Abstract
Background
Follicular lymphoma (FL), the most common indolent non-Hodgkin’s Lymphoma, is a heterogeneous disease and a paradigm of the contribution of immune tumor microenvironment to disease onset, progression, and therapy resistance. Patient-derived models are scarce and fail to reproduce immune phenotypes and therapeutic responses.
Methods
To capture disease heterogeneity and microenvironment cues, we developed a patient-derived lymphoma spheroid (FL-PDLS) model culturing FL cells from lymph nodes (LN) with an optimized cytokine cocktail that mimics LN stimuli and maintains tumor cell viability.
Results
FL-PDLS, mainly composed of tumor B cells (60% on average) and autologous T cells (13% CD4 and 3% CD8 on average, respectively), rapidly organizes into patient-specific three-dimensional (3D) structures of three different morphotypes according to 3D imaging analysis. RNAseq analysis indicates that FL-PDLS reproduces FL hallmarks with the overexpression of cell cycle, BCR, or mTOR signaling related gene sets. FL-PDLS also recapitulates the exhausted immune phenotype typical of FL-LN, including expression of BTLA, TIGIT, PD-1, TIM-3, CD39 and CD73 on CD3+ T cells. These features render FL-PDLS an amenable system for immunotherapy testing. With this aim, we demonstrate that the combination of obinutuzumab (anti-CD20) and nivolumab (anti-PD1) reduces tumor load in a significant proportion of FL-PDLS. Interestingly, B cell depletion inversely correlates with the percentage of CD8+ cells positive for PD-1 and TIM-3.
Conclusions
In summary, FL-PDLS is a robust patient-derived 3D system that can be used as a tool to mimic FL pathology and to test novel immunotherapeutic approaches in a context of personalized medicine.
Keywords: Immunotherapy, Hematologic Neoplasms, Tumor Microenvironment, Immune Checkpoint Inhibitors
WHAT IS ALREADY KNOWN ON THIS TOPIC
Follicular lymphoma-tumor microenvironment (FL-TME) is characterized by a rich infiltrate of T cells expressing immune checkpoint (ICP) involved in the incurability of the disease.
Developments over the last decade leading to targeted therapeutics and novel immunotherapeutic strategies effective against FL were performed and the evaluation of their efficacy is currently underway.
However, easy to handle relevant preclinical FL models that can predict drug response are very few and full characterization including two-dimensional and three-dimensional imaging, immunophenotyping and immune cell identification, ICP expression, immune checkpoint inhibitor response, have not been studied on the same models at the same time.
WHAT THIS STUDY ADDS
This study provides a method to establish and characterize a new preclinical FL model that recapitulates FL fundamental hallmarks and immune exhaustion profiles thus allowing a better understanding of the pathology.
FL-patient-derived lymphoma spheroid (FL-PDLS) is a relevant model to determine and predict response to therapy. Indeed, we present evidence that the percentage of PD-1 and TIM-3 expressing CD8+ cells negatively correlates with the sensitivity to anti-CD20 and anti-PD1 treatment, thus confirming clinical studies.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study provides a complete workflow and methods allowing the establishment and the full characterization of preclinical FL models.
In the era of personalized medicine, FL-PDLS appears as a promising model that can help to predict patient response, discover new therapeutic targets and identify new mechanisms of resistance. This system can evolve toward full FL-TME reconstituted organoids, and therefore, become powerful theragnostic biomaterials that combine therapy with diagnosis necessary for individualized therapies for patients.
Background
B cell non-Hodgkin’s lymphoma (B-NHL) is a highly heterogeneous disease that is well characterized by the contribution of somatic mutations and the tumor microenvironment (TME).1 In follicular lymphoma (FL), the TME is a rich intricate of immune CD4+ T cells comprizing CD4+ T follicular helper (TFh) cells, CD4+ T follicular regulatory (TFr) cells, CD4+ T regulatory cells (Treg), CD8+ cytotoxic T cells, follicular dendritic cells (FDC), stromal cells (mesenchymal stromal cells and tumor-associated macrophages) but also cytokines, proangiogenic factors and extracellular matrix (ECM) components that can support or regulate survival, proliferation and migration of tumorous B cells.2–7 FL is characterized by a rich infiltrate of T cells that exhibit an exhausted phenotype characterized by the expression of immune checkpoints (ICPs).8 9 This rich, well-interconnected and supportive network may account for the incurability of this indolent lymphoma. Despite remission following chemoimmunotherapy, a significant proportion of patients relapse leading to reduced overall survival (OS).2
Thus, a better understanding of this pathology with relevant in vitro models is essential to identify therapeutic targets and perform preclinical studies. In the era of personalized medicine, our aim was to develop a reliable and representative model of both intratumor and intertumor heterogeneity. The culture of lymphoma B cells in suspension is not representative of lymph node (LN) spatial organization and architecture, rendering drug efficacy results difficult to interpret. Indeed, these models do not mimic the neoplastic heterogeneity and drug response of the original tumor. Three-dimensional (3D) cell cultures are largely used and studied for solid cancers and their advantages have been recognized for over 50 years.10 11 In contrast to two-dimensional (2D) cultures, cell–cell and cell–matrix interactions, spatial organization, mechanical constraints, nutrients and O2 gradients, are well-known parameters that influence disease biology and response to treatments and are reproduced in 3D models. They also offer useful properties for drug screening.12–15 In contrast to solid cancers, relevant 3D models for B-NHL are poorly described.16 Among them, spheroids/organoids from DLBCL cell lines or fresh samples from patients are the most developed.17–21 We are pioneers in the development of 3D cultures established from FL cell lines demonstrating that conventional 2D culture of lymphoma B cells does not reproduce FL signaling pathways22–26 while a 3D hanging-drop (HD) system that we term HD-MALC (multicellular aggregates of lymphoma cells) recapitulates FL transcriptomic profiles including the overexpression of gene families involved in survival pathways (ie, NF-κB pathway, cell cycle regulation, hypoxia). MALC can be cocultured with NK or gamma delta (γδ) T cells to study therapeutic monoclonal antibody responses, immune escape (ie,) mechanisms, drug penetration and immune cell infiltration.25–27 More recently, we improved the 3D FL model using B-NHL cell lines.28 Based on a scaffold-free technique, cells are directly seeded in ultra-low attachment plates (ULA), which allows multiplexing and facilitates the spheroid imaging.11 29 Although ULA-MALCs are similar in gene expression or mutational profiles to HD-MALC, they are not representative of FL heterogeneity. Lamaison and collaborators recently established a 3D model including ECM, using tonsil stromal cells and FL /DLBCL cells or primary FL B cells (n=2) to study the dynamic relationship between lymphoma B cells and their microenvironment.21 These models, including mechanical constraints, bring new interesting perspectives. However, they are artificially reconstituted and do not include immune TME, a key component in the era of immunotherapeutic strategies and personalized medicine, particularly in FL.
Here, we present a new 3D model of patient-derived lymphoma spheroids (FL-PDLS) established from a whole FL LN biopsy that recreates in vitro FL immune TME and provides a suitable preclinical platform for drug testing and discovery of new therapeutic targets. In this model, no scaffold or matrix are introduced in order to attribute the results obtained to the FL-PDLS per se and not to the component used to maintain the primary cells in 3D. Maintaining primary FL cells in vitro is known to be challenging. Here, we establish a simple, robust and reproducible workflow allowing the maintenance of viable cells isolated from FL biopsies. We characterized FL-PDLS both by 2D and 3D imaging and by multiparametric flow cytometry analyses, and evaluated their sensitivity toward two monoclonal antibodies (mAbs) (anti-CD20 and anti-PD-1). Altogether, our results, that were obtained by Faria et al (unpublished PhD thesis manuscript), demonstrate that FL-PDLS constitutes a representative preclinical FL model integrating patient immune TME and patient heterogeneity. Our results represent a proof-of-concept to implement these models in personalized medicine research.
Methods
mAbs
Obinutuzumab (GA101) an anti-CD20 mAb was provided by Roche (Basel, Switzerland). Anti-PD-1 mAb of class IgG4 (Nivolumab) was obtained from the department of pharmacy at the IUCT (Toulouse).
Patient samples
LN was collected and processed at the CRB Cancer des Hôpitaux de Toulouse following ethics guidelines (Declaration of Helsinki), and written informed consent was obtained from each patient diagnosed between 2018 and 2021 with FL (grade II according to the WHO classification30) at the department of hematology (IUC, Toulouse-Oncopole, France). CRB collection was declared to the Ministry of Research (DC-2009-989) and a transfer agreement (AC-2008-820) was obtained after approval from the appropriate ethics committees. Patient characteristics are listed table 1 and cells from FL-LN were collected as reported in online supplemental methods.
Table 1.
FL patient characteristics
| Patient No | Ann Arbor stage | Flipi | GELF | B symptoms | BM involvement | Treatment response | Primary treatment | Date of diagnostic | Last follow-up | Relapse/transformation |
| #1 | IV | 4 | >1 | No | Yes | PR | EPI-R-CHOP | 23 Dec 2020 | 2022-02-09 | No |
| #2 | IV | 1 | 1 | No | No | CR | GA101-CHOP+GA maintenance | 13 Apr 2020 | 2022-01-19 | No |
| #3 | I–II | 0 | 0 | No | No | NA | Surveillance | 21 May 2019 | 2020-05 | Yes (tFL) |
| #4 | III | 2 | 0 | No | No | CR | GA101-CVP+GA maintenance | 24 Sep 2019 | 2022-01-20 | No |
| #5 | III | 3 | >1 | No | No | Ongoing | GA101-CVP | 05 Sep 2019 | 2022-03-04 | No |
| #6 | IV | 3 | 1 | No | NA | CR | R-Chemotherapy+R maintenance | 04 Jan 2019 | 2022-02-09 | No |
| #7 | IV | 3 | 0 | No | No | NA | Surveillance | 20 Mar 2021 | 2022-02-25 | No |
| #8 | IV | 1 | Yes | Yes | CR | EPI-R-CHOP | 27 Mar 2021 | 2022-02-08 | No | |
| #9 | IV | 3 | 3 | Yes | Yes | CR | EPI-R-CHOP | 11 May 2021 | 2022-01-21 | No |
| #10 | IV | 4 | 2 | No | No | CR | 4 GA101-CHOP+2 R-CHOP+R maintenance | 30 Jan 2019 | 2021-11-17 | No |
C, cyclophosphamide; CR, complete response; EPI-R-CHOP protocol, R-CHOP+TAZEMETOSTAT + 2 RTX + RTX maintenance; GA, GA101/Obinutuzumab; GELF, The Groupe d'Etude des Lymphomes Folliculaires ; H, hydroxyadriamycin; NA, not applicable; O, oncovin; P, prednisone; PR, partial response; R, rituximab; tFL, transformed follicular lymphoma; V, vincristine.
jitc-2023-007156supp008.pdf (216.6KB, pdf)
FL-PDLS generation
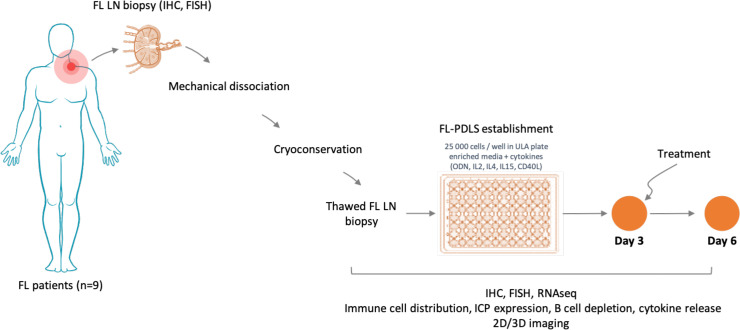
A total of 25,000 cells were seeded in 96-well round bottom ULA plates (Corning, Samois sur Seine, France) and cultured in an enriched medium as detailed in online supplemental methods and figure 1.
Figure 1.
Workflow for PDLS establishment and characterization. 2D, two-dimensional; 3D, three-dimensional; FISH, fluorescence in situ hybridization; FL, follicular lymphoma; IHC, immunohistochemistry; ICP, immune checkpoint; ODN, oligodeoxynucleotide; PDLS, patient-derived lymphoma spheroid; ULA, ultra-low attachment.
Immunohistochemistry
FL-PDLS was fixed with 4% PFA (Alfa Aesar, Haverhill, Massachusetts, USA), embedded in 1% low-melting agarose (Life Technologies, Villebon sur Yvette, France) and included in paraffin before performing sections (3 µm). The antibodies used for IHC labeling are listed in online supplemental table 1 and details for IHC in online supplemental methods.
jitc-2023-007156supp004.pdf (154.7KB, pdf)
2D imaging
FL-PDLS was visualized by brightfield illumination (BF) using a high throughput microplate imager for high-content analyses device, equipped with a ×5 objective (Operetta, Perkin Elmer, Villebon sur Yvette, France). Morphological parameters (BF area, roundness) were analyzed using the associated Columbus software.
Live-cell imaging
After centrifugation in ULA plates, FL-PDLS was imaged from day 0 to day 6 with Incucyte S3 Live-Cell Analysis System (Sartorius, Göttingen, Germany), placed in a standard tissue culture incubator to automatically acquire phase images at ×4 magnification.
3D imaging
FL-PDLS was fixed with 4% PFA, embedded in 1% low-melting agarose (Life Technologies) and cleared before analysis with a 880 confocal microscope (Zeiss, Oberkochen, Germany). For details, see online supplemental methods.
Flow cytometry analyses
Immune cell composition of FL samples and FL-PDLS, as well as ICP expression, B cell depletion and cytokine release were determined by flow cytometry as detailed in online supplemental methods. The antibodies used for flow cytometry are listed in online supplemental table 2.
jitc-2023-007156supp005.pdf (312.1KB, pdf)
3’mRNA sequencing
Libraries were prepared with 500 ng of RNA using the QuantSeq 3’mRNA-Seq Library Prep Kit-FWD (Lexogen, Vienna, Austria) and UMI Second Strand Synthesis Module (Lexogen) following the manufacturers’ instructions. The libraries were quantified using the Qubit dsDNA HS Assay Kit (Invitrogen, Life Technologies) and sequenced on single read 75 pb run, on a NextSeq550DX instrument (Illumina). Online supplemental methods detail the data analysis procedure.
RNAseq analyses
Sequencing reads were trimmed using cutadapt (3) and aligned with STAR (10.1093/bioinformatics/bts635). Gene counts were generated using HTSeq (doi:10.1093/bioinformatics/btu638) with GRCh38.p13 reference annotation. Differential expression was computed using DESeq2 (10.18129/B9.bioc.DESeq2), and Gene signatures were determined with GSEA V.4.3.2 (Broad Institute, Cambridge, Massachusetts, USA) using the hallmark gene sets, the C2 curated gene sets, the C5 gene ontology gene sets and the C6 oncogenic signatures (Molecular Signature Database V.2.5). A two-class analysis with 1000 permutations of gene sets and a weighted metric was used. Expression heatmaps were created using Morpheus software (https://software.broadinstitute.org/morpheus/, Broad Institute).
Fold-changes of FL-PDLS versus 2D models and p value criteria were represented in a volcano plot. Data were visualized with the Multiplot Studio V.1.5.20 Software from GenePattern archive (http://gparc.org/).
Correlogram and correlation curves
Correlogram was obtained with Open source Rstudio (RStudio Team (2020), PBC, Boston, Massachusetts, USA URL http://www.rstudio.com/) and corrplot package. For details, see online supplemental methods.
Statistics
t-tests were applied and all tests were performed with GraphPad Prism software.
Results
FL-PDLS organize in patient-specific 3D structures
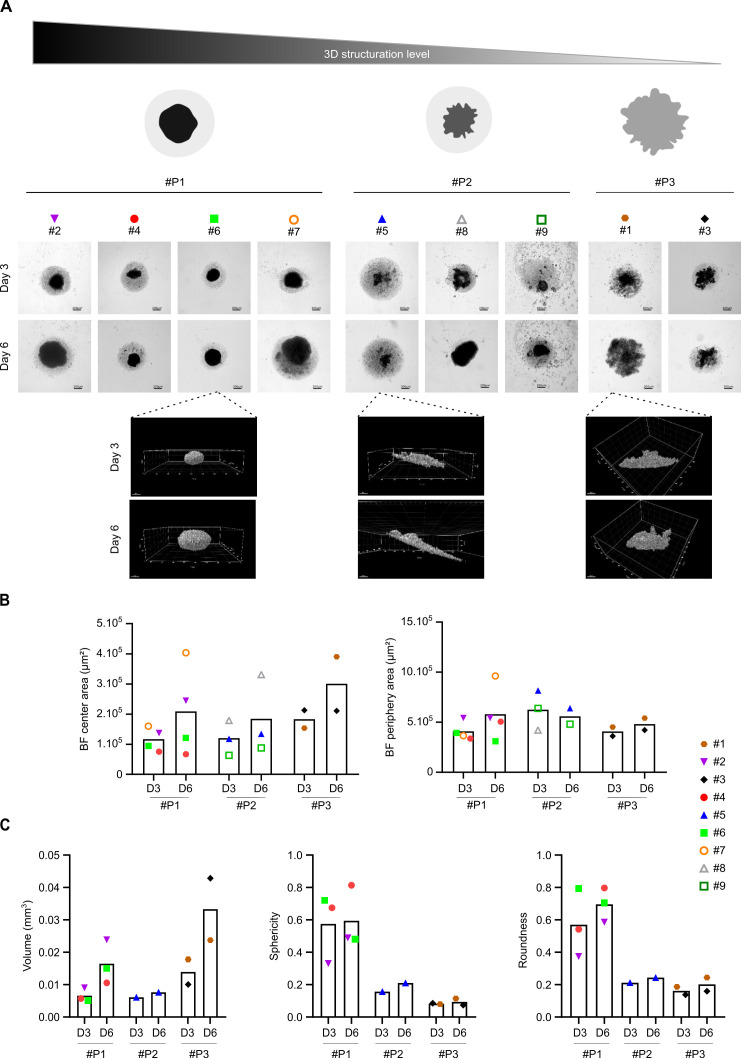
Different tests of cell seeding densities and different media were necessary to establish viable FL-PDLS with FL LN and the most favorable conditions to obtain spheroids with a viability of approximately 60% was achieved at 25,000 cells per well (data not shown). Global morphology was examined by 2D imaging with a high throughput imaging confocal system and by 3D imaging with a confocal microscope (figure 2). We observed different FL-PDLS morphotypes (figure 2A, upper panel) exhibiting (1) a thin monocellular layer and a round and dense cell aggregate in the clearly delineated center (pattern 1, #P1), (2) a more or less enlarged thin layer and a less structured central aggregate (pattern 2, #P2) and (3) a reduced and destructured thin layer and a large and disorganized central structure with no clear delimitation between monocellular layer and aggregated parts (pattern 3, #P3). In all cases, cell aggregation occurs in the first few hours (online supplemental video). After 6 days of culture, we observed a significant global increase of the FL-PDLS size for most patients (six out of nine) without strong modification of morphotype except for patient #8. These morphological patterns were confirmed by 3D imaging (figure 2A, lower panel) and morphological parameters were measured based on 2D imaging. First, areas of FL-PDLS’s periphery (composed mainly of dead cells, data not shown), or center (composed of viable cells) were determined (figure 2B). Most FL-PDLS presented an increase in the central area between day 3 and day 6 ranging from 13% to 152% (patient #5 and #7, respectively) (figure 2B). For the periphery areas, three different profiles were observed (figure 2B) with an increase (patients #1, #3, #4 and #7), a decrease (patients #5, #6 and #9) or no variation (patient #2).
Figure 2.
FL-PDLS characterization by 2D and 3D imaging. (A) Global morphology observed by brightfield (Operetta, 5X, scale: 200 µm) of FL-PDLS from nine patients or by confocal microscopy (×10) for patients #1, #5, and #6 in untreated condition at D3 and D6. #P represents patterns based on the morphology. (B) Histograms representing center/core and periphery areas (µm2) measured after 2D imaging with Columbus software and (C) volume (mm3), sphericity and roundness quantification after 3D imaging for all patients clustered according to their morphotypes (patterns, #P). 2D, two-dimensional; 3D, three-dimensional; FL-PDLS, follicular lymphoma-patient-derived lymphoma spheroid.
jitc-2023-007156supp010.m4v (7.3MB, m4v)
Real volume as well as sphericity and roundness were then analyzed by 3D imaging (figure 2C). Volume increased between day 3 and day 6 with the highest difference observed for patient #3. In contrast, sphericity and roundness did not mirror volume variation, but both exhibited same profiles and range of values, with an increase observed in 4 out of 6 patients (#1, #2, #4, #5). For patient #3, no variation in sphericity and roundness was observed and for patient #6, both sphericity and roundness were decreased. Moreover, we noticed a large scale of volume variation (0.0045 mm3–0.05 mm3), sphericity (0.004–0.83) and roundness (0.03–0.88).
Thus, we show that cells from FL biopsies can be cultured in 3D with an enriched medium maintaining a high viability. 2D and 3D imaging analyses highlighted the interpatient variability in terms of aggregation dynamics, morphology and behavior during the time of culture.
FL-PDLS recapitulates FL hallmarks
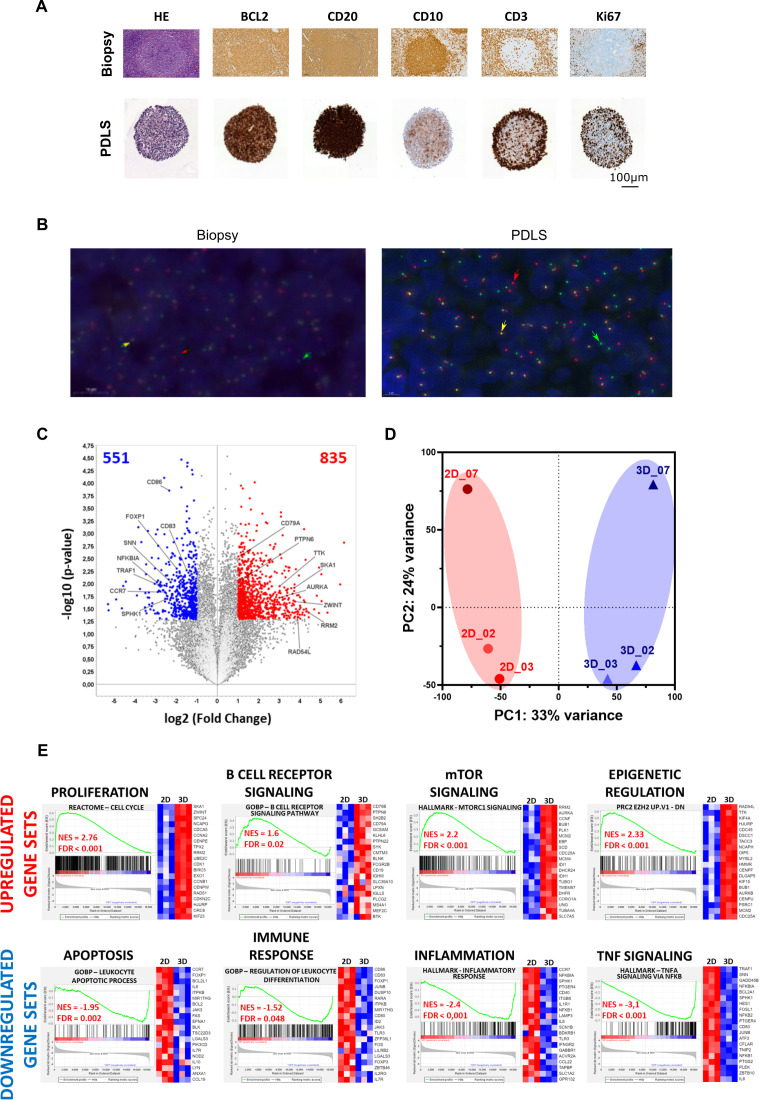
Immune cell type distribution in FL-PDLS was then determined by IHC (figure 3A) and t(14;18) translocation by fluorescence in situ hybridization FISH (figure 3B). At the FL-PDLS center, a clustering of CD20+, CD79a+ B cells was identified whereas CD3+ T cells were localized at the surface, an architecture reminiscent of FL neoplastic follicles. In addition, Ki-67 staining confirmed the presence of proliferative T cells. No FDC was observed due to the non-enzymatic dissociation and freezing cycle. A FISH experiment was performed with a “break apart” probe to assess BCL2 rearrangements in FL-PDLS. Figure 3B indicates the presence of both normal B cells by yellow arrows and lymphomatous B cells, characterized by a BCL2 split, by green and red arrows. Altogether, these results confirm that the FL-PDLS model mimics the spatial organization of FL, with follicles composed mainly of tumor B cells coexpressing CD10 and BCL2, surrounded by a microenvironment rich in T cells with little or no penetration of the follicle.
Figure 3.
Global FL-PDLS characterization by different approaches. (A) Representative IHC on sliced FL (LN biopsy and PDLS at day 3 of culture from patient #4). (B) Representative FISH on sliced FL-PDLS #4. (C) Volcano plot of differentially expressed genes (DEG) contrasting 3D–2D models. After application of thresholds, 835 genes were found upexpressed (red genes) and 551 downexpressed (blue genes), respectively. Top modulated genes are indicated. (D) PCA analysis using expression values obtained from RNA-seq data in 3D and 2D models. (E) Gene set enrichment analysis. Most significant upregulated and downregulated gene sets filtered by FDR<0.05 and NES>1.5 or <−1.5. Heatmaps correspond to top 20 genes. 2D, two-dimensional; 3D, three-dimensional; FDR, false discovery rate; FISH, fluorescence in situ hybridization; FL-PDLS, follicular lymphoma-patient-derived lymphoma spheroid; NES, Normalized Enrichment Score; PCA, principal component analysis.
To further characterize FL-PDLS, we performed 3’ RNA sequencing to compare cells in FL biopsies (2D) to cells grown for 3 days in FL-PDLS (3D). Cells from patient #2, #3 and #7 were used in this comparison. Differential expression analyses of paired samples contrasting 3D–2D models indicated that 551 genes were upregulated and 835 downregulated in the 3D models (figure 3C) highlighting a significant transcriptome modulation. Furthermore, 3D samples clearly clustered in the principal component analysis (PCA) generated with the RNA-seq gene expression (figure 3D) indicating common features in gene expression among the 3D samples.
Next, we proceeded with a GSEA analysis using the hallmark gene sets, C2 curated gene sets, C5 gene ontology gene sets and C6 oncogenic signatures (MSigDB V.2023.1Hs) which identified upregulated gene sets related with cell proliferation like cell cycle regulation and DNA replication as the most significant. In addition, we observed an enrichment in our 3D models of survival pathways such as mTOR signaling or DNA repair. Moreover, GSEA analysis highlighted two other upregulated FL classical pathways in 3D models including B cell receptor signaling and epigenetic regulation. In contrast, GSEA analysis also revealed downregulation of gene sets involved in the regulation of apoptosis, immune response, inflammation and TNF signaling pathways in 3D models. The top 20 genes of representative signatures from GSEA plots are presented in heatmaps (figure 3E). The summary of gene sets overrepresented in FL-PDLS compared with 2D cultures is listed in online supplemental table 3 and a more complete table with other interesting gene sets identified by GSEA analysis such as cell adhesion, cell migration or angiogenesis that appeared downregulated in 3D models is listed in online supplemental figure 2 and online supplemental table 4.
jitc-2023-007156supp006.pdf (177.5KB, pdf)
jitc-2023-007156supp002.pdf (1.8MB, pdf)
jitc-2023-007156supp007.pdf (284.2KB, pdf)
Taken together, these data suggest that the favorable culture conditions of our 3D model (FL-PDLS) maintain viability and minimizes cell death, and upregulates FL hallmarks as observed in FL patients such as survival, proliferation and downregulation in apoptosis or immune response.31
FL-PDLS immune microenvironment composition
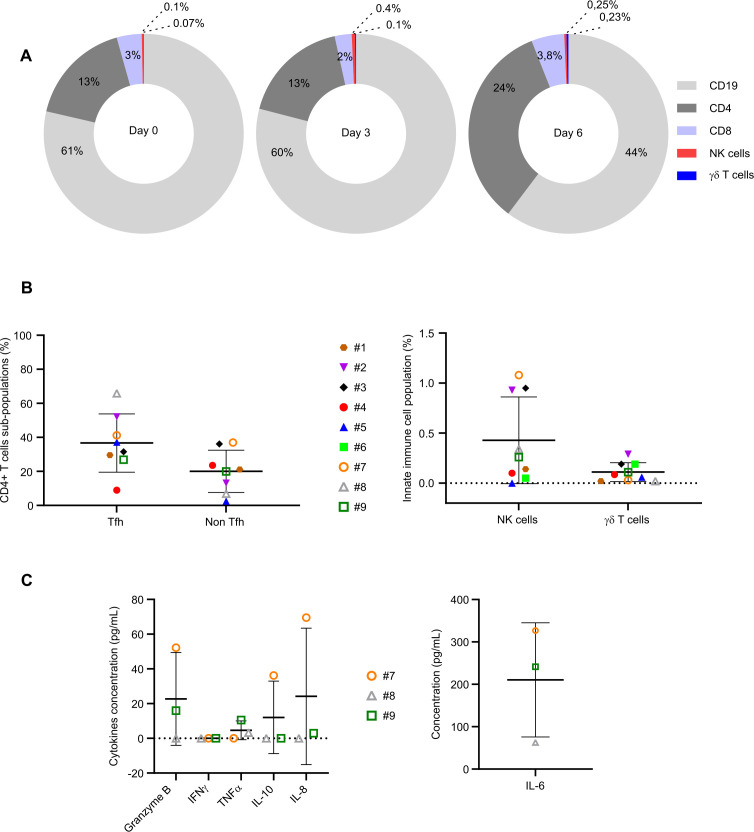
FL immune TME is a rich intricate of different cellular subtypes mainly composed of tumorous CD10+ B cells, CD4+ T, CD8+ T, γδT and NK cells.2 3 7 26 32 Thus, we performed multiparametric flow cytometry analyses to determine the composition of FL-PDLS and compared them to the initial biopsies. On average at day 0 of culture, B lymphocytes (60% of total cells) were the prominent population composing the thawed samples, followed by CD4+ (13%), CD8+ (3%), NK (0.1%) and γδT (0.07%) cells (figure 4A). Importantly, 3D cultures did not affect the percentage of each cell subtype at day 3 and only a slight decrease was observed for B cells with 44% of CD19+ cells at day 6 (Figure 4A).
Figure 4.
Immune cell distribution and cytokine release. (A) Percentage of immune cells evaluated by flow cytometry at day 0, day 3 and day 6 of 3D culture. (B) Percentage of TFh (CD3+CD4+CXCR5+ICOS+), non-TFh (CD3+CD4+CXCR5−ICOS−), NK (CD3-CD56+) and gamma delta T cells (CD3+TCRgamma9+) in FL-PDLS at day 3 evaluated by flow cytometry. (C) Cytokine release (granzyme B, IFNγ, TNFα, IL-10, IL-8, IL-6) was evaluated by flow cytometry at day 6 of culture. 3D, three-dimensional; FL-PDLS, follicular lymphoma-patient-derived lymphoma spheroid.
Phenotyping of CD4+ T cells revealed that 40% of CD4+ T cells were TFh and 20% were non-TFh (figure 4B left). Individually, we distinguished patients with a high (> 50%, patients #2 and #8), medium (around 30 %, patients #1, #3, #5, #7 and #9) and low percentage of TFh (<10%, patient #4). Regarding non-TFh cells, high (around 36%, patients #3 and #7), medium (around 20%, patients #1, #2, #4, #9) and low percentages (<10%, patients #5 and #8) were observed (for more details, see online supplemental results).
jitc-2023-007156supp009.pdf (172KB, pdf)
The presence of innate immune cells in the FL-PDLS was very low (NK<1% and γδT<0.1 %) (figure 4B right). Here again, we were able to distinguish patients exhibiting a higher percentage of NK cells (#2,#3,#7) and patients with a very low percentage.
Finally, FL-PDLS exhibited high (patient #7), intermediate (patient #9) or very low (patient #8) basal levels of T cell activation as attested by the measurement of secreted cytokines in the culture medium (figure 4C, for more details see online supplemental results).
Altogether, these results highlight FL-PDLS as a relevant model exhibiting similar features to FL patients such as proportion of B/T cells and variability of immune cell composition. Moreover, these results show that FL-PDLS culture conditions maintain the viability of cells composing the immune TME during cell culture.
FL-PDLS maintain FL immune escape features
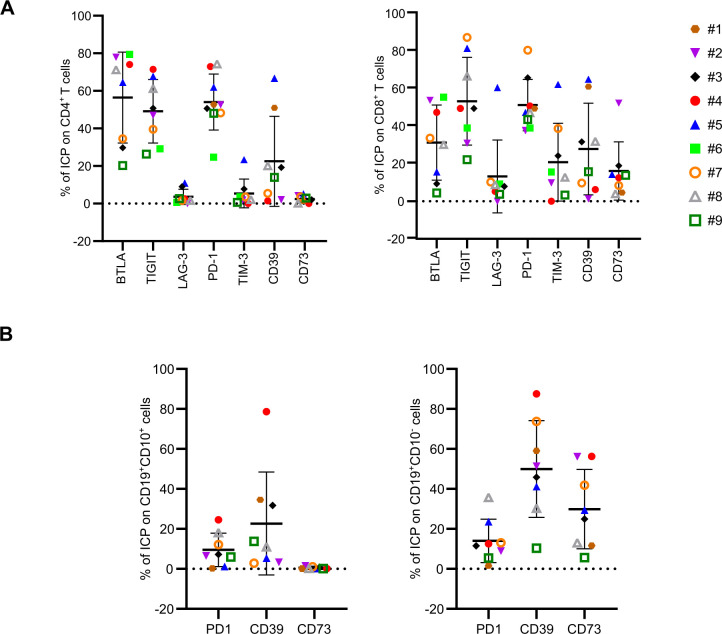
To further characterize FL-PDLS, we investigated the profile of ICP expression described in FL.33–40 BTLA, TIGIT, LAG-3, PD-1, TIM-3, CD39 and CD73 expression was determined at the cell surface of CD4+ T and CD8+ T cells after thawing and at day 3 of 3D culture (figure 5A and online supplemental figure 1 for day 0). A high proportion (50%–55%) of CD4+ cells expressed BTLA, TIGIT and PD-1, a medium (roughly 20%) percentage expressed CD39 and less than 10% expressed LAG-3, TIM-3 and CD73. On CD8+ lymphocytes, TIGIT and PD-1 were expressed by approximately 50% of cells, TIM-3, CD39 and BTLA by roughly 20%–30% and less than 20% expressed LAG-3 and CD73. We also determined ICP expression on tumorous B cells (CD19+CD10+) and observed PD-1 expression in fewer than 10% of cells and CD39 in roughly 20%. No CD73, PDL1 and PDL2 expression was detected. In healthy B cells (CD19+CD10−), the percentage of CD39 and CD73 was higher with approximately 50% and 30%, respectively (figure 5B).
Figure 5.
FL-PDLS immune checkpoint characterization. 10 FL-PDLS from nine different FL patients at day 3 of culture were pooled and the percentage of ICP was analyzed by flow cytometry. (A) Percentage of BTLA, TIGIT, LAG-3, PD-1, TIM-3, CD39, CD73 on CD4+ and CD8+ T cells. (B) Percentage of PD-1, CD39, CD73 on tumorous cells (CD10+CD19+) and healthy B cells (CD10−CD19+). FL-PDLS, follicular lymphoma-patient-derived lymphoma spheroid; ICP, immune checkpoint.
jitc-2023-007156supp001.pdf (29.3KB, pdf)
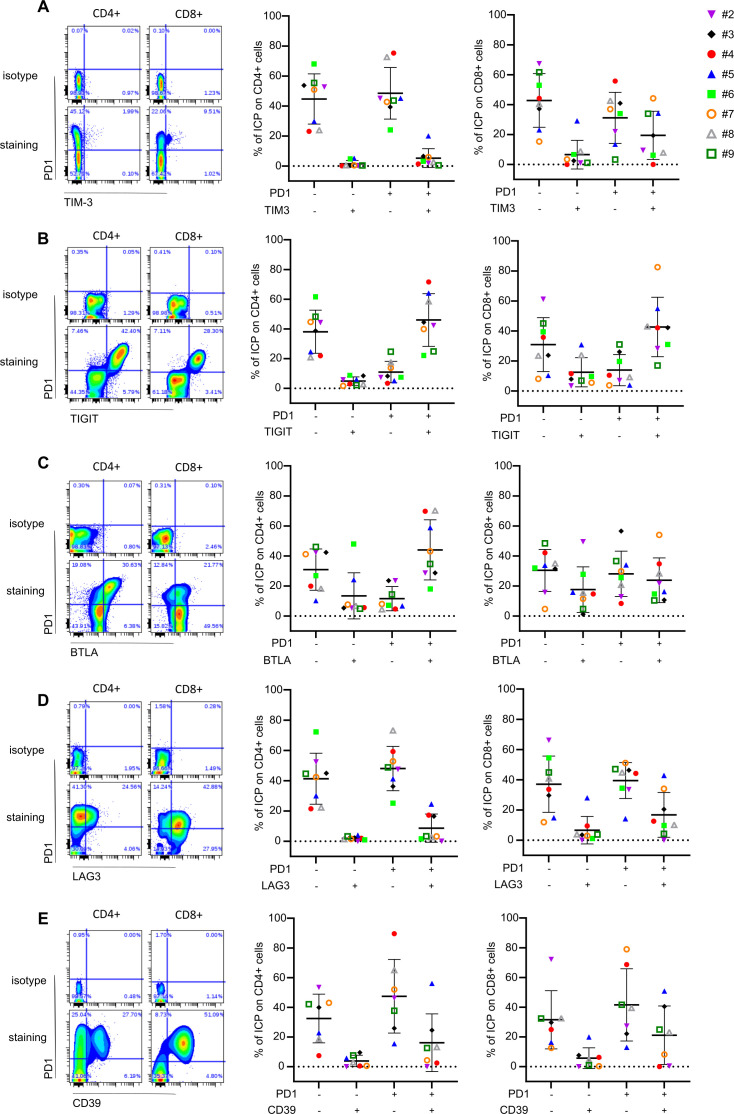
FL is characterized by coexpression of ICP markers such as PD-1/TIM-3,41 PD-1/TIGIT,42 PD-1/LAG-3,43 PD-1/BLTA,44 which predicts patient outcome. There is currently a special interest in the development of bispecific antibodies against ICP. Thus, we determined the coexpression of these ICP in both CD4+ and CD8+ cells by applying different gating strategies (figure 6A–E). We observed that globally, CD4+ were mainly PD-1+TIM-3− (48%) (figure 6A), PD-1+TIGIT+ (46%) (figure 6B), PD-1+BTLA+ (44%) (figure 6C) and PD-1+LAG-3− (48%) (figure 6D). CD8+ were mostly PD-1-TIM-3− (42%) (figure 6A) and PD-1+TIGIT+ (42%) (figure 6B). The percentage of PD-1−BTLA−, PD-1+BTLA− or PD-1+BTLA+ CD8+ cells were quite similar (30, 28 and 23%, respectively) (figure 6C). We also observed 37% of PD-1−LAG-3− and 39% of PD-1+LAG-3− CD8+cells (figure 6D). CD39 is a relevant marker in NHL TME33–35 and its coexpression with PD-1 has been recently described in exhausted TIL in epithelial malignancies.45 Thus, we determined the level of PD-1/CD39 in the FL-PDLS analyses and revealed that CD39+CD4+ or CD8+ cells were mainly PD-1+ with 16 and 21%, respectively, compared with 3.8 and 5.6% for PD-1−CD4+ and CD8+ cells (figure 6E).
Figure 6.
ICP coexpression in CD4+ and CD8+ T cells from FL-PDLS. Ten FL-PDLS from eight different FL patients at day 3 of culture were pooled and the percentage of ICP was analyzed by flow cytometry. Left panels represent gating strategies. Right panels represent percentage of CD4+ and CD8+ T cells expressing double ICP. PD-1/TIM-3 (A), PD-1/TIGIT (B), PD-1/BTLA (C), PD-1/LAG-3 (D) an PD-1/CD39 (E). FL-PDLS, follicular lymphoma-patient-derived lymphoma spheroid; ICP, immune checkpoint.
Altogether, our results indicate that the FL-PDLS model reproduces interpatient variability and immune suppression observed in FL LNs. Importantly, most CD4+ and CD8+ T cells are PD-1+TIGIT+. Moreover, CD39+PD-1+ TILs also appears as an interesting population infiltrating FL tumors. Thus, these results strongly support the use of FL-PDLS for testing immunotherapy in a context of personalized medicine.
FL-PDLS as tool for immunotherapy screening in FL
Finally, we aimed to determine whether FL-PDLS could represent preclinical models for immunotherapy screening. For this purpose, we tested the efficacy of an anti-CD20 mAb used for FL therapy, obinutuzumab (GA101) combined or not with the anti-PD-1 mAb, nivolumab. We established a workflow adapted for medium throughput screening in 96-well plates and developed specific tools to study the effect of therapies on FL-PDLS morphology and behavior as well as B cell depletion. 2D imaging allowed a global characterization of FL-PDLS and the observation of different patterns after drug treatment without any potent variation after immunotherapy (online supplemental results and online supplemental figure 3).
jitc-2023-007156supp003.pdf (884.8KB, pdf)
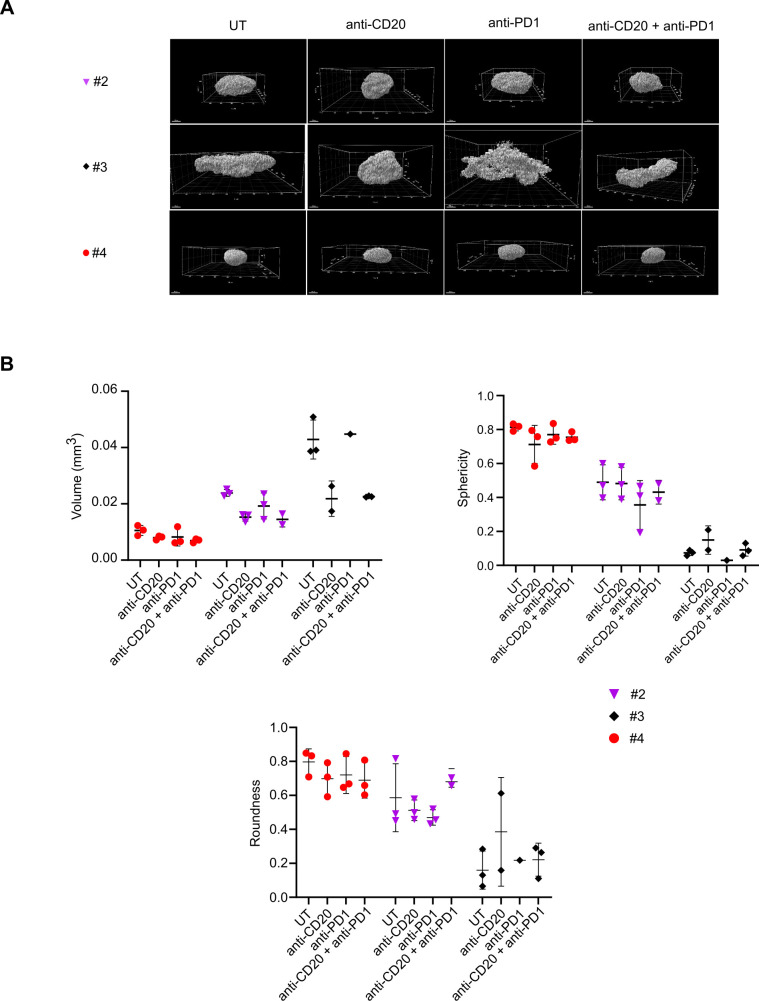
Therefore, we investigated the effect of these therapeutic mAbs by 3D imaging (figure 7A). First, we observed by 3D confocal imaging the already described 3D shape differences between patients in untreated condition (UT). Anti-CD20 alone or in combination with anti-PD-1 seemed to strongly modify #2 and #3 FL-PDLS morphology. As for figure 2C, volume, sphericity and roundness extracted from 3D imaging were determined by specific algorithms developed for ULA-MALC28 and FL-PDLS (figure 7B).
Figure 7.
Drug effect on FL-PDLS 3D morphology and volume. (A) 3D reconstruction by IMARIS from 880 confocal acquisitions at ×10 magnification of FL-PDLS labeled with PI and cleared by BABB for patients #2, #3 and #4 at D6 in untreated and treated conditions. (B) Volume and morphology 3D quantification. Based on 3D acquisitions (A), volume (mm3), roundness and sphericity were calculated for each patient and each condition and represented as graphs. Mean±SD of up to three replicates per condition. 3D, three-dimensional; BABB, methanol-benzyl alcohol/benzyl benzoate; FL-PDLS, follicular lymphoma-patient-derived lymphoma spheroid; PI, propidium iodide.
We observed a decrease of FL-PDLS volume in the 3 patients tested after anti-CD20 treatment and two of them were also sensitive to anti-PD-1 treatment (#2 and #4). Combination seemed to enhance the volume decrease in patients sensitive to single drugs. Sphericity and roundness were also analyzed but exhibited high interpatient variation. For patient #4, both sphericity and roundness decreased after anti-CD20 or anti-PD-1 mAbs treatment and combination did not enhance these effects. For patient #2, anti-PD-1 mAb alone affected sphericity, whereas roundness was decreased by both single drugs, but surprisingly not with the combination where an increase was observed. Finally, for patient #3, which was the less spherical model (sphericity=0.07 in untreated condition), anti-CD20 mAb increased the sphericity and roundness, whereas anti-PD-1 mAb decreased sphericity and increased roundness. The combination did not modify the effect of single drugs on sphericity or roundness (figure 7B).
Altogether, these results show the ability to assess the effect of immunotherapies on the volume and morphology of FL-PDLS and the interpatient variability. They also underline the importance of studying therapeutic efficacy beyond 2D imaging.
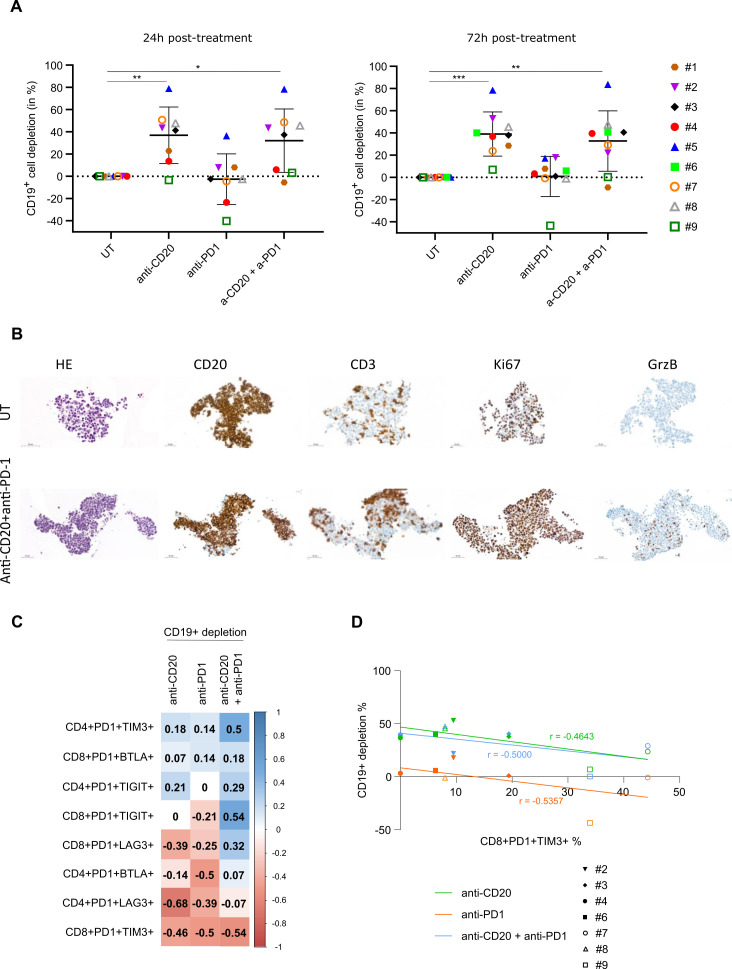
Flow cytometry analyses were then performed on dissociated FL-PDLS to evaluate the effect of treatment on target B cells. Thus, we observed that in 8 out of 9 FL-PDLS tested, anti-CD20 mAb induced a potent CD19+ B cell depletion (up to 80%) in a similar fashion at 24 hours and 72 hours of treatment (figure 8A). However only three out of nine FL-PDLS responded to anti-PD1 treatment as a single agent. Interestingly, in FL-PDLS from patients #2, #3 and #4, 3D volume variation (figure 7B) was correlated with the B cell depletion observed in response to treatments (figure 8A). Moreover, we were able to observe by IHC an increase of T cell infiltration and activation, as attested by GrB labeling after anti-CD20/anti-PD-1 combination (figure 8B).
Figure 8.
B cell depletion and T cell activation on anti-CD20 and anti-PD-1 mAb treatments. (A) Percentage of depletion of CD19+ B cells at 24 hours and 72 hours post-treatments (10 FL-PDLSs were pooled from 6 to 9 different FL patients after treatment with anti-CD20 (GA101, 10 µg/mL) and/or anti-PD-1 (10 µg/mL). (B) Representative IHC of sliced PDLS (patient #3) treated by anti-CD20 (GA101, 10 µg/mL) and anti-PD-1 (10 µg/mL) or not during 72 hours. (C) Values from FL-PDLS (seven different FL patients) gathered for all the parameters listed: percentage of PD-1+/TIM3+, PD-1+/BTLA+, PD-1+/TIGIT+, PD-1+/LAG3+ CD4+ and CD8+ cells and percentage of B cell depletion (normalized by untreated condition) after 72 hours of treatment by anti-CD20, anti-PD-1 and combination. Matrix of correlation based on correlation coefficients (non-parametric Spearman’s correlation) of side-by-side represented as a graph. Correlation coefficients represented by squares where values and color were determined according to correlation coefficient values. (D) Correlation curves of percentage of PD1+TIM3+ expressing CD8+cells (x-axis) and percentage of B cell depletion after 72 hours of treatment (y-axis) with correlation coefficients (r) extracted from correlogram in B. FL-PDLS, follicular lymphoma-patient-derived lymphoma spheroid.
Finally, we assessed the correlation between B cell depletion and ICP expression on CD4+ and CD8+ populations. To do so, double positive (PD-1+TIM-3+, PD-1+TIGIT+, PD-1+BTLA+, PD-1+LAG-3+) population percentage and B cell depletion percentage obtained after 72 hours of treatment with anti-CD20 and anti-PD-1 alone or in combination were used to generate a matrix of correlation represented as a correlogram (figure 8C). With this side-by-side comparison, we observed that the percentage of PD-1+BTLA+ CD4+ T cells negatively correlated with B cell depletion induced by anti-PD-1. Finally, the percentage of CD4+ or CD8+ T cells co-expressing PD-1+LAG-3+ negatively correlated with B cell depletion in response to the two single treatments, with a higher score for CD4+ populations after treatment with anti-CD20 mAb (−0.68) (figure 8C). Very interestingly, the percentage of PD-1+TIM-3+ CD8+ T cells negatively correlated with the B cell depletion induced by anti-PD-1, anti-CD20 or their combination (figure 8D).
Altogether, these correlations suggest a potential link between the expression of ICP markers and the response to anti-PD-1 and anti-CD20 mAbs and reveal PD-1+TIM-3+ expression on CD8+ cells as a marker of lower response to both single or combo treatment.
Discussion/conclusion
Here, we present a novel patient-derived 3D model for FL that may be of particular relevance to understand disease biology and immune cell distribution. Indeed, IHC analyses reveal that the 3D FL model reproduces the same pattern of expression as observed in biopsies with CD79a, BCL2, CD10 and CD20 labeling and peripheral distribution of T cells. 3D imaging and specific analyses,28 show an interpatient variability of shape with three different patterns of aggregation and different profiles of response to treatment. Noteworthy, FL-PDLS from patient #3, which was not well structured and presented the least roundness and sphericity, exhibited the highest responses to anti-CD20 and anti-PD1 mAbs. This could be explained by a better mAb penetration within the 3D structure. Although the number of samples could not allow us to draw definitive conclusions, these results highlight another application based on the use of FL-PDLS as in vitro models to characterize the mechanisms of action of anti-lymphomatous drugs and model volume, aggregation, ECM production or mechanical constraints and parameters influencing drug sensitivity. Obviously, we cannot exclude that FL-PDLS, like any preclinical model, remains a study model that does not exactly recapitulate the FL LN.
Despite the absence of stromal cells, FL-PDLS represents a robust tool to recapitulate FL biology for a number of reasons:
First, both tumor B cells and autologous T cells maintain good viability for at least 1 week, a window which enables us to analyze the efficacy of most therapeutic agents.
Second, FL-PDLS recapitulates the fundamental hallmarks of FL in terms of tumorous cell markers (BCL2, CD79b, CD20+, and CD10+), Bcl2 translocation, and overexpression of cell cycle, mTOR, DNA replication, BCR signaling, and epigenetic-related gene sets. However, we cannot overlook our findings regarding the downregulation of immune response, inflammation, and TNF signaling gene sets. Several hypotheses, mainly focusing on technical issues, can be considered: (1) One could speculate that the enriched medium used to maintain viable cells could influence gene expression to some extent. (2) Since the processes for biopsy dissociation/conservation/thawing unfortunately discard certain companion FL-B cells, such as macrophages or stromal cells, we cannot exclude the possibility that the absence of these subpopulations might interfere with certain transcriptomic programs in FL-B cells. (3) Although FL-PDLS exhibits a decrease in certain pathways when comparing 3D–2D cultures, the question of how it compares to reactive LNs cultured under the same conditions remains open. All of these issues are currently under investigation in the laboratory.
Third, FL-PDLS recapitulates the immune exhaustion profile typical of FL LN, preserving inter and intrapatient T cell heterogeneity as seen in single cell studies.36 37
Fourth, FL-PDLS represents an affordable 96/well format for drug screening, including ICP inhibitors (such as anti-PD-1) thanks to the presence of autologous T cells capable to experience activation (as demonstrated cytokine secretion in FL-PDLS supernatants).
The clinical development of effective immunotherapies needs the identification of tumor environmental features that could predict sensitivity to ICP blockade. While impressive efficacy was observed in Hodgkin and primary mediastinal-B cell lymphomas, disappointing results were obtained in FL or DLBCL.38 NHL are categorized as inflamed or non-inflamed tumors.8 38 Large-scale microarray profiling revealed four stages of IE in B-NHL that were correlated with OS.8 This immune landscape of lymphoma is a critical point to predict the response to immunotherapy and to design new therapeutic approaches.39 FL, of which 73% FL exhibit a stage III (immunogenic tumors with IE) or IV (fully immune-edited tumors),8 is an indolent lymphoma with abundant levels of PD-1+ infiltrating T-cells40 that can coexpress other exhausted markers such as TIM-3,41 TIGIT,42 46 LAG-3,43 or BTLA44 that predict patient outcome. Thus, it is becoming clear that FL specific features must be taken into account in preclinical studies to predict patient response. In this regard, there is an urgent need of patient-derived immunocompetent systems. Here, we present FL-PDLS as a model exhibiting similar inter-patient variability as that observed in the biopsy from which they originate. Indeed, the immune cell population characterization reveals not only that FL-PDLS are mainly composed, as expected, of B cells and T cells (CD8+ T cells, CD4+ T cells, TFh, non-TFh, NK and γδT cells) in variable proportions among patients, but they also exhibit similar ICP expression profiles after 3 days of culture compared with day 0 (online supplemental figure 1). Interestingly, we observed that the coexpression of PD-1 and TIM-3 on CD8+ cells negatively correlates with the sensitivity to treatments corroborating results from Yang’s clinical study where a high percentage of PD-1lowTIM-3+CD8+ cells is associated with a poor outcome.41 These results set the rational basis for novel ICP inhibitor combinatorial approaches that could be tested in FL-PDLS advancing toward personalized immunotherapies.
Altogether, we present evidence that FL-PDLS is a relevant pre-clinical FL model that can be used to better characterize FL pathology and predict patient response drug testing, new target discovery and characterization of mechanisms of action and/or resistance to antilymphomatous drugs. Although the FL-PDLS model does not recapitulate intra and inter LN variability, nor clonal evolution (which predictably affects TME and response to therapy) over time, we firmly believe that FL-PDLS must evolve by integrating other TME components (stromal, myeloid and endothelial cells) to recreate in vitro, a complete FL TME. This is currently under development.47 Moreover, integration of these tumoroids in vascularized systems could be of particular interest to study immune cell recruitment and/or drug delivery to the tumor site. This is especially true in the current context of the development of chimeric antigen receptor-T cells for refractory/relapsed FL. Thus, by combining tumor modeling of each patient with medical imaging and bioinformatic tools to analyze genomic data, it should be possible to provide a full ID card of each patient and propose personalized therapies in a disease that remains incurable.
jitc-2023-007156supp011.pdf (192.1KB, pdf)
Acknowledgments
We are grateful to our healthcare professionals for their boundless investment during the COVID-19 crisis. Authors thank Christian Klein from Roche Pharma Research & Early Development, Schlieren, Switzerland for providing Obinutuzumab; Julie Bordenave from Inserm CRCT, Toulouse, France for technical help on initial experiments on FL biopsies; L Ligat, M Farcé and M Tosoloni from the Pôle technologique du CRCT for their advices on flow cytometry, 2D imaging, 2D imaging analyses; all members of JJ Fournié/ C Laurent’s team (CRCT, Toulouse, France) for their stimulating comments; Pascale Bernes-Lasserre, Aurélie Gomez, Marine Norlund and Valérie Lobjois from IMACTIV-3D (Toulouse) and Laetitia Pieruccioni and Jacques Rouquette from RESTORE Research Center (Toulouse) for their fruitful discussions and advices on 3D imaging. Stéphanie Grenard, Annie Alloy and François-Xavier Frenois from Imag’IN Platform (Toulouse) for IHC experiments on FL-PDLS. Authors also thank, Anne-Marie Benot, Stephanie Nevouet and Muriel Bouas for administrative support and Cathy Greenland for english proofreading.
Footnotes
Twitter: @jgv1982, @@perezgalan_p
Contributors: FG, CF and CB designed the experimental strategy, organized the experiments and collected and analyzed the data. CF, FG, PG, CD-L, JGV and CR performed in vitro experiments. CF performed and analyzed the experiments of flow cytometry and on the operetta system. CQ performed 3’mRNA sequencing and JGV and GJ the analyses. FG performed experiments for SPIM and confocal imaging. RM generated specific algorithms for 3D image processing. FG, RM and J-ML analyzed 3D imaging analyses. PG, NVA and CL developed IHC and FISH specifically for FL-PDLS and analyzed labelling. LY and CL selected FL samples and provided patients clinical data. CB and PP-G co-supervised and discussed the experiments. CF, FG and CB wrote the manuscript. All authors discussed and approved the manuscript. CB is a acting as guarantor.
Funding: This work was part of an Interreg POCTEFA program (IMLINFO EFA281/16/). This study was also funded by the Institut Claudius Regaud CLCC (CIEL, R20027BB), Labex TOUCAN (G20000BB), institutional grants from INSERM, Université Paul Sabatier and CNRS. PG was supported by the CALYM Carnot Institute and FG by Labex TOUCAN (G20000BB). CD-L was supported by a personal FPI fellowship from the Ministry of Economy and competitiveness (PRE2018-083797) associated to the project SAF2017-88275-R lead by PPG (granted by Ministerio de Economia y Competitividad- MINECO).
Competing interests: RM and J-ML are employees of Imactiv3D.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
References
- 1. Scott DW, Gascoyne RD. The tumour Microenvironment in B cell Lymphomas. Nat Rev Cancer 2014;14:517–34. 10.1038/nrc3774 [DOI] [PubMed] [Google Scholar]
- 2. Carbone A, Roulland S, Gloghini A, et al. Follicular lymphoma. Nat Rev Dis Primers 2019;5:83. 10.1038/s41572-019-0132-x [DOI] [PubMed] [Google Scholar]
- 3. Amé-Thomas P, Tarte K. The Yin and the Yang of follicular lymphoma cell niches: role of Microenvironment heterogeneity and plasticity. Semin Cancer Biol 2014;24:23–32. 10.1016/j.semcancer.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 4. Chraa D, Naim A, Olive D, et al. T lymphocyte Subsets in cancer immunity: friends or foes. J Leukoc Biol 2019;105:243–55. 10.1002/JLB.MR0318-097R [DOI] [PubMed] [Google Scholar]
- 5. Dobaño-López C, Araujo-Ayala F, Serrat N, et al. Follicular lymphoma Microenvironment: an intricate network ready for therapeutic intervention. Cancers (Basel) 2021;13:641. 10.3390/cancers13040641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valero JG, Matas-Céspedes A, Arenas F, et al. The receptor of the colony-stimulating Factor-1 (CSF-1R) is a novel Prognostic factor and therapeutic target in follicular lymphoma. Leukemia 2021;35:2635–49. 10.1038/s41375-021-01201-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Watanabe T. The tumor Microenvironment in follicular lymphoma: its pro-malignancy role with therapeutic potential. Int J Mol Sci 2021;22:5352. 10.3390/ijms22105352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tosolini M, Algans C, Pont F, et al. Large-scale microarray profiling reveals four stages of immune escape in non-Hodgkin lymphomas. OncoImmunology 2016;5:e1188246. 10.1080/2162402X.2016.1188246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Armengol M, Santos JC, Fernández-Serrano M, et al. Immune-Checkpoint inhibitors in B-cell lymphoma. Cancers (Basel) 2021;13:214. 10.3390/cancers13020214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sutherland RM, McCredie JA, Inch WR. Growth of Multicell Spheroids in tissue culture as a model of nodular Carcinomas. J Natl Cancer Inst 1971;46:113–20. [PubMed] [Google Scholar]
- 11. Bissell MJ. Goodbye flat biology - time for the 3RD and the 4TH dimensions. J Cell Sci 2017;130:3–5. 10.1242/jcs.200550 [DOI] [PubMed] [Google Scholar]
- 12. Kunz-Schughart LA, Freyer JP, Hofstaedter F, et al. The use of 3-D cultures for high-throughput screening: the Multicellular Spheroid model. J Biomol Screen 2004;9:273–85. 10.1177/1087057104265040 [DOI] [PubMed] [Google Scholar]
- 13. Friedrich J, Seidel C, Ebner R, et al. Spheroid-based drug screen: considerations and practical approach. Nat Protoc 2009;4:309–24. 10.1038/nprot.2008.226 [DOI] [PubMed] [Google Scholar]
- 14. Pampaloni F, Stelzer E. Three-dimensional cell cultures in toxicology. Biotechnol Genet Eng Rev 2010;26:117–38. 10.5661/bger-26-117 [DOI] [PubMed] [Google Scholar]
- 15. Sirenko O, Mitlo T, Hesley J, et al. High-content assays for characterizing the viability and morphology of 3d cancer Spheroid cultures. Assay Drug Dev Technol 2015;13:402–14. 10.1089/adt.2015.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rossi M, Alviano F, Righi S, et al. Three-dimensional models: a novel approach for lymphoma research. J Cancer Res Clin Oncol 2022;148:753–65. 10.1007/s00432-021-03897-9 [DOI] [PubMed] [Google Scholar]
- 17. Tian YF, Ahn H, Schneider RS, et al. Integrin-specific Hydrogels as adaptable tumor Organoids for malignant B and T cells. Biomaterials 2015;73:110–9. 10.1016/j.biomaterials.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mannino RG, Santiago-Miranda AN, Pradhan P, et al. 3d Microvascular model Recapitulates the diffuse large B-cell lymphoma tumor Microenvironment in vitro. Lab Chip 2017;17:407–14. 10.1039/c6lc01204c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sabhachandani P, Sarkar S, Mckenney S, et al. Microfluidic assembly of Hydrogel-based Immunogenic tumor Spheroids for evaluation of anticancer therapies and biomarker release. J Control Release 2019;295:21–30. 10.1016/j.jconrel.2018.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Foxall R, Narang P, Glaysher B, et al. Developing a 3D B cell lymphoma culture system to model antibody therapy. Front Immunol 2020;11:605231. 10.3389/fimmu.2020.605231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lamaison C, Latour S, Hélaine N, et al. A novel 3D culture model Recapitulates primary FL B-cell features and promotes their survival. Blood Adv 2021;5:5372–86. 10.1182/bloodadvances.2020003949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gravelle P, Jean C, Valleron W, et al. Innate predisposition to immune escape in follicular lymphoma cells. Oncoimmunology 2012;1:555–6. 10.4161/onci.19365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Decaup E, Jean C, Laurent C, et al. Anti-tumor activity of Obinutuzumab and Rituximab in a follicular lymphoma 3D model. Blood Cancer J 2013;3:e131. 10.1038/bcj.2013.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gravelle P, Jean C, Familiades J, et al. Cell growth in aggregates determines gene expression, proliferation, survival, Chemoresistance, and sensitivity to immune effectors in follicular lymphoma. Am J Pathol 2014;184:282–95. 10.1016/j.ajpath.2013.09.018 [DOI] [PubMed] [Google Scholar]
- 25. Decaup E, Rossi C, Gravelle P, et al. A Tridimensional model for NK cell-mediated ADCC of follicular lymphoma. Front Immunol 1943;10:10. 10.3389/fimmu.2019.01943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossi C, Gravelle P, Decaup E, et al. Boosting Γδ T cell-mediated antibody-dependent cellular cytotoxicity by PD-1 blockade in follicular lymphoma. Oncoimmunology 2019;8:1554175. 10.1080/2162402X.2018.1554175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vidal-Crespo A, Matas-Céspedes A, Rodriguez V, et al. Daratumumab displays in vitro and in vivo anti-tumor activity in models of B-cell non-Hodgkin lymphoma and improves responses to standard Chemo-Immunotherapy regimens. Haematologica 2020;105:1032–41. 10.3324/haematol.2018.211904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gava F, Faria C, Gravelle P, et al. 3d model characterization by 2d and 3d imaging in T(14;18)-Positive B-NHL: perspectives for in vitro drug screens in follicular lymphoma. Cancers 2021;13:1490. 10.3390/cancers13071490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horvath P, Aulner N, Bickle M, et al. Screening out irrelevant cell-based models of disease. Nat Rev Drug Discov 2016;15:751–69. 10.1038/nrd.2016.175 [DOI] [PubMed] [Google Scholar]
- 30. Jaffe ES. The 2008 WHO classification of Lymphomas: implications for clinical practice and Translational research. Hematology Am Soc Hematol Educ Program 2009:523–31. 10.1182/asheducation-2009.1.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huet S, Sujobert P, Salles G. From Genetics to the clinic: a Translational perspective on follicular lymphoma. Nat Rev Cancer 2018;18:224–39. 10.1038/nrc.2017.127 [DOI] [PubMed] [Google Scholar]
- 32. Tosolini M, Pont F, Poupot M. Assessment of tumor-infiltrating TCRVγ9Vδ2 Γδ lymphocyte abundance by Deconvolution of human cancers microarrays. Oncoimmunology 2017;6:e1284723. 10.1080/2162402X.2017.1284723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hilchey SP, Kobie JJ, Cochran MR, et al. Human follicular lymphoma CD39+-Infiltrating T cells contribute to adenosine-mediated T cell Hyporesponsiveness. J Immunol 2009;183:6157–66. 10.4049/jimmunol.0900475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cardoso CC, Auat M, Santos-Pirath IM, et al. The importance of CD39, CD43, CD81, and CD95 expression for differentiating B cell lymphoma by flow Cytometry: CD39, CD43, CD81, AND CD95 FOR LYMPHOMA DIAGNOSIS. Cytometry B Clin Cytom 2018;94:451–8. 10.1002/cyto.b.21533 [DOI] [PubMed] [Google Scholar]
- 35. Casey M, Segawa K, Law SC, et al. Inhibition of CD39 Unleashes macrophage antibody-dependent cellular Phagocytosis against B-cell lymphoma. Leukemia 2023;37:379–87. 10.1038/s41375-022-01794-9 [DOI] [PubMed] [Google Scholar]
- 36. Andor N, Simonds EF, Czerwinski DK, et al. Single-cell RNA-Seq of follicular lymphoma reveals malignant B-cell types and Coexpression of T-cell immune checkpoints. Blood 2019;133:1119–29. 10.1182/blood-2018-08-862292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han G, Deng Q, Marques-Piubelli ML, et al. Follicular lymphoma Microenvironment characteristics associated with tumor cell mutations and MHC class II expression. Blood Cancer Discov 2022;3:428–43. 10.1158/2643-3230.BCD-21-0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laurent C, Charmpi K, Gravelle P, et al. Several immune escape patterns in non-Hodgkin's Lymphomas. OncoImmunology 2015;4:e1026530. 10.1080/2162402X.2015.1026530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kline J, Godfrey J, Ansell SM. The immune landscape and response to immune Checkpoint blockade therapy in lymphoma. Blood 2020;135:523–33. 10.1182/blood.2019000847 [DOI] [PubMed] [Google Scholar]
- 40. Richendollar BG, Pohlman B, Elson P, et al. Follicular programmed death 1-positive lymphocytes in the tumor Microenvironment are an independent Prognostic factor in follicular lymphoma. Hum Pathol 2011;42:552–7. 10.1016/j.humpath.2010.08.015 [DOI] [PubMed] [Google Scholar]
- 41. Yang Z-Z, Grote DM, Ziesmer SC, et al. PD-1 expression defines two distinct T-cell sub-populations in follicular lymphoma that Differentially impact patient survival. Blood Cancer J 2015;5:e281. 10.1038/bcj.2015.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Josefsson SE, Beiske K, Blaker YN, et al. TIGIT and PD-1 mark Intratumoral T cells with reduced Effector function in B-cell non-Hodgkin lymphoma. Cancer Immunol Res 2019;7:355–62. 10.1158/2326-6066.CIR-18-0351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Z-Z, Kim HJ, Villasboas JC, et al. Expression of LAG-3 defines exhaustion of Intratumoral PD-1+ T cells and correlates with poor outcome in follicular lymphoma. Oncotarget 2017;8:61425–39. 10.18632/oncotarget.18251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carreras J, Lopez-Guillermo A, Kikuti YY, et al. High TNFRSF14 and low BTLA are associated with poor prognosis in follicular lymphoma and in diffuse large B-cell lymphoma transformation. J Clin Exp Hematop 2019;59:1–16. 10.3960/jslrt.19003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balança C-C, Salvioni A, Scarlata C-M, et al. PD-1 blockade restores helper activity of tumor-infiltrating, exhausted PD-1HiCD39+ CD4 T cells. JCI Insight 2021;6:e142513. 10.1172/jci.insight.142513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Josefsson SE, Huse K, Kolstad A, et al. T cells expressing Checkpoint receptor TIGIT are enriched in follicular lymphoma tumors and characterized by reversible suppression of T-cell receptor signaling. Clin Cancer Res 2018;24:870–81. 10.1158/1078-0432.CCR-17-2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dobaño-López C, Valero JG, Araujo-Ayala F, et al. Immunocompetent 3d follicular lymphoma model: A Preclinical tool to design tailored Immunotherapies. Blood 2022;140(Supplement 1):9279–80. 10.1182/blood-2022-166131 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007156supp008.pdf (216.6KB, pdf)
jitc-2023-007156supp004.pdf (154.7KB, pdf)
jitc-2023-007156supp005.pdf (312.1KB, pdf)
jitc-2023-007156supp010.m4v (7.3MB, m4v)
jitc-2023-007156supp006.pdf (177.5KB, pdf)
jitc-2023-007156supp002.pdf (1.8MB, pdf)
jitc-2023-007156supp007.pdf (284.2KB, pdf)
jitc-2023-007156supp009.pdf (172KB, pdf)
jitc-2023-007156supp001.pdf (29.3KB, pdf)
jitc-2023-007156supp003.pdf (884.8KB, pdf)
jitc-2023-007156supp011.pdf (192.1KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information.