Abstract
Conventional genome engineering with CRISPR–Cas9 creates double-strand breaks (DSBs) that lead to undesirable byproducts and reduce product purity. Here we report an approach for programmable integration of large DNA sequences in human cells that avoids the generation of DSBs by using Type I-F CRISPR-associated transposases (CASTs). We optimized DNA targeting by the QCascade complex through protein design and developed potent transcriptional activators by exploiting the multi-valent recruitment of the AAA+ ATPase TnsC to genomic sites targeted by QCascade. After initial detection of plasmid-based integration, we screened 15 additional CAST systems from a wide range of bacterial hosts, identified a homolog from Pseudoalteromonas that exhibits improved activity and further increased integration efficiencies. Finally, we discovered that bacterial ClpX enhances genomic integration by multiple orders of magnitude, likely by promoting active disassembly of the post-integration CAST complex, akin to its known role in Mu transposition. Our work highlights the ability to reconstitute complex, multi-component machineries in human cells and establishes a strong foundation to exploit CRISPR-associated transposases for eukaryotic genome engineering.
RNA-guided DNA endonucleases encoded by CRISPR–Cas systems offer ease of programmability and high-efficiency activity in a wide range of cells and organisms and have, therefore, experienced widespread adoption for basic research, agricultural applications and human therapeutics1,2. In mammalian cells, Cas9-mediated double-strand breaks (DSBs) are primarily repaired in one of two ways–non-homologous end joining (NHEJ) and homology-directed repair (HDR)–with the efficiency of NHEJ typically exceeding that of HDR by at least an order of magnitude3. Although improved methods of HDR-based insertion have begun to emerge4-7, precise modifications necessitating larger cargo sizes remain inefficient and difficult to generate, particularly in cell types that do not express sufficient levels of recombination machinery8-10.
Recent studies have further highlighted the range of undesirable (and previously undetected) byproducts of DSB-based genome editing, including large-scale genomic deletions, chromosomal translocations and chromothripsis11-14, which confound results and pose serious safety concerns. Next-generation editing reagents, including base editors and prime editors, exploit CRISPR–Cas9 for programmable RNA-guided DNA targeting while leveraging fused effector domains to perform site-specific chemistry on the genome, enabling precise, DSB-independent modifications15-17. However, both approaches have traditionally been restricted to edits ranging from single to ~50 base pairs (bp), rendering larger insertions inaccessible. Recently developed tools that combine prime editors with serine integrases, such as TwinPE and PASTE, have been shown to enable larger DNA insertions with efficiencies up to ~25%, but these methods still require resolution of complex DNA intermediates, can generate undesired indels at the target site and produce incomplete modifications when the multiple enzymatic events do not occur in concert18,19.
Lentiviral vectors are a highly used gene delivery vehicle for biotechnological applications because they integrate with high efficiency across diverse cell types, although they may exhibit promiscuous specificity, offer little control over copy number, present limitations in cargo capacity and design and require numerous manufacturing steps20. Transposases such as Sleeping Beauty and piggyBac also integrate DNA without relying on host recombination and can better accommodate large sequences for insertion, but they lack specificity and copy number control21-23. In contrast to these approaches, recombinases such as Cre and Bxb1 offer excellent specificity and product purity but are not programmable and, thus, require researchers to first generate engineered cells containing the obligate recombination site24,25. The ideal DNA integration technology would function in a single step and avoid generating DSBs or indels while retaining the programmability afforded by RNA-guided DNA targeting.
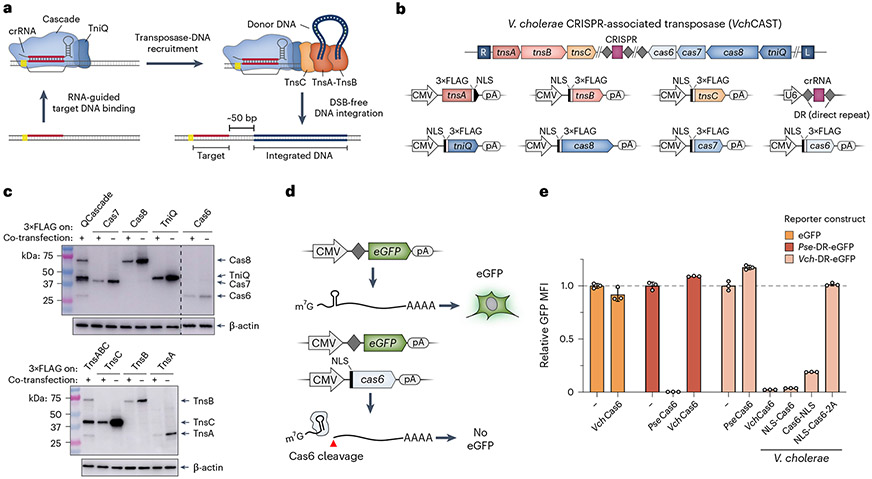
Recent studies have attempted to engineer RNA-guided transposases by fusing Cas9 to various transposase domains, but these efforts have remained reliant on DSBs and/or failed to achieve stringent specificity control26-29. In contrast, bacterial CRISPR-associated transposases (CASTs) catalyze insertion of large DNA sequences in a targeted manner without DSBs. Using a Type I-F CAST system derived from Vibrio cholerae Tn6677, we recently reported DSB-free DNA insertions in multiple bacterial species and demonstrated that this approach exhibited exquisite genome-wide specificity and could be easily reprogrammed to user-defined sites with single-bp accuracy30,31. We, therefore, sought to leverage RNA-guided transposases for targeted DNA integration in mammalian cells, despite the obstacle of reconstituting a complex, multi-component pathway that depends on a donor DNA, guide CRISPR RNA (crRNA) and assembly of seven distinct proteins, many of which function in an oligomeric state (Fig. 1a,b).
Fig. 1 ∣. Reconstitution of protein–RNA CAST components in human cells.
a, Schematic detailing DNA integration using RNA-guided transposases. b, Type I-F CRISPR-associated transposons encode the CRISPR RNA (crRNA) and seven proteins needed for DNA integration (top). Mammalian expression vectors used for heterologous reconstitution in human cells are shown at the bottom. c, Western blotting with anti-FLAG antibody demonstrates robust protein expression upon individual (−) or multi-plasmid (+) co-transfection of HEK293T cells. Co-transfections contained all VchCAST components, with the FLAG-tagged subunit(s) indicated. β-actin was used as a loading control. Western blots were repeated in biological duplicates with similar results. d, Schematic of eGFP knockdown assay to monitor crRNA processing by Cas6 in HEK293T cells. Cleavage of the CRISPR direct repeat (DR)-encoded stem-loop severs the 5′ cap from the ORF and polyA (pA) tail, leading to a loss of eGFP fluorescence (bottom). e, Transposon-encoded VchCas6 (Type I-F3) exhibits efficient RNA cleavage and eGFP knockdown, as measured by flow cytometry. Knockdown was similar to PseCas6 from a canonical CRISPR–Cas system (Type I-E)41, was absent with a non-cognate DR substrate and was sensitive to C-terminal tagging. To control for overexpression, data were normalized to negative control conditions (−), in which dCas9 was co-transfected with the reporter. Data are shown as the mean ± s.d. for n = 3 biologically independent samples. Uncropped western blots are shown in Source Data Fig. 1.
Here we report mammalian CAST activity using two diverse systems from V. cholerae and Pseudoalteromonas32, demonstrating that the same molecular determinants of RNA-guided transposition hold true in bacteria and eukaryotes. Integration efficiencies were initially much lower at endogenous target sites compared to episomal plasmid substrates, which led us to identify bacterial ClpX as a critical accessory factor that enhanced genomic integration by more than two orders of magnitude. During our engineering efforts, we also developed a strategy for targeted recruitment of an oligomeric transposase component, TnsC, which we harnessed to achieve potent transcriptional activation at levels similar to conventional dCas9-based reagents. Taken together with recent studies harnessing alternative Type I CRISPR–Cas systems for eukaryotic genome and transcriptome engineering33-37, our work challenges the reliance on single-effector editing reagents and provides a strong starting point for genome engineering using RNA-guided, CRISPR-associated transposases.
Results
Heterologous expression of CAST components in human cells
Bacterial Tn7-like transposases have co-opted at least three distinct types of nuclease-deficient CRISPR–Cas systems for RNA-guided transposition (I-B, I-F and V-K)30,38,39, with each exhibiting unique features. We carefully reviewed fidelity and programmability parameters for experimentally characterized CAST systems, alongside recently described Cas9–transposase fusion approaches27-29, and opted to focus our efforts on the Type I-F V. cholerae CAST (VchCAST; previously also referred to as VchINTEGRATE) because of its optimal integration efficiency, specificity and absence of co-integrates30,31,38,40. Within this system, a ribonucleoprotein complex comprising TniQ and Cascade (VchQCascade) performs RNA-guided DNA targeting, thereby defining sites for transposon DNA insertion30,41. Excision and integration reactions are catalyzed by the heteromeric TnsA–TnsB transposase after prior recruitment of the AAA+ ATPase, TnsC42,43. Integrated DNA payloads must be flanked by transposon left and right end sequences, which encode TnsB binding sites and define boundaries of the mobile element.
We adopted a methodical, bottom-up approach to port VchCAST into human cells. To first establish whether the component parts were efficiently expressed, we cloned each protein-coding gene onto a standard mammalian expression vector with an N-terminal or C-terminal nuclear localization signal (NLS) and 3×FLAG epitope tag (Fig. 1b). Using western blotting, we showed robust heterologous protein expression, both individually and when all CAST proteins were co-expressed (Fig. 1c). Cellular fractionation provided evidence of nuclear trafficking, and we also demonstrated efficient expression and trafficking of an engineered TnsAB fusion protein (TnsABf) that we previously showed retains wild-type activity (Supplementary Fig. 1)40. However, initial attempts to reconstitute RNA-guided DNA integration in HEK293T cells proved unsuccessful, even after exploring numerous strategies to enrich rare events through both positive and negative selection. We, therefore, decided to separately assess guide RNA (gRNA) expression by adapting a previously developed approach34 to monitor crRNA biogenesis within the 5′ untranslated region (UTR) of a GFP-encoding mRNA. Cas6 is a ribonuclease subunit of Cascade that cleaves the CRISPR repeat sequence in most Type I CRISPR–Cas systems44, which, in our assay, would sever the 5′ cap from the GFP open reading frame (ORF) and, thus, lead to fluorescence knockdown (Fig. 1d). Accordingly, we observed near-total loss of GFP fluorescence when the reporter plasmid was co-transfected with cognate VchCas6 but not when the reporter encoded a non-cognate CRISPR repeat or lacked a repeat altogether (Fig. 1e). Interestingly, GFP knockdown was substantially reduced when Cas6 contained a C-terminal NLS or 2A peptide (Fig. 1e), indicating a sensitivity to terminal tagging that could not be easily explained by the cryogenic electron microscopy (cryoEM) structure (see below)41. Collectively, these experiments verified expression of all protein and RNA components from VchCAST, leading us to next focus on functional reconstitution of RNA-guided DNA targeting by QCascade.
QCascade and TnsC function as transcriptional activators
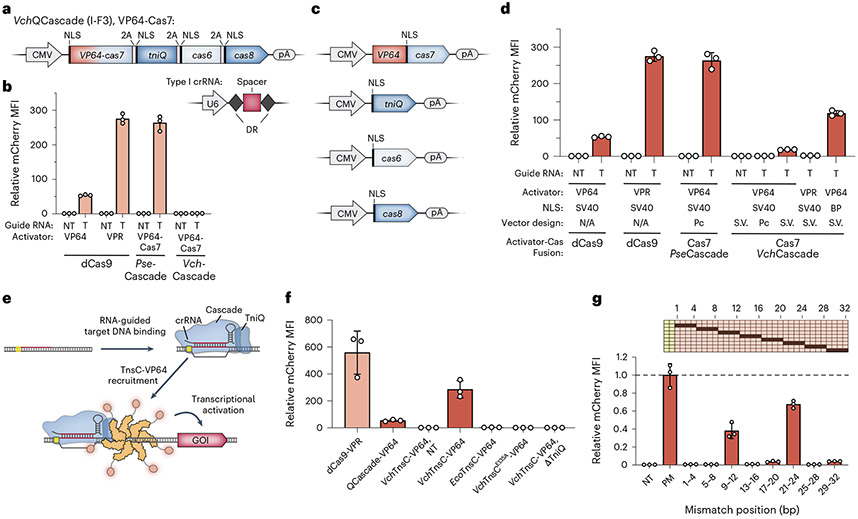
Unlike most Type II and V CRISPR–Cas systems, which encode single effector proteins that function as RNA-guided DNA nucleases (Cas9 and Cas12, respectively), the Cascade complex encoded by Type I systems does not possess DNA cleavage activity and, instead, exhibits long-lived target DNA binding upon R-loop formation, analogously to catalytically inactive Cas9 (dCas9)45. We decided to leverage this activity for transcriptional activation of an mCherry reporter gene by fusing transcriptional activators to QCascade, as recently done for other Type I systems34,35,37, thereby converting DNA binding into a detectable signal that would allow facile troubleshooting and optimization of QCascade function (Supplementary Fig. 2a).
We first constructed activators using a Type I-E Cascade unrelated to transposases from Pseudomonas sp. S-6–2 (PseCascade_IE), which we previously exploited for genome engineering in human cells33. We fused VP64 to the hexameric Cas7 subunit and concatenated all five cas genes within a single polycistronic vector downstream of a cytomegalovirus (CMV) promoter, by linking them together with virally derived 2A ‘skipping’ peptides; the crRNA was separately expressed from a U6 promoter (Supplementary Fig. 2a). The resulting expression plasmids yielded ~260-fold mCherry activation when co-transfected with the reporter plasmid, similar to levels achieved with dCas9–VPR, and the effect was ablated in the presence of a non-targeting crRNA (Fig. 2b). Surprisingly, when we tested nearly identical designs using the transposon-encoded Type I-F QCascade homolog from V. cholerae, we did not detect any activation (Fig. 2a,b).
Fig. 2 ∣. Development of QCascade-based and TnsC-based transcriptional activators to monitor DNA targeting.
a, Design of mammalian expression vectors encoding transposon-encoded Type I-F3 systems (VchQCascade). Cascade subunits are concatenated on a single polycistronic vector and connected by virally derived 2A peptides, as described previously33. b, Normalized mCherry fluorescence levels for the indicated experimental conditions, measured by flow cytometry. Whereas PseCascade stimulated robust activation, VchQCascade was inactive under these conditions. NT, non-targeting sgRNA/crRNA; T, targeting sgRNA/crRNA. c, Design of separately encoded VchQCascade mammalian expression vectors with optimized NLS tag placement. d, VchQCascade mediates transcriptional activation when encoded by re-engineered expression vectors, as measured by flow cytometry. mCherry expression is further enhanced when replacing mono-partite (SV40) NLS tags with BP NLS tags. Pc, polycistronic; S.V., single vectors; NT, non-targeting; T, targeting. e, Schematic of transcriptional activation assay, in which DNA targeting by VchQCascade leads to multi-valent recruitment of VchTnsC–VP64. The assembly mechanism is based on our recent biochemical, structural and functional data41. f, Normalized mCherry fluorescence levels for the indicated experimental conditions, measured by flow cytometry. VchTnsC-based activation requires cognate protein–protein interactions, is strictly dependent on the presence of TniQ and involves ATP-dependent oligomer formation, which is eliminated with the E135A mutation. Several controls are shown for comparison, and gRNAs target the same sites shown in Supplementary Fig. 3a. NT, non-targeting crRNA. g, Transcriptional activation shows strong sensitivity to RNA–DNA mismatches within both the PAM-proximal seed sequence and a PAM-distal region implicated in TnsC recruitment. Data are shown as in f, and the schematic at the top displays the mismatched positions that were tested. Data were normalized to the perfectly matching (PM) crRNA. Data in b, d, f and g are shown as the mean ± s.d. for n = 3 biologically independent samples.
We suspected that the presence of N-terminal NLS tags, C-terminal 2A tags or both might be inhibiting QCascade assembly and/or RNA-guided DNA targeting, despite the fact that all termini appeared to be solvent-accessible in our experimentally determined VchQCascade structure (Supplementary Fig. 2b)41. To systematically investigate this possibility, we cloned peptide tags onto the termini of all VchCAST components and tested their impact in Escherichia coli transposition assays. Whereas some tags had little effect on activity, others led to a severe reduction or complete loss of targeted DNA integration (Supplementary Fig. 2c), highlighting the sensitivity of this system to minor perturbations. The transposase components were particularly vulnerable, with an N-terminal tag on TnsA and C-terminal tags on TnsB and TnsC being largely prohibitive. Within the context of QCascade, C-terminal 2A tags on TniQ and Cas7 each reduced integration by more than 90%, which could explain the lack of transcriptional activation that we observed using polycistronic vector designs. We also screened multiple components for activator fusions and found that the N-terminus of Cas7 was amenable to both VP64 and VPR fusions in bacteria (Supplementary Fig. 2d).
With these data in hand, we retested QCascade–VP64 in human cells using individual expression vectors with optimized NLS tag locations for each component and detected mCherry activation for two distinct crRNAs, evidencing successful assembly and target binding in human cells (Fig. 2c,d and Supplementary Fig. 2e). Activation levels were further increased by replacing all mono-partite SV40 NLS tags with bi-partite (BP) NLS tags, and this activity was strictly dependent on the simultaneous expression of Cas8, Cas7, Cas6 and a targeting crRNA (Fig. 2d and Supplementary Fig. 2e,f). Interestingly, although Cas7 tolerated a VPR fusion in bacteria, we were unable to detect transcriptional activation in mammalian cells using VPR–Cas7 (Fig. 2d and Supplementary Fig. 2d,e). These results highlighted the importance of carefully dissecting the effects of all sequence modifications being introduced to VchCAST components, even those appearing innocuous, and emphasized the value of fluorescence reporter assays in debugging molecular events upstream of DNA integration.
Early dCas9-based transcriptional activators revolved around recruitment of an activator domain co-valently linked to a single dCas946-48, whereas later methods have exploited strategies for multi-valent recruitment of one or more effector domains49,50. In the case of CAST systems, recent experiments have demonstrated that TnsC forms large ATP-dependent oligomers that assemble onto double-stranded DNA (dsDNA) and are specifically recruited to DNA-bound QCascade with high genome-wide specificity in E. coli43,51,52. We, thus, hypothesized that these properties could be leveraged for multi-valent assembly of TnsC to increase the potency of transcriptional activation in mammalian cells while also demonstrating recruitment of a critical transposase component in a QCascade-dependent fashion (Fig. 2e).
We fused VP64 to either the N-terminus or C-terminus of TnsC, targeted seven candidate sites upstream of our mCherry reporter gene (Supplementary Fig. 3a) and investigated the potential for TnsC to stimulate transcriptional activation. Strikingly, TnsC–VP64 activators drove substantially higher levels of mCherry activation than QCascade alone, and activation levels could be further improved by optimizing the relative amount of each expression plasmid used during transfection (Fig. 2f and Supplementary Fig. 3b). This effect was absent when TniQ was omitted or an E. coli TnsC homolog was substituted, confirming the importance of cognate TniQ–TnsC interactions. Furthermore, a TnsC ATPase mutant that prevents oligomer formation (E135A)43 also abolished transcriptional activation, suggesting that the observed signal requires protein oligomerization on DNA (Fig. 2f). TnsC homologs from Type V-K CAST systems form filaments non-specifically on dsDNA51,52, and we were, therefore, keen to investigate the fidelity of VchTnsC-mediated activation. Non-targeting controls generated undetectable mCherry mean fluorescence intensity (MFI) above background levels, demonstrating the specificity of potential TnsC filamentation in Type I-F CASTs (Fig. 2f). When probing the specificity of QCascade DNA binding, intermediate levels of transcriptional activation were retained when mismatches were tiled within the middle of the 32-bp target site, but there was a strict requirement for cognate pairing in the seed (positions 1–8) and PAM-distal (positions 25–32) regions (Fig. 2g).
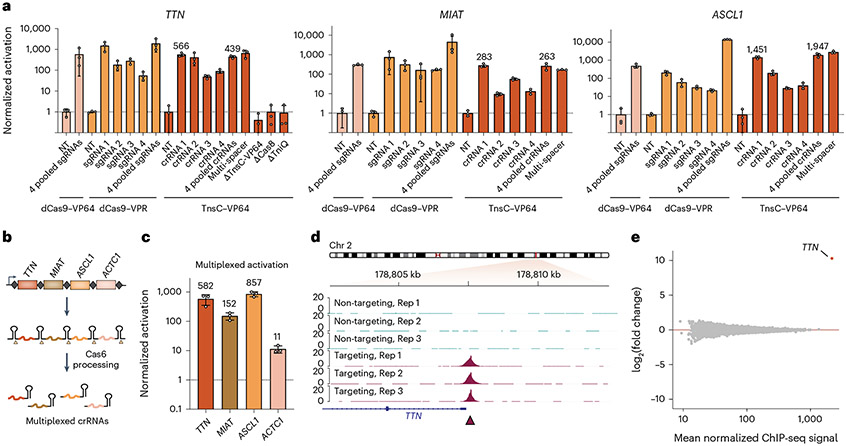
Having demonstrated the ability of TnsC-based activation to potently induce expression of a reporter gene, we targeted four endogenous genes in the human genome (TTN, MIAT, ASCL1 and ACTC1), which have been previously targeted with CRISPRa using dCas9–VPR53. We designed three or four distinct crRNAs tiled upstream of the transcription start site (TSS) and delivered them by transfecting a single crRNA expression plasmid, co-transfecting multiple crRNA expression plasmids or transfecting a single crRNA expression plasmid containing a four-spacer CRISPR array (Fig. 3a and Supplementary Fig. 3c,d). TTN induction by TnsC–VP64 was similar to dCas9–VP64 and dCas9–VPR activation, and, consistent with our model, the presence of Cas8 and TniQ were strictly required (Fig. 3a). Potent activation was seen on other genomic targets ranging from 200-fold (MIAT) to more than 1,000-fold (ASCL1), highlighting the programmability of our multi-meric system (Fig. 3a), although other sites showed more moderate activation (Supplementary Fig. 3e). Furthermore, we demonstrated the ability to use a multiplexed CRISPR array containing four spacers that each targeted a different gene to achieve robust transcriptional activation of all four genes (TTN, MIAT, ASCL1 and ACTC1) in the same cell population at levels similar to activation achieved by single-spacer CRISPR arrays (Fig. 3b,c).
Fig. 3 ∣. Potent genomic transcriptional activation via RNA-guided recruitment of the AAA+ ATPase, TnsC.
a, TnsC–VP64 directs efficient transcriptional activation of endogenous human gene expression, as measured by RT–qPCR. Four distinct crRNAs were combined for each condition and were delivered individually, as a pool or as a single multi-spacer multiplexed CRISPR array. The dCas9–VP64 and dCas9–VPR comparisons used four distinct sgRNAs encoded on separate plasmids. NT, non-targeting; T, targeting. b, Schematic demonstrating Cas6’s ability to process CRISPR arrays in vivo, thus allowing for the use of multiplexed CRISPR arrays to target multiple sites concurrently. c, Multiplexed activation of four distinct genes in the same cell pool. d, A 10-kb viewing window of ChIP-seq signal at the TTN promoter corresponding to TTN guide 1. e, Differential binding analysis plot. Across consensus peaks for each condition, the only region exhibiting significantly different ChIP enrichment (FDR < 0.05) between targeting and non-targeting conditions was the peak at the TTN promoter. Data in a and c are shown as the mean ± s.d. for n = 3 biologically independent samples. Viewing windows in d are shown for three biologically independent targeting and non-targeting samples, and ChIP-seq signal is visualized as SPMR. Data in e are shown as the mean for n = 3 biologically independent samples for each condition on the y axis and the mean for all n = 6 biologically independent samples on the x axis, irrespective of condition.
We next investigated the fidelity of TnsC recruitment by performing chromatin immunoprecipitation followed by sequencing (ChIP-seq) after co-transfecting plasmids encoding FLAG-tagged TnsC, protein components of QCascade and a TTN-specific crRNA. Analysis of the resulting data revealed a sharp peak directly upstream of the TTN TSS at the expected target site, which was absent in non-targeting samples transfected with a crRNA containing a spacer not found in the human genome (Fig. 3d and Supplementary Fig. 4a,b). To assess off-target binding, we analyzed all peaks in both targeting and non-targeting conditions across three biological replicates and performed differential binding analysis, revealing only a single region at the TTN promoter that exhibited significantly different binding affinity between both conditions (false discovery rate (FDR) < 0.05)54, highlighting the specificity of Type I-F CAST assembly (Supplementary Fig. 4c and Fig. 3e). Heat map analysis of additional peaks that were called in either targeting or non-targeting conditions revealed low enrichment values, and a further manual inspection of five potential off-target sites that exhibited high similarity to the TTN spacer sequence lacked any detectable signal enrichment in the ChIP-seq datasets (Supplementary Fig. 4d-g). Together with our recent study of VchCAST factor recruitment in E. coli43, these results indicate that TnsC binds target sites marked by QCascade with high fidelity and that the intrinsic ability of TnsC to form ATP-dependent oligomers enables multiple copies of an effector protein to be delivered to genomic sites targeted by a crRNA.
This programmable, multi-valent recruitment represents an exciting opportunity to further develop genome and transcriptome engineering tools that benefit from RNA-guided DNA binding of an effector ATPase. In the context of efforts to reconstitute CAST systems, TnsC-mediated transcriptional activation provided compelling evidence that both CRISPR-associated and transposon-associated protein components can be functionally assembled at plasmid and genomic target sites in a highly specific and programmable manner, encouraging our efforts to next probe for RNA-guided DNA integration.
RNA-guided episomal DNA integration in human cells
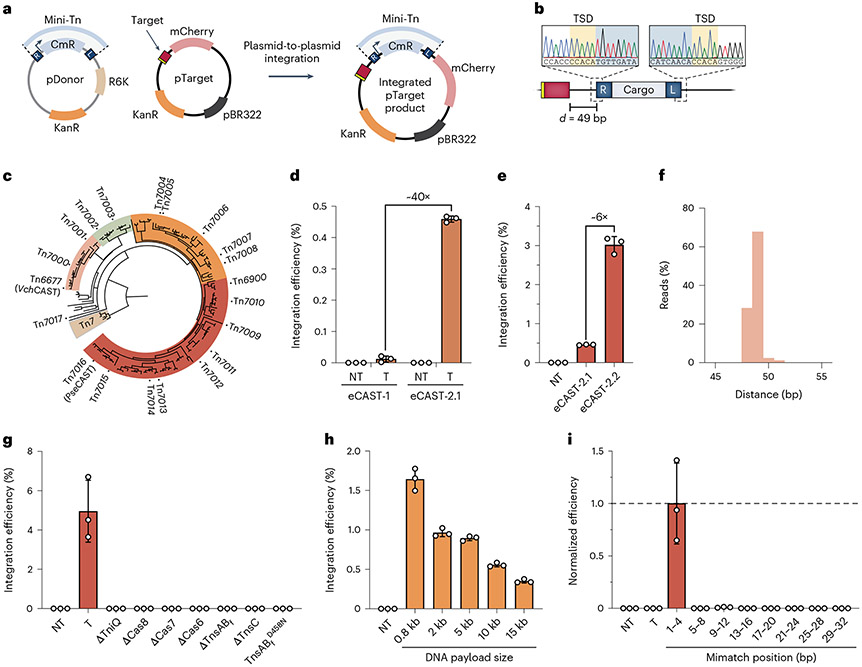
We reasoned that the baseline efficiency of RNA-guided transposition might be low before optimization, and, therefore, we sought to develop a sensitive assay that would enrich integration products. We cloned a promoter-driven chloramphenicol resistance cassette (CmR) within the mini-transposon of a donor plasmid (pDonor) and then targeted the same sequence on the mCherry reporter plasmid (pTarget) that was used in transcriptional activation experiments. Upon successful transposition in HEK293T cells, integrated pTarget products will carry both CmR and kanamycin-resistance (KanR) drug markers and can, thus, be selected for by transforming E. coli with plasmid DNA isolated from transfected cells (Fig. 4a). Notably, in these experiments, we used a pDonor backbone that cannot be replicated in standard E. coli strains, reducing background from unreacted plasmids. We also opted to use a TnsAB fusion protein (TnsABf)40 that contains an internal BP NLS and maintains wild-type activity in E. coli (Supplementary Fig. 1c), thereby reducing the number of unique protein components; this modified system is hereafter referred to as engineered CAST-1 (eCAST-1).
Fig. 4 ∣. Plasmid-based RNA-guided DNA integration in human cells using diverse CASTs.
a, Schematic of plasmid-to-plasmid transposition assay in human cells. b, Sanger sequencing confirmation of targeted integration products after plasmid isolation from human cells and selection in E. coli (a), showing the expected insertion site position and presence of target site duplication. c, Phylogenetic tree of Type I-F CRISPR-associated transposon systems adapted from previous work in the lab32, with labels of the homologs that were tested in human cells. d, Comparison of plasmid-to-plasmid integration efficiencies with eCAST-1 (VchCAST) and eCAST-2.1 (PseCAST), as measured by qPCR. Efficiencies are calculated by comparing Cq values between the integration junction product and a reference sequence located elsewhere on pTarget, as described in Methods. e, Optimization of eCAST-2 (PseCAST) integration efficiencis by varying NLS placement and plasmid stoichiometries, as described in Supplementary Fig. 7, yielded an approximate six-fold increase in integration efficiencies. f, Amplicon sequencing reveals a strong preference for integration 49 bp downstream of the 3′ edge of the site targeted by the crRNA in T-RL integrants. g, Deletion experiments confirm the obligate requirement of each protein component, a targeting crRNA and intact transposase active site (D220N mutation in TnsB, D458N mutation in TnsABf) for successful integration. h, RNA-guided DNA integration functions with genetic payloads spanning 1–15 kb in size, transfected based on molar amount. i, RNA-guided DNA integration shows a strong sensitivity to mismatches across the entire 32-bp target site. Data were normalized to the perfectly matching (PM) crRNA, which exhibited an efficiency of 4.7 ± 1.8%. Data in d, e and g–i are shown as the mean ± s.d. for n = 3 biologically independent samples. Data in d, e and g–i were determined by qPCR.
After transfecting HEK293T cells with pDonor, pTarget and all protein–RNA expression plasmids, purifying the plasmid mixture from cells and using the mixture to transform E. coli, we observed the emergence of colonies that were chloramphenicol resistant, which outnumbered the corresponding colonies obtained from experiments using a non-targeting crRNA that did not match pTarget (Supplementary Fig. 5a). Encouraged by this result, we performed junction polymerase chain reaction (PCR) on select colonies and obtained bands of the expected size, which subsequent Sanger sequencing confirmed were integration products arising from DNA transposition 49 bp downstream of the target site (Fig. 4b), as expected from our bacterial studies30. Further analyses of individual clones revealed the expected junction sequences across both the transposon left and right ends (Supplementary Fig. 5b). Next, we showed that the same products could be detected by nested PCR directly from HEK293T cell lysates (Supplementary Fig. 5c), and we developed a sensitive TaqMan probe-based quantitative (qPCR) strategy to quantify integration events from lysates by detecting site-specific, plasmid–transposon junctions (Supplementary Fig. 5d). Using this approach, we performed an initial optimization screen by varying the relative amounts of expression and pDonor plasmids and found that efficiencies were greatest with low levels of pTnsC and high levels of pTnsABf and pDonor (Supplementary Fig. 5e). Nevertheless, absolute efficiencies of plasmid-to-plasmid integration with this eCAST-1 system from V. cholerae remained less than 0.1%, leading us to pursue other avenues for improved activity (Supplementary Fig. 5e).
We recently described the bioinformatic mining and experimental characterization of 18 new Type I-F CRISPR-associated transposons (denoted Tn7000–Tn7017), many of which exhibited high-efficiency and high-fidelity RNA-guided DNA integration in E. coli (Fig. 4c)32. We hypothesized that sampling from this diversity would uncover variants with improved activity in human cells and, thus, embarked on a hierarchical screening approach to concentrate our efforts on the most promising systems (Supplementary Fig. 6a). In brief, our scheme involved filtering based on robust activity in three key areas: (1) crRNA biogenesis by Cas6, assessed using our GFP knockdown assay; (2) transposon DNA binding by TnsB, assessed using a tdTomato reporter assay; and (3) transcriptional activation by TnsC–VP64, assessed using our mCherry reporter assay. In all cases, genes were human codon optimized, which was often necessary to achieve strong expression (Supplementary Fig. 6b), and tagged with NLS sequences on the same termini as for Tn6677 (VchCAST). We found that most systems exhibited efficient crRNA biogenesis and transposon DNA binding activity that was similar to that observed with Tn6677 (Supplementary Fig. 6c,d). Interestingly, of those systems selected for testing in transcriptional activation experiments, only Tn7016 showed reproducible induction of mCherry expression, albeit at levels ~8-fold lower than Tn6677 (Supplementary Fig. 6e). We, therefore, decided to focus on Tn7016––a 31-kilobase (kb) transposon from Pseudoalteromonas sp. S983 (PseCAST)––and next investigated its RNA-guided DNA integration activity.
After verifying that fusing TnsA and TnsB from PseCAST with an internal NLS retained function, and optimizing the length of left and right transposon ends (Supplementary Fig. 7a,b), we repeated plasmid-to-plasmid transposition assays in HEK293T cells. Strikingly, the engineered Pseudoalteromonas CAST (eCAST-2.1) was ~40-fold more active than eCAST-1 when tested under un-optimized conditions (Fig. 4d and Supplementary Fig. 7c). To further improve integration efficiencies, we systematically varied the design of the crRNA, location of NLS tags and relative amounts of each expression plasmid; the resulting eCAST-2.2 yielded a further ~6-fold improvement to reach levels of 3–5% integration, and PCR followed by Sanger or Illumina sequencing analysis confirmed the expected site of integration 49 bp downstream of the target (Fig. 4e,f and Supplementary Fig. 7d-h). Of note, these efficiencies were similar to integration efficiencies achieved with BxbI under similar plasmid-to-plasmid conditions (Supplementary Fig. 7i). Peak integration occurred 4–6 days after transfection, with the efficiency exhibiting sensitivity to both cell density and the choice of cationic lipid delivery method55 (Supplementary Fig. 8a-c). We also found that the observed integration efficiency was increased by >5-fold by co-transfection with a GFP marker and separately analyzing sorted cells exhibiting high GFP fluorescence levels, suggesting that activity was dependent not only on the stoichiometry of the transfected plasmids but also on the plasmid dosage across the population of cells (Supplementary Fig. 8d,e).
Next, we sought to confirm the genetic requirements for RNA-guided DNA integration and further investigate specificity. Integration was strictly dependent on a targeting crRNA and the presence of all protein components, including an intact TnsB active site (Fig. 4g), and functioned with genetic payloads spanning 1–15 kb in size, albeit with a ~3-fold decrease in efficiency with larger payloads (Fig. 4h). We generated a panel of mismatched crRNAs in which mutations were tiled along the length of the 32-nucleotide (nt) guide and found that activity was ablated regardless of the location (Fig. 4i), indicating a greater degree of discrimination than that observed in activation experiments using VchCAST in activation experiments or in E. coli30. We used an alternative qPCR approach to confirm that integration orientation for eCAST-2.2 was highly biased toward T-RL, as expected from prior bacterial integration data32 (Supplementary Fig. 9a). Finally, we used an NGS-based amplicon sequencing approach to quantify all integration events at the expected insertion site (Supplementary Fig. 9b,c) and performed droplet digital PCR (ddPCR) to further corroborate the quantitative data obtained from TaqMan qPCR (Supplementary Fig. 9d).
RNA-guided DNA integration into the human genome
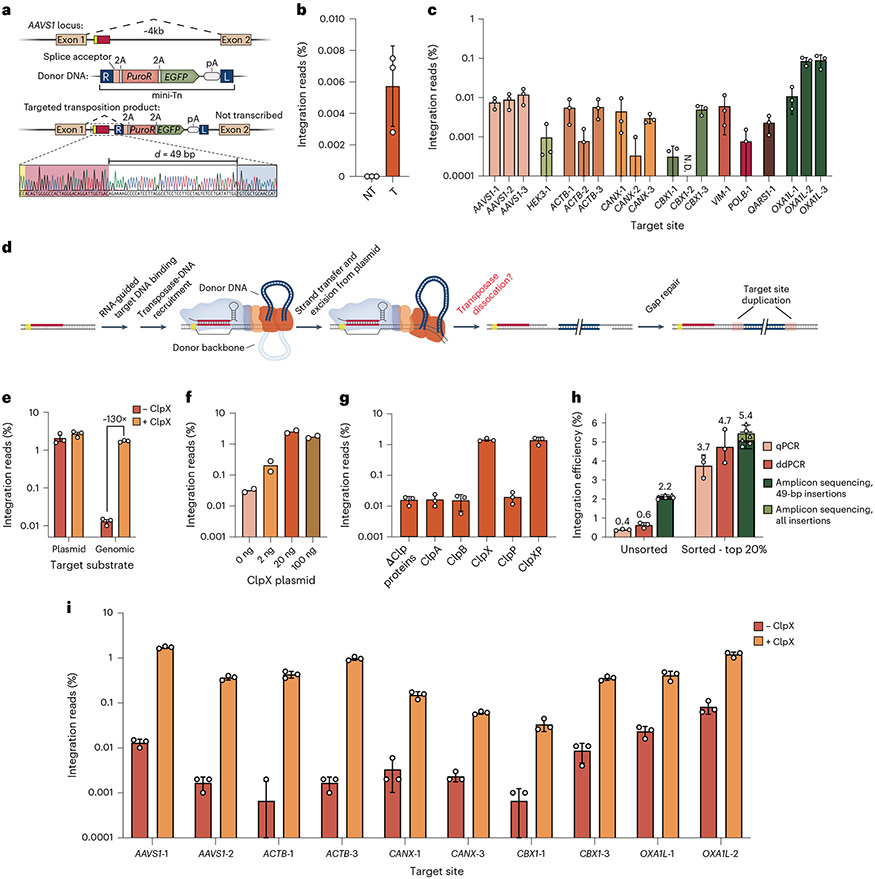
After optimization efforts on episomal plasmid DNA integration with eCAST-2.2, we next turned our attention to reconstituting RNA-guided integration into endogenous genomic sites. We first screened a panel of guide sequences targeting the AAVS1 safe harbor locus via a plasmid-to-plasmid integration assay, in which we cloned 32-bp target sites derived from AAVS1 into pTarget and leveraged existing assays to identify two active crRNAs that outperformed our original plasmid-specific crRNA (Supplementary Fig. 10a). When we tested the AAVS1 locus for genomic integration using a nested PCR strategy, we identified RNA-guided DNA integration products that again maintained the expected 49-bp distance dependence from the target site (Fig. 5a). However, detection was often not consistent across biological replicates, suggesting that integration efficiencies flirted with our limit of detection. We, therefore, applied an NGS-based amplicon sequencing method established in our prior plasmid-based assays, yielding reproducible efficiencies on the order of ~0.005% (Fig. 5b and Supplementary Fig. 9b).
Fig. 5 ∣. ClpX-mediated enhancement of genomic DNA integration with eCAST-3.
a, Sanger sequencing of nested PCR of genomic lysates In which eCAST-2.2 targeted the AAVS1 genome and showed a junction product 49 bp downstream of the target site targeted by crRNA12 (AAVS1-1), one of the optimal crRNAs screened in Supplementary Fig. 10a. b, Initial quantifications of genomic integration efficiencies at AAVS1-1. c, Integration efficiencies across multiple loci within human genome showed broadly limited efficiencies. Quantified integration efficiencies less than 0.0001% were not plotted, and ‘N.D.’ represents a target site in which no integration events were detected across three biological replicates. d, Proposed steps required for successful targeted integration, including the downstream gap repair needed for complete resolution of the integration product. e, Co-transfection of EcoClpX specifically improves genomic, but not plasmid, integration efficiencies in human cells. f, Co-transfecting EcoClpX at varied amounts directly impacts genomic integration efficiencies in human cells. g, Investigating the impact of various Clp proteins from E. coli on genomic integration efficiencies in human cells. h, Integration efficiencies for samples before and after FACS of a fluorescent transfection marker to select for the top 20% brightest cells. Sorting enriched integration efficiencies, as measured by qPCR, ddPCR and amplicon sequencing (Supplementary Fig. 9b). For amplicon sequencing samples, triangle data points represent all insertions characterized, whereas circle data points represent only 49-bp insertions. i, Integration efficiencies were investigated across multiple loci within the human genome with and without EcoClpX. Quantified integration efficiencies less than 0.0001% were not plotted. Data in b, c, e and g–i are shown as the mean ± s.d. for n = 3 biologically independent samples. Data in f are shown as the mean for n = 2 biologically independent samples. Data in b, c, e, f, g and i are quantified by amplicon sequencing. FACS, fluorescence-activated cell sorting.
We next targeted an additional eight sites across the genome, with 1–3 crRNAs per locus, and detected integration at efficiencies that varied but were generally ~0.01% (Fig. 5c). Attempts to increase the efficiency further through simplified delivery of a polycistronic QCascade expression vector, serial additions of extra NLS sequences, constitutive expression of the targeting machinery, inclusion of bacterial IHFa/b56 or phenotypic drug selection to enrich for integration events (Supplementary Fig. 10b-f) did not reduce the large, 100–1,000× discrepancy between observed integration efficiencies at plasmid and genomic target sites. Although differences in chromatinization remained a distinct possibility, we hypothesized that the discrepancy might be due to potential toxicity of genomic integration intermediate products.
TnsB performs trans-esterification reactions to join the two ends of the transposon DNA to both strands of the target DNA with a 5-bp offset, leading to the generation of an initial product characterized by 5-nt gaps on either strand of the integrated DNA57. Subsequent gap repair involves gap fill-in by DNA polymerase and DNA ligase, resulting in the hallmark 5-bp target site duplication (TSD), but these reactions require prior dissociation of the transpososome (Fig. 5d). We questioned whether incomplete dissociation of the post-transposition CAST complex might limit observed frequencies of genomic integration, perhaps leading to stalled replication forks or DNA repair pathways akin to crosslink-induced replication fork stalling58-60. Notably, these effects would likely be less deleterious on plasmid DNA substrates, because pTarget does not undergo active DNA replication and is not critical for cellular fitness. Our hypothesis was bolstered by previous studies demonstrating the extreme stability of the analogous post-transposition complex (PTC) in Tn7 and Mu transposons61-63 and the requirement for an additional factor—ClpX—in active Mu PTC disassembly, gap repair and phage propagation61-65. ClpX is a sequence-specific AAA+ ATPase that unfolds protein substrates by denaturing and translocating them through a central hexameric pore, and it recognizes degron tags that are often exposed only under certain conditions, allowing for sensitive regulation of protein unfolding and degradation66. In the case of MuA, ClpX recognizes a specific C-terminal motif, and the PTC undergoes a conformational rearrangement to expose additional MuA residues that enable more efficient ClpX binding, resulting in targeted unfolding and destabilization of the PTC65,67. We hypothesized that CAST systems might also require bacterial ClpX, or some other accessory factor, for active mechanical disassembly of the PTC.
To test this, we co-transfected human cells with eCAST-2.2 components and a plasmid expressing NLS-tagged E. coli ClpX (EcoClpX), collectively referred to as eCAST-3. Remarkably, genomic integration efficiencies increased by ~100× in a ClpX dose-responsive manner, albeit with observable ClpX-induced cellular toxicity, whereas plasmid integration efficiencies were unaffected (Fig. 5e,f). To investigate if the effect was specific to ClpX, we tested other bacterial unfoldases, including ClpA and ClpB, and found that ClpX was the only tested ATPase that enhanced genomic integration. ClpP, which functions as the peptidase component within the ClpXP protease complex68, had no effect on integration, either alone or in combination with ClpX, suggesting that protein unfolding—but not protein degradation—is necessary (Fig. 5g). When we introduced point mutations that ablate ATP hydrolysis (E185Q or R370K)69,70 or substrate engagement (Y153A)71, ClpX failed to enhance genomic integration (Supplementary Fig. 11a), further supporting the mechanistic link between ATPase-driven protein unfolding and PTC disassembly. ClpX is highly conserved across bacterial species, and the homolog from Pseudoalteromonas (80% amino acid identity) also stimulated integration, albeit to a slightly lesser extent that EcoClpX (Supplementary Fig. 11b); NLS-tagged human ClpX, which normally functions in the mitochondria, had no effect on integration (Supplementary Fig. 11c). Interestingly, genomic integration with eCAST-1 (VchCAST) was reproducibly detectable in the presence of EcoClpX or VchClpX but not in its absence, indicating a consistent effect across Type I-F CAST systems, although lower intrinsic activity of VchCAST was observed similar to plasmid-to-plasmid integration assays (Supplementary Fig. 11b). Collectively, these results suggest that PTC disassembly may be a critical bottleneck limiting integration into genomic target sites and identify ClpX as an accessory factor that acts to unfold one or more components within the CAST transpososome (Supplementary Fig. 11d). Future experiments will be needed to further dissect mechanistic details of this pathway.
Single-digit genomic integration efficiencies at the AAVS1 locus allowed us to explore other parameters of eCAST-3 design and delivery. We found that crRNAs functioned best with 33-nt spacers on both plasmid and genomic targets (Supplementary Fig. 12a,b) and that transfections could be simplified by placing the U6-driven crRNA cassette directly on pDonor without an adverse effect on activity (Supplementary Fig. 12c). Integration could be further improved with the appropriate selection of cationic lipid formulation (Supplementary Fig. 12d) or by selecting/sorting cells that were co-transfected with either a drug or fluorescent marker, with efficiencies reaching ~5% as measured by amplicon sequencing and ddPCR (Fig. 5h and Supplementary Fig. 12e,f). Notably, we also carefully inspected our next-generation sequencing (NGS) data to assess product purity at genomic sites of integration, looking specifically at whether unedited alleles showed any evidence of mutations and whether edited alleles containing a transposon insertion harbored unexpected modifications. These analyses revealed an absence of indels above background (~0.04% sequencing error) at unedited target sites and an absence of detectable mutations surrounding genome–transposon junctions (Supplementary Fig. 12g-i), suggesting that CAST systems are less prone to the range of byproducts common to Cas9 nuclease and nickase-based approaches18,19.
Lastly, we revisited previously targeted sites across the human genome and assessed integration efficiency to test the generalizability of ClpX enhancement (Supplementary Fig. 13a). Strikingly, we observed a 10–600-fold increase in integration efficiencies across all tested loci (Fig. 5i), with a consistent preference for insertions ~49 bp downstream of the crRNA-matching target site (Supplementary Fig. 13b), as first reported in our E. coli studies30,31.
Discussion
Here we describe successful implementation of CAST systems for RNA-guided DNA integration into endogenous sites in the human genome. Recent reports described preliminary evidence of Type V-K CASTs functioning on plasmid substrates in human cells72,73, albeit at efficiencies below 0.05%, similar to the upper-end efficiencies of eCAST-1. Our iterative engineering yielded single-digit genomic integration efficiencies with eCAST-3, showcasing the ability of CASTs to insert large genetic sequences without generating DNA DSBs, despite their molecular complexity. Advances with TwinPE and PASTE technologies have achieved up to ~25% DNA integration efficiencies with similar delivery methods and show promising results from in vivo delivery. However, unlike CAST systems, these approaches require multiple independent editing steps, produce low levels of indels, insert the entire vector via recombination and require extensive prime editing guide RNA (pegRNA) optimization18,19. Although further CAST improvements will be necessary for broad use in research and therapeutic applications, our results pinpoint specific features that would circumvent the drawbacks of TwinPE and PASTE. More generally, this work demonstrates the feasibility of reconstituting multi-component editing pathways in human cells and highlights a robust pipeline to engineer promising candidates for continued development.
We established functional assays to carefully assess each modular component of the V. cholerae Type I-F CAST system (eCAST-1), which revealed specific terminal tagging modifications that severely reduced or, in some cases, altogether eliminated activity (Supplementary Fig. 2c). Accordingly, the integration experiments in this study relied on transient delivery of multiple protein and RNA expression plasmids alongside the donor plasmid in a single co-transfection. Given the sensitivity of CAST systems to protein/complex stoichiometry, this approach reduces the fraction of cells that receive optimal distributions of each component. Moving forward, the increased amenability of eCAST-2 (that is, PseCAST) components to N-terminal and C-terminal tagging, as compared to eCAST-1 (Supplementary Fig. 7e), together with structure-guided engineering and recent examples of naturally fused class 1 complexes74, provide strong support for further streamlining the system into fewer molecular components while retaining its intrinsic properties. In addition, direct delivery of purified protein, RNA and DNA components offers a particularly promising area of investigation, and electroporation of pre-assembled transpososomes comprising the transposon DNA and TnsABf may improve trafficking and/or co-localization of the donor genetic payload and transposase to the target site.
When we screened homologous systems, we observed a wide range of relative activities for crRNA maturation, transposon DNA binding and TnsC-based transcriptional activation, indicating that each molecular step of the pathway may require independent optimization. For example, although components derived from Tn6677 (VchCAST) exhibited the strongest levels of activation, our assay for transposon DNA binding by TnsB revealed that homologs from Tn7005and Tn7010 exhibited more than 200-fold activation in human cells (Supplementary Fig. 6). Although we cannot exclude the possibility that the specific fusion constructs and reporter assay designs developed for this experiment fail to faithfully reflect TnsB activity, our results nevertheless suggest that none of the systems currently tested combines optimal activities in human cells across each molecular component.
In addition to the potential that RNA-guided transposases offer for DNA integration applications, we were excited to find that recruitment of the AAA+ ATPase TnsC, when fused with VP64 domains, stimulated robust levels of transcriptional activation at both plasmid and genomic target sites that were similar to levels achieved with dCas9–VPR fusion proteins. Recent structural and functional studies have demonstrated that TnsC homologs form ATP-dependent oligomers that assemble around dsDNA51,52,75, and we showed that TnsC is recruited to genomic loci in eukaryotic cells with high fidelity in a QCascade-dependent manner (Fig. 3), similar to recent experiments performed in E. coli43. Thus, combining these molecular components, while foregoing the heteromeric transposase itself, reveals a potent strategy to assemble an intrinsically multi-meric protein at user-defined target sites, for applications where multi-valency offers a considerable benefit. In addition to fusing TnsC to other activation or repression domains for control over gene expression levels, similar to existing CRISPRa and CRISPRi tools, we propose tethering epigenetic modifiers for DNA and/or histone modifications or fluorescent proteins for higher signal-to-noise ratios for chromosomal loci imaging assays without requiring arrays of gRNAs76,77 Furthermore, by also leveraging the multi-subunit nature of the QCascade complex, one could access more elaborate scaffolding approaches to recruit multiple functionalities to individual target sites simultaneously, such as by fusing effector domains to Cas8, Cas7 and/or TnsC in various combinatorial fashions.
Perhaps the most notable outcome of our study was the identification of bacterial ClpX as a novel accessory protein involved in CRISPR RNA-guided transposition (Fig. 5 and Supplementary Fig. 11). The disparity that we observed between integration efficiencies into episomal plasmid substrates versus genomic targets inspired us to more carefully consider the importance of CAST transpososome disassembly in exposing integration product intermediates for gap fill-in and repair. Based on a careful review of the Tn7 and Mu transposon literature, we hypothesized that protein unfoldases might facilitate active dissociation of one or more CAST components. Subsequent experiments revealed that ClpX enhanced genomic integration activity by two orders of magnitude, reaching single-digit efficiencies across multiple target sites (Fig. 5). Heterologous expression of bacterial ClpX did show evidence of CAST-independent cellular toxicity, suggesting deleterious effects on protein homeostasis that will require further investigation. However, we envision focusing future engineering efforts on alternative strategies to stimulate transpososome disassembly without the need for additional factors, informed by a better understanding of the underlying molecular mechanism. Alongside recent studies that uncovered the unexpected roles of ribosomal protein S15 and integration host factor (IHF) in select Type V-K and I-F CAST systems56,78, our ClpX finding indicates that CAST systems may be more reliant on host proteins than previously appreciated and that all chemical steps in the transposition pathway need to be critically evaluated.
CRISPR-based genome engineering tools have largely focused on single-protein effectors over the past decade, including Cas9, Cas12 and Cas13, because of the straightforward design of expression vectors, ease of viral delivery and perceived simplicity in reconstitution. However, recent studies highlight the feasibility of transplanting more complex CRISPR–Cas effectors into eukaryotic cells while retaining the ability to achieve high editing efficiencies and exploit novel enzymatic functionalities33-36,79. Our work extends this paradigm further while leveraging a class of transposases that offers the promise of single-step insertion of large, multi-kilobase genetic payloads with the programmability afforded by RNA-guided CRISPR–Cas systems.
Methods
Plasmid construction
Genes were human codon optimized and synthesized by GenScript, and plasmids were generated using a combination of restriction digestion, ligation, Gibson assembly and inverted (around-the-horn) PCR. All PCR fragments for cloning were generated using Q5 DNA Polymerase (New England Biolabs (NEB)).
The CRISPR array sequence (repeat-spacer-repeat) for VchCAST is as follows:
5′–GTGAACTGCCGAGTAGGTAGCTGATAAC–N32–GTGAACTGCCGAGTAGGTAGCTGATAAC–3′ where N32 represents the 32-nt guide region. The sequence of the mature crRNA is as follows:
5′–CUGAUAAC–N32–GUGAACUGCCGAGUAGGUAG–3′
The CRISPR array sequence (repeat-spacer-repeat) for PseCAST is as follows:
5′–GTGACCTGCCGTATAGGCAGCTGAAAAT–N32–GTGACCTGCCGTATAGGCAGCTGAAAAT–3′ where N32 represents the 32-nt guide region. The sequence of the mature crRNA is as follows:
5′–CUGAAAAU–N32–GUGACCUGCCGUAUAGGCAG–3′
We also used ‘atypical’ repeats32,80 for PseCAST (unless otherwise mentioned) to reduce the likelihood of recombination during cloning. For these variant CRISPR arrays, the repeat-spacer-repeat sequence is as follows:
5′–GTGACCTGCCGTATAGGCAGCTGAAGAT–N32–TAATTCTGCCGAAAAGGCAGTGAGTAGT–3′ where N32 represents the 32-nt guide region. The sequence of the mature crRNA is as follows:
5′–CUGAAGAU–N32–UAAUUCUGCCGAAAAGGCAG–3′. Where noted, we modified the 32-nt guide region to have varying lengths. The repeat sequences flanking the guide region were not modified in these experiments.
Clp proteins from the E. coli genome were PCR amplified from BL21 DE3 cells with primers that specifically amplified the ORF of the indicated protein and cloned into pcDNA3.1 expression vectors with an N-terminal BP NLS tag. ClpX sequences from E. coli, Pseudoalteromonas sp. and V. cholerae were then codon optimized by GenScript and ordered as Twist fragments to be cloned into pcDNA3.1 expression vectors with an N-terminal BP NLS tag.
E. coli culturing and general transposition assays
Chemically competent E. coli BL21(DE3) cells carrying pDonor, pDonor and pTnsABC, or pDonor and pQCascade, were prepared and transformed with 150–250 ng of pEffector, pQCascade or pTnsABC, respectively. Transformations were plated on agar plates with the appropriate antibiotics (100 μg ml−1 spectinomycin, 100 μg ml−1 carbenicillin, 50 μg ml−1 kanamycin) and 0.1 mM IPTG. For bacterial transposition assays investigating PseCAST activity, cells were co-transformed with pEffector and pDonor. Cells were incubated for 18–20 hours at 37 °C and typically grew as densely spaced colonies, before being scraped, resuspended in LB medium and prepared for subsequent analysis. A full list of all plasmids used for transposition experiments is provided in Supplementary Table 1, and a list of crRNAs used is provided in Supplementary Table 2.
E. coli qPCR analysis of transposition products
The optical density (OD) of resuspended colonies from our transposition assays was measured at 600 nm, and approximately 3.2 × 108 cells (the equivalent of 200 μl of OD600 = 2.0) were pelleted by centrifugation at 4,000g for 5 minutes. The cell pellets were resuspended in 80 μl of water before being lysed by incubating at 95 °C for 10 minutes in a thermal cycler. The cell debris was pelleted by centrifugation at 4,000g for 5 minutes, and 5 μl of lysate supernatant was removed and serially diluted in water to generate 20-fold and 500-fold lysate dilutions for qPCR analysis.
Integration in the T-RL orientation was measured by qPCR by comparing Cq values of a T-RL-specific primer pair (one transposon-specific primer and one genome-specific primer) to a genome-specific primer pair that amplifies an E. coli reference gene (rssA). Transposition efficiency was then calculated as 2ΔCq, in which ΔCq is the Cq difference between the experimental reaction and the reference reaction. A full list of oligos used for integration efficiency measurements is provided in Supplementary Table 3. qPCR reactions (10 μl) contained 5 μl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), 1 μl of water, 2 μl of 2.5 μM primers and 2 μl of 500-fold diluted cell lysate. Reactions were prepared in 384-well white PCR plates (Bio-Rad), and measurements were performed on a CFX384 Real-Time PCR Detection System (Bio-Rad) using the following thermal cycling parameters: polymerase activation and DNA denaturation (98 °C for 3 minutes) and 35 cycles of amplification (98 °C for 10 seconds, 59 °C for 1 minute).
Mammalian cell culture and transfections
HEK293T cells were cultured at 37 °C and 5% CO2. Cells were maintained in DMEM media with 10% FBS and 100 U ml−1 of penicillin and streptomycin (Thermo Fisher Scientific). The cell line was authenticated by the supplier and tested negative for mycoplasma.
Cells were typically seeded at approximately 100,000 cells per well in a 24-well plate (Eppendorf or Thermo Fisher Scientific) coated with poly-d-lysine (Thermo Fisher Scientific) 24 hours before transfection. Cells were transfected with DNA mixtures and 2 μl of Lipofectamine 2000 (Thermo Fisher Scientific) per the manufacturer’s instructions. Transfection reactions typically contained between 1 μg and 1.5 μg of total DNA. For detailed transfection parameters specific to distinct assays, refer to the sections below.
Western immunoblotting and nuclear/cytoplasmic fractionation
Cells were transfected with epitope-tagged protein expression plasmids. Approximately 72 hours after transfection, cells were washed with PBS and harvested using Cell Lysis Buffer (150 mM NaCl, 0.1% Triton X-100, 50 mM Tris-HCl pH 8.0, protease inhibitor (Sigma Aldrich)). For nuclear and cytoplasmic fractionation experiments, cells were harvested using Cell Lysis Buffer (Thermo Fisher Scientific) per the manufacturer’s instructions. Proteins were separated by SDS-PAGE and transferred to a PVDF membrane (Thermo Fisher Scientific). The membrane was then washed with TBS-T (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.1% Tween 20) and blocked with blocking buffer (TBS-T with 5% w/v BSA). Membranes were then incubated with primary antibodies overnight at 4 °C in blocking buffer. Membranes were then washed and incubated with secondary antibodies at room temperature for 1 hour. All antibodies (both primary and secondary) were diluted 1:10,000 in blocking buffer. Membranes were again washed and then developed with SuperSignal West Dura (Thermo Fisher Scientific). Antibodies used for immunostaining are listed in Supplementary Table 4.
HEK293T fluorescent reporter assays and flow cytometry analysis and sorting
HEK293T cells were seeded at approximately 50,000 cells per well in a 24-well plate coated with poly-d-lysine 24 hours before transfection. For Cas6-mediated RNA processing assays, cells were co-transfected with 300 ng of GFP reporter plasmid, 300 ng of pCas6 and 10 ng of an mCherry expression plasmid (as a transfection marker). In negative control experiments, cells were transfected with 300 ng of a pdCas9 instead of a pCas6 to control for possible expression burden or squelching. For transcriptional activation assays, cells were co-transfected with 60 ng of reporter plasmid, 20 ng of a plasmid encoding an orthogonal fluorescent protein (as a transfection marker) and the additional indicated plasmids. Unless otherwise noted, transcriptional activation assays utilized crRNA1 as the pCRISPR. In separate wells, cells were transfected with 100 ng of Cas9-based transcriptional activators and 50 ng of either a non-targeting or a targeting sgRNA as positive controls81. Representative flow cytometry analysis can be seen in Supplementary Fig. 15.
DNA mixtures were transfected using 2 μl of Lipofectamine 2000 (Thermo Fisher Scientific) per the manufacturer’s instructions. Approximately 72–96 hours after transfection, cells were collected for assay by flow cytometry. Transfected cells were analyzed by gating based on fluorescent intensity of the transfection marker relative to a negative control, as previously described81. For assays that involved cell sorting, cells were transfected with a GFP expression plasmid and collected 4 days after transfection. A BD FACSAria flow cytometer was used to sort cells and obtain flow cytometry data. Cells with the top 20% brightest GFP fluorescence were sorted by 5% increments into four bins. Cells were immediately harvested after sorting, as detailed below.
HEK293T genomic activation and RT–qPCR analysis
HEK293T cells were seeded at approximately 50,000 cells per well in a 24-well plate coated with poly-d-lysine 24 hours before transfection. Cells were co-transfected as described above, with the following VchCAST components: 100 ng of pTnsABf, 50 ng of pTnsC-VP64, 50 ng of pTniQ, 50 ng of pCas6, 250 ng of pCas7, 50 ng of pCas8 and 62.5 ng each of four targeting crRNAs for TTN, MIAT and ASCL1 (or 83.3 ng each of three targeting crRNAs for ACTC1) (pCRISPR). In control experiments, cells were co-transfected with 100 ng of either pdCas9–VP64 or pdCas9–VPR plasmid, 62.5 ng each of four targeting sgRNAs for TTN (psgRNA) and a pUC19 plasmid to standardize transfected DNA amounts; see Supplementary Table 2 for crRNAs and sgRNAs used. Cells were harvested 72 hours after transfection using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was subsequently synthesized using the iScript cDNA Synthesis Kit (Bio-Rad) using 1,000 ng of RNA in a 20-μl reaction. Gene-specific qPCR primers53 were designed to amplify an approximately 180–250-bp fragment to quantify the RNA expression of each gene, and a separate pair of primers was designed to amplify ACTB (β-actin) reference gene for normalization purposes. A comprehensive list of oligonucleotides used in the study is available in Supplementary Table 3.
qPCR reactions (10 μl) contained 5 μl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), 2 μl of water, 1μl of 5 μM primer pair and 2 μl of cDNA diluted 1:4 in water. Reactions were prepared in 384-well white PCR plates (Bio-Rad), and measurements were performed on a CFX384 Real-Time PCR Detection System (Bio-Rad) using the following thermal cycling parameters: polymerase activation and DNA denaturation (98 °C for 2 minutes), 40 cycles of amplification (95 °C for 10 seconds, 60 °C for 30 seconds) and terminal melt curve analysis (65 °C–95 °C in 0.5 °C-per-5-second increments). Each condition was analyzed using three biological replicates, and two technical replicates were run per sample. Normalized gene activation was calculated as the ratio of the 2−ΔCq of the targeting samples to the non-targeting samples, in which ΔCq is the Cq difference between the experimental gene primer pair and the reference gene primer pair.
ChIP
For ChIP-seq analysis experiments, HEK293T cells were seeded at approximately 1,500,000 cells per well in a 10-cm dish coated with poly-d-lysine 24 hours before transfection. Cells were co-transfected as described above with the following eCAST-1 components: 1.5 μg of p3×FLAG-TnsC, 1.5 μg of pTniQ, 1.5 μg of pCas6, 7.5 μg of pCas7, 1.5 μg of pCas8 and 3 μg of either a targeting (TTN crRNA 1) or a non-targeting crRNA. See Supplementary Table 2 for crRNAs used. Seventy-two hours after transfection, cells were harvested and pelleted by centrifugation at 300g for 5 minutes, and the supernatant was aspirated. The pellets were processed as described previously43,82,83. In brief, pellets were resuspended in 1% freshly made formaldehyde (Thermo Fisher Scientific) in DPBS and shaken gently for 10 minutes. Fixation was quenched by adding 2.5 M glycine, for a final concentration of 125 mM glycine, and rotating cells for 5 minutes. Cells were pelleted, washed with cold DPBS, pelleted, resuspended in DPBS and 1× cOmplete EDTA-free protease inhibitors (Sigma-Aldrich), pelleted, flash frozen in liquid nitrogen and stored at −80 °C.
On the day of sonication, the cross-linked pellets were resuspended in 1 ml of Lysis Buffer 1 (50 mM HEPES-KOH, 140 mM NaCl,1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) and 1× protease inhibitors and rotated for 10 minutes. Cells were pelleted at 1,350g for 5 minutes. Pellets were resuspended in 1 ml of Lysis Buffer 2 (10 mM Tris-HCl, 200 mM, NaCl, 1 mM EDTA, 0.5 mM EGTA) and 1× protease inhibitors and rotated for 10 minutes before being pelleted at 1,350g for 5 minutes. Pellets were resuspended in 900 μl of Lysis Buffer 3 (10 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine), 100 μl of 10% Triton X-100 and 1× protease inhibitors. All steps took place at 4 °C.
The resuspended cells were transferred to a 1-ml milliTUBE AFA Fiber (Covaris) and sonicated on an M220 Focused-ultrasonicator (Covaris) under the following SonoLab 7.2 settings: minimum temperature 4 °C, set point 6 °C, maximum temperature 7 °C, Peak Power 75.0, Duty Factor 10.0, Cycles/Burst 200 and sonication time 490 seconds. Sonicated cell lysate was centrifuged at 20,000g for 10 minutes at 4 °C. The supernatant was transferred to a new tube, and 5% was saved as the input sample. The remaining supernatant was incubated with Dynabeads Protein G (Thermo Fisher Scientific) that were bound to the monoclonal anti-Flag M2 antibody at a 1:8 dilution (Sigma-Aldrich) the day before sonication by overnight rotating at 4 °C, and the lysate–Dynabeads mixture was rotated overnight at 4°C.
The samples were washed three times each with low salt buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1 mM EDTA, 50 mM Tris HCl); high salt buffer (550 mM NaCl, 0.1% SDS, 1% Triton X-100, 1 mM EDTA, 50 mM Tris HCl); and LiCl buffer (150 mM LiCl, 0.5% Na-deoxycholate, 0.1% SDS, 1% Nonidet P-40, 1 mM EDTA, 50 mM Tris HCl) on a magnetic stand at 4 °C. The samples were washed with 1 ml of TE buffer (1 mM EDTA, 10 mM Tris HCl) with 50 mM NaCl and centrifuged at 960g for 3 minutes at 4 °C. The supernatant was aspirated, and 210 μl of elution buffer (1% SDS, 50 mM Tris HCl, 10 mM EDTA, 200 mM NaCl) was added to samples and incubated for 30 minutes at 65 °C. Samples were centrifuged for 1 minute at 16,000g at room temperature, and 200 μl of supernatant was incubated overnight at 65 °C. The input sample was diluted in 150 μl of elution buffer and also incubated overnight at 65 °C. Then, 0.5 μl of 10 mg ml−1 RNase was added, and samples were incubated for 1 hour at 37 °C. Next, 2μl of 20 mg ml−1 proteinase K was added, and samples were incubated for 1 hour at 55 °C. The DNA was recovered by the QiaQUICK PCR Purification Kit (Qiagen), and DNA was eluted in 50 μl of water for downstream analysis.
ChIP-seq sample preparation
Sample DNA concentration was determined by the DeNovix dsDNA High Sensitivity Kit. Illumina libraries were generated using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB), as described previously43. Sample concentrations were normalized such that 12 ng of DNA in each condition was used for library preparation. The concentration of DNA was determined for pooling using the DeNovix dsDNA High Sensitivity Kit. Illumina libraries were sequenced in paired-end mode on the Illumina NextSeq platform with automated demultiplexing and adaptor trimming. For each ChIP-seq sample, 75-bp paired-end reads were obtained, and between 9.5 million and 18.9 million uniquely mapped fragments were analyzed.
ChIP-seq analysis
ChIP-seq data were processed using CoBRA version 2.0 (ref. 84) with modifications as follows. Each experimental condition (TnsC with TTN-targeting gRNA or TnsC with non-targeting gRNA) was processed with three biological replicate ChIP samples and one corresponding non-immunoprecipitated input sample. Reads were aligned to the hg38 human reference genome using BWA-MEM with default settings. Reads were sorted and indexed using SAMtools85, and multi-mapping reads with a MAPQ score <1 were removed using the SAMtools view command. Peaks were called using MACS2 version 2.2.6 (ref. 86). The callpeak function was executed in paired-end mode with the following parameters: −g 2.7e9 −q 0.0001–keep-dup auto–nomodel. Input samples were used as controls for peak calling. bedGraph files for each sample with pileup information in signal per million reads (SPMR) were generated with the –SPMR and –B subcommands of MACS2 callpeak and were converted to bigWig files using bedGraphToBigWig. ChIP-seq signal at individual genomic loci was visualized with Integrative Genomics Viewer (IGV)87. Reads mapping to the Y chromosome or the mitochondrial genome were removed before downstream analysis.
A consensus list of peaks for each experimental condition was identified using BEDTools version 2.30.0 (ref. 88). First, peak files for the three replicates were concatenated and sorted, and overlapping peaks were merged. Then, peaks appearing in fewer than three replicates were removed. Blacklisted regions of the genome defined by the ENCODE Consortium were also removed89. The consensus lists for the conditions were then intersected to identify peaks exclusive to either condition (BEDTools intersect –v) or peaks shared by both conditions (BEDTools intersect –u). Differential binding analysis was performed using DiffBind version 3.6.5 (ref. 90) to compare ChIP-seq read density between the two conditions in the regions defined by their consensus peak lists. Reads were counted using dba.count with the following arguments: summits = F, bUseSummarizeOverlaps = T, bRemoveDuplicates = F, bSubControl = F. Read counts were normalized to account for differences in sequencing depth between samples. Normalized read counts were passed to DESeq2 to calculate the mean across conditions, as well as fold change and q value (using the Benjamini–Hochberg procedure) between conditions, for each peak. The result of differential binding analysis was visualized using ggplot2.
Heat maps of ChIP-seq signal intensity over peaks exclusive to the TTN gRNA condition were plotted using deepTools version 3.3.2 (ref. 91). Score matrices were generated using computeMatrix in reference-point mode. Peaks were sorted in descending order by mean signal over 2-kb windows around peak centers before plotting using plotHeatmap.
For manual inspection of potential off-target sites, a custom script was used to identify genomic loci with high similarity to the TTN spacer sequence. Other than the TTN locus itself, no loci with fewer than five mismatches were identified. TnsC ChIP-seq signal at the five most similar loci was visualized with IGV.
HEK293T integration assays
For assays in which plasmids were isolated and used to transform bacteria, HEK293T cells were transfected with requisite eCAST-1 expression plasmids, a pDonor that contained a non-replicative origin of replication (R6K), a pTarget plasmid and a crRNA expression plasmid (pCRISPR) that encoded either a non-targeting crRNA or a crRNA targeting pTarget. Seventy-two hours after transfection, cells were washed with PBS, harvested using TrypLE (Thermo Fisher Scientific), neutralized with culture media and pelleted. After removal of supernatant, transfected plasmids were harvested using Qiagen Miniprep columns per the manufacturer’s instructions, and further concentrated using the Qiagen MinElute column. Of this final purified plasmid mixture, 1 μl was used to electroporate NEB 10-beta electrocompetent E. coli cells (NEB) per the manufacturer’s instructions. After recovery at 37 °C, cells were plated onto LB agar plates containing chloramphenicol. Chloramphenicol-resistant colonies were then replated onto LB agar plates containing both chloramphenicol and kanamycin, and doubly-resistant colonies were harvested for genotypic analyses.
For all other integration assays, HEK293T cells were counted using a Countess 3 Cell Counter and seeded at 20,000 cells per well, unless otherwise specified, in a 24-well plate coated with poly-d-lysine 24 hours before transfection. Cells were transfected using plasmid DNA mixtures and 2 μl of Lipofectamine 2000 per the manufacturer’s instructions. For eCAST-1 transposition assays, HEK293T cells were transfected with the following optimized VchCAST components, unless otherwise stated: 300 ng of pTnsABf, 25 ng of pTnsC, 100 ng each of pTniQ, pCas6, pCas7 and pCas8, 200 ng of pDonor, 100 ng of pTarget and 100 ng of a targeting or non-targeting crRNA (pCRISPR). For eCAST-2 transposition assays, HEK293T cells were transfected with the following PseCAST components, unless otherwise specified: 200 ng of pTnsABf, 50 ng each of pTnsC, pTniQ, pCas6, pCas7 and pCas8, 200 ng of pDonor and 100 ng of pTarget and a targeting or non-targeting crRNA (pCRISPR). When a QCascade polycistronic expression vector was used (pQCas), 75 ng was transfected. For eCAST-3 transposition assays, eCAST-2 conditions were used with pQCas, and 20 ng of pClpX was co-transfected as well (unless otherwise noted). All eCAST-3 transposition assays used puromycin selection (unless otherwise noted; see below for puromycin conditions), as constitutive ClpX expression led to visible toxicity independent of CAST machineries. A full list of plasmids and crRNAs used is available in Supplementary Table 1 and Supplementary Table 2, respectively. Unless otherwise stated, cells were cultured for 4 days after transfection. Cells were washed with DPBS with no calcium or magnesium (Thermo Fisher Scientific), harvested using TrypLE (Thermo Fisher Scientific) and neutralized with culture media. Twenty percent of the resuspended cells were pelleted by centrifugation at 300g for 5 minutes, and the supernatant was aspirated. Cell pellets were resuspended in 50 μl of Quick Extract (Lucigen), and genomic DNA was prepared per the manufacturer’s instructions.
For assays that used puromycin selection, HEK293T cells were transfected as described above with the addition of 20 ng of puromycin resistance expression plasmid as a transfection marker. Media were changed 24 hours after transfection, and selection with 1 μg ml−1 of puromycin was started. Cells were harvested using Quick Extract (Lucigen) per the manufacturer’s instructions, either 4 days after transfection or, for time-course experiments, beginning at 2 days after transfection until 6 days after transfection, with or without puromycin selection. For plasmid-based assays that used cell sorting, HEK293T cells were transfected with eCAST-2 components as described above with an additional 5 ng of GFP expression plasmid as a transfection marker. Four days after transfection, the GFP-positive cells with the brightest MFI were sorted in four bins of 5% increments to encompass the 20% brightest cells and were immediately harvested as described above. For genomic assays that used cell sorting, HEK293T cells were seeded at approximately 100,000 cells in six-well plates coated with poly-d-lysine 24 hours before transfection. Cells were transfected with the following eCAST-3 components: 1,000 ng each of pTnsABf and pDonor, 250 ng of pTnsC, 375 ng of polycistronic pCas7-Cas8-Cas6-TniQ, 20 ng of pGFP, 100 ng of pClpX and 500 ng of a targeting crRNA (pCRISPR). Four days after transfection, the top 20% of GFP-positive cells with the brightest MFI were sorted and immediately harvested, as described above. For genomic integration assays, cells were harvested by previously described assays, using 100 μl of freshly prepared lysis buffer (10 mM Tris-HCl pH 7.5, 0.05% SDS, 25 μg ml−1 proteinase K (Thermo Fisher Scientific)) directly into each well of the tissue culture plate. The genomic DNA mixture was incubated at 37 °C for 1–2 hours, followed by an 80 °C enzyme inactivation step for 30 minutes17.
For assays that used cargo sizes ranging from 798 bp to 15 kb, HEK293T cells were transfected as described above with eCAST-2 component plasmids, except the 5-kb, 10-kb and 15-kb pDonor plasmids were transfected in molar equivalents to the 798-bp pDonor (~406 fmol), to account for the size difference between donor plasmids. For assays that used amplicon deep sequencing, HEK293T cells were transfected as described above, with a pDonor plasmid that contained a primer binding site immediately downstream of the right transposon end that matched a primer binding site present in the unedited pTarget plasmid. Cells were harvested 4 days after transfection.
Nested PCR analysis of transposition assays
DNA amplification was performed by PCR using Q5 Hot Start High-Fidelity DNA Polymerase (NEB) following the manufacturer’s protocol. In brief, for PCR-1, 1 μl of cell lysate was added to a 25-αl PCR reaction. Thermocycling conditions were as follows: 98 °C for 45 seconds, 98 °C for 15 seconds, 66 °C for 15 seconds, 72 °C for 10 seconds and 72 °C for 2 minutes, with steps 2–4 repeated 24 times. The annealing temperature was adjusted depending on primers used. One microliter of the first PCR reaction served as the template for PCR-2, a 25-μl PCR reaction that was run under the same thermocycling conditions. Primer pairs contained one target-specific primer and one transposon-specific primer, and the primers used in the second PCR reaction generated a smaller amplicon than the first reaction (see Supplementary Table 3 for oligonucleotides used in this study). PCR amplicons were resolved by 1–2% agarose gel electrophoresis and visualized by staining with SYBR Safe (Thermo Fisher Scientific). Negative control samples were always analyzed in parallel with experimental samples to identify mis-priming products, some of which presumably result from the analysis being performed on crude cell lysates that still contain the pDonor and target site DNA.
qPCR analysis of plasmid-to-plasmid and genomic integration products
Transposition-specific qPCR primers were designed to amplify a ~140-bp fragment to quantify integration efficiency. Primer pairs were designed to span the integration junction, with the forward primer annealing to pTarget, or the genome, and the reverse primer annealing within the transposon. Additionally, a custom 5′ FAM-labeled, ZEN/3′ IBFQ probe (Integrated DNA Technologies) was designed to anneal to each unique integration junction. A separate pair of primers and a SUN-labeled, ZEN/3′ IBFQ probe (Integrated DNA Technologies) were designed to amplify a distinct reference sequence in the target plasmid or the human genome, for efficiency calculation purposes. For a full list of oligonucleotides used in qPCR, refer to Supplementary Table 3.
Probe-based qPCR reactions (10 μl) contained 5 μl of TaqMan Fast Advanced Master Mix, 0.5 μl of each 18 μM primer pair, 0.5 μl of each 5 μM probe, 1 μl of water and 2 μl of ten-fold diluted cell lysate for plasmid-based transposition samples or 2 μl of five-fold diluted cell lysate for genomic transposition samples. Reactions were prepared in 384-well white PCR plates (Bio-Rad), and measurements were performed on a CFX384 Real-Time PCR Detection System (Bio-Rad) using the following thermal cycling parameters: polymerase activation (95°C for 10 minutes) and 50 cycles of amplification (95 °C for 15 seconds, 59.5 °C for 1 minute). Each condition was analyzed using either two or three biological replicates, and two technical replicates were run per sample. Baseline threshold ratios were manually adjusted to be 1:1 for the reference primer pair to the transposition primer pair. Integration efficiency was calculated as a percentage as 2−ΔCq times 100, in which ΔCq is the Cq difference between the reference primer pair and the transposition primer pair.
To analyze the frequency of left–right insertion (T-LR) versus right–left insertion (T-RL) of the PseCAST transposon in plasmid-based assays, integration-specific qPCR primers were designed to span the T-LR integration junction, in addition to the primer pairs used for T-RL integration and the reference amplicon in the probe-based qPCR analysis described above. qPCR reactions (10 μl) contained 5 μl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), 2 μl of water, 1 μl of 5 μM primer pair and 2 μl of ten-fold diluted cell lysate. Reactions were prepared in 384-well white PCR plates (Bio-Rad), and measurements were performed on a CFX384 Real-Time PCR Detection System (Bio-Rad) using the following thermal cycling parameters: polymerase activation and DNA denaturation (98 °C for 2 minutes), 50 cycles of amplification (95 °C for 10 seconds, 59.5 °C for 20 seconds) and terminal melt curve analysis (65 °C–95 °C in 0.5 °C-per-5-second increments). Each condition was analyzed using three biological replicates, and two technical replicates were run per sample.
ddPCR analysis of integration products
During harvesting of HEK293T plasmid-based integration assays, 50% of the resuspended cells were reserved during lysate generation. Then, 500 μl of resuspended cells were pelleted by centrifugation at 300g for 5 minutes. The supernatant was aspirated, and DNA was extracted from cell pellets using the Qiagen DNeasy Blood and Tissue Kit (Qiagen). DNA was eluted in water and diluted to a concentration of 2.5 ng μl−1. For genomic integration assays, crude cell lysate, generated as described above, was purified using two-sided AMPure XP beads (Beckman Coulter) as follows43: 45 μl of AMPure XP beads were added to 20–80 μl of genomic lysate and incubated for 5 minutes before being placed on a magnetic PCR rack for 5 minutes. The supernatant was aspirated, and the beads were washed twice with 80% ethanol. The beads were dried for 5 minutes, and then 25 μl of water was added to resuspend the beads. The suspension was incubated for 10 minutes off the magnetic rack and then placed back on the rack for 5 minutes. The supernatant was transferred to a new tube.
ddPCR was performed with the same primers and probes as detailed above for plasmid-to-plasmid integration analysis and genomic integration assays with the exception of the OXA1L-2 target site, which was not quantified via qPCR. For a full list of oligonucleotides used in ddPCR, refer to Supplementary Table 3. Plasmid-based ddPCR reactions (20 μl) contained 10 μl of ddPCR Supermix for Probes (Bio-Rad), 1 μl of each 5 μM probe, 1 μl of each 18 μM primer pair, 5 U of HindIII (NEB), 4.13 μl of water and 2 μl of 2.5 ng μl−1 DNA. Genomic ddPCR reactions (20 μl) contained 10 μl of ddPCR Supermix for Probes (Bio-Rad), 1 μl of each 5 μM probe, 1 μl of each 18 μM primer pair, 5 U of HindIII (NEB) and 6.33 μl of purified DNA, ranging from ~6 ng to ~500 ng. Reactions were assembled at room temperature, and droplets were generated using the Bio-Rad QX200 Droplet Generator according to the manufacturer’s instructions. Thermocycling was performed on a Bio-Rad C1000 Touch Thermocycler with the following parameters: enzyme activation (95 °C for 10 minutes), 40 cycles of amplification (94 °C for 30 seconds, 61.5 °C for 1 minute) and enzyme deactivation (98 °C for 10 minutes). After thermocycling, droplets were hardened at 4 °C for 2 hours. Droplets were analyzed using the QX200 Droplet Reader according to the manufacturer’s instructions. Integration percentages were calculated as the number of FAM-positive molecules divided by the number of SUN/VIC-positive molecules times 100.
Amplicon sequencing strategy to quantify integration efficiencies
To improve sensitivity of genomic integration assays in human cells, we designed an NGS-based approach in which both unedited sites and integration products are simultaneously amplified in a single PCR (Supplementary Fig. 9b). PCR-1 products were generated as described for PCR-1 in the nested PCR analyses, except primers contained universal Illumina adaptors as 5′ overhangs (Supplementary Table 3), and the cycle number was reduced to 15 for plasmid-to-plasmid integration assays and 25 for genomic integration assays. Additionally, up to five degenerate nucleotides were placed between the primer binding site and the Illumina adaptor 5′ overhang to improve library diversity when sequencing. One microliter of lysate per 10 μl of overall PCR reaction was used; plasmid-to-plasmid integration assays were 20-μl PCR reactions, whereas genomic integration assays were 250-μl PCR reactions to sample sufficient alleles. These products were then diluted 20-fold into a fresh PCR (PCR-2) containing indexed p5/p7 primers and subjected to ten additional thermal cycles using an annealing temperature of 65 °C. After verifying amplification by analytical gel electrophoresis, barcoded reactions were pooled and resolved by 2% agarose gel electrophoresis; DNA was isolated by Gel Extraction Kit (Qiagen); and NGS libraries were quantified by qPCR using the NEBNext Library Quant Kit (NEB). Illumina sequencing was performed using the NextSeq platform with automated demultiplexing and adaptor trimming (Illumina).
To determine integration efficiencies and distributions, reads were filtered that contained the expected 10-bp sequence immediately downstream of the forward primer, to verify that they derived from the target site. Next, reads containing a 10-bp transposon end sequence were counted as ‘integration reads’, and the integration distance was calculated from the start of the transposon end to the PAM-distal end of the target sequence. Reads that instead contained a 10-bp sequence from the unedited site at the end of the read were counted as ‘unedited reads’. The integration efficiency, or ‘integration reads (%)’, as marked in Fig. 5b-g, was calculated as the number of ‘integration reads’ divided by the sum of both ‘integration reads’ and ‘unedited reads’, converted to a percentage. Histograms of integration distances were plotted by compiling distances across all reads within a given sample.
Supplementary Material
Acknowledgements
We thank N. Jaber and S. R. Pesari for laboratory support, L. F. Landweber for qPCR instrument access, A. Santiago-Frangos for helpful ClpX discussions, the Columbia University Institute for Genomic Medicine for ddPCR instrument access, W. Frankel for thermocycler instrument access, the Columbia Stem Cell Initiative Flow Cytometry Core, M. Kissner and R. Gordon-Schneider for assistance in cell sorting and flow cytometry analysis, and the JP Sulzberger Columbia Genome Center for NGS support. We thank C. Lu and V. Sahu for cell sonicator instrument access and valuable discussions on ChIP-seq analysis. R.T.K. is supported by NIH grant 1F31HL167530-01 from the National Heart, Lung, and Blood Institute. A.C. is supported by a Career Award for Medical Scientists from the Burroughs Wellcome Fund and NIH grant HG011855-01 from the National Human Genome Research Institute. S.H.S. is supported by NIH grant DP2HG011650, a Pew Biomedical Scholarship, a Sloan Research Fellowship, an Irma T. Hirschl Career Scientist Award, and a generous startup package from the Columbia University Irving Medical Center Dean’s Office and the Vagelos Precision Medicine Fund.
Footnotes
Competing interests
Columbia University has filed a patent application related to this work. S.E.K., P.L.H.V., A.C. and S.H.S. are inventors on other patents and patent applications related to CRISPR–Cas systems and uses thereof. A.C. is a consultant for Vor Biopharma and an equity holder and scientific advisor for Cellgorithmics. S.H.S. is a co-founder of and scientific advisor to Dahlia Biosciences, a scientific advisor to Prime Medicine and CrisprBits and an equity holder in Dahlia Biosciences and CrisprBits. The remaining authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41587-023-01748-1.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587-023-01748-1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Sequencing data have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive under Gene Expression Omnibus accession number GSE223174 (ref. 92). Source data are provided with this paper.
References
- 1.Pickar-Oliver A & Gersbach CA The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol 20, 490–507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knott GJ & Doudna JA CRISPR–Cas guides the future of genetic engineering. Science 361, 866–869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol 38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Maruyama T. et al. Inhibition of non-homologous end joining increases the efficiency of CRISPR/Cas9-mediated precise genome editing. Nature 33, 538–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakade S. et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun 5, 5560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu VT et al. Increasing the efficiency of homology-directed repair for CRISPR–Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol 33, 543–548 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Yeh CD, Richardson CD & Corn JE Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol 21, 1468–1478 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Heyer W-D, Ehmesn KT & Liu J Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet 44, 113–139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moynahan ME & Jasin M Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol 11, 196–207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin S, Staahl BT, Alla RK & Doudna JA Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuccaro MV et al. Allele-specific chromosome removal after Cas9 cleavage in human embryos. Cell 183, 1650–1654 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Adikusuma F. et al. Large deletions induced by Cas9 cleavage. Nature 560, E8–E9 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol 36, 765–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leibowitz ML et al. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet 53, 895–905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol 35, 371–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anzalone AV et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol 40, 731–740 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yarnall MTN et al. Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases. Nat. Biotechnol 10.1038/s41587-022-01527-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naldini L, Trono D & Verma IM Lentiviral vectors, two decades later. Science 353, 1101–1102 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Querques I. et al. A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nat. Biotechnol 37, 1502–1512 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yusa K, Zhou L, Li MA, Bradley A & Craig NL A hyperactive piggyBac transposase for mammalian applications. Proc. Natl Acad. Sci. USA 108, 1531–1536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tipanee J, Vandendriessche T & Chuah MK Transposons: moving forward from preclinical studies to clinical trials. Hum. Gene Ther 28, 1087–1104 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Gaidukov L. et al. A multi-landing pad DNA integration platform for mammalian cell engineering. Nucleic Acids Res. 46, 4072–4086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durrant MG et al. Systematic discovery of recombinases for efficient integration of large DNA sequences into the human genome. Nat. Biotechnol 10.1038/s41587-022-01494-w (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hew BE, Sato R, Mauro D, Stoytchev I & Owens JB RNA-guided piggyBac transposition in human cells. Synth. Biol 4, ysz018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovač A. et al. RNA-guided retargeting of Sleeping Beauty transposition in human cells. eLife 9, e53868 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo W et al. Comparative analysis of chimeric ZFP-, TALE- and Cas9-piggyBac transposases for integration into a single locus in human cells. Nucleic Acids Res. 45, 8411–8422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen SP & Wang HH An engineered Cas-Transposon system for programmable and site-directed DNA transpositions. CRISPR J. 2, 376–394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klompe SE, Vo PLH, Halpin-Healy TS & Sternberg SH Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Vo PLH et al. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol 39, 480–489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klompe SE et al. Evolutionary and mechanistic diversity of type I-F CRISPR-associated transposons. Mol. Cell 82, 616–628 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cameron P. et al. Harnessing type I CRISPR–Cas systems for genome engineering in human cells. Nat. Biotechnol 37, 1471–1477 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Chen Y. et al. Repurposing type I-F CRISPR–Cas system as a transcriptional activation tool in human cells. Nat. Commun 11, 3136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickar-Oliver A. et al. Targeted transcriptional modulation with type I CRISPR–Cas systems in human cells. Nat. Biotechnol 37, 1493–1501 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolan AE et al. Introducing a spectrum of long-range genomic deletions in human embryonic stem cells using type I CRISPR–Cas. Mol. Cell 74, 936–950 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young JK et al. The repurposing of type I-E CRISPR-Cascade for gene activation in plants. Commun. Biol 2, 383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strecker J et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 364, 48–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito M et al. Dual modes of CRISPR-associated transposon homing. Cell 184, 2441–2453.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vo PLH, Acree C, Smith ML & Sternberg SH Unbiased profiling of CRISPR RNA-guided transposition products by long-read sequencing. Mob. DNA 12, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halpin-Healy TS, Klompe SE, Sternberg SH & Fernández IS Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature 577, 271–274 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Peters JE Tn7. Microbiol. Spectr 10.1128/microbiolspec.MDNA3-0010-2014 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Hoffmann FT et al. Selective TnsC recruitment enhances the fidelity of RNA-guided transposition. Nature 609, 384–393 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Behler J & Hess WR Approaches to study CRISPR RNA biogenesis and the key players involved. Methods 172, 12–26 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G & Karvelis T Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl Acad. Sci. USA 111, 9798–9803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mali P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol 31, 833–838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez-pinera P. et al. RNA-guided gene activation by CRISPR–Cas9-based transcription factors. Nat. Methods 10, 973–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS & Vale RD A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JU et al. Structural basis for target site selection in RNA-guided DNA transposition systems. Science 373, 768–774 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Querques I, Schmitz M, Oberli S, Chanez C & Jinek M Target site selection and remodelling by type V CRISPR-transposon systems. Nature 599, 49–502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chavez A. et al. Highly-efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–3228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thakore PI et al. Highly specific epigenome editing by CRISPR–Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang T, Larcher LM, Ma L & Veedu RN Systematic screening of commonly used commercial transfection reagents towards efficient transfection of single-stranded oligonucleotides. Molecules 23, 2564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker MWG, Klompe SE, Zhang DJ & Sternberg SH Transposon mutagenesis libraries reveal novel molecular requirements during CRISPR RNA-guided DNA integration. Preprint at https://www.biorxiv.org/content/10.1101/2023.01.19.524723v1 (2023). [Google Scholar]
- 57.Sarnovsky RJ, May EW & Craig NL The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 15, 6348–6361 (1996). [PMC free article] [PubMed] [Google Scholar]
- 58.North SH & Nakai H Host factors that promote transpososome disassembly and the PriA-PriC pathway for restart primosome assembly. Mol. Microbiol 56, 1601–1616 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Adeyemi RO et al. The Protexin complex counters resection on stalled forks to promote homologous recombination and crosslink repair. Mol. Cell 81, 4440–4456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ciccia A & Elledge SJ The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holder JW & Craig NL Architecture of the Tn7 posttransposition complex: an elaborate nucleoprotein structure. J. Mol. Biol 401, 167–181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levchenko I, Luo L & Baker TA Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 9, 2399–2408 (1995). [DOI] [PubMed] [Google Scholar]
- 63.Kruklitis R, Welty DJ & Nakai H ClpX protein of Escherichia coli activates bacteriophage Mu transposase in the strand transfer complex for initiation of Mu DNA synthesis. EMBO J. 15, 935–944 (1996). [PMC free article] [PubMed] [Google Scholar]
- 64.Mhammedi-Alaoul A, Pato M & Gama M-J & Toussaint A A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol. Microbiol 11, 1109–1116 (1994). [DOI] [PubMed] [Google Scholar]
- 65.Abdelhakim AH, Oakes EC, Sauer RT & Baker TA Unique contacts direct high-priority recognition of the tetrameric Mu transposase-DNA complex by the AAA+ unfoldase ClpX. Mol. Cell 30, 39–50 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sauer RT & Baker TA AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem 80, 587–612 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Levchenko I, Yamauchi M & Baker TA ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 11, 1561–1572 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Baker TA & Sauer RT ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta Mol. Cell Res 1823, 15–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hersch GL, Burton RE, Bolon DN, Baker TA & Sauer RT Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell 121, 1017–1027 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Joshi SA, Hersch GL, Baker TA & Sauer RT Communication between ClpX and ClpP during substrate processing and degradation. Nat. Struct. Mol. Biol 11, 404–411 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Siddiqui SM, Sauer RT & Baker TA Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 18, 369–374 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strecker J, Zhang F & Ladha A CRISPR-associated transposase systems and methods of use thereof. https://patents.google.com/patent/WO2020131862A1/en (2020). [Google Scholar]
- 73.Tou CJ, Orr B & Kleinstiver BP Precise cut-and-paste DNA insertion using engineered type V-K CRISPR-associated transposases. Nat. Biotechnol 10.1038/s41587-022-01574-x (2023). [DOI] [PubMed] [Google Scholar]
- 74.Özcan A. et al. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature 597, 720–725 (2021). [DOI] [PubMed] [Google Scholar]
- 75.Shen Y et al. Structural basis for DNA targeting by the Tn7 transposon. Nat. Struct. Mol. Biol 29, 143–151 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Gu B. et al. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359, 1050–1055 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen B, Guan J & Huang B Imaging specific genomic DNA in living cells. Annu. Rev. Biophys 45, 1–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmitz M, Querques I, Oberli S, Chanez C & Jinek M Structural basis for the assembly of the type V CRISPR-associated transposon complex. Cell 185, 4999–5010 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fricke T et al. Targeted RNA knockdown by a type III CRISPR–Cas complex in zebrafish. CRISPR J. 3, 299–313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petassi MT, Hsieh S & Peters JE Guide RNA categorization enables target site choice in Tn7-CRISPR-Cas transposons. Cell 183, 1757–1771 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yeo NC et al. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods 15, 611–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee TI, Johnstone SE & Young RA Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc 1, 729–748 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinberg DN et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qiu X. et al. CoBRA: Containerized Bioinformatics Workflow for Reproducible ChIP/ATAC-seq Analysis. Genomics Proteomics Bioinformatics 19, 652–661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson JT et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Amemiya HM, Kundaje A & Boyle AP The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep 9, 9354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stark R & Brown G DiffBind: differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf 1–29 (2011). [Google Scholar]
- 91.Ramírez F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lampe GD et al. Integration in human cells without double-strand breaks using CRISPR RNA-guided transposases. Gene Expression Omnibus https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223174 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive under Gene Expression Omnibus accession number GSE223174 (ref. 92). Source data are provided with this paper.