum adenocarcinoma; PRAD, prostate adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PAAD, pancreatic adenocarcinoma; OV, ovarian serous cystadenocarcinoma; MESO, mesothelioma; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma; LIHC, liver hepatocellular carcinoma; LGG, brain lower grade glioma; KIRC, kidney renal clear cell carcinoma; KICH, kidney chromophobe; HNSC, head and neck cancer; HPV, human papillomavirus; GBM, glioblastoma multiforme; ESCA, esophageal carcinoma; DLBC, lymphoid neoplasm diffuse large b-cell lymphoma; COAD, colon adenocarcinoma; CHOL, cholangiocarcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; BRCA, breast invasive carcinoma; BLCA, bladder urothelial carcinoma; ACC, adrenocortical carcinoma.
To further explore the biological functions of PEX13 in PAAD, the enrichment of PEX13 positively and negatively related genes and their enrichment pathways in PAAD were analyzed using the Linkedomics portal. Fig. S3A shows the heat maps of 50 positively and 50 negatively related genes of PEX13 in PAAD. Fig. S3B-E showed the GO-BP and KEGG enrichment analysis of 100 positively and 100 negatively related genes of PEX13, respectively. The results showed that GO-BP was mainly enriched in positive regulation of drug response, small GTPase mediated signal transduction, regulation of epidermal growth factor-activated receptor activity and ERBB signaling pathway among the positively related genes of PEX13 and in the regulation of glycogen (starch) synthase activity, intracellular protein transport, intracellular protein transport and autophagosome maturation among the negatively related genes of PEX13 (Fig. S3B and C). For the KEGG enrichment analysis, PEX13-positive related genes were mainly enriched in Rap1 signaling pathway, ErbB signaling pathway, EGFR tyrosine kinase inhibitor resistance, Neurotrophin signaling pathway and AMPK signaling pathway and PEX13-negatively related genes were mainly enriched in Vitamin B6 metabolism, Apelin and Insulin signaling pathway (Fig. S3D-E). These findings show that PEX13 could regulate mRNA processing, RNA splicing, cell cycle and the response to drugs in cancers. In PAAD, PEX13 expression was related to protein transport, tumor cell response to drugs, multiple cancer pathways and immune pathways.
Analysis of WGCNA in PAAD
The TCGA-PAAD cohort was analyzed via WGCNA to identify hub genes strongly associated with PEX13 expression in PAAD. Using the average linkage approach and Pearson's correlation method, the samples of TCGA-PAAD data were first clustered (Fig. 5A-C). With the help of the average linkage hierarchical clustering, a total of 23 modules were obtained (Fig. 5D). According to the expression level of PEX13, we analyzed the differences among 23 modules, among which darkgrey module had the strongest association with PEX13 expression (Fig. 5E-F). The darkgrey module was selected as the significant module for further analysis. Fig. 5G shows the hub genes of darkgrey module and GO-BP and KEGG enrichment analyses were used to analyze these hub genes. In GO-BP analysis, hub genes mainly enriched in cell migration, regulation of ERK1 and ERK2 cascade, wound healing, ephrin receptor signaling pathway, ERBB2-EGFR signaling pathway, positive regulation of MAP kinase activity and signal transduction (Fig. 5H). In KEGG enrichment analysis, hub genes mainly enriched in PI3K-Akt signaling pathway, extracellular matrix (ECM)-receptor interaction, focal adhesion, pathways in cancer and MAPK signaling pathway (Fig. 5I). These results suggested that the hub genes of darkgrey module with different PEX13 expression were mainly involved in cell migration, wound healing, PI3K-Akt and MAPK signaling pathway.
Figure 5.
Weighted gene co-expression network analysis of PAAD associated with PEX13 expression. (A) Analysis of the scale-free fit index for various soft-thresholding powers (β). (B) Analysis of the mean connectivity for various soft-thresholding powers. (C) Clustering dendrogram of PAAD samples. (D) Module feature vector clustering. (E) Dendrogram of all differentially expressed genes clustered based on a dissimilarity measure. (F) Heatmap of the correlation between module eigengenes and PEX13 expression level of PAAD. (G) The hub genes diagram of darkgrey module. The GO-BP (H) and KEGG (I) enrichment analyses of hub genes of darkgrey module in PAAD. GO-BP, Gene Ontology-Biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes; PEX, peroxisomal biogenesis factor; PAAD, pancreatic adenocarcinoma.
Construction of protein interaction and ncRNA-miRNA-PEX13 regulatory network
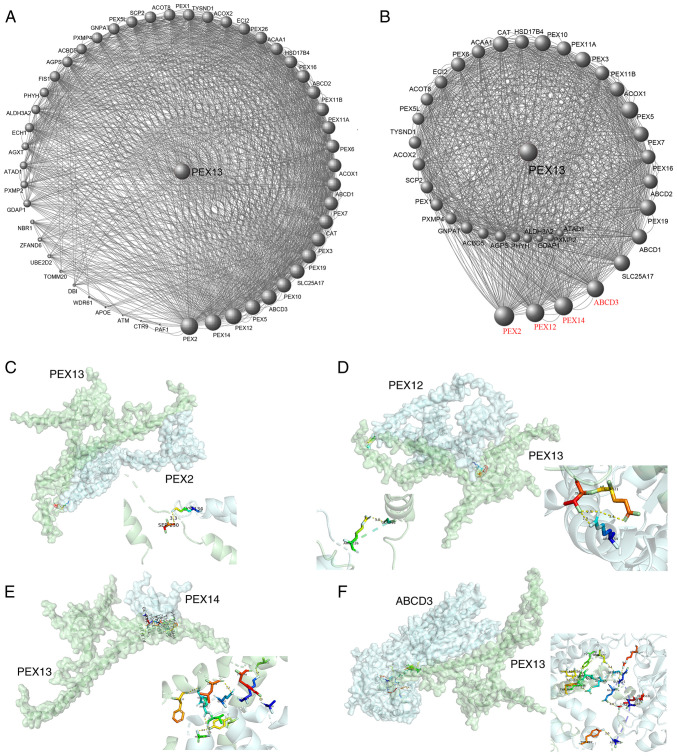
The 50 PEX13 interacting proteins were identified using STRING database and the PEX13 interacting protein network map was generated in Cytoscape (Fig. 6A). In this protein interaction network, key protein interaction network of PEX13 was extracted using Cytoscape, as shown in Fig. 6B. The PEX13 interacting proteins were scored in Cytoscape and a larger volume indicated higher interacting score of these proteins in the network diagram. Subsequently, four proteins had the highest interaction scores with PEX13, including PEX2, PEX12, PEX14 and ABCD3 (Fig. 6B). Molecular docking technique was applied to forecast the binding sites between PEX13 and PEX2, PEX12, PEX14 and ABCD3 (Fig. 6C-F). The results showed that PEX13 may interact with PEX2, PEX12, PEX14 and ABCD3 to influence the biological functions and acted as a significant role in various tumors.
Figure 6.
Protein-protein interaction analysis of PEX13. (A) Protein-protein interaction network of PEX13 by STRING portal. (B) Key protein-protein interaction network of PEX13 using STRING and Cytoscape tools. The binding sites between PEX13 and PEX2 (C), PEX12 (D), PEX14 (E) and ABCD3 (F) via molecular docking technique. PEX, peroxisomal biogenesis factor; ABCD3, TP binding cassette subfamily d member 3.
In addition, numerous studies found that lncRNA and circRNA regulated the expression of downstream mRNA by adsorbing target miRNAs, which acted as a crucial factor in tumorigenesis and the progression of tumors (30,31). Therefore, the present study explored the lncRNA/circRNA-miRNA network that may regulate mRNA expression in multiple cancers. LncRNA-miRNA regulatory networks of PEX13 were constructed to investigate the underlying molecular mechanisms of PEX13 in tumors. Through the miRabel, miRDB, TargetScan and ENCORI databases, 16 miRNAs that might target PEX13 mRNA were identified (Fig. 7A). Subsequently, four miRNAs, including hsa-miR-4465, hsa-miR-16-5p, hsa-miR-195-5p and hsa-miR-6838-5p, were screened and selected using miRabel, miRDB and miRWalk. Using the ENCORI online tool, the lncRNAs and circRNAs that targeted their corresponding miRNAs were predicted and Cytoscape was used to visualize the lncRNA-miRNA and circRNA-miRNA regulatory networks (Fig. 7B and C). These findings suggested the upstream ncRNA-miRNA regulatory networks that may control the aberrant expression of PEX13 in cancers.
Figure 7.
Construction of lncRNA/circRNA/miRNA-PEX13 network. (A) Venn diagram showed the 16 miRNAs that might target PEX13 mRNA by the miRabel, miRDB, TargetScan and ENCORI databases and 4 miRNAs were eventually identified via miRabel, miRDB and miRWalk databases. (B) Construction of lncRNA-miRNA-PEX13 network through Cytoscape tool. (C) Construction of circRNA-miRNA-PEX13 network through Cytoscape tool. LncRNA, long non-coding RNA; miRNA, microRNA; circRNA, circular RNA; PEX, peroxisomal biogenesis factor.
PEX13 regulated tumor immunity in multiple tumors
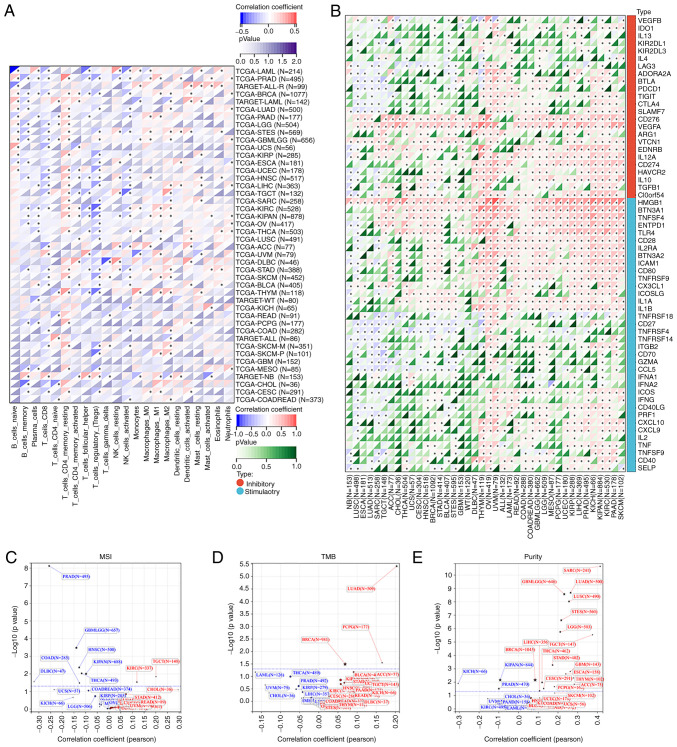
The relationships between PEX13 and the tumor immune microenvironment (TIM) were further analyzed in a variety of tumors, and the correlation between PEX13 and the composition of tumor infiltrating immune cells (TIICs) was investigated in multiple cancers. Studies showed that TIICs were a significant component of the tumor microenvironment and regulated the occurrence, development and metastasis of tumors (32,33). The Sangerbox database was applied to investigate the correlation between PEX13 and TIIC levels in diverse tumors. The expression of PEX13 in LAML, PRAD, BRCA, PAAD, STES, HNSC, LIHC, KIRC, THCA, STAD, SKCM, THYM and CESC was remarkably related to various immune cells, including T_cells_CD4_memory_resting, B_cell_memory, T_cells_CD8, macrophages_M1, macrophages_M2, Tregs, NK_cells_activated, dendritic_cells_activated and neutrophils (Fig. 8A). Fig. S4A demonstrated the relationship between PEX13 expression and B cell infiltration levels in different tumors. In PAAD, the expression level of PEX13 under different algorithms was mainly negatively related to the infiltration level of B cells (Fig. S4B). Fig. S4C presents a heat map of the relationship between PEX13 expression levels and macrophage infiltration levels in various tumors. PEX13 expression level was also negatively related to the infiltration level of macrophages under different algorithms in PAAD (Fig. S4D).
Figure 8.
Correlations between PEX13 expression and immunity in pan-cancers. (A) Association between PEX13 expression level and immune cell infiltration level in pan-cancers via Sangerbox. (B) The correlations between PEX13 expression and immune checkpoint genes in multiple tumors. The correlations between PEX13 expression and microsatellite instability (C), tumor mutation burden (D) and tumor purity (E) in various tumors. *P<0.05. PEX, peroxisomal biogenesis factor; MSI, microsatellite instability; TMB, tumor mutation burden; ICP, immune checkpoint.
Furthermore, the correlation between PEX13 expression level and ICP genes, MSI, TMB and tumor purity were analyzed via SangerBox. The occurrence of MSI is caused by the functional defect of DNA mismatch repair in tumor tissues (34). The presence of MSI and ineffective DNA mismatch repair is a crucial clinical tumor marker (35). TMB has a strong relationship to the effectiveness of PD-1/PD-L1 inhibitors and can forecast their benefits in terms of efficacy (36). The present study discovered that PEX13 expression was positively linked with ICP genes in several tumor types (Fig. 8B). PEX13 expression was positively correlated with MSI in TGCT and KIRC and negatively correlated with MSI in GBMLGG, HNSC, PRAD, KIPAN, COAD, DLBC and THCA (Fig. 8C). In LUAD and BRCA and pheochromocytoma and paraganglioma, PEX13 expression was linked favorably with TMB (Fig. 8D). In tumor purity, PEX13 expression was negatively related to purity in KICH, KIPAN and PRAD and positively related to purity in GBMLGG, SARC, LUAD, LUSC, STES, LGG, STAD, BRCA, ESCA, etc. (Fig. 8E). Moreover, it was evaluated the relationship between the ESTIMATE score and PEX13 expression level in various cancers. ImmuneScore reflects the proportion of infiltrating immune cells in cancer tissues. The proportion of stromal cells in tumor tissue was reflected by the Stromal score (StromalScore). The ESTIMATE score (ESTIMATEScore) is the sum of immune and stromal scores and reflects the state of the TIM and tumor purity (18,37). The present study found significant relationships between PEX13 levels and StromalScore, ImmuneScore and ESTIMATEScore across multiple tumors, especially in LUAD, STES, SARC and LUSC (Fig. S5A-C). These findings indicated that PEX13 expression regulated the sensitivity and resistance of certain cancers to immunotherapy. Thus, PEX13 emerged from this analysis as a potential immunotherapy biomarker and predictor of tumor immunotherapy response.
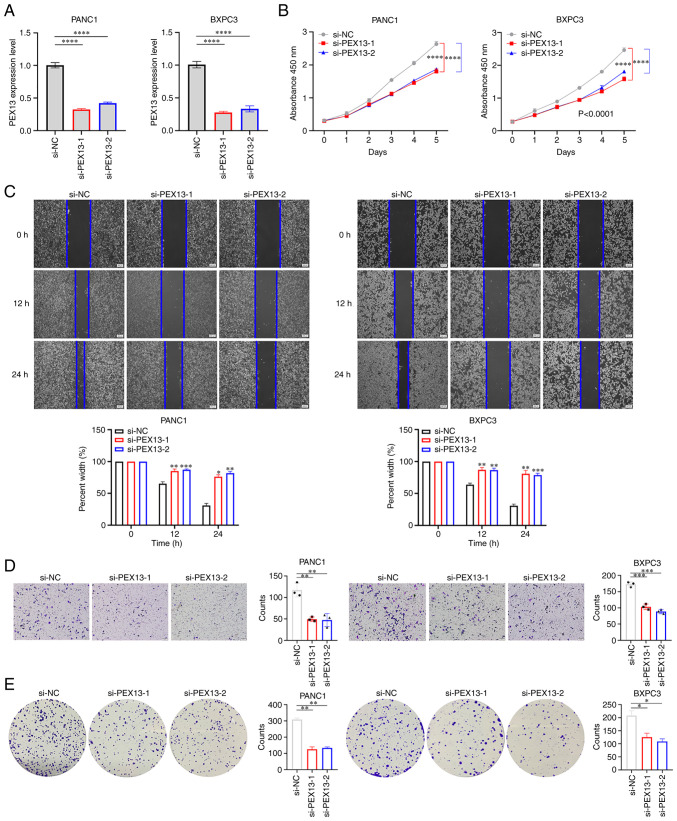
PEX13 promoted cell proliferation, migration, invasion and colony formation in PAAD
PANC1 and BxPC3 cell lines were used to verify the biological functions of PEX13. The knockdown of PEX13 expression in PANC1 and BxPC3 cells transfected with PEX13 siRNAs was confirmed using RT-qPCR (Fig. 9A). The CCK-8 assay suggested that PEX13 knockdown inhibited the cell proliferation of PANC1 and BxPC3 cells in vitro (Fig. 9B). In addition, wound healing and Transwell experiments demonstrated that PEX13 knockdown suppressed the cell migration and invasion of PANC1 and BxPC3 cells in vitro (Fig. 9C-D). The colony formation assay suggested that PEX13 knockdown could inhibit the cell colony formation ability of PANC1 and BxPC3 cells in vitro (Fig. 9E). These findings showed that PEX13 could increase cell proliferation, migration, invasion and colony formation abilities of PANC1 and BxPC3 cell lines in vitro.
Figure 9.
Biological functions of PEX13 in PAAD. (A) Reverse transcription-quantitative PCR assay verified the knockdown efficiency of PEX13 small interfering RNAs. (B) Cell Counting Kit-8 assay measured the cell proliferation after PEX13 knockdown in PANC1 and BxPC3 cells. (C) Wound healing, (D) Transwell and (E) colony formation assays measured cell migration, invasion and colony formation abilities after PEX13 knockdown in PANC1 and BxPC3 cells. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. si-, small interfering RNA; NC, negative control; PEX, peroxisomal biogenesis factor; si-PEX13, siRNA targeting PEX13.
Discussion
Metabolic reprogramming is one of the hallmarks of cancer and has a selective advantage in the occurrence and progression of cancer cells (38). Metabolites ingested through cell membranes are mostly initially processed in the cytoplasm and then typically transported to intracellular organelles for further processing, such as peroxisome and mitochondria (39). Although mitochondria are key metabolic hubs of cancer cells, the metabolic functions of other organelles in cancer cells are rarely studied and need further exploration.
In mammals, the peroxisome, a single membrane organelle, participates in >50 separate enzymatic functions (40). Defects in genes encoding peroxisome proteins are associated with peroxisome disease. Lipid metabolism is also critical to tumorigenicity (8). Lipid synthesis pathways (such as the production of lipogenic triglycerides and the synthesis of phospholipids and cholesterol) are critical for energy storage, cell membrane structure and cell signaling regulation. For lipid synthesis and activation of the corresponding signaling network to be coordinated in rapidly growing cancer cells, a significant volume of cell membrane formation is required (41). High levels of ether phospholipid in certain cancers suggest that elevated lipid synthesis of peroxisome is related to tumor progression (42). PEX13 is an indispensable key molecule in the occurrence of peroxisome, but, to the best of our knowledge, its functions and effects in tumors have not been reported in the literature. Therefore, the present study aimed to investigate the biological functions and clinical significance of PEX13 in various tumors, especially in PAAD.
The present study first revealed that PEX13 was upregulated in several types of tumors compared with the corresponding normal tissues, including GBM, LGG, GBMLGG, UCEC, CESC, BRCA, ESCA, LUAD, STES, COADREAD, COAD, PRAD, STAD, LUSC, HNSC, WT, LIHC, THCA, BLCA, PAAD, UCS, ALL, LAML and ACC. In ACC, KICH, LGG, LIHC and PAAD, patients with high PEX13 mRNA expression had significantly lower OS, and patients with high PEX13 mRNA expression had significantly higher OS in KIRC. Cox regression analysis indicated that PEX13 mRNA was related to OS, DFI, PFI and DSS in various tumors. In addition, high expression of PEX13 was associated with poorer prognosis in patients with PAAD. Subsequently, genetic alteration analysis of PEX13 showed the presence of PEX13 gene mutations, amplification and deletion in multiple tumors. The PAAD tissues displayed different somatic mutations and CNVs depending on the level of PEX13 expression, indicating that the PEX13 gene alteration may mediate the occurrence and progression of multiple tumors, especially in PAAD. The GO-BP and KEGG enrichment analyses of PEX13-related genes found that PEX13 expression was significantly correlated with endocytosis, mRNA surveillance pathway, nucleocytoplasmic transport, T cell receptor signaling pathway and hippo signaling pathway in cancers. In PAAD, GO-BP and KEGG enrichment analyses of PEX13-related genes revealed that PEX13 expression was significantly associated with Rap1, ErbB, Neurotrophin, AMPK, Apelin and Insulin signaling pathways. WGCNA analysis suggested that PEX13-related hub genes were mainly enriched in PI3K-Akt signaling pathway, ECM-receptor interaction, focal adhesion and MAPK signaling pathway via KEGG enrichment analysis. Moreover, the present study discovered that PEX13 may interact with PEX2, PEX12, PEX14 and ABCD3 to influence the biological functions and serve as a significant factor in pan-cancers and the upstream lncRNA/circRNA-miRNA regulatory networks may regulate the aberrant expression of PEX13 in various cancers.
Furthermore, PEX13-related genes were mainly enriched in immune and cancer-related pathways, and the correlation between PEX13 and tumor immunity was further analyzed. As an important component of the immune microenvironment, TIICs act as a key factor in the occurrence, progression and treatment of tumors. PEX13 mRNA levels were associated with TIICs in multiple tumors, including PAAD. The present study evaluated the effect of PEX13 on immunotherapy sensitivity and resistance in patients with cancer. The ICP, MSI, TMB and tumor purity analyses suggested that PEX13 may be a promising target for the treatment of patients with certain tumors, especially in immunotherapy. In vitro assays also demonstrated that PEX13 knockdown could significantly inhibit the proliferation, migration, invasion and colony formation of PAAD cells.
In conclusion, the present results demonstrated that PEX13 may be an invaluable prognostic indicator and a possible predictor of immunotherapy sensitivity and resistance in patients with malignant tumors, especially in PAAD. However, the present study has limitations because it only uses TCGA database, while further databases should be used for corroboration, and only verifies the relationship between PEX13 and PAAD. The possible mechanisms by which PEX13 functions in PAAD were not further explored.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ACC
adrenocortical carcinoma
- BLCA
bladder urothelial carcinoma
- BRCA
breast invasive carcinoma
- CESC
cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
cholangiocarcinoma
- CNA
copy number alteration
- COAD
colon adenocarcinoma
- DLBC
lymphoid neoplasm diffuse large b-cell lymphoma
- ESCA
esophageal carcinoma
- GTEx
Genotype-Tissue Expression
- GBM
glioblastoma multiforme
- GEPIA2
Gene Expression Profiling Interactive Analysis 2
- HGG
brain higher grade glioma
- HNSC
head and neck cancer
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KICH
kidney chromophobe
- KIRC
kidney renal clear cell carcinoma
- KIPAN
Pan-kidney cohort
- ICP
immune checkpoint
- LAML
acute myeloid leukemia
- LGG
brain lower grade glioma
- LIHC
liver hepatocellular carcinoma
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MESO
mesothelioma
- MSI
microsatellite instability
- OS
overall survival
- OV
ovarian serous cystadenocarcinoma
- PAAD
pancreatic adenocarcinoma
- PCPG
pheochromocytoma and paraganglioma
- PEX
peroxisomal biogenesis factor
- PRAD
prostate adenocarcinoma
- READ
rectum adenocarcinoma
- SARC
sarcoma
- SKCM
skin cutaneous melanoma
- STAD
stomach adenocarcinoma
- TCGA
The Cancer Genome Atlas
- TGCT
testicular germ cell tumors
- THCA
thyroid carcinoma
- THYM
thymoma
- TMB
tumor mutation burden
- UCEC
uterine corpus endometrial carcinoma
- UCS
uterine carcinosarcoma
- UVM
uveal melanoma
- WGCNA
weighted gene co-expression network analysis
Funding Statement
The present study was supported by the Natural Science Foundation of Shanxi Province (grant no. 202103021223011).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JS designed the study. PD and XD performed the bioinformatics analyses and experiments. TY and DL analyzed the results. YW and YD performed data analysis and wrote the manuscript. JS and PD confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fujiki Y, Okumoto K, Mukai S, Honsho M, Tamura S. Peroxisome biogenesis in mammalian cells. Front Physiol. 2014;5:307. doi: 10.3389/fphys.2014.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee MY, Sumpter RJ, Zou Z, Sirasanagandla S, Wei Y, Mishra P, Rosewich H, Crane DI, Levine B. Peroxisomal protein PEX13 functions in selective autophagy. EMBO Rep. 2017;18:48–60. doi: 10.15252/embr.201642443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugiura A, Mattie S, Prudent J, Mcbride HM. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature. 2017;542:251–254. doi: 10.1038/nature21375. [DOI] [PubMed] [Google Scholar]
- 6.Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain IH, Calvo SE, Markhard AL, Skinner OS, To TL, Ast T, Mootha VK. Genetic screen for cell fitness in high or low oxygen highlights mitochondrial and lipid metabolism. Cell. 2020;181:716–727. doi: 10.1016/j.cell.2020.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q, Luo Q, Halim A, Song G. Targeting lipid metabolism of cancer cells: A promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Di Cara F, Savary S, Kovacs WJ, Kim P, Rachubinski RA. The peroxisome: An up-and-coming organelle in immunometabolism. Trends Cell Biol. 2023;33:70–86. doi: 10.1016/j.tcb.2022.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Diskin C, Zotta A, Corcoran SE, Tyrrell VJ, Zaslona Z, O'Donnell VB, O'Neill L. 4-Octyl-Itaconate and Dimethyl Fumarate inhibit COX2 expression and prostaglandin production in macrophages. J Immunol. 2021;207:2561–2569. doi: 10.4049/jimmunol.2100488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 12.Ding L, Sun W, Balaz M, He A, Klug M, Wieland S, Caiazzo R, Raverdy V, Pattou F, Lefebvre P, et al. Peroxisomal beta-oxidation acts as a sensor for intracellular fatty acids and regulates lipolysis. Nat Metab. 2021;3:1648–1661. doi: 10.1038/s42255-021-00489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JY, Plakidas A, Lee WH, Heikkinen A, Chanmugam P, Bray G, Hwang DH. Differential modulation of Toll-like receptors by fatty acids: Preferential inhibition by n-3 polyunsaturated fatty acids. J Lipid Res. 2003;44:479–486. doi: 10.1194/jlr.M200361-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Moreno-Fernandez ME, Giles DA, Stankiewicz TE, Sheridan R, Karns R, Cappelletti M, Lampe K, Mukherjee R, Sina C, Sallese A, et al. Peroxisomal beta-oxidation regulates whole body metabolism, inflammatory vigor, and pathogenesis of nonalcoholic fatty liver disease. JCI Insight. 2018;3:e93626. doi: 10.1172/jci.insight.93626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demers ND, Riccio V, Jo DS, Bhandari S, Law KB, Liao W, Kim C, Mcquibban GA, Choe SK, Cho DH, et al. PEX13 prevents pexophagy by regulating ubiquitinated PEX5 and peroxisomal ROS. Autophagy. 2023;19:1781–1802. doi: 10.1080/15548627.2022.2160566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen XF, Tian MX, Sun RQ, Zhang ML, Zhou LS, Jin L, Chen LL, Zhou WJ, Duan KL, Chen YJ, et al. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. Embo Rep. 2018;19:e45124. doi: 10.15252/embr.201745124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn YH, Yang Y, Gibbons DL, Creighton CJ, Yang F, Wistuba, Lin W, Thilaganathan N, Alvarez CA, Roybal J, et al. Map2k4 functions as a tumor suppressor in lung adenocarcinoma and inhibits tumor cell invasion by decreasing peroxisome proliferator-activated receptor gamma2 expression. Mol Cell Biol. 2011;31:4270–4285. doi: 10.1128/MCB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei C, Wang B, Peng D, Zhang X, Li Z, Luo L, He Y, Liang H, Du X, Li S, et al. Pan-cancer analysis shows that ALKBH5 is a potential prognostic and immunotherapeutic biomarker for multiple cancer types including gliomas. Front Immunol. 2022;13:849592. doi: 10.3389/fimmu.2022.849592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navani S. Manual evaluation of tissue microarrays in a high-throughput research project: The contribution of Indian surgical pathology to the Human Protein Atlas (HPA) project. Proteomics. 2016;16:1266–1270. doi: 10.1002/pmic.201500409. [DOI] [PubMed] [Google Scholar]
- 21.Wu P, Heins ZJ, Muller JT, Katsnelson L, de Bruijn I, Abeshouse AA, Schultz N, Fenyo D, Gao J. Integration and analysis of CPTAC proteomics data in the context of cancer genomics in the cBioPortal. Mol Cell Proteomics. 2019;18:1893–1898. doi: 10.1074/mcp.TIR119.001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liefeld T, Reich M, Gould J, Zhang P, Tamayo P, Mesirov JP. GeneCruiser: A web service for the annotation of microarray data. Bioinformatics. 2005;21:3681–3682. doi: 10.1093/bioinformatics/bti587. [DOI] [PubMed] [Google Scholar]
- 23.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45:W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei C, Zhang X, Peng D, Zhang X, Guo H, Lu Y, Luo L, Wang B, Li Z, He Y, et al. LncRNA HOXA11-AS promotes glioma malignant phenotypes and reduces its sensitivity to ROS via Tpl2-MEK1/2-ERK1/2 pathway. Cell Death Dis. 2022;13:942. doi: 10.1038/s41419-022-05393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR, Campbell PJ. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Y, Zheng S, Wu Q, Wu J, Zhou R, Wang C, Wu Z, Rong X, Huang N, Sun L, et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy. 2021;17:4083–4101. doi: 10.1080/15548627.2021.1901204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Y, Lu C, Zhou P, Zhao L, Lyu X, Yin J, Shi Z, You Y. EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neuro Oncol. 2021;23:611–624. doi: 10.1093/neuonc/noaa214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baxevanis CN, Fortis SP, Perez SA. The balance between breast cancer and the immune system: Challenges for prognosis and clinical benefit from immunotherapies. Semin Cancer Biol. 2021;72:76–89. doi: 10.1016/j.semcancer.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Jia K, Sun Y, Zhang C, Li Y, Zhang L, Chen Z, Zhang J, Hu Y, Yuan J, et al. Predicting response to immunotherapy in gastric cancer via multi-dimensional analyses of the tumour immune microenvironment. Nat Commun. 2022;13:4851. doi: 10.1038/s41467-022-32570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baretti M, Le DT. DNA mismatch repair in cancer. Pharmacol Ther. 2018;189:45–62. doi: 10.1016/j.pharmthera.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Lin A, Zhang J, Luo P. Crosstalk between the MSI status and tumor microenvironment in colorectal cancer. Front Immunol. 2020;11:2039. doi: 10.3389/fimmu.2020.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luchini C, Bibeau F, Ligtenberg M, Singh N, Nottegar A, Bosse T, Miller R, Riaz N, Douillard JY, Andre F, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: A systematic review-based approach. Ann Oncol. 2019;30:1232–1243. doi: 10.1093/annonc/mdz116. [DOI] [PubMed] [Google Scholar]
- 37.Wu W, Wang X, Le W, Lu C, Li H, Zhu Y, Chen X, An W, Xu C, Wu Q, et al. Immune microenvironment infiltration landscape and immune-related subtypes in prostate cancer. Front Immunol. 2022;13:1001297. doi: 10.3389/fimmu.2022.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deberardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porporato PE, Filigheddu N, Pedro J, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28:265–280. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 41.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dean JM, Lodhi IJ. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.