Abstract
Coexpression of di-α-globin and β-globin in Escherichia coli in the presence of exogenous heme yielded high levels of soluble, functional recombinant human hemoglobin (rHb1.1). High-level expression of rHb1.1 provides a good model for measuring mistranslation in heterologous proteins. rHb1.1 does not contain isoleucine; therefore, any isoleucine present could be attributed to mistranslation, most likely mistranslation of one or more of the 200 codons that differ from an isoleucine codon by 1 bp. Sensitive amino acid analysis of highly purified rHb1.1 typically revealed ≤0.2 mol of isoleucine per mol of hemoglobin. This corresponds to a translation error rate of ≤0.001, which is not different from typical translation error rates found for E. coli proteins. Two different expression systems that resulted in accumulation of globin proteins to levels equivalent to ∼20% of the level of E. coli soluble proteins also resulted in equivalent translational fidelity.
Protein synthesis in living organisms, the ultimate step in biological information transfer, is also the most error-prone step. In Escherichia coli, 2 to 20 translation errors typically occur in every 10,000 codons translated (15). Errors can occur through incorrect charging of tRNAs (misaminoacylation), incorrect pairing of noncognate tRNAs with codons (missense errors), frameshifting and drop-offs (processivity errors), misreading of sense codons as terminators (false termination), or misreading of terminators as sense codons (termination readthrough) (13; reviewed in references 2, 5, 8, and 15). Growth conditions, particularly starvation for one or more amino acids, can significantly increase the translation error rates (15, 16). The rates at which many of these errors occur are also exacerbated by the presence of codons whose cognate tRNAs are rare in E. coli (reviewed in reference 6). For this reason, most heterologous proteins are overexpressed in E. coli from genes whose codon bias has been altered to conform to that typical of E. coli (6).
Missense errors resulting from incorrect pairing of noncognate tRNAs with mRNA codons are the most frequent translation errors under general growth conditions (8). These errors are attributed to recognition of a codon by a tRNA containing a less-than-perfect anticodon nucleotide match, as described by the wobble hypothesis and the two-out-of-three hypothesis (reviewed in reference 2). Missense errors have been measured at specific sites, including in recombinant methionyl human granulocyte colony-stimulating factor, which contained Gln substitutions for His (12), or in general throughout the protein, including in mouse epidermal growth factor (mEGF) (18). The general missense errors were measured by analyzing mEGF for the presence of phenylalanine not encoded by the gene (18). Radiolabeled Phe and Leu were incorporated into mEGF, and the ratio of these amino acids indicated that there was a missense error rate of 2 × 10−2. This translation error rate is 1 to 2 orders of magnitude higher than the rates typical for E. coli protein synthesis. However, no other studies of missense errors in intact overexpressed proteins have confirmed that this is a general phenomenon for highly expressed heterologous proteins in E. coli.
Functional recombinant human hemoglobin (rHb1.1) is produced in E. coli as a heterotrimer when there is concomitant expression from a plasmid-borne operon of di-α and β subunits and exogenous heme is provided (10). Di-α-globin is a pair of α-globin molecules that are genetically linked to prevent αβ dimerization and thereby reduce renal filtration and extend the intravascular half-life (11). Low-level mistranslation of rHb1.1 by misaminoacylation resulting in norvaline substitution for leucine has been observed (1). In this study, we examined the missense error rate by a technique which we call absent amino acid analysis (AAAA). Using this technique, we detected trace amounts of isoleucine, which is not encoded by the rHb1.1 genes. Therefore, the isoleucine content of the purified hemoglobin reflected a specific class of missense errors. The study described here had two advantages over previous studies: (i) very highly purified protein samples were used, so contributions of contaminants were negligible; and (ii) a sensitive direct method was used to measure traces of isoleucine by AAAA rather than radiolabeling.
MATERIALS AND METHODS
Hemoglobin expression and purification.
E. coli SGE1662 (19) contains pSGE705, a medium-copy-number pBR-based plasmid with di-alpha- and beta-globin genes in which most codons conform to the E. coli bias. These genes are in an operon whose transcription is dependent upon the tac promoter (11) and express rHb1.1. A similar high-copy-number plasmid containing the pUC origin of replication was used to express rHb1.1 in SGE1464 (19).
Fermentations were performed at 28 to 30°C in defined medium in 15-, 600-, and 1,500-liter fermentors generally as described by Looker et al. (10) and Weickert et al. (20). Expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a concentration of either 55 μM (SGE1662) or 100 μM (SGE1464). IPTG was added when the optical density at 600 nm was approximately one-third the expected final cell density. Incubation was continued for 10 to 16 h postinduction, and hemin was added to a concentration of ∼0.2 g/liter (SGE1662) or ∼0.4 g/liter (SGE1464). One-milliliter samples were withdrawn, placed in 1.7-ml Eppendorf tubes, and centrifuged for 3 min, and the supernatants were removed. The pellets were stored at −80°C until they were assayed for soluble and/or insoluble rHb1.1, as described by Weickert and Curry (19) and Weickert et al. (20). rHb1.1 was purified as described by Plomer et al. (17). The functionality of the purified rHb1.1 samples was determined as described by Hoffman et al. (4). Purified protein was characterized by trypsin mapping performed as described by Lippincott et al. (9), and a liquid chromatography-mass spectrometry analysis was performed as described by Apostol et al. (1).
Measurement of E. coli proteins.
The amounts of E. coli protein contaminants in purified rHb1.1 samples were determined by a proprietary enzyme-linked immunosorbent assay (ELISA) by using antibodies developed at Somatogen. After identical fermentation of an equivalent strain containing plasmids from which the rHb1.1 genes were removed, E. coli proteins were recovered by purification of a lysate. This produced antibodies that were more sensitive than commercial anti-E. coli protein reagents and that could be used to detect protein contaminants in an rHb1.1 sample which were present at levels too low to be visualized by gel electrophoresis.
Amino acid analysis.
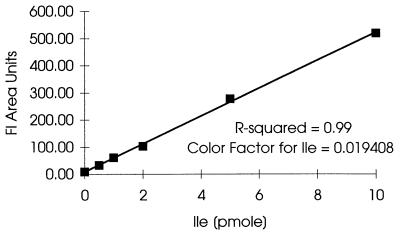
Purified rHb1.1 samples were subjected to gas phase hydrolysis at 165°C for 1.5 h in the presence of 6 N HCl containing 0.1% phenol with a Savant AminoPrep model AP100 hydrolyzer. Levels of norvaline substitution were determined by a modified ortho-phthaldehyde method as described in Apostol et al. (1). Other amino acids were analyzed by precolumn derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC). The standard AccQ-Tag method was used to derivatize amino acids. AQC derivatives of amino acids were separated on a Zorbax XDB C8 column (2 by 150 mm) by using a model HP 1090IIM chromatograph. Separation was monitored by examining fluorescent signals (excitation at 254 nm, emission at 395 nm) and UV signals (254 nm). The UV signal was used to determine the concentration of all amino acids except Ile, and the amplified fluorescent signal was used to determine the isoleucine content (Fig. 1). One level of calibration at 125 pmol/injection was used for the UV signal. Solvent A was diluted AccQ-Tag eluant A from Waters (1,000 ml of water, 85 ml of concentrated eluant A). Solvent B was 48% acetonitrile–12% isopropanol in high-performance liquid chromatography grade water. Separation was accomplished by using the following gradient conditions: 1% solvent B at zero time, 4% solvent B at 1 min, 20% solvent B at 16 min, 37% solvent B at 40 min, 100% solvent B from 42 to 46 min, and no solvent B at 47 min. A hydrolysate of hemoglobin from sample 79 (Table 1) was used to calibrate the assay and to determine the degree to which low levels of isoleucine added to the sample were recovered. This hydrolysate was used as the matrix, and different levels of amino acid standards, including Ile, were added. The areas under Ile peaks (fluorescent signal) were plotted versus the amounts of Ile added (Fig. 2). The levels of recovery of small amounts of added Ile (0.5 and 1.0 pmol) exceeded 100% (130 and 118%, respectively) because of the small amount of Ile already present in the hydrolysate. This small amount of native isoleucine was not significant when the larger amounts (2.0, 5.0, and 10.0 pmol) of Ile were added; under these conditions the average level of recovery was 102%. The area under the Ile peak was proportional to the amount added, with an R2 value of 0.9987 and a P value of 1.1 × 10−6. The inverse of the slope represented the color factor (extinction coefficient) used to determine low levels of isoleucine in a hemoglobin matrix. The level of Ile in an unknown sample was calculated by multiplying the area under the Ile peak by the color factor.
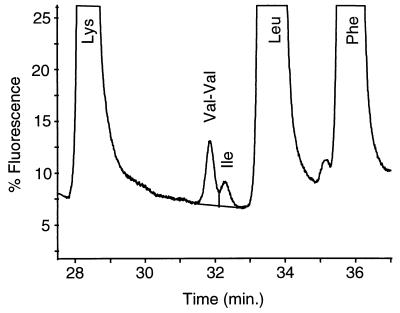
FIG. 1.
Magnified portion of chromatogram from an amino acid analysis (sample 94-3A) (Table 2), showing resolution of Ile and Val-Val dipeptide. The lines used to establish a baseline and to separate the Val-Val and Ile peaks in order to enable peak integration and Ile quantitation are shown.
TABLE 1.
Attributes of purified rHb1.1 samplesa
| Sample | rHb1.1 functionality
|
E. coli protein concn (ppm)b | mol of norvaline/mol of rHb1.1 | Overall yield (%) | |
|---|---|---|---|---|---|
| P50 | Hill max | ||||
| 74 | 28.8 | 2.4 | <0.16 | 0.10 | 31 |
| 75 | 29.4 | 2.4 | 1.30 | <0.04 | 20 |
| 76 | 28.7 | 2.4 | 0.42 | 0.35 | 35 |
| 77 | 31.1 | 2.4 | 0.39 | 0.09 | 27 |
| 78 | 31.6 | 2.5 | <0.16 | 0.19 | 35 |
| 79 | 30.7 | 2.4 | <0.16 | 0.20 | 36 |
| 80 | 31.8 | 2.5 | <0.16 | 0.32 | 33 |
| 81 | 31.6 | 2.4 | 0.20 | 0.08 | 31 |
| 94 | 30.2 | 2.5 | <0.16 | 0.36 | 20 |
The RSDs for P50, Hill max, E. coli protein concentration, and proportion of norvaline were 2.3, 2.1, 25, and 8.9%, respectively.
As determined by the ELISA.
FIG. 2.
Calibration curve for Ile. Hydrolysate of sample 79 was used as the matrix, and different levels of amino acid standards, including Ile, were added. Areas under the Ile peak were plotted versus the amount of added Ile. The inverse of the slope represents the color factor for a low level of isoleucine in a hemoglobin matrix (R-squared = 0.9987 and P value = 1.1 × 10−6). Fl, fluorescence.
Sample 94 (Table 1) was used as a reference sample for determining the precision and consistency of the Ile analyses. Aliquots of sample 94 were hydrolyzed and analyzed multiple times. The molar content of Ile in hemoglobin was calculated by using the three most stable amino acids, Leu, Val, and Ala, as the references. The value reported for each experiment was the mean number of moles of Ile per mole of rHb1.1 calculated by using these three amino acids. On average, 0.102 mol of Ile/mol of rHb1.1 was found in sample 94, and the standard deviation was 0.036 mol (relative standard deviation [RSD], 35%) (Table 2). In low-level amino acid analyses factors such as foreign particles, cross contamination, traces of glove powder, etc. may contribute to relatively large errors.
TABLE 2.
Consistency and precision of the Ile assay
| Sample | Amt (pmol) of amino acids as determined by AAAAa
|
Proportions of Ile compared with other amino acids
|
||||||
|---|---|---|---|---|---|---|---|---|
| Ile | Leu | Val | Ala | Ile/Leub | Ile/Valc | Ile/Alad | Avge | |
| 94.1A | 0.775 | 441.72 | 366.64 | 446.37 | 0.126 | 0.125 | 0.125 | 0.125 |
| 94-1B | 0.969 | 433.16 | 357.89 | 434.71 | 0.161 | 0.160 | 0.161 | 0.160 |
| 94-1C | 0.527 | 431.99 | 358.57 | 435.29 | 0.088 | 0.087 | 0.087 | 0.087 |
| 94-2A | 0.407 | 426.67 | 357.40 | 436.13 | 0.069 | 0.067 | 0.067 | 0.068 |
| 94-3A | 0.633 | 452.58 | 374.31 | 446.11 | 0.101 | 0.100 | 0.102 | 0.101 |
| 94-3B | 0.400 | 411.50 | 344.01 | 418.50 | 0.070 | 0.069 | 0.069 | 0.069 |
Each rHb1.1 molecule contains 72 leucine residues, 59 valine residues, and 72 alanine residues.
Calculated as follows: (picomoles of Ile/picomoles of Leu) × 72.
Calculated as follows: (picomoles of Ile/picomoles of Val) × 59.
Calculated as follows: (picomoles of Ile/picomoles of Ala) × 72.
The overall average ± standard deviation was 0.102 ± 0.036, and the RSD was 35%.
RESULTS
Expression and purification of rHb1.1.
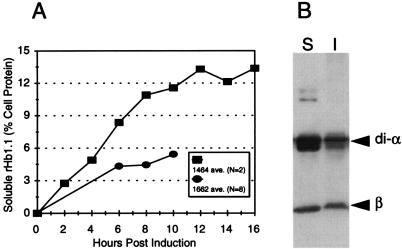
rHb1.1 was produced in E. coli with medium-copy-number (SGE1662) and high-copy-number (SGE1464) plasmids containing the di-alpha-globin and beta-globin genes in an operon under control of the tac promoter (4, 10, 19, 20). In eight fermentations of SGE1662 approximately 5.4% of the soluble E. coli protein was soluble rHb1.1 (Fig. 3A). In two fermentations of SGE1464 the average level of soluble rHb1.1 accumulated was 13.4% of the soluble E. coli proteins (Fig. 3A). Western blot assays of the insoluble and soluble fractions of recombinant hemoglobins from SGE1464 lysates indicated that approximately one-third (37% ± 3%, as determined by densitometry scanning) of the globins were insoluble (Fig. 3B). In SGE1464, this corresponded to a maximum accumulation of total hemoglobin equivalent to approximately 20% of the E. coli proteins, which is consistent with previous observations (20).
FIG. 3.
rHb1.1 expression. (A) Accumulation of soluble rHb1.1 during fermentation in E. coli SGE1662 and SGE1464. N is the number of fermentation preparations from which samples were taken. Except for the last time point, most but not all preparations were assayed for each time point. (B) Western blot of soluble (lane S) and insoluble (lane I) globin from an SGE1464 fermentation cell lysate.
Soluble rHb1.1 was recovered and rigorously purified and the final yield was 20 to 36% of the starting material (Table 1). The level of E. coli proteins was measured by the ELISA, and in the majority of samples the levels of E. coli proteins were below the level of detection (<0.16 ppm relative to rHb1.1) (Table 1). The presence of very pure rHb1.1 was confirmed by trypsin mapping and reversed-phase chromatography along with mass spectrometry (9). The functionality of the purified rHb1.1 was also assessed, and the values for all samples were within the expected range of values for oxygen affinity and cooperativity (Table 1). Rigorous purification of rHb1.1 allowed us to assess the amino acid composition without worrying about potential contamination of the signal by E. coli proteins (Table 1).
Measurement of the isoleucine content of rHb1.1.
An analysis to determine the absent amino acid was the analytical method used to determine the isoleucine content in several samples of hemoglobin produced by two different strains (Table 3). Highly purified rHb1.1 samples were subjected to hydrolysis followed by amino acid analysis as described above. AQC derivatives of amino acids were separated on a reversed-phase column, and the concentrations of all amino acids except isoleucine were determined from the UV signal. The highly amplified fluorescent signal was used only to determine isoleucine content (Fig. 1). Special care was taken to resolve the residual Val-Val dipeptide from Ile to avoid interference. The Val-Val dipeptide results from incomplete hydrolysis of the peptide bond (3), which occurs twice in β-globin.
TABLE 3.
Isoleucine contents of different lots of rHb1.1 and estimated translation errors
| Sample | Strain | mol of Ile/mol of rHb1.1 | Translation error rate |
|---|---|---|---|
| 44 | SGE1662 | BLOQa | ≤0.0002 |
| 46 | SGE1662 | BLOQ | ≤0.0002 |
| 47 | SGE1662 | BLOQ | ≤0.0002 |
| 51 | SGE1662 | 0.18 | 0.0009 |
| 74 | SGE1662 | 0.17 | 0.0008 |
| 75 | SGE1662 | 0.20 | 0.0010 |
| 76 | SGE1662 | 0.15 | 0.0007 |
| 77 | SGE1662 | 0.18 | 0.0009 |
| 78 | SGE1662 | BLOQ | ≤0.0002 |
| 79 | SGE1662 | BLOQ | ≤0.0002 |
| 80 | SGE1662 | 0.09 | 0.0004 |
| 81 | SGE1662 | 0.07 | 0.0004 |
| 94 | SGE1662 | 0.18 | 0.0010 |
| 151 | SGE1464 | 0.05 | ≤0.0002 |
| 601 | SGE1464 | 0.22 | 0.0011 |
BLOQ, below limit of quantitation (<0.05 mol of Ile/mol of rHb1.1).
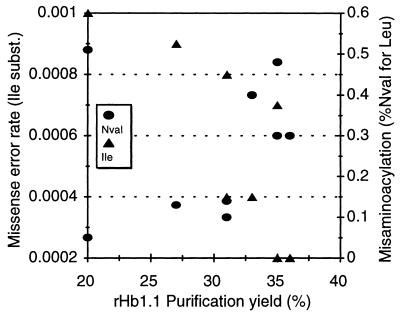
The number of moles of isoleucine in 1 mol of rHb1.1 was divided by the number of codons in rHb1.1 that differed from the Ile codons by one base to calculate the frequency of this class of mistranslation error. A total of 200 of the 575 codons in the sequence of recombinant hemoglobin met this criterion. Three of these codons encode initiating methionines (one for di-α subunits of rHb1.1, two for β subunits of rHb1.1), and they typically could be excluded, except that initiation of heterologous protein translation by isoleucine has been observed in E. coli (7). Exclusion of these three amino acid codons should have had little effect on the calculated rate of translation error. The calculated translation error rate did not exceeded 0.0011, and one-third of the samples had a translation error rate of ≤0.0002 (Table 2). It was estimated that the lower limit of quantitation for Ile determinations was 0.05 mol of Ile per mol of rHb1.1, which corresponds to a translation error rate of 0.0002. This is equivalent to two translation errors in every 10,000 translations of rHb1.1 codons that differ from isoleucine codons by one nucleotide. The observed levels of translation error did not correlate with hemoglobin functionality measurements for oxygen affinity (P50) and cooperativity (Hill max) (Tables 1 and 3). There was a considerable distribution of error rates among the various purification yields (Fig. 4).
FIG. 4.
Relationship between purification yield and amino acid misincorporation. Misaminoacylation produced norvaline (Nval), shown as the percentage of norvaline substituted for leucine, and missense substitutions (subst.) introduced isoleucine, shown as the error rate per codon translated.
Measurement of the norvaline content of rHb1.1.
Another type of mistranslation has been observed in rHb1.1; this type of mistranslation, substitution of norvaline for leucine, arises by misaminoacylation and depends on conditions during cell growth (1). Norvaline substitutions in rHb1.1 occur late in fermentations after most cell protein mass has already accumulated (1); thus, norvaline is not typically found in E. coli proteins. Highly purified rHb1.1 samples were analyzed for the frequency of this substitution. The level of norvaline measured in purified rHb1.1 varied by at least 1 order of magnitude (Table 1). The frequency of this substitution was not correlated with the purification yield, with the level of isoleucine misincorporation (Fig. 4), or with hemoglobin functionality (P50, Hill max) (Table 1).
Another advantage of examining norvaline substitution is that since norvaline is not typically found in E. coli proteins, contaminants should not interfere with measurements of norvaline in rHb1.1 in crude samples. We studied norvaline substitution throughout purification to see if bias was introduced into the final pure sample by removal of the substituted population. Several additional fermentations were performed under conditions likely to generate relatively high levels of norvaline substitution (1). The norvaline substitution levels in crude rHb1.1 from two early purification steps were compared to norvaline levels in final, highly purified rHb1.1 (Table 4). Purification did not significantly influence the quantity of norvaline in the samples. The norvaline level in highly purified rHb1.1 was 104% ± 15% of the level found in crude rHb1.1 pools.
TABLE 4.
Norvaline levels during rHb1.1 purification
| Prepn | mol of norvaline/mol of rHb1.1 ina:
|
||
|---|---|---|---|
| Sample 88 | Sample 90 | Sample 91 | |
| Crude prepn I (before heat treatment) | 0.48 | 0.24 | 0.23 |
| Crude prepn II (after heat treatment) | 0.54 | 0.30 | 0.29 |
| Final pure rHb1.1 prepn | 0.40 | 0.27 | 0.27 |
The final pure preparation/crude preparation I ratios for samples 88, 90, and 91 were 0.833:1, 1.125:1, and 1.174:1, respectively.
DISCUSSION
A serious dilemma confronts investigators attempting to determine translation error in specific proteins. A crude protein preparation represents a large sample of the population of protein molecules of interest, but contaminants can produce misleading signals; however, a highly purified protein preparation could be biased if the purification procedure selectively eliminates the mistranslated members of the population. In addition, many of the proteins studied contain at least one residue of each amino acid. Therefore, researchers analyzing the inappropriate presence of absent amino acids frequently rely on only a fragment of the protein and extrapolate mistranslation of the whole protein from the measurement obtained for this fragment (2, 15). Our approach examines mistranslation in the whole protein and avoids signals arising from contaminants. Several lines of evidence indicate that rigorous sample purification is unlikely to bias our results significantly. We examined translational fidelity during overexpression of recombinant hemoglobin by taking advantage of the absence of isoleucine in the amino acids of rHb1.1. Therefore, most of the isoleucine found in purified rHb1.1 samples could be attributed to mistranslation. Misacylation could also have accounted for some Ile in rHb1.1 samples, but the fidelity of aminoacylation is generally greater than ribosome fidelity by at least 1 order of magnitude (5), so this class of translation errors probably contributed little to the Ile detected in these experiments.
Many (33%) of our samples had no quantifiable translation error (error rate, ≤0.0002). The average number of errors for rHb1.1 translation was 6 in 10,000 codons (Table 3). This is within the normal range of the error rates measured for homologous proteins in E. coli. When the samples with error rates of ≤2 × 10−4 were not included in the analysis, the upper estimate of the error rate was 8 × 10−4. There was no significant difference between the two strains studied, even though the level of rHb1.1 accumulation was much higher in SGE1464. The average error rate for SGE1662 was 0.0005 ± 0.0003 (n = 13), while the average error rate for SGE1464 was 0.0007 ± 0.0005 (n = 2), suggesting that higher levels of soluble and/or total globin expression did not increase mistranslation in highly purified samples. The average error rate for purified soluble rHb1.1 (≤6 × 10−4) was at least 2 orders of magnitude lower than the average error rate for another highly expressed recombinant protein, recombinant mEGF (18).
Purification can affect the measurement of translation error in two ways; first, contaminating E. coli protein can contribute an isoleucine signal in AAAA, and second, the purified sample may be biased by the purification techniques if they eliminate a higher proportion of the mistranslated protein. Since the level of E. coli protein can result in an overestimate of Ile, we measured the E. coli protein contents of our samples. Only one rHb1.1 sample contained more than 1 ppm of E. coli proteins, and most E. coli protein levels were below the limit of quantitation (<0.16 ppm). A sample containing 0.1 mol of Ile per mol of rHb1.1 would require about 0.4% E. coli protein contamination, assuming that E. coli proteins contain 5% Ile, which is typical (Table 5). The levels of E. coli proteins in our samples were at least 3 to 4 orders of magnitude lower than this and therefore could not significantly contribute to the Ile signal measured.
TABLE 5.
Amino acid compositions of rHb1.1 and typical E. coli proteins
| Amino acid | No. of residues in rHb1.1 | μmol/g of rHb1.1 | μmol/g of E. coli protein (avg)a | % Change in distributionb |
|---|---|---|---|---|
| Ala | 72 | 1,159 | 887 | 8 |
| Arg | 12 | 193 | 511 | −16 |
| Asn | 18c | 290 | 416 | −8 |
| Asp | 30 | 483 | 416 | 4 |
| Cys | 6 | 97 | 158 | −10 |
| Glu | 24 | 386 | 455 | −4 |
| Gln | 8 | 129 | 455 | −18 |
| Gly | 41 | 660 | 1,058 | −9 |
| His | 38 | 612 | 164 | 68 |
| Ile | 0 | 0 | 502 | −25 |
| Leu | 72c | 1,159 | 778 | 12 |
| Lys | 46c | 741 | 593 | 6 |
| Met | 9c | 145 | 265 | −11 |
| Phe | 30c | 483 | 320 | 13 |
| Pro | 28 | 451 | 382 | 5 |
| Ser | 32 | 515 | 373 | 10 |
| Thr | 32c | 515 | 438 | 4 |
| Trp | 6 | 97 | 98 | −0 |
| Tyr | 12 | 193 | 238 | −5 |
| Val | 59c | 950 | 731 | 7 |
| Total | 575 | 9,258 | 9,238 | 0 |
Calculated by using the data of Neidhardt and Umbarger (14).
Percent change in amino acid distribution in the total protein of E. coli cells when the level of rHb1.1 expression was 25%.
Some but not necessarily all of the codons for this amino acid are identical to an Ile codon at two of the three positions; 200 of the 575 codons tested were identical to an Ile codon at two of the three positions.
Purification bias might be introduced if it selectively removes molecules containing mistranslations, resulting in a virtually error-free product. The scatter plot of the Ile mistranslation levels distributed among the various purification yields (Fig. 4) did not reveal any purification bias that affected our results. We simultaneously measured a second, independent mistranslation event, norvaline substitution for leucine, which is due to misaminoacylation (1). We reasoned that the levels of amino acid misincorporation, whether occurring by mistranslation or misaminoacylation, should be similarly affected by the protein purification procedure. Again there was no correlation between the purification yield and the level of norvaline substitution (Fig. 4) or between norvaline misincorporation and isoleucine misincorporation (Fig. 4). In addition, norvaline is not typically found in native E. coli proteins, which allowed us to easily measure norvaline substitution in upstream (crude) samples. The proportions of this mistranslation were the same for crude rHb1.1 and for highly purified rHb1.1, indicating that purification did not affect norvaline mistranslation measurements. We also saw no evidence that the rate of either type of mistranslation correlated with rHb1.1 functionality (Tables 1 and 3).
Although highly purified, our samples represented a large portion of the soluble rHb1.1, averaging about 30% of the starting material in the experiments. Since the soluble rHb1.1 accounts for at least 60 to 65% of the total globin protein, purified samples included 20 to 30% of the total globin. Therefore, the measured rate of translation error was the average rate for a large fraction of the total globin population. It may have represented the lower limit of mistranslation for globin, but was unlikely to differ a great deal from the rate for the entire globin population, assuming that translation errors are normally distributed among proteins and Ile-substituted proteins behave like norvaline-substituted proteins during purification. In rHb1.1, most of the globin codons have been optimized for E. coli expression (4, 10). Since translation errors are strongly context dependent (15), codon optimization may have removed the opportunities for mistranslation at a rate other than the rate typified by E. coli proteins.
It has been proposed that high-level overexpression of a heterologous protein having a distribution of amino acids atypical for E. coli could result in overutilization of some amino acids. This could result in amino acid starvation or other perturbations in E. coli amino acid availability. Starvation or depletion of particular amino acid pools could stimulate a compensatory increase in the mistranslation error rate, especially for those codons involving cognate tRNAs usually charged with amino acids made scarce by overutilization (reviewed in reference 8). We calculated the effect of rHb1.1 overexpression on E. coli amino acid requirements and found it to be surprisingly small. Assuming that accumulation of rHb1.1 accounted for about 25% of the E. coli protein, a value slightly greater than the value measured in SGE1464 fermentations, the requirement for histidine increased by 68%, but no other amino acid requirement increased by more than 13% (Table 5). It appears that the level of histidine utilization did not affect E. coli growth to a degree indicative of amino acid starvation, since no significant difference was observed in the growth rates and cell densities of SGE1464 cultures in which rHb1.1 synthesis was induced or not induced (18a). This suggests that the amino acid requirements in E. coli do not appear to be seriously perturbed by rHb1.1 overexpression. However, our method was not suitable to address the potential for His substitutions.
We quantified translation missense errors in highly purified samples representing 20 to 30% of the total population of globin molecules by using AAAA. The fidelity of translation of this portion of rHb1.1 in E. coli is indistinguishable from the fidelity of translation of homologous proteins. This may indicate that when native E. coli codons are used, even expression of globin proteins accounting for up to ∼20% of the total protein can be accommodated without seriously compromising the quality of the purified fraction of this large, complex protein. It appears that overexpression of heterologous proteins in E. coli is not necessarily deleterious to the fidelity of translation, which is especially reassuring for those proteins destined for therapeutic use, like rHb1.1.
ACKNOWLEDGMENTS
We sincerely thank Maria Pagratis for technical assistance in the insoluble globin analysis and Mary Simonson, Julie MacGregor, Chris Tapparo, Bruce Whitesel, Jared Rowe, and Daryl Ogden for soluble hemoglobin measurements and functionality testing. We also greatly appreciate the assistance of a group, too large to name individually, that was responsible for fermentation and purification of rHb1.1. We also thank Doug Looker, Dan Doherty, Jeff Etter, and Spencer Anthony-Cahill for their comments on the manuscript.
REFERENCES
- 1.Apostol I, Levine J, Lippincott J, Leach J, Hess E, Glascock C R, Weickert M J, Blackmore R. Incorporation of norvaline at leucine positions in recombinant human hemoglobin expressed in Escherichia coli. J Biol Chem. 1997;272:28980–28988. doi: 10.1074/jbc.272.46.28980. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham R H, Grosjean H. The accuracy of mRNA-tRNA recognition. In: Kirkwood T B L, Rosenberger R F, Galas J D, editors. Accuracy in molecular processes. New York, N.Y: Chapman and Hall; 1986. pp. 83–126. [Google Scholar]
- 3.Glazer A N, Delange R J, Sigman D S. Chemical modification of proteins: selected methods and analytical procedures. New York, N.Y: Elsevier Biomedical; 1987. [Google Scholar]
- 4.Hoffman S J, Looker D L, Roehrich J M, Cozart P E, Durfee S L, Tedesco J L, Stetler G L. Expression of fully functional tetrameric human hemoglobin in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:8521–8525. doi: 10.1073/pnas.87.21.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakubowski H, Goldman E. Editing of errors in selection of amino acids for protein synthesis. Microbiol Rev. 1992;56:412–429. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kane J. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 7.Köpke A K E, Leggatt P A. Initiation of translation at an AUA codon for an archeabacterial protein expressed in E. coli. Nucleic Acids Res. 1991;19:5169–5172. doi: 10.1093/nar/19.19.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurland C, Gallant J. Errors of heterologous protein expression. Curr Opin Biotechnol. 1996;7:489–493. doi: 10.1016/s0958-1669(96)80050-4. [DOI] [PubMed] [Google Scholar]
- 9.Lippincott J, Hess E, Apostol I. Mapping of recombinant hemoglobin using immobilized trypsin cartridges. Anal Biochem. 1997;252:314–325. doi: 10.1006/abio.1997.2334. [DOI] [PubMed] [Google Scholar]
- 10.Looker D, Mathews A J, Neway J O, Stetler G L. Expression of recombinant human hemoglobin in Escherichia coli. Methods Enzymol. 1994;231:364–374. doi: 10.1016/0076-6879(94)31025-4. [DOI] [PubMed] [Google Scholar]
- 11.Looker D L, Abbott-Brown D, Cozart P, Durfee S, Hoffman S, Mathews A J, Miller-Roehrich J, Shoemaker S, Trimble S, Fermi G, Komiyama N H, Nagai K, Stetler G L. A human recombinant haemoglobin designed for use as a blood substitute. Nature. 1992;356:258–260. doi: 10.1038/356258a0. [DOI] [PubMed] [Google Scholar]
- 12.Lu H S, Fausset P R, Sotos L S, Clogston C L, Rohde M F, Stoney K S, Herman A C. Isolation and characterization of three recombinant human granulocyte colony stimulating factor His→Gln isoforms produced in Escherichia coli. Protein Expr Purif. 1993;4:465–472. doi: 10.1006/prep.1993.1061. [DOI] [PubMed] [Google Scholar]
- 13.Lu K V, Rohde M F, Thomason A R, Kenney W C, Lu H S. Mistranslation of a TGA termination codon as tryptophan in recombinant platelet-derived growth factor expressed in Escherichia coli. Biochem J. 1995;309:411–417. doi: 10.1042/bj3090411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neidhardt F C, Umbarger H E. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 13–16. [Google Scholar]
- 15.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker J, Precup J, Fu C. Misreading of the argL message in Escherichia coli. FEMS Microbiol Lett. 1992;79:141–145. doi: 10.1111/j.1574-6968.1992.tb14032.x. [DOI] [PubMed] [Google Scholar]
- 17.Plomer, J. J., J. R. Ryland, M.-A. H. Matthews, D. Traylor, E. E. Milne, S. L. Durfee, A. J. Mathews, and J. O. Neway. May 1996. International patent application PCT WO96/15151 (purification of hemoglobin).
- 18.Scorer C A, Carrier M J, Rosenberger R F. Amino acid misincorporation during high-level expression of mouse epidermal growth factor in Escherichia coli. Nucleic Acids Res. 1991;19:3511–3516. doi: 10.1093/nar/19.13.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Weickert, M. J. Unpublished data.
- 19.Weickert M J, Curry S R. Turnover of recombinant human hemoglobin in Escherichia coli occurs rapidly for insoluble and slowly for soluble globin. Arch Biochem Biophys. 1997;348:337–346. doi: 10.1006/abbi.1997.0410. [DOI] [PubMed] [Google Scholar]
- 20.Weickert M J, Pagratis M, Curry S R, Blackmore R. Low temperature improves accumulation of soluble recombinant hemoglobin in Escherichia coli by stabilization of apo-globin. Appl Environ Microbiol. 1997;63:4313–4320. doi: 10.1128/aem.63.11.4313-4320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]