Abstract
Background:
Very young premenopausal women diagnosed with hormone receptor-positive, human epidermal growth factor receptor 2-negative (HR+HER−) early breast cancer (EBC) have higher rates of recurrence and death for reasons that remain largely unexplained.
Patients and methods:
Genomic sequencing was applied to HR+HER2− tumours from patients enrolled in the Suppression of Ovarian Function Trial (SOFT) to determine genomic drivers that are enriched in young premenopausal women. Genomic alterations were characterised using next-generation sequencing from a subset of 1276 patients (deep targeted sequencing, n = 1258; whole-exome sequencing in a young-age, case-control subsample, n = 82). We defined copy number (CN) subgroups and assessed for features suggestive of homologous recombination deficiency (HRD). Genomic alteration frequencies were compared between young premenopausal women (<40 years) and older premenopausal women (≥40 years), and assessed for associations with distant recurrence-free interval (DRFI) and overall survival (OS).
Results:
Younger women (<40 years, n = 359) compared with older women (≥40 years, n = 917) had significantly higher frequencies of mutations in GATA3 (19% versus 16%) and CN amplifications (CNAs) (47% versus 26%), but significantly lower frequencies of mutations in PIK3CA (32% versus 47%), CDH1 (3% versus 9%), and MAP3K1 (7% versus 12%). Additionally, they had significantly higher frequencies of features suggestive of HRD (27% versus 21%) and a higher proportion of PIK3CA mutations with concurrent CNAs (23% versus 11%). Genomic features suggestive of HRD, PIK3CA mutations with CNAs, and CNAs were associated with significantly worse DRFI and OS compared with those without these features. These poor prognostic features were enriched in younger patients: present in 72% of patients aged <35 years, 54% aged 35–39 years, and 40% aged ≥40 years. Poor prognostic features [n = 584 (46%)] versus none [n = 692 (54%)] had an 8-year DRFI of 84% versus 94% and OS of 88% versus 96%. Younger women (<40 years) had the poorest outcomes: 8-year DRFI 74% versus 85% and OS 80% versus 93%, respectively.
Conclusion:
These results provide insights into genomic alterations that are enriched in young women with HR+HER2− EBC, provide rationale for genomic subgrouping, and highlight priority molecular targets for future clinical trials.
Keywords: breast cancer, hormone receptor positive, young women, genomics, prognosis
INTRODUCTION
Younger premenopausal women diagnosed with early breast cancer (EBC) have a significantly higher riskof recurrence and death than older women for reasons that remain largely unexplained. This is particularly true for hormone receptor-positive (HR+), luminal breast cancers.1,2 In order to address this clinical need, the pivotal Suppression of Ovarian Function Trial (SOFT) was developed. SOFT was a multicenter, open-label, phase III trial for premenopausal women diagnosed with HR+ early-stage breast cancer.3 The study demonstrated a significant improvement in disease-free survival and overall survival (OS) with the addition of ovarian function suppression (OFS) to tamoxifen.4,5 These practice-changing results led to the standard incorporation of OFS into adjuvant therapy for high-risk premenopausal patients diagnosed with early-stage HR+/human epidermal growth factor receptor 2-negative (HER2−) breast cancers. Despite the addition of OFS therapy, the prognosis of women aged <35 years remained significantly worse than that of older women, with a 5-yearbreastcancer-freeintervalofonly 77% versus 91.5%, respectively.6
The biology underlying the poor prognosis observed in very young women with HR+HER2− breast cancer remains incompletely understood. In a large pooled analysis of 71 studies investigating gene expression assays, young women aged ≤40 years had higher proportions of intermediate- to high-risk tumours by EndoPredict, MammaPrint, and Oncotype DX assays compared with older women; however this analysis was not stratified by breast cancer subtype.7 This is also consistent with data demonstrating that young women with HR+ breast cancers have higher proportions of luminal B tumours and lower proportions of luminal A tumours than older women.1 Analysis of The Cancer Genome Atlas (TCGA)-Breast dataset using broader gene expression data showed increases in gene expression signatures indicating proliferation, stem cell features, and endocrine resistance in young patients compared with older patients; however, this analysis was also not stratified by breast cancer subtype.8
DNA-based genomic analyses between HR+HER2− tumours in young women and older women involve far fewer patients. There are limited numbers of HR+HER2− breast cancer samples from young women represented in publicly available databases (Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2023.01.009). Germline cancer predisposition alterations such as with BRCA1, BRCA2, and PALB2 are known to occur in a higher frequency in young women but this is observed to a greater degree in patients with triple-negative breast cancer rather than HR+HER2− breast cancers.9,10 The previously discussed TCGA-Breast analysis demonstrated higher frequencies of GATA3 mutations and chromosome 6q27 deletions in younger patients compared with older patients, but again this was not stratified by breast cancer subtype.8 A recently reported study investigating patients aged ≤35 years at diagnosis described genomic findings in 92 patients, of which 49 had HR+HER2− breast cancer. They identified higher rates of alterations in GATA3 and ARID1A, and lower rates of PIK3CA mutations in young patients compared with older patients, particularly in luminal A tumours.10
This study was designed to investigate genomic features that may be enriched in young premenopausal women diagnosed with HR+HER2− EBC, using samples obtained from patients enrolled in the SOFT clinical trial who received contemporary treatment. Current treatment strategies for very young patients remain similar to older premenopausal women and postmenopausal women. We hypothesised that uniquely designed prospective trials may be required for this at-risk population.
PATIENTS AND METHODS
Patients
SOFT (ClinicalTrials.gov, NCT00066690) is a randomised, phase III trial in which 3066 premenopausal women with HR+ early-stage breast cancer were randomly assigned to adjuvant endocrine therapy with tamoxifen alone, tamoxifen plus ovarian suppression, or exemestane plus ovarian suppression for 5 years. Details of the study have been previously reported.3–5 Tumour and normal formalin-fixed paraffin-embedded tissue blocks were prospectively collected for translational research purposes. The IBCSG Biological Project Working Group approved this investigation.
The study workflow is shown in Supplementary Figure S1, available at https://doi.org/10.1016/j.annonc.2023.01.009, including reasons for sample exclusion. There were 1509 tumour samples selected for targeted DNA sequencing after exclusions. A smaller young-age, case-control subsample was selected for whole-exome sequencing (WES). Case-control patient selection for the subsample is described in the Supplementary Methods, available at https://doi.org/10.1016/j.annonc.2023.01.009.
Next-generation sequencing
Targeted DNA sequencing was carried out using a customised hybridisation capture panel designed specifically for recurrent breast cancer genes. A list of target genes is shown at https://breast-cancer-research.biomedcentral.com/articles/10.1186/s13058-020-01328-0#Sec2. 11After exclusions, 1258 samples were evaluable for the analysis. WES was successfully completed in 82 patients from the case-control subsample, of which 73 (78%) had tumour/ normal pairs successfully sequenced, and 9 (10%) had tumour samples successfully sequenced but without the normal sample. Details on variant calling, copy number (CN) calling, integrated cluster estimation, analysis of homologous recombination deficiency (HRD) using the previously developed HRDetect model12 adapted for WES, analysis of HRD-related genes, and other genomic features are available in the Supplementary Methods, available at https://doi.org/10.1016/j.annonc.2023.01.009. Publicly available clinical and genomic data from the TCGA-Breast and a secondary analysis of the BIG 1–98 clinical trial were used for validation purposes.13,14
Statistical analysis
The combined sequencing cohort includes all patients with tumour samples who successfully underwent DNA sequencing of any type (n=1276). The young-age, case-control subsample includes only patients aged <45 years who underwent successful WES of both tumour/normal or only tumour (n = 82); 64 patients had both targeted sequencing and WES. Luminal-like status was defined using previously published St. Gallen consensus guidelines using centrally determined estrogen receptor (ER), progesterone receptor, and Ki-67 expression levels by immunohistochemistry.15,16
Comparisons between patient subgroups were carried out using chi-square tests (categorical variables), and t-tests and Mann—Whitney Wilcoxon tests (normally and non-normally distributed continuous variables, respectively). For comparisons in driver alteration frequency between subgroups, multiple testing correction using the false discovery method was used. For analysis of time-to-event endpoints, the primary endpoint was distant recurrence-free interval (DRFI), defined as the time from randomisation to recurrence at a distant site. In patients without a distant recurrence, censoring occurred at the date of last follow-up or death. The secondary endpoint was OS. Cox proportional hazards regression models were used to analyse associations with time-to-event endpoints [stratified by nodal status and (neo)adjuvant chemotherapy receipt, and adjusted by treatment assignment]. For association of endpoints with: (i) driver alterations, patients with the presence of the driver alteration were compared with patients without the driver alteration; (ii) CN-altered subgroups, each CN-altered subgroup was compared with the subgroup classified as ‘amplification-devoid’. For association with prognostic genomic subgroupings, each genomic subgroup was compared with the subgroup with ‘no poor prognostic features’. Hazard ratios (HRs) with 95% confidence intervals (CIs) were estimated and Wald tests were used, with a two-sided value of P < 0.05 deemed to be statistically significant. To analyse for association of eHRDetect score (positive versus negative) with DRFI in the young-age, case-control subsample, a conditional logistic regression model was used, stratified by the case-control matching.
RESULTS
We successfully sequenced tumour samples from 1276 (n = 1276/1509, 85%) premenopausal women with HR+HER2− EBC who were randomised in the SOFT study (Figure 1A). Given the age of these archival tumour samples, we applied a deep targeted sequencing approach for the majority of samples (n = 1258). The clinical characteristics of the combined sequencing cohort were similar to those of the overall SOFT study population: the median age was 43 years (range 24–58 years) and median follow-up was 8 years (Supplementary Table S2, available at https://doi.org/10.1016/j.annonc.2023.01.009). In order to focus on the very young, we additionally carried out WES on a matched case-control subsample of 82 women, 42 with and 40 without a distant recurrence event. The matched case-control subsample had a median age of 38 years (Supplementary Table S3, available at https://doi.org/10.1016/j.annonc.2023.01.009).
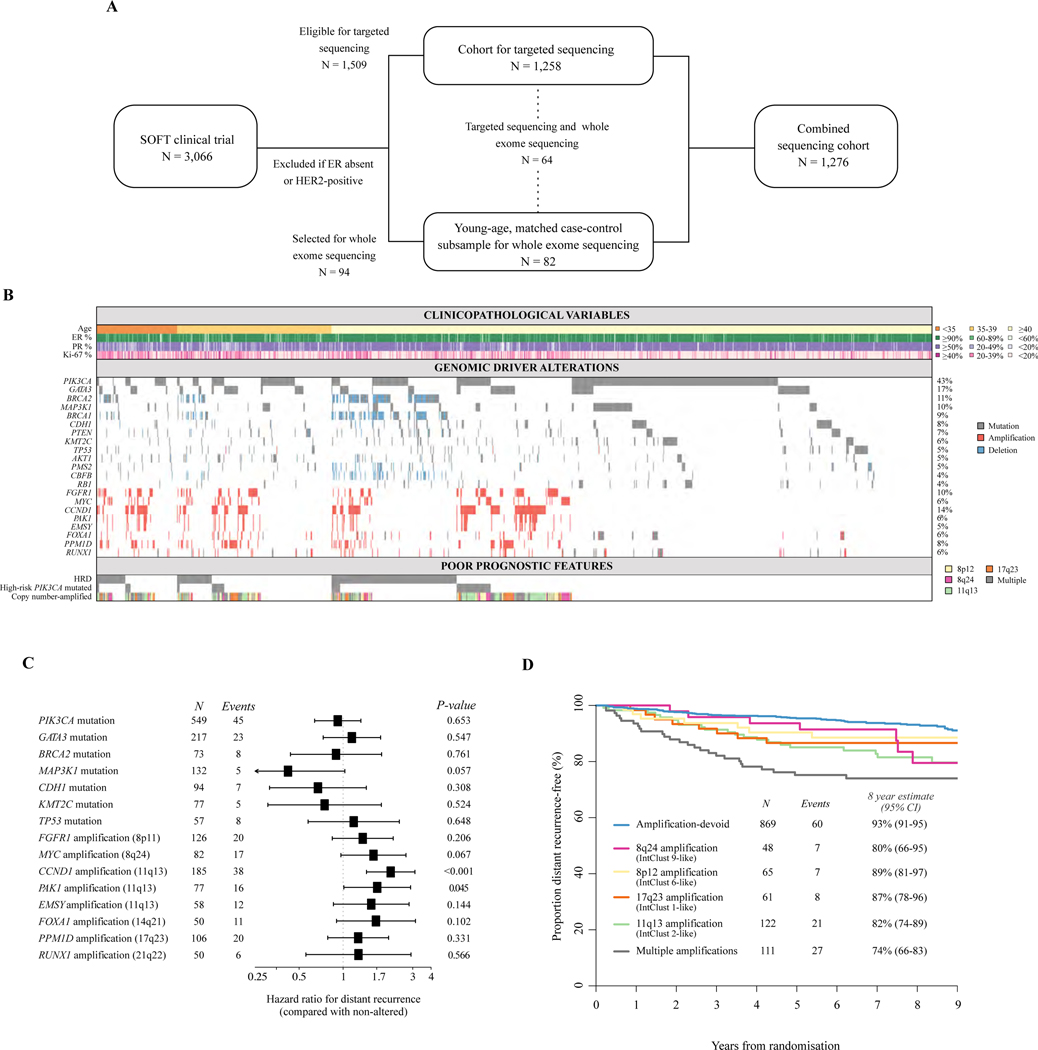
Figure 1. The genomic landscape of premenopausal HRDHER2−L breast cancer in SOFT.
(A) Summary of the sequencing cohorts derived from the SOFT clinical trial, including overlapping cases. Further details on patient selection are shown in Supplementary Figure S1, available at https://doi.org/10.1016/j.annonc.2023.01.009. (B) Plot demonstrating the frequencies and co-existence of clinicopathological variables (top panel), genomic driver alterations (middle panel), and poor prognostic genomic features (bottom panel) in the SOFT combined sequencing cohort (n = 1276). (C) Hazard ratio estimates (boxes) and 95% confidence intervals (lines) derived from Cox proportional hazards regression models comparing patients with tumours that harbour the driver alteration with patients with tumours that do not harbour the driver alteration for the endpoint of distant recurrence-free interval in the SOFT combined sequencing cohort (n = 1276). Only driver alterations with ≥5 events are included. (D) Kaplan–Meier plot estimating the rate of freedom from distant recurrence based on the copy number-amplified subgrouping in the SOFT combined sequencing cohort (n = 1276).
CI, confidence interval; ER, estrogen receptor; HER2−, human epidermal growth factor receptor 2-negative; HR+, hormone receptor-positive; PR, progesterone receptor; SOFT, Suppression of Ovarian Function Trial.
Genomic landscape of premenopausal HR+HER2− breast cancer
The landscape of frequent somatic driver alterations in the combined sequencing cohort of 1276 is shown in Figure 1B. There was a mean number of 3 driver alterations identified per tumour (range 0–22). No driver alteration was identified in 96 tumour samples (7.5%). Twenty-two genes harboured oncogenic drivers in at least 50 patients (≥4%), of which 9 were predominantly mutational (PIK3CA, GATA3, MAP3K1, CDH1, PTEN, KMT2C, TP53, AKT1, CBFB), 4 were tumour suppressors which were either mutated and/or CN deleted (BRCA2, BRCA1, PMS2, RB1), and the remainder were within recurrent amplicons on chromosomes 11q13 (CCND1, PAK1, EMSY), 8p11 (FGFR1), 8q24 (MYC), 17q23 (PPM1D), 14q21 (FOXA1), and 21q22 (RUNX1). ERBB2 mutations were uncommon (n = 23, 2%). We detected no ESR1 mutations, consistent with previous data indicating that these arise following selective pressure from endocrine therapies.17 As per individual tumour sample, double or multiple genomic alterations were observed in well-described tumour suppressors such as BRCA2, BRCA1, MAP3K1, and PTEN; however, this was infrequent (Supplementary Figure S2, available at https://doi.org/10.1016/j.annonc.2023.01.009). Recently described double or multiple PIK3CA mutations occurred in 47 patients (9% of PIK3CA-mutated tumours).18 PIK3CA amplifications were rare, but nearly all co-existed with PIK3CA activating mutations (six patients with amplifications, five with co-existing hotspot PIK3CA mutations). Frequencies of genomic driver alterations in our premenopausal cohort were overall similar to those in postmenopausal women enrolled in the BIG 1–98 (n = 538) clinical trial and in TCGA-Breast (HR+HER2− n = 451) (Supplementary Figure S3, available at https://doi.org/10.1016/j.annonc.2023.01.009),14 with the exception of numerically less TP53 mutations (5% versus 15%−19%) and CDH1 mutations (7% versus 12%−19%) in premenopausal women versus postmenopausal women, respectively.
We next investigated the association of each frequent somatic driver with the risk of distant recurrence, compared with its non-altered counterpart (Figure 1C). Consistent with previous reports, key gene-level CN amplifications (CNAs) on chromosomes 11q13, 8q24, 14q21, 8p11 17q23, and 21q22 dominated driver associations with increased risk of distant recurrence.19,20 Similar findings were previously reported for postmenopausal BIG 1–98 trial patients.14 By contrast, MAP3K1 mutations were associated with a numerically lower risk of distant recurrence. MAP3K1 mutations have been previously reported to be associated with a less proliferative and more endocrine-sensitive luminal A phenotype.21 PIK3CA mutations were associated with decreased risk of distant recurrence, similar to a recent large meta-analysis;22 however, this was not statistically significant. Associations with OS demonstrated similar findings (Supplementary Table S4, available at https://doi.org/10.1016/j.annonc.2023.01.009).
A recent integrated analysis of EBCs combining CN segmentation and gene expression data identified 10 clusters termed IntClusts, each with unique CN and prognostic profiles.19,20 Most of these clusters are defined by characteristic gene-level CN aberrations.20,23 For HR+HER2− breast cancers, the key IntClust subgroups were defined by characteristic CNAs. In the absence of gene expression data, we evaluated this classification framework using a simplified system based solely on IntClust-specific CNAs. We first compared tumours that harboured each of these IntClust-specific CNAs with tumours that were CNA-devoid. Similar to the integrated cluster data, each CNA group was associated with a significantly higher risk of distant recurrence than the CNA-devoid group, with the exception of the group of patients with amplifications on chromosome 17q23 (Supplementary Table S5, available at https://doi.org/10.1016/j.annonc.2023.01.009). Similar findings were observed for CNA associations with OS (Supplementary Figure S6 and Supplementary Table S5, available at https://doi.org/10.1016/j.annonc.2023.01.009).
As a number of tumours harboured concurrent CNAs in multiple IntClust-characteristic amplicons, we grouped the tumours that contained amplifications in multiple amplicons together as ‘multiple amplifications’ (Supplementary Figure S4, available at https://doi.org/10.1016/j.annonc.2023.01.009). Frequencies of the CNA subgroups in our premenopausal cohort were similar to frequencies observed in postmenopausal women enrolled in the BIG 1–98 clinical trial and in TCGA-Breast (Supplementary Figure S5, available at https://doi.org/10.1016/j.annonc.2023.01.009). The CNA-devoid group had the best prognosis with an estimated rate of freedom from distant recurrence of 93% (95% CI 91% to 95%) and OS of 95% (95% CI 93% to 97%) at 8 years (Figure 1D, Supplementary Figure S5, available at https://doi.org/10.1016/j.annonc.2023.01.009). Notably, patients classified with multiple amplifications had the poorest prognosis with an estimated freedom from distant recurrence rate of 74% (95% CI 66% to 83%) and OS of 79% (95% CI 71% to 87%) at 8 years.
Genomic characteristics of breast cancer in premenopausal patients aged <40 years
The higher risk of recurrence in very young compared with older premenopausal patients enrolled in the SOFT trial has been previously reported.3,6,24 Other studies have also demonstrated this even after stratification by intrinsic subtype, despite different ‘young’ age cut-off values.1,2 Similarly, after adjustment for clinicopathological variables and treatment assignment, in our targeted sequencing cohort patients aged <40 years had greater risk of distant recurrence than patients aged ≥40 years in the combined sequencing cohort (HR 1.57, 95% CI 1.08–2.28), with an estimated 8-year freedom from distant recurrence rate of 78% (95% CI 74% to 83%) versus 94% (95% CI 92% to 95%), respectively (Figure 2A). Consistent with prior literature,1,2 the poor prognostic association of age <40 years was observed in both luminal A-like [estimated 8-year freedom from distant recurrence rate of 82% (95% CI 71% to 95%) versus 97% (95% CI 95% to 99%)] and luminal B-like populations [estimated 8-year freedom from distant recurrence rate of 78% (95% CI 73% to 84%) versus 91% (95% CI 89% to 94%)] (Supplementary Figure S7, available at https://doi.org/10.1016/j.annonc.2023.01.009).
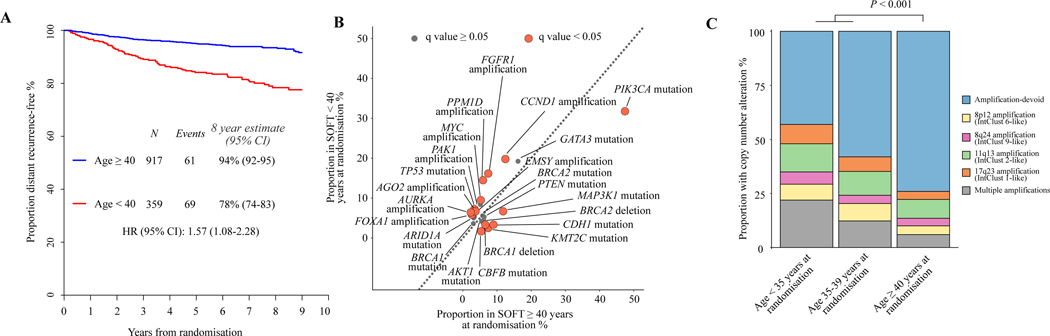
Figure 2. Genomic drivers of poor prognosis breast cancer in the very young.
(A) Kaplan–Meier plot estimating the rate of freedom from distant recurrence according to age at randomisation in the SOFT combined sequencing cohort (n = 1276). (B) A comparison of genomic driver alteration frequencies of patients in the SOFT combined sequencing cohort (n = 1276) aged <40 years at randomisation with those of patients aged ≥40 years at randomisation. The dotted line provides demonstration of the plot points that represent equal frequencies between the groups. Points in burgundy demonstrated significantly different frequencies after adjustment for multiple testing using the false discovery method. (C) A comparison of the frequencies of copy number-altered subgroups according to patients’ age at randomisation in the SOFT combined sequencing cohort (n = 1276).
CI, confidence interval; HR, hazard ratio; SOFT, Suppression of Ovarian Function Trial.
We next assessed for differences in the frequency of oncogenic drivers in younger patients compared with older premenopausal patients. Patients aged <40 years had significantly lower frequencies of mutations in PIK3CA (32% versus 47%, q < 0.001), CDH1 (3% versus 9%, q = 0.002), and MAP3K1 (7% versus 12%, q = 0.014), but higher frequencies of mutations in TP53 (7% versus 3%, q < 0.010) than patients aged ≥40 years (Figure 2B). In comparison with the postmenopausal cohorts (BIG 1–98 and TCGA-Breast), patients aged <40 years had similarly lower frequencies of mutations in PIK3CA, CDH1, and MAP3K1, but not TP53 (Supplementary Figures S8 and S9, available at https://doi.org/10.1016/j.annonc.2023.01.009), whereas higher frequencies of mutations in BRCA2 and GATA3 were observed. Given the reported poorer prognosis of very young patients with luminal A subtype EBCs,25 we further compared genomic drivers by luminal breast cancer subtype (Supplementary Figure S10, available at https://doi.org/10.1016/j.annonc.2023.01.009). Interestingly, the difference in frequency of GATA3 and CDH1 mutations appeared to be confined to the luminal A subtype.
Younger patients <40 years of age also had significantly higher frequencies of CNAs compared with older premenopausal patients (47% versus 26%, q < 0.001). This increase was observed for all assessed CNA subgroups; however, the largest increase was observed in the ‘multiple amplifications’ subgroup (16% versus 6%, q < 0.001) (Figure 2C). This effect was even more pronounced as age decreased, with patients aged <35 years (n = 123) having very high frequencies of CNAs (57%), but low frequencies of PIK3CA mutation (24%). We found similar findings when comparing with postmenopausal cohorts, with the greatest magnitude of difference observed in very young patients aged <35 years (Supplementary Figure S11, available at https://doi.org/10.1016/j.annonc.2023.01.009).
Whole-exome sequencing in the very young case-control subsample
We next carried out tumour-normal WES in a matched case-control subsample (successful 73/82, 89%) in order to investigate if there were unique genomic features specific to younger women. A summary of genomic findings is shown in Figure 3. In this case-control subsample, CCND1 amplifications were again significantly associated with higher risk of distant recurrence (Supplementary Figure S12, available at https://doi.org/10.1016/j.annonc.2023.01.009). Whilst we found no significant correlation between somatic mutation number and the whole-genome integrity index (wGII), a significantly higher number of mutations were observed in tumours where whole-genome doubling was present versus absent (P < 0.001) (Supplementary Figure S13, available at https://doi.org/10.1016/j.annonc.2023.01.009). Higher wGII scores were significantly associated with higher risk of distant recurrence; however, higher mutation burden was not (Supplementary Figure S13, available at https://doi.org/10.1016/j.annonc.2023.01.009). There was no correlation between number of somatic mutations and stromal tumour-infiltrating lymphocytes’ quantity (Spearman’s correlation 0.08), or age (Spearman’s correlation 0.06) (Supplementary Figure S13 and S14, available at https://doi.org/10.1016/j.annonc.2023.01.009). However, there was a moderate positive correlation between number of somatic mutations and Ki-67 levels (Spearman’s correlation 0.25) (Supplementary Figure S14, available at https://doi.org/10.1016/j.annonc.2023.01.009).
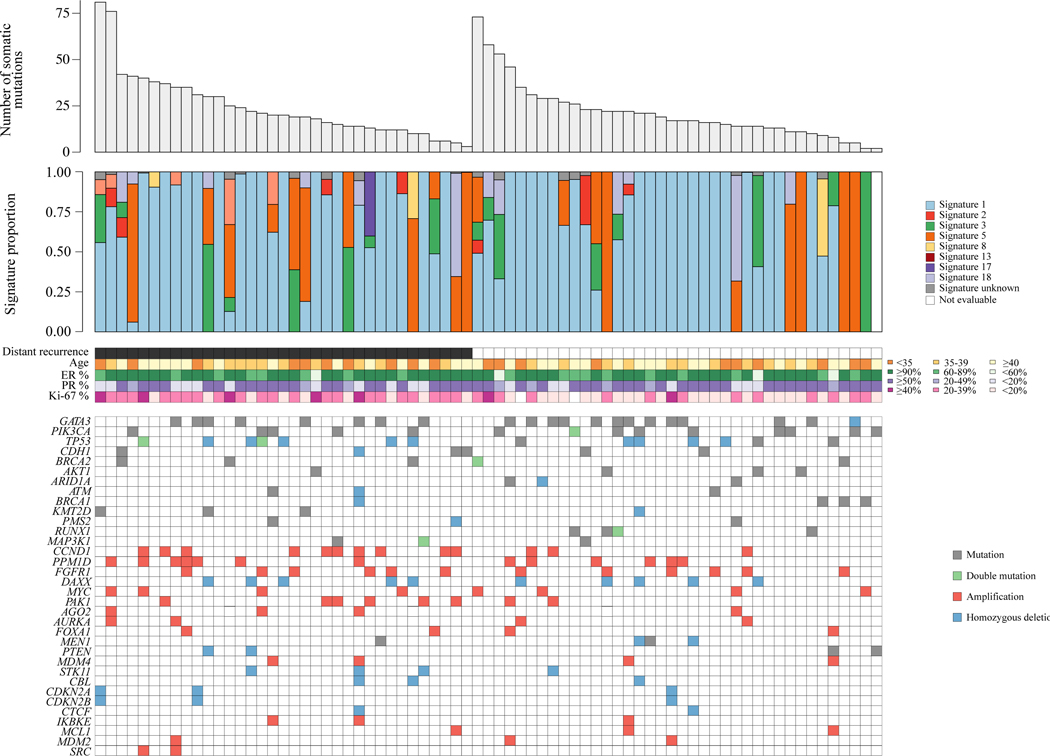
Figure 3. Genomic features of the very young, high-risk subsample.
Combined plot demonstrating the number of somatic mutations, mutational signature proportions, clinicopathological variables, and genomic driver alterations in the SOFT matched case-control subsample that had paired tumour-normal whole-exome sequencing (n = 73). Mutational signature 1 is associated with spontaneous deamination of 5-methylcytosine, signature 3 is associated with HRD, and signatures 2 and 13 are associated with APOBEC mutagenesis.
ER, estrogen receptor; HRD, homologous recombination deficiency; PR, progesterone receptor; SOFT, Suppression of Ovarian Function Trial.
We evaluated the distribution of mutational signatures in this subsample, acknowledging WES for this purpose has limitations compared with whole-genome sequencing. Signature 1 was the most common predominant mutational signature amongst the assessed tumours (72%) (Supplementary Figure S15, available at https://doi.org/10.1016/j.annonc.2023.01.009). This signature represents spontaneous deamination of 5-methylcytosine and usually is associated with increasing patient age in other tumour types. Notably, signatures reflective of APOBEC mutagenesis (Supplementary Figures S2 and S13, available at https://doi.org/10.1016/j.annonc.2023.01.009) were uncommon here compared with previous reports.26 Interestingly, Signature 3, previously characterised in tumours with defective homologous recombination DNA repair, was present in 22% of tumours.
Genomic features of homologous recombination deficiency
We went on to further investigate the above findings using other algorithms that are designed to detect functional HRD. Deleterious alterations in genes involved in homologous recombination repair, such as germline BRCA1 and BRCA2 mutations, are a known risk factor for the development of early-onset breast cancer. Furthermore, recent research has suggested that estrogen signalling itself can suppress DNA damage response pathways, potentiating the development of homologous recombination-deficient, ER-positive breast cancers.27 Homologous recombination-deficient tumours are potentially amenable to therapeutic strategies that include poly (ADP-ribose) polymerase (PARP) inhibitors28,29 or platinum chemotherapy.30
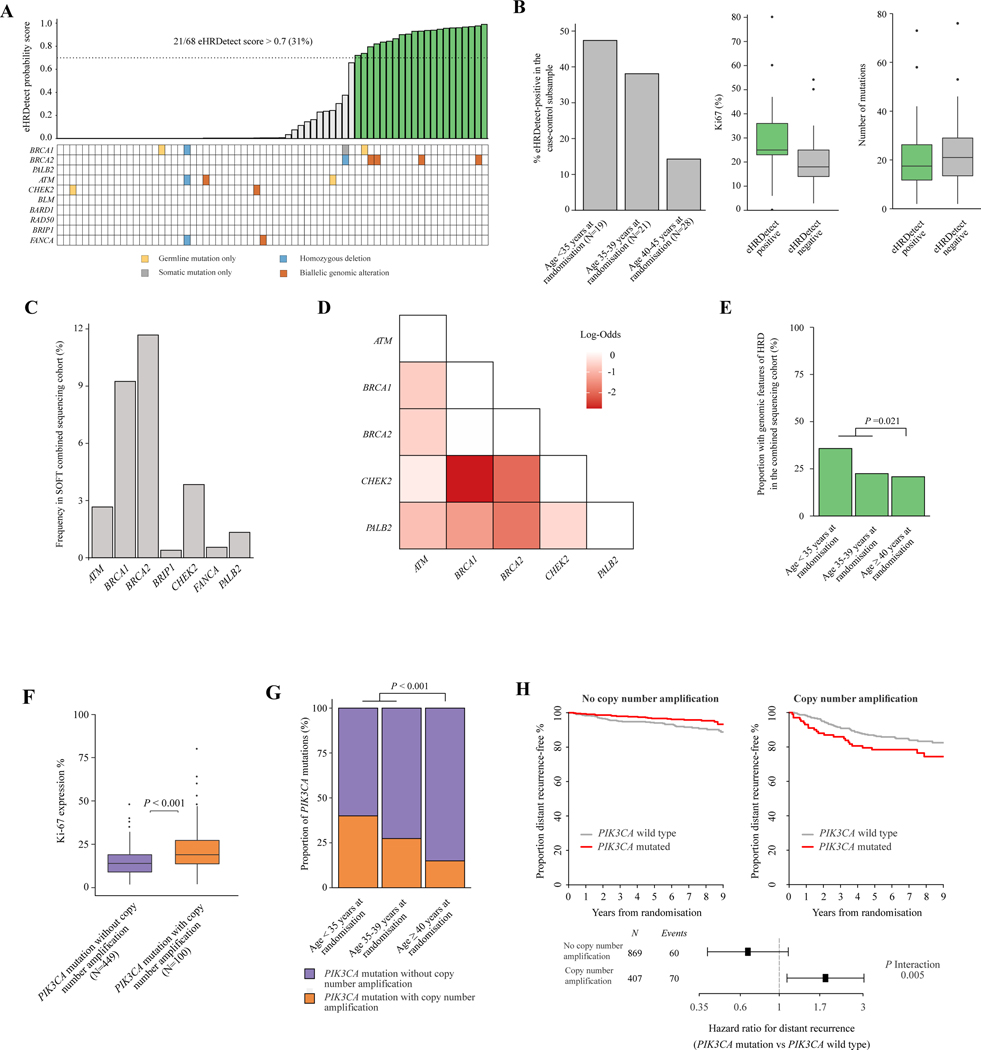
To analyse for the presence of features of HRD, we first investigated the matched case-control subsample with paired tumour-normal WES (n = 73). Using the ‘HRDetect’ algorithm modified for WES (here termed eHRDetect), 21 of 68 evaluable (31%) tumours tested positive using the pre-defined cut-off of 0.7.12 Of the 21 patients with eHRDetect-positive tumours, only 4 had germline BRCA2 mutations and 1 had a germline BRCA1 mutation (total n = 5/21, 24%). There were no patients with PALB2, RAD51C, or RAD51D germline mutations (Figure 4A). To further confirm this finding, we analysed the WES mutational signatures using SigMA,31 a published tool that is optimised to detect mutational signature 3 (the signature associated with HRD) in exomes. SigMA findings were concordant with eHRDetect in 57 of 60 (95%) samples (Supplementary Figure S15, available at https://doi.org/10.1016/j.annonc.2023.01.009). Tumours that were eHRDetect positive had a median Ki-67 level of 25% [interquartile range (IQR) 23%−36%] compared with tumours that were eHRDetect negative with a median Ki-67 level of 18% (IQR 14%−25%) (Figure 4B). There was no significant difference in the number of somatic mutations (P = 0.56) or stromal tumour-infiltrating lymphocytes (P = 0.38) by HRDetect status (Figure 4B, Supplementary Figure S15, available at https://doi.org/10.1016/j.annonc.2023.01.009). Notably, eHRDetect-positive tumours had higher rates of distant recurrence compared with eHRDetect-negative tumours (62% versus 40%, odds ratio 6.5, 95% CI 0.80–53.33, P = 0.080, respectively). We further observed that there was increasing frequency of eHRDetect-positive tumours with decreasing age: 41% in patients <40 years of age, and 47% in patients <35 years of age compared with 14% in patients 40–45 years of age (Figure 4B).
Figure 4. Genomic features of HRD and high-risk PIK3CA mutations in young patients.
(A) Plot demonstrating eHRDetect probability score (top panel) in 68 assessable patients in the case-control subsample who underwent paired tumour-normal whole-exome sequencing. Green bars indicate eHRDetect score above the 0.7 probability score threshold, termed eHRDetect positive. Genomic alterations in HRD-related genes (bottom panel) are also shown. (B) Plots demonstrating the associations between eHRDetect-positive score with age at randomisation, Ki-67 expression level, and number of somatic mutations in the case-control subsample who were assessable for eHRDetect (n = 68). (C) Bar plot demonstrating the frequency of patients with genomic features of HRD according to age at randomisation in the SOFT combined sequencing cohort (n = 1276). Genomic features of HRD included eHRDetect positivity and/or genetic alterations in HRD-related genes. (D) Pairwise analysis of HRD-related genes in the SOFT combined sequencing cohort (n = 1276). Only genes with ≥10 patients with genetic alterations were included. Fisher’s exact test was applied to each genetic alteration pair. Multiple testing correction using false discovery rate was applied. Only log-odds with a false discovery rate of <0.2 are displayed. Burgundy colour indicates an association with mutual exclusivity. (E) Bar plot demonstrating the proportion of tumours with genomic features of HRD according to age at randomisation in the SOFT combined sequencing cohort (n = 1276). Genomic features of HRD included eHRDetect positivity and/ or genetic alterations in HRD-related genes. (F) Boxplot demonstrating the association between concurrent copy number amplification status with PIK3CA mutations and Ki-67 expression levels in the SOFT combined sequencing cohort (n = 1276). (G) Bar plot demonstrating the frequency of patients with a PIK3CA mutation and concurrent copy number amplification according to age at randomisation in the SOFT combined sequencing cohort (n = 1276). (H) Kaplan–Meier plots and forest plots estimating the rate of freedom from distant recurrence according to PIK3CA mutation status and copy number amplification status in the SOFT combined sequencing cohort (n = 1276).
HRD, homologous recombination deficiency; SOFT, Suppression of Ovarian Function Trial.
We next evaluated for genomic alterations in HRD-related genes in the 1276 patients in the combined sequencing cohort. There were 274 patients (21%) with a genomic alteration in at least one HRD-related gene, with BRCA2 (12%) and BRCA1 (9%) alterations being the most prevalent (Figure 4C). Of the 274 patients, 68 (24.8%) had a genomic alteration in at least one HRD-related gene that was predicted to be germline in origin (Supplementary Figure S16, available at https://doi.org/10.1016/j.annonc.2023.01.009). Significant pairwise mutual exclusivity between these genes suggested functional redundancy (Figure 4D).
Taken together, HRD genomic features were present in 288 (23%) of the SOFT premenopausal combined sequencing cohort including tumours with eHRDetect positivity and/or genetic alterations in HRD-related genes. Again, we observed a significantly higher frequency of HRD genomic features in patients <40 years of age compared with patients ≥40 years of age (27% versus 21%, P = 0.021) (Figure 4E), with frequency increasing in patients <35 years of age at randomisation (n = 44, 36%). These findings suggest that genomic features of HRD are significantly enriched with decreasing age in young premenopausal patients with HR+HER2− EBC. The underlying biological reasons for this are unknown.
Heterogeneous outcomes for PIK3CA-mutated breast cancer
We next assessed whether oncogenic driver alterations had significantly different prognostic associations in women aged <40 years as compared with those aged ≥40 years (Supplementary Table S6, available at https://doi.org/10.1016/j.annonc.2023.01.009). PIK3CA mutations were the only oncogenic driver to demonstrate significantly different prognostic associations in women aged <40 years (HR 1.78, 95% CI 1.08–2.92) compared with women aged ≥40 years (HR 0.58, 95% CI 0.33–0.99, P interaction = 0.002). Heterogeneous disease outcome associations of those breast cancers with PIK3CA mutations have been previously reported, with better prognosis observed in a large meta-analysis of early-stage disease,22 and contrastingly poor prognosis observed in metastatic disease.32 We hypothesised that concurrent genomic features might explain the heterogeneous outcomes. Double or multiple PIK3CA mutations have recently been reported to enable hyper-activated PI3-kinase signalling and enhanced proliferation.18 Multiple PIK3CA mutations were present in 47 patients; however, these were not associated with higher Ki-67 levels, poorer outcomes, or enrichment in patients <40 years of age in this study (Supplementary Figure S17, available at https://doi.org/10.1016/j.annonc.2023.01.009). On the other hand, PIK3CA mutations with co-existing CNAs, while uncommon (n = 100, 8% of the combined sequencing cohort, 18% of patients with a PIK3CA mutation), demonstrated significantly higher Ki-67 levels (median 19% versus 14%, P < 0.001), enrichment in patients <40 years of age (31% versus 15%, P < 0.001), and a higher risk of distant recurrence (HR 3.37, 95% CI 1.86–6.10, P < 0.001) than PIK3CA mutations without co-existing CNAs (Figure 4F–H). This validates a similar previously reported finding of prognostic associations of PIK3CA mutations between different IntClust groups.33 The number of patients with a PIK3CA mutation within each IntClust group was small, limiting our ability to analyse each group independently; however, the poor prognostic effect was observed in the PIK3CA-mutated tumours with multiple amplifications, and amplifications on 11q13, 8p12, and 17q23 (Supplementary Figure S18, available at https://doi.org/10.1016/j.annonc.2023.01.009).
Rationale for prognostic genomic subgrouping of premenopausal breast cancer
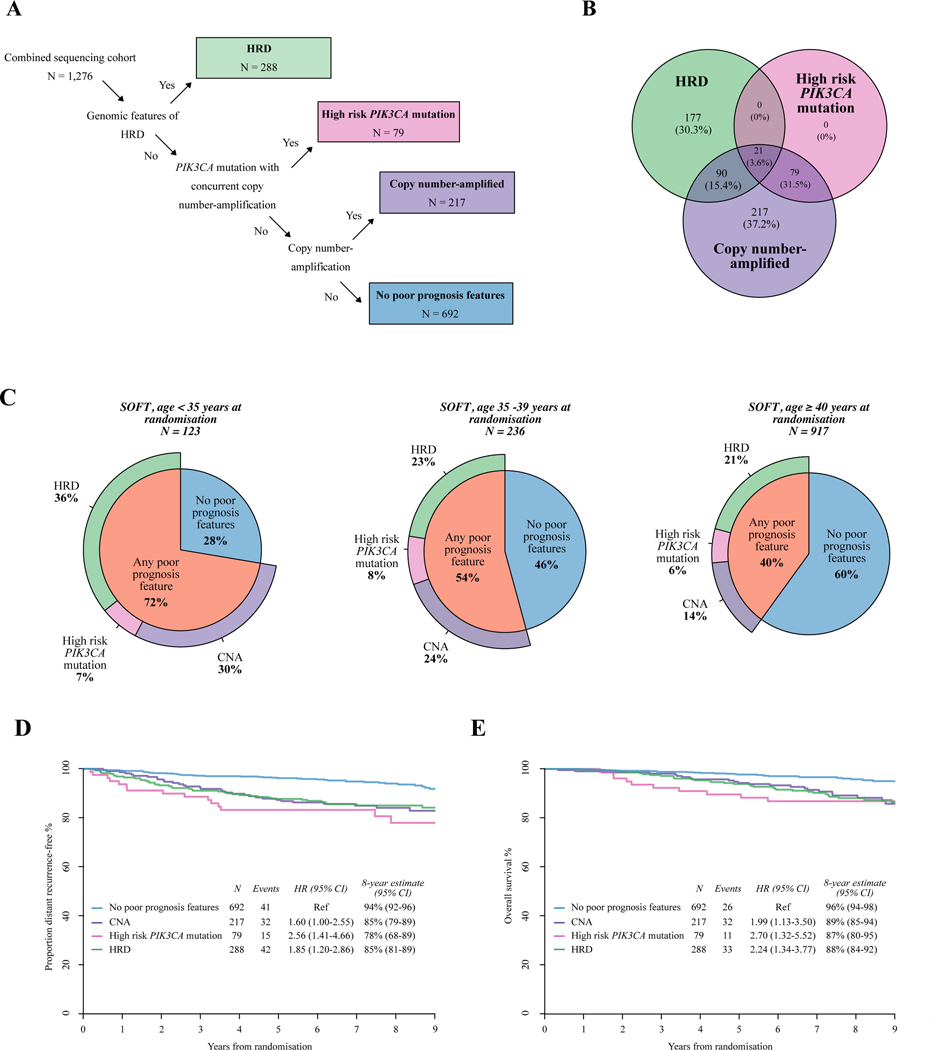
We have identified a number of key genomic features in premenopausal HR+HER2− EBC that associate with distinct prognostic profiles in this heterogeneous breast cancer subtype. These features may help define potential therapeutic targets that could be addressed in clinical trial settings. This provides rationale for genomic subgrouping of premenopausal breast cancer in order to develop new treatment strategies. An example of such a proposed genomic subgrouping framework is presented (Figure 5A). These genomic subgroups are not entirely mutually exclusive, but are based on their priority as molecular targets (Figure 5B). As we have described above, poor prognostic genomic features are enriched in younger patients: present in 72% of patients aged <35 years, 54% in patients aged 35–39 years, and 40% in patients aged ≥40 years (Figure 5C).
Figure 5. Framework for genomic subgrouping of premenopausal HRDHER2− breast cancer.
(A) Proposed framework for genomic subgrouping for premenopausal patients with HR+HER2− early breast cancers, and number of patients with each feature in the SOFT combined sequencing cohort (n = 1276). (B) Venn diagram demonstrating the number and proportion of tumours assigned to each poor prognosis genomic subgroup in the SOFT combined sequencing cohort (n = 1276) using the proposed framework. (C) Pie charts demonstrating the frequencies of the proposed genomic subgroups according to age at randomisation in the SOFT combined sequencing cohort (n = 1276). (D) Kaplan–Meier plot estimating the rate of freedom from distant recurrence according to the proposed genomic subgroups in the SOFT combined sequencing cohort (n = 1276). (E) Kaplan–Meier plot estimating the overall survival according to the proposed genomic subgroups in the SOFT combined sequencing cohort (n = 1276).
CI, confidence interval; CNA, copy number amplification; ER, estrogen receptor; HER2−, human epidermal growth factor receptor 2-negative; HR, hazard ratio; HR+, hormone receptor-positive; HRD, homologous recombination deficiency; SOFT, Suppression of Ovarian Function Trial.
Collectively, these poor prognostic genomic features are associated with a higher risk of distant recurrence (HR 1.85, 95% CI 1.27–2.69; 8-year rate of freedom from distant recurrence 84% versus 94%) and poorer OS (HR 2.20, 95% CI 1.40–3.48; 8-year OS 88% versus 96%) when compared with those with no poor prognostic features, suggesting they may define breast cancer subsets with aggressive disease biology resistant to standard adjuvant treatments (Figure 5D and E). The genomic subgrouping also provided significant additional prognostic information after adjustment by luminal-like status, age, and other prognostic factors for both DRFI (P = 0.011) and OS (P = 0.016) (Supplementary Table S7 and Figures S19–S21, available at https://doi.org/10.1016/j.annonc.2023.01.009 https://doi.org/10.1016/j.annonc.2023.01.009). Similar prognostic associations were observed in the BIG 1–98 postmenopausal population (Supplementary Figure S22, available at https://doi.org/10.1016/j.annonc.2023.01.009). By contrast, the subgroup with no poor prognostic features (n = 692, 53%) demonstrated excellent 8-year rates of freedom from distant recurrence (94%, 95% CI 92% to 96%) and OS (96%, 95% CI 94% to 98%), even in the luminal B phenotype. Finally, as an exploratory analysis, we investigated for OFS treatment interactions between the different genomic subgroups for risk of distant recurrence. Whilst numbers in each subgroup are too small for significant interactions, the point estimates for the HRD subgroup did not suggest additional benefit from the addition of OFS, hence the need to investigate other therapeutic approaches (Supplementary Figure S23, available at https://doi.org/10.1016/j.annonc.2023.01.009).
DISCUSSION
The results from this dataset highlight the genomic alterations in HR+HER2− EBCs arising in premenopausal women, focusing on women aged <40 years. We have demonstrated age-related differences in genomic profiles with enrichment of genomic features associated with poor prognosis in these younger premenopausal women compared with older premenopausal and postmenopausal women. We validated the prognostic associations of previously described CNA drivers described in IntClusts,19,20 and demonstrate that poor prognosis IntClusts are enriched in patients aged <40 years. Furthermore, we have identified a number of possible therapeutic targets that are enriched in patients aged <40 years, highlighting the potential for age-focussed treatment strategies. The strength of our work is the use of a unique patient dataset, with tumour samples from those enrolled in a landmark clinical trial of premenopausal women, which has well-annotated clinical data and long and accurate survival follow-up. To our knowledge this is the largest such cohort of young, premenopausal women with HR+HER2− EBC. Crucially, these findings are in the context of patients who have received contemporary and standard-of-care treatments including OFS.
Genomic features of HRD, present in up to 36% in the very young (age <35 years), and 23% of the combined sequencing cohort, present as a potential molecular target given drugs affecting this pathway are already well established. It should be noted that patients with germline BRCA1 or BRCA2 mutations may be underrepresented in the SOFT clinical trial as patients who had already had bilateral oophorectomy or were planned oophorectomy within 5 years were excluded. Notably, from the 21 patients with eHRDetect-positive tumours, only 4 had biallelic BRCA2 alterations. Our data support that broader genomic strategies for detecting features of HRD or testing for genomic alterations in HRD-related genes other than solely germline BRCA1 and BRCA2 mutation may have clinical utility in this setting. We acknowledge that HRDetect is likely to be more accurate using a whole-genome sequencing approach as this provides a greater number of mutations and improved clarity on structural genomic alterations for down-stream analysis (rather than WES). Prediction of sensitivity to molecularly targeted approaches with PARP inhibitors in these tumours still remains an unanswered important clinical question. In support of this notion, the recently reported GeparOLA randomised clinical trial demonstrated strikingly high pathological complete response rates in patients treated with the combination of chemotherapy with olaparib in HR+, HRD-positive tumours (52.6%), as well as in patients aged <40 years (76.2%).34 Given the paucity of data demonstrating PARP inhibitor monotherapy efficacy beyond patients with germline BRCA1 and BRCA2 mutations, combination strategies with PARP inhibitors (e.g. with chemotherapy) seem to be required.
We also identified a subgroup of PIK3CA-mutated tumours with CNAs that were associated with a higher risk of distant recurrence enriched in the very young. Notably, this included CNAs in recurrent amplicons rather than PIK3CA itself. A recent report has additionally highlighted that PIK3CA-mutated tumours harbouring a concurrent PIK3CA gain are also associated with significantly worse disease outcomes;35 however, in our cohort of EBCs, PIK3CA mutations with concurrent PIK3CA gain were rare (0.5%). PI3-kinase inhibitors have recently demonstrated significant clinical efficacy in the metastatic setting;36 however, further biomarkers beyond the presence of PIK3CA mutations are needed. PIK3CA mutations in HR+ breast cancer are reported to have heterogeneous clinical outcomes, with recent data suggesting a possible biological reason for this in that the number of mutant alleles was found to be important in mediating increased pathway activation.37 Given the poor prognosis associated with CN-amplified, PIK3CA-mutated tumours that was observed in our dataset, this subgroup may have the potential to benefit from escalation of therapeutic strategies that include PI3-kinase or AKT inhibitors in the early-stage setting as they could have higher levels of pathway activation. This hypothesis could be further validated experimentally.
We note here some limitations to our study. The majority of tumours underwent deep targeted sequencing capturing known breast cancer genes, with a subsample of tumours undergoing WES. Thus, we believe whole-genome sequencing in a larger cohort of younger women with HR+HER2− EBC will be important for validation of our findings, but was not possible for this dataset due to the age of our archival tumour samples. Mutational signatures derived from whole-genome sequencing could also offer a means to study aetiology and pathophysiology of the disease arising in the very young. In addition to this, combination with gene expression data would allow for even more refinement of our data: for example, incorporation of prognostic gene assays, more accurate calling of previously described IntClusts, as well as elucidating transcriptional PI3K pathway activation in PIK3CA-mutant tumours according to CN status. We recognise that age is a continuum; however, we have used a 40-year age cut-off as our main threshold. Whilst this is largely arbitrary, both age cut-offs of 40 years and 35 years have been shown many times to be associated with poorer outcomes in this breast cancer subtype. Finally, we present genomic subgroupings not as a definitive stratification, but rather as a proposal for a genomic-based prognostic algorithm for use in future clinical trials using an age-focussed approach. We believe this strategy could be refined for use in the clinic with a single, optimised assay.
The recently reported RxPONDER trial demonstrated a significant benefit for adjuvant chemotherapy in early-stage, HR+, node-positive breast cancer (1–3 nodes positive) in premenopausal women, but not in postmenopausal women, further supporting the notion that HR+ breast cancer may have different biological characteristics depending on age at diagnosis.38 Our study demonstrates key genomic features that are particularly enriched with younger age rather than menopausal status per se. We propose that prospective trials in young women with HR+HER2− EBC addressing these specific molecular pathways will be pivotal to improving their clinical outcomes.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to the women, physicians, nurses, trial coordinators, and data managers who participated in the SOFT clinical trial and the many pathologists who submitted tumour and normal tissue blocks for research purposes; to the International Breast Cancer Study Group (IBCSG) for the design of the trial, coordination, data management, medical review, and statistical support; and to the IBCSG Central Pathology Office for tumour block collection and processing. The trials were coordinated by the International Breast Cancer Study Group (IBCSG), in collaboration with the Breast International Group (BIG), BIG cooperative groups, and United States National Cancer Institute National Clinical Trials Network cooperative groups. SJL was supported by the University of Melbourne (2016–2019). SL is supported by Breast Cancer Research Foundation (BCRF) NY, the John Colebatch Cancer Council Victoria Clinical fellowship (2014–2018), and the National Breast Cancer Foundation (NBCF) of Australia.
FUNDING
This work was supported by a Susan G. Komen for the Cure Promise Grant [grant number CCR14299387 to SL], the National Health and Research Council (NHMRC) Australia [grant number 1084531], the BCRF, NBCF Australia, and with the support from the family of Judy Eisman in Australia. The SOFT clinical trial receives financial support for trial conduct from Pfizer, the International Breast Cancer Study Group, the United States National Cancer Institute, the Breast Cancer Research Foundation (16–185, 17–187, 18–003, 19–011, 20–011), Ipsen, TerSera, and AstraZeneca. Pfizer and Ipsen provided drug supply. Support for central pathology included Susan G. Komen for the Cure Promise Grant [grant number KG080081 to GV, MMR] and the Breast Cancer Research Foundation. Support for the coordinating group, IBCSG: Frontier Science and Technology Research Foundation, Swiss Group for Clinical Cancer Research (SAKK), Cancer Research Switzerland, Oncosuisse, Cancer League Switzerland, Foundation for Clinical Cancer Research of Eastern Switzerland (OSKK), United States National Cancer Institute [United States NIH grant number CA075362]. Grant support of cooperative groups: Breast Cancer Trials Australia & New Zealand [NHMRC grant numbers 351161, 510788, 1105058]; Institute of Cancer Research Clinical Trials and Statistics Unit (ICR-CTSU) on behalf of the National Cancer Research Institute Breast Clinical Studies Group United Kingdom (NCRI-BCSG–ICR-CTSU Partnership) [Cancer Research UK grant numbers CRUKE/03/022, CRUKE/03/023, A15955]; National Institute for Health Research Royal Marsden/Institute of Cancer Research Biomedical Research Centre; and National Institute for Health Research/Cambridge Biomedical Research Centre; Alliance for Clinical Trials in Oncology [United States NIH grant number U10CA180821]; SWOG [United States NIH grant number U10CA180888]; ECOG-ACRIN Cancer Research Group [United States NIH grant number U10CACA180820, UG1CA233328]; NRG Oncology [United States NIH grant numbers U10CA180868, U10CA180822, UG1CA189867]; Canadian Cancer Trials Group [United States NIH grant number U10CA180863]; and Canadian Cancer Society [grant number 704970].
DISCLOSURE
GV has received honoraria (outside of this submitted study) from MSD Oncology, Roche, Pfizer, Novartis, Bayer, Daiichi Sankyo, and Dako Agilent. SNZ declares multiple patents on mutational signature-based algorithms including HRDetect. PS has acted as an uncompensated consultant for Roche-Genentech. EC declares consulting or advisory role for Lilly, Novartis, MSD, AstraZeneca, Pfizer, Roche; speakers’ bureau for Lilly, Roche, Pfizer; travel accommodation and expenses from Pfizer and Roche. PK has research contracts with PFS genomics and Prelude DX. MCl declares honoraria from BMS, Astellas, Janssen, MSD, Sanofi, Bayer, Roche, Pfizer, Novartis, Ipsen; consultation for BMS, MSD, Bayer, EUNSA, Pfizer, Roche, Janssen, Pierre Fabre, Ipsen; travel funding from Janssen, Astellas, Roche, Ipsen, MSD. CG declares travel expenses from Genentech, Roche, Daiichi-Sankyo, AstraZeneca; medical writing assistance from Roche and Abbvie; uncompensated advisory boards with Genentech, Roche, Daiichi-Sankyo, Seattle Genetics; compensated advisory boards with Exact Sciences; uncompensated consulting with Daiichi-Sankyo; and compensated consulting with Athenex. RC has received honoraria (outside of this study) from Amgen, Astra Zeneca, ITM, Novartis, and Scancell. BT declares stocks with Novartis; consultation fees from Eli Lilly and AstraZeneca. MCo declares research funding from Roche. PF declares travel funding from Novartis, Ipsen. MMR declares institutional research funding and/or provision of drug supply for clinical trials from Novartis, Pfizer, AstraZeneca, Roche, TerSera, Ipsen; institutional research funding from Bayer, Bristol-Myers Squibb; institutional advisory role from Ipsen; advisory role and honoraria from Bristol-Myers Squibb, Tolmar. SL receives research funding to her institution from Novartis, BMS, Merck, Roche-Genentech, Puma Biotechnology and Pfizer; consultant (not compensated) for Seattle Genetics, Pfizer, Novartis, BMS, Merck, and Roche-Genentech.
Footnotes
DISCLOSURE
All other authors have declared no conflicts of interest.
DATA SHARING
The datasets generated during and/or analysed during the current study are available on controlled access. Applications are reviewed and are approved by the IBCSG. Data access will be provided on approval to any party able to comply with the necessary agreements. This process is to comply with the ethics, legal, and data privacy obligations approved by the site ethical committees. To request access to data, contact the IBCSG Statistical Center (stat_center@ibcsg.org).
REFERENCES
- 1.Azim HA Jr, Michiels S, Bedard PL, et al. Elucidating prognosis and biology of breast cancer arising in young women using gene expression profiling. Clin Cancer Res. 2012;18(5):1341–1351. [DOI] [PubMed] [Google Scholar]
- 2.Partridge AH, Hughes ME, Warner ET, et al. Subtype-dependent relationship between young age at diagnosis and breast cancer survival. J Clin Oncol. 2016;34(27):3308–3314. [DOI] [PubMed] [Google Scholar]
- 3.Francis PA, Regan MM, Fleming GF, et al. Adjuvant ovarian suppression in premenopausal breast cancer. N Engl J Med. 2015;372(5):436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis PA, Pagani O, Fleming GF, et al. Tailoring adjuvant endocrine therapy for premenopausal breast cancer. N Engl J Med. 2018;379:122–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis PA, Fleming GF, Lang I, et al. Adjuvant endocrine therapy in premenopausal breast cancer: 12-year results from SOFT. J Clin Oncol. 2022. 10.1200/JCO.22.01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saha P, Regan MM, Pagani O, et al. Treatment efficacy, adherence, and quality of life among women younger than 35 years in the International Breast Cancer Study Group TEXT and SOFT adjuvant endocrine therapy trials. J Clin Oncol. 2017;35:3113–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villarreal-Garza C, Ferrigno AS, De la Garza-Ramos C, Barragan-Carrillo R, Lambertini M, Azim HA Jr. Clinical utility of genomic signatures in young breast cancer patients: a systematic review. NPJ Breast Cancer. 2020;6:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azim HA Jr, Nguyen B, Brohee S, Zoppoli G, Sotiriou C. Genomic aberrations in young and elderly breast cancer patients. BMC Med. 2015;13:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copson ER, Maishman TC, Tapper WJ, et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): a prospective cohort study. Lancet Oncol. 2018;19(2):169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waks AG, Kim D, Jain E, et al. Somatic and germline genomic alterations in very young women with breast cancer. Clin Cancer Res. 2022;28:2339–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Geelen CT, Savas P, Teo ZL, et al. Clinical implications of prospective genomic profiling of metastatic breast cancer patients. Breast Cancer Res. 2020;22(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23(4):517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luen SJ, Asher R, Lee CK, et al. Association of somatic driver alterations with prognosis in postmenopausal, hormone receptor-positive, HER2−negative early breast cancer: a secondary analysis of the BIG 1–98 randomized clinical trial. JAMA Oncol. 2018;4:1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24(9):2206–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regan MM, Pagani O, Francis PA, et al. Predictive value and clinical utility of centrally assessed ER, PgR, and Ki-67 to select adjuvant endocrine therapy for premenopausal women with hormone receptor-positive, HER2−-negative early breast cancer: TEXT and SOFT trials. Breast Cancer Res Treat. 2015;154(2):275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson DR,Wu YM,Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasan N, Razavi P, Johnson JL, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Ka inhibitors. Science. 2019;366(6466):714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rueda OM, Sammut SJ, Seoane JA, et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature. 2019;567(7748):399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zardavas D, Te Marvelde L, Milne RL, et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J Clin Oncol. 2018;36(10):981–990. [DOI] [PubMed] [Google Scholar]
- 23.Dawson SJ, Rueda OM, Aparicio S, Caldas C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 2013;32(5):617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pagani O, Francis PA, Fleming GF, et al. Absolute improvements in freedom from distant recurrence to tailor adjuvant endocrine therapies for premenopausal women: results from TEXT and SOFT. J Clin Oncol. 2020;38(12):1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HJ, Kim S, Freedman RA, Partridge AH. The impact of young age at diagnosis (age <40 years) on prognosis varies by breast cancer subtype: a U.S. SEER database analysis. Breast. 2021;61:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caldon CE. Estrogen signaling and the DNA damage response in hormone dependent breast cancers. Front Oncol. 2014;4:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA-mutation. N Engl J Med.2017;377:523–533. [DOI] [PubMed] [Google Scholar]
- 29.Litton JK, Rugo HS, Ettl J, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tutt A, Tovey H, Cheang MCU, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. 2018;24(5):628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulhan DC, Lee JJ, Melloni GEM, Cortés-Ciriano I, Park PJ. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat Genet. 2019;51(5):912–919. [DOI] [PubMed] [Google Scholar]
- 32.Bertucci F, Ng CKY, Patsouris A, et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569(7757):560–564. [DOI] [PubMed] [Google Scholar]
- 33.Pereira B, Chin SF, Rueda OM, et al. The somatic mutation profiles of 2, 433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fasching PA, Link T, Hauke J, et al. Neoadjuvant paclitaxel/olaparib in comparison to paclitaxel/carboplatinum in patients with HER2−negative breast cancer and homologous recombination deficiency (GeparOLA study). Ann Oncol. 2021;32(1):49–57. [DOI] [PubMed] [Google Scholar]
- 35.Migliaccio I, Paoli M, Risi E, et al. PIK3CA co-occurring mutations and copy-number gain in hormone receptor positive and HER2− negative breast cancer. NPJ Breast Cancer. 2022;8(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andre F, Ciruelos E, Rubovszky G, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380(20):1929–1940. [DOI] [PubMed] [Google Scholar]
- 37.Madsen RR, Erickson EC, Rueda OM, et al. Positive correlation between transcriptomic stemness and PI3K/AKT/mTOR signaling scores in breast cancer, and a counterintuitive relationship with PIK3CA genotype. PLoS Genet. 2021;17(11):e1009876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalinsky K, Barlow WE, Meric-Bernstam F, et al. First results from a phase III randomized clinical trial of standard adjuvant endocrine therapy (ET) +/− chemotherapy (CT) in patients (pts) with 1–3 positive nodes, hormone receptor-positive (HR+) and HER2−-negative (HER2−) breast cancer (BC) with recurrence score (RS) <25: SWOG S1007 (RxPonder). Paper presented at the 2020 San Antonio Breast Cancer Symposium. December 8–11, 2020; San Antonio, Texas. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available on controlled access. Applications are reviewed and are approved by the IBCSG. Data access will be provided on approval to any party able to comply with the necessary agreements. This process is to comply with the ethics, legal, and data privacy obligations approved by the site ethical committees. To request access to data, contact the IBCSG Statistical Center (stat_center@ibcsg.org).