Abstract
Changes in gene regulation are thought to underlie most phenotypic differences between species. For subterranean rodents such as the naked mole-rat, proposed phenotypic adaptations include hypoxia tolerance, metabolic changes, and cancer resistance. However, it is largely unknown what regulatory changes may associate with these phenotypic traits, and whether these are unique to the naked mole-rat, the mole-rat clade, or are also present in other mammals. Here, we investigate regulatory evolution in the heart and liver from two African mole-rat species and two rodent outgroups using genome-wide epigenomic profiling. First, we adapted and applied a phylogenetic modeling approach to quantitatively compare epigenomic signals at orthologous regulatory elements and identified thousands of promoter and enhancer regions with differential epigenomic activity in mole-rats. These elements associate with known mole-rat adaptations in metabolic and functional pathways and suggest candidate genetic loci that may underlie mole-rat innovations. Second, we evaluated ancestral and species-specific regulatory changes in the study phylogeny and report several candidate pathways experiencing stepwise remodeling during the evolution of mole-rats, such as the insulin and hypoxia response pathways. Third, we report nonorthologous regulatory elements overlap with lineage-specific repetitive elements and appear to modify metabolic pathways by rewiring of HNF4 and RAR/RXR transcription factor binding sites in mole-rats. These comparative analyses reveal how mole-rat regulatory evolution informs previously reported phenotypic adaptations. Moreover, the phylogenetic modeling framework we propose here improves upon the state of the art by addressing known limitations of inter-species comparisons of epigenomic profiles and has broad implications in the field of comparative functional genomics.
Most phenotypic changes across mammals are thought to arise from differences in gene regulation. African mole-rats are a group of rodents displaying unusual longevity (Emmrich et al. 2022) and evolutionary adaptations to their subterranean environment (for reviews, see Park and Reznick 2022; Wong et al. 2022), including cooperative behavior (Zöttl et al. 2016; Bennett et al. 2018), loss of vision (Partha et al. 2017), resistance to hypoxia (Larson and Park 2009; Park et al. 2021; Cheng et al. 2022), anoxia (Park et al. 2017) and hypercapnia (Park et al. 2021), metabolic adaptations (Park et al. 2017; Faulkes et al. 2019), and pain insensitivity (Smith et al. 2011). These unusual traits have prompted genome sequencing of both the naked mole-rat (Heterocephalus glaber) (Kim et al. 2011) and the Damaraland mole-rat (Fukomys damarensis) (Fang et al. 2014), as well as genomic investigations on species-specific changes in protein sequences and signatures of positive selection (Davies et al. 2015; Sahm et al. 2018). Recent work has applied proteomic (Heinze et al. 2018) and metabolomic (Ma et al. 2015; Faulkes et al. 2019) approaches to the study of mole-rat traits, yet the extent to which mole-rat-specific changes in gene regulation may underlie these adaptations remains unexplored. Moreover, the significance and uniqueness of mole-rat phenotypic traits have been subject to debate (Buffenstein et al. 2022), in part because of many observations being limited to comparisons between the naked mole-rat and mouse.
Mammalian gene regulation is largely enacted by collections of noncoding promoter and enhancer regions, known to bind hundreds of transcription factors combinatorially (Spitz and Furlong 2012; Moorthy et al. 2017; Andersson and Sandelin 2020). Previous studies have extensively documented the evolution of mammalian regulatory elements (Cotney et al. 2013; Vierstra et al. 2014; Reilly et al. 2015; Villar et al. 2015), which is especially fast for enhancers. Comparative analyses across species, tissues, and developmental stages have suggested a greater functional relevance for conserved regulatory activity (Arnold et al. 2014; Castelijns et al. 2020; VanOudenhove et al. 2020; Wong et al. 2020). In contrast, lineage-specific elements appear partly compensatory of proximally lost events (Arnold et al. 2014; Berthelot et al. 2018) and typically arise in genomic regions with pre-existing regulatory activity (Cotney et al. 2013; Emera et al. 2016). Although mammalian adaptations in gene regulation have been linked to the lineage-specific expansion of repetitive elements (Lynch et al. 2011; Trizzino et al. 2017; Roller et al. 2021), identifying the subsets of rapidly evolving noncoding elements associated with lineage-specific shifts in regulatory activity has proved more challenging, in part because of limitations in comparative approaches for functional genomics data (Dunn et al. 2018; Price et al. 2022).
Here, we applied a phylogeny-aware approach to quantitatively compare epigenomically defined promoters and enhancers across two tissues from four rodents, including two mole-rat species and two rodent outgroups. Our study design aimed to identify mole-rat-specific shifts in promoter and enhancer activities and to inform the gene regulatory component of mole-rat adaptations. Last, we investigated the contribution of repetitive elements to lineage-specific gene regulation in the mole-rat clade.
Results
Comparative epigenomics of liver and heart regulatory activities in mole-rats
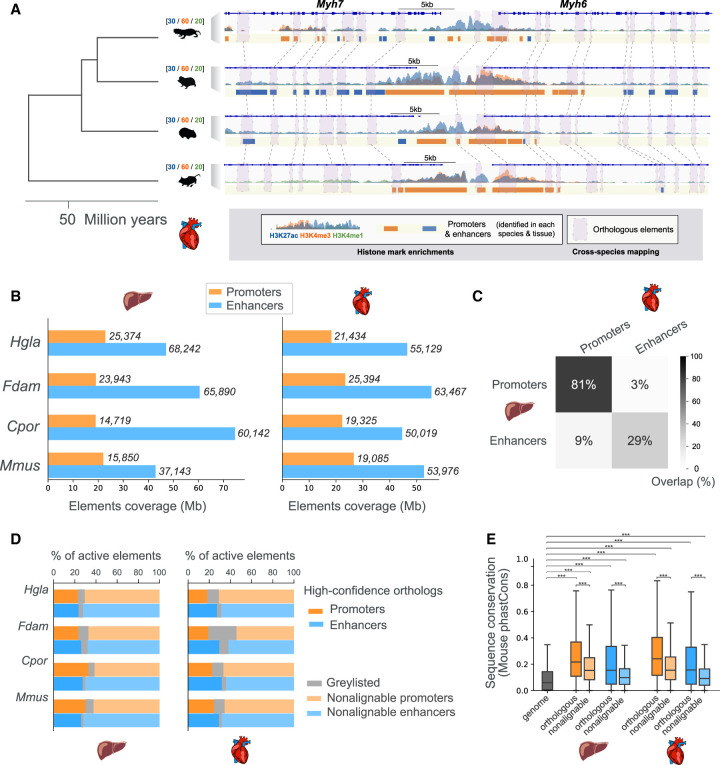
To investigate the contribution of enhancer evolution to tissue-specific gene regulation in mole-rats, we generated histone mark chromatin immunoprecipitation (ChIP)-sequencing data to identify promoters and enhancers active in two somatic tissues with distinct metabolic and functional roles (liver and heart) from the naked mole-rat, Damaraland mole-rat, and two outgroup rodents (guinea pig and mouse) (Fig. 1A; Supplemental Fig. S1). Using two to six replicates for each combination of species, histone mark, and tissue (Supplemental Table S1), we obtained an average of around 20,000 promoters and 50,000 enhancers reproducibly detected across individuals (Methods) (Fig. 1B). In the four study species, promoters show >80% commonality across tissues (Fig. 1C; Supplemental Fig. S1), in line with their general association with gene expression (Vierstra et al. 2014; Young et al. 2015). In contrast, liver and heart enhancers in each of the four rodent species are largely distinct, with just under 30% being typically common to both tissues (Fig. 1C; Supplemental Fig. S1), consistent with their role as tissue-specific cis-regulators (Castelijns et al. 2020; Roller et al. 2021).
Figure 1.
Comparative epigenomics of mole-rat promoters and enhancers in the heart and liver. (A) Epigenomic profiling and cross-mapping approach exemplified by the Myh6/Myh7 heart locus. H3K27ac (blue), H3K4me3 (orange), and H3K4me1 (green) histone mark enrichment in the heart are shown for each of the four species: (Hgla) naked-mole rat, (Fdam) Damaraland mole-rat, (Cpor) guinea pig, and (Mmus) mouse. Scales above species names indicate fold enrichment over input (averaged across replicates). Note that we use the same scale across species to facilitate direct visual comparison of histone mark enrichments. Identified promoters and enhancers are represented by orange and blue boxes, respectively. Purple boxes connected by dashed lines correspond to orthologous promoters and enhancers in each species. (B) Coverage and numbers of promoters and enhancers by species and tissue. Bars correspond to total genomic coverage; the number of elements are indicated at the right of bars. (C) Overlap between promoters and enhancers across the liver and heart tissues, averaged over the four species. Percentages represent the fraction of overlapping elements in each category. (D) Percentage of promoter and enhancer elements containing high-confidence orthologous regions across the four species (dark orange and blue bars). Lighter shade bars indicate promoters and enhancers with no high-confidence orthologs (nonalignable). Graylisted regulatory elements correspond to elements with elevated read density in input ChIP (Methods). (E). Sequence conservation of orthologous and nonalignable promoters and enhancers in the mouse. Orthologous mouse elements have significantly higher phastCons sequence conservation scores than those of nonalignable elements. Both orthologous and nonalignable mouse elements have sequences significantly more conserved than the genomic background: Mann–Whitney U tests, with Benjamini–Hochberg correction for multiple testing, (***) corrected P-values < 0.001. See also Supplemental Figures S1 and S2 and Supplemental Table S1.
To compare regulatory regions across the four study species, we used pairwise whole-genome alignments to identify promoters and enhancers for which orthologous regions can be confidently assigned across all species (Methods) (Fig. 1A). Specifically, we identified subregions of any promoter and enhancer with a reciprocal alignment between the mouse genome and each of the three other rodents (Methods). To illustrate this process, we show regulatory elements in each species around the heart-specific genes Myh6 and Myh7, for which a number of orthologous elements were identified (Fig. 1A, solid boxes). Using this approach, we can compare regulatory activities at orthologous locations within a substantial fraction of promoters and enhancers across the four study species (shaded boxes and connecting lines in Fig. 1A; Supplemental Fig. S2). Overall, and as expected for nonchromosomal genome assemblies, we identify four-way orthologous regions within 20%–30% promoters and enhancers in each species (Fig. 1D). We define these elements as orthologous promoters and enhancers. In agreement with previous studies (Cotney et al. 2013; Vierstra et al. 2014; Villar et al. 2015), orthologous promoters are mostly active across all study species (70%), whereas orthologous enhancers are largely active in only one species (55%) (Supplemental Fig. S2). Last, orthologous elements in either tissue show significantly higher levels of sequence conservation in the mouse compared with nonalignable elements (Fig. 1E). These results closely agree with those reported in recent similar data sets (Trizzino et al. 2017; Roller et al. 2021) and indicate that across the four species, we capture promoters and enhancers corresponding to both conserved and fast-evolving sequences. As such, the orthologous and nonalignable regulatory elements we defined constitute a genome-wide collection to study regulatory evolution in mole-rats.
Phylogenetic modeling of regulatory activity identifies promoter and enhancer shifts in mole-rats
We next focused on orthologous promoters and enhancers to identify mole-rat-specific changes in gene regulation. We adapted the EVE phylogenetic modeling method, initially developed for transcriptomic data (Rohlfs and Nielsen 2015), to identify promoters and enhancers showing shifts in regulatory activity along mole-rat branches (Fig. 2A). This approach is based on an Ornstein–Uhlenbeck model with two optima and allows statistical assessment of branch-specific shifts in regulatory activity while accounting for inter-species variation (Methods). Because EVE had not been applied to ChIP-seq data previously, we extensively evaluated the performance of phylogenetic modeling with simulated data (Methods) (Supplemental Fig. S3), and selected suitable data normalization and statistical thresholds (Supplemental Fig. S3; Supplemental Table S2). Here, we focus on results with H3K27ac data, as they are broadly distributed across promoters and enhancers and show a large dynamic range of normalized read densities and good phylogenetic signal across our study species (Fig. 2B; Supplemental Fig. S3).
Figure 2.
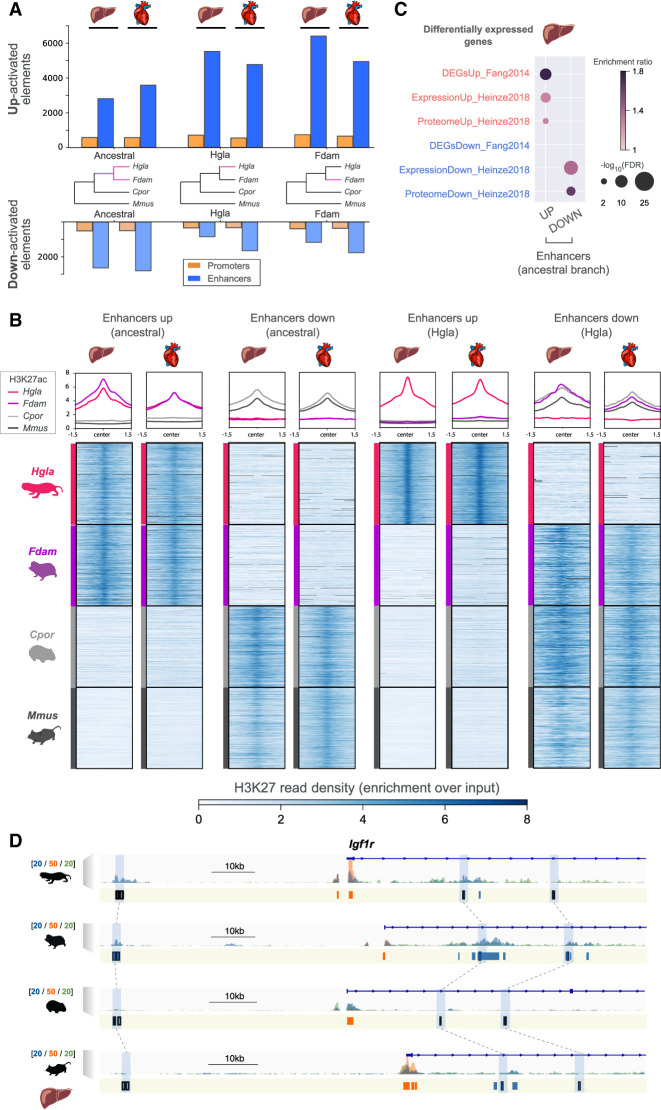
Identification of promoters and enhancers with an activity shift in mole-rats. (A) Number of regulatory elements identified via phylogenetic modeling with an increase (up) or decrease (down) in activity in the indicated branches of the species tree: mole-rat ancestral (ancestral), naked mole-rat (Hgla), and Damaraland mole-rat (Fdam). (B) H3K27ac read density heatmaps and profile plots for up and down enhancers in the ancestral and naked mole-rat branches. Reads densities are presented as fold enrichment over input, averaged across biological replicates. Profile plots show distributions in a 3-kb window. (C) Liver enhancers with an ancestral activity shift in mole-rats are significantly associated with previously identified differentially expressed genes in mole-rats liver (GREAT enrichment tests; Methods). (D) Example of liver enhancers up-regulated in the ancestral mole-rat branch and associated with the Igf1r insulin response locus. H3K27ac (blue), H3K4me3 (orange), and H3K4me1 (green) histone mark enrichment in the liver around Igf1r; scale units are as in Figure 1A. Promoters are shown in orange; enhancers, in blue. Up enhancers are identified by black boxes and linked by light blue boxes and dashed lines across species. See also Supplemental Figures S3 through S7 and Supplemental Tables S2 and S3.
Using this approach, we identified shifts in regulatory activity across three branches of the study phylogeny (Fig. 2A,B): the ancestral branch leading to mole-rats (Ancestral) and the single-species branches leading to the naked-mole rat (Hgla) and the Damaraland mole-rat branch (Fdam). Across both ancestral and single-species branches, phylogenetic modeling accurately identified orthologous regions presenting increased (Up) or decreased (Down) H3K27ac read densities in mole-rat species compared with outgroup rodents (Fig. 2B). For each tested branch, phylogenetic modeling identified approximately 400 promoters and 3000–6000 enhancers with increased regulatory activity in mole-rat branches (Fig. 2A, top bars), which we define as Up elements. Conversely, we obtained approximately 200–500 promoters and approximately 1000–2500 enhancers with reduced regulatory activity (Fig. 2B, bottom bars), which we define as Down promoters and enhancers. We additionally confirmed that Up enhancers have significantly fewer 3D chromatin contacts in the mouse than do Down enhancers (Fisher's exact test: P-value < 0.05 for liver, P-value < 10−5 for heart; Methods) (Chapski et al. 2018), which is consistent with activity gains in mole-rats.
We next compared the locations and properties of Up and Down elements identified with phylogenetic modeling (Supplemental Table S3) to regulatory changes inferred by state-of-the-art methods, such as parsimony (Roller et al. 2021) and differential binding analysis (Supplemental Figs. S4, S5; Ross-Innes et al. 2012). Phylogenetic modeling recovered regulatory shifts with read density patterns clearly consistent with each tested branch (Fig. 2B; Supplemental Figs. S4, S5). Moreover, and to assess the relative significance of enhancer shifts inferred by the three methods, we overlapped their nearby genes with previously reported differentially expressed genes and proteins in the mole-rat liver (Methods) (Fang et al. 2014; Heinze et al. 2018). In this analysis, enhancer shifts identified by phylogenetic modeling are significantly associated with mole-rat differentially expressed genes (Fig. 2C). In agreement with this observation, we found genes flanking Up and Down promoter and enhancer shifts include a number of previously reported loci with differential gene expression between mole-rats and other rodents, such as the up-regulated insulin response gene Igf1r (Fig. 2D; Fang et al. 2014) or the down-regulated urate oxidase gene Uox (GREAT genes-to-regions association; Methods) (Supplemental Table S3; Ma et al. 2015). For both genes, we measured (1) chromatin interactions between promoters and Up or Down enhancers (Supplemental Fig. S6) and (2) downstream expression levels (Supplemental Fig. S7). We detected, consistent with mole-rat enhancer shifts in the Igf1r locus (Fig. 2D), significant interactions between the Igf1r promoter and several Up enhancers in the mole-rat heart but not in the mouse heart (Supplemental Fig. S6A,B) and observed increased Igf1r expression in mole-rat tissues (Supplemental Fig. S7). Similarly, Uox promoter interactions and gene expression levels were consistent with enhancer losses at this locus (Supplemental Table S3; Supplemental Figs. S6, S7).
Enhancer shifts in the ancestral mole-rat branch enrich for tissue-specific metabolic and morphological adaptations
To investigate gene regulatory pathways associated with mole-rat enhancer shifts, we initially focused on the ancestral branch leading to both mole-rat species. On this branch, the top enriched Gene Ontologies (GOs) across enhancer shifts in liver and heart tissue provided a summary map of gene regulatory changes in mole-rats (Fig 3A; Supplemental Tables S4–S7). In the heart, Up enhancers were associated with heart contraction, energy metabolism, and cellular respiration GOs. Previous studies have reported low heart rate and resting cardiac contractility observed in the naked mole-rat heart (Grimes et al. 2017), which our results can inform from a gene regulation perspective. In the liver, Up enhancers were linked to miRNA transcription, liver development, and lipid metabolism, in line with reported adaptations in fatty acid utilization in both mole-rat species (Heinze et al. 2018; Farhat et al. 2020; Yap et al. 2022). For Down enhancers, top enrichments were observed for migration and angiogenic processes in the heart and for hematopoietic and purine catabolism in the liver (Fig. 3A); the latter aligns with previous reports (Ma et al. 2015). In sum, although some of the associations observed above may reflect the biology of liver and heart tissues, several enrichments suggest specific connections to known adaptations seen in the mole-rat clade and not observed in control analyses on the guinea pig branch (Supplemental Table S8).
Figure 3.
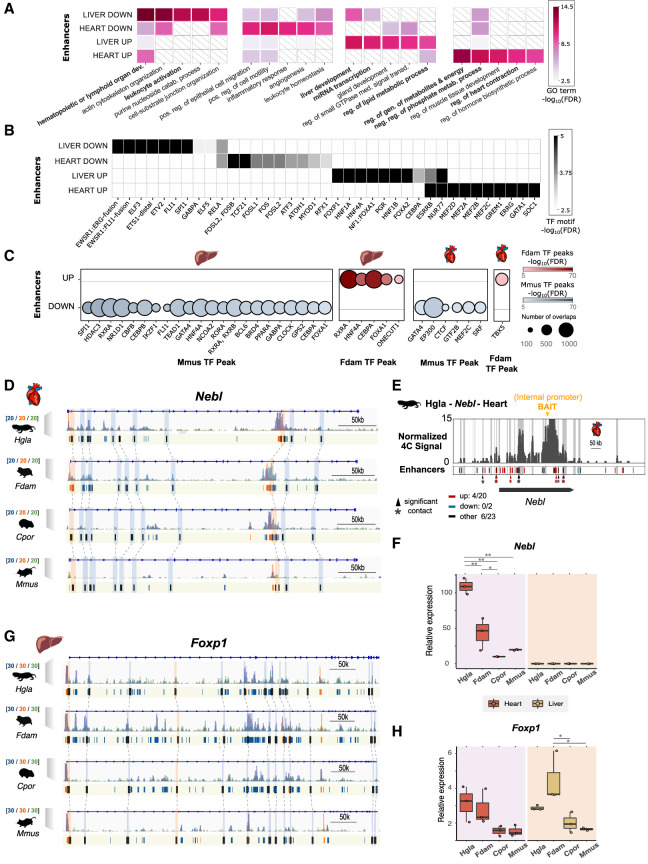
Gene Ontologies (GOs) and transcription factor binding sites enriched in enhancers with an activity shift on the ancestral mole-rat branch. (A) Top GO terms enriched in enhancers with an activity shift on the ancestral mole-rat branch. The top five associated GO terms (GREAT enrichment tests, corrected P-values < 0.05; Methods) are shown for each set. Colors indicate P-value significance levels (−log10 FDR) of the GO term in each set, and white barred boxes mark sets in which the term is not enriched (not in the top 100). The complete list of enriched terms is available in Supplemental Tables S4 through S6. (B) Transcription factor binding motifs enriched in mole-rat enhancers with an activity shift on the ancestral branch. Enriched motifs are represented as a heatmap, with color scale proportional to P-value significance levels (−log10 FDR) of the motif in enrichment tests (HOMER enrichment tests; Methods). The complete list of enriched TF motifs is available in Supplemental Table S7. (C) ChIP-seq transcription factor binding signals enriched in mole-rat enhancers with an activity shift on the ancestral branch. Circle plots show enrichments with size proportional to the number of overlapping peaks, and darker colors denote lower FDR significance. Enrichments are shown for either tissue and each tested transcription factor in mouse (Mmus) and Damaraland mole-rat (Fdam) ChIP-sequencing data sets. (D) Example of heart promoters and enhancers with an activity shift in mole rats and associated to the nebulette (Nebl) locus. Representation as in Figure 2D: H3K27ac (blue), H3K4me3 (orange), and H3K4me1 (green) histone marks enrichment in heart displayed around Nebl. Promoters are shown in orange; enhancers, in blue. Orthologous up enhancers and promoters are identified by black boxes and are linked by light blue or orange boxes (respectively) and dashed lines across species. (E) Chromatin interactions between the Nebl internal promoter (bait; orange arrow) and genomic elements in the same locus in naked mole-rat heart. Gray bars denote significant interactions (4C peaks; Methods), and asterisks denote significant contacts with a candidate enhancer (enhancers overlapping a 4C peak, bars in track below; red for “up,” blue for “down,” and black for “other”). (F) Relative Nebl mRNA expression across the heart and liver samples from the four species. (*) P < 0.05, (**) P < 0.001 (one-way ANOVA, Tukey post-hoc test). (G) Example of Up promoters and enhancers at the Foxp1 locus in liver. Representation as in D. (H) Relative Foxp1 mRNA expression. Representation as in F. See also Supplemental Tables S4 through S9 and Supplemental Figures S6 through S9.
To further explore the relationship between enriched GOs and tissue-specific gene regulation, we identified transcription factor binding motifs enriched in Up and Down enhancers in either tissue (Fig. 3B; Supplemental Table S9). Up enhancer shifts were enriched for binding motifs of tissue-specific transcription factors, such as MEF family members in the heart and FOX transcription factors in the liver. In contrast, Down enhancer shifts associated with binding motifs of broadly expressed transcription factors, such as FOS members in the heart and ETS proteins in the liver. To determine whether the observed motif enrichments correspond to in vivo TF binding, we measured the enrichment of TF binding signals detected in ChIP experiments from mouse (Hammal et al. 2022) or mole-rat samples (Methods) (Fig. 3C). In agreement with their histone mark enrichments (Fig. 2B), Up enhancers in either tissue were bound by TFs in the mole-rat but not in the mouse, whereas TF binding in Down enhancers was only observed in the mouse. Moreover, we detected specific pairs of motif/TF peaks and ontology terms significantly associated with each set of enhancers (Supplemental Fig. S8), suggesting regulatory rewiring of tissue-specific processes. As an example, for Up enhancers in the heart, binding motifs for MEF family members associate with the precursor metabolites and energy ontology category, which includes genomic loci such as the transcriptional coactivator Ppargc1a, a regulator of mitochondrial biogenesis and respiratory capacity (Austin and St-Pierre 2012) with context-dependent effects in the heart (Zhu et al. 2019; Oka et al. 2020). In the mole-rat heart, we observed increased Ppargc1a expression (Supplemental Fig. S7) and interactions between Up enhancers and the gene promoter (Supplemental Fig. S6). However, further research is required to determine the impact of Ppargc1a dosage in mole-rats.
Taken together, these observations also suggest specific responses and transcriptional pathways with altered gene regulation in mole-rats (Supplemental Table S9), such as the Nebl heart contraction locus (Fig. 3D–F). In this region, we detect a large number of Up promoters and enhancers, some of which also contain MEF transcription factor binding motifs. In liver, FOXP1 binding motifs were enriched in Up enhancer shifts (Fig. 3B), and we also found a concordant recruitment of Up promoters and enhancers at the Foxp1 gene locus (Fig. 3G; Supplemental Tables S3, S9), suggesting a coordinated up-regulation of this pathway in mole-rats. In the mole-rat liver, we observed increased Foxp1 expression (Fig. 3H). FOXP1 is a transcriptional repressor regulating hepatic glucose homeostasis (Zou et al. 2015). However, we did not find an accompanying reduction in expression for genes in the gluconeogenesis pathway in the mole-rat liver. Thus, the effects of increased Foxp1 expression may be compensated by other transcription factors such as Foxo1 (Supplemental Fig. S7) or may be apparent in response to specific stimuli.
Enhancer shifts along mole-rat evolution identify temporal rewiring of gene regulation
Our study phylogeny allows for temporal investigation of regulatory evolution in mole-rats. We thus asked whether ancestral and species-specific enhancer shifts associate with common or divergent molecular processes along the evolution of mole-rats. To this end, we clustered related GOs detected in enhancer shifts across branches (Methods) (Fig. 4A; Supplemental Table S10), and we focused on gene regulatory pathways found across several branches in the phylogeny (Fig. 4A).
Figure 4.
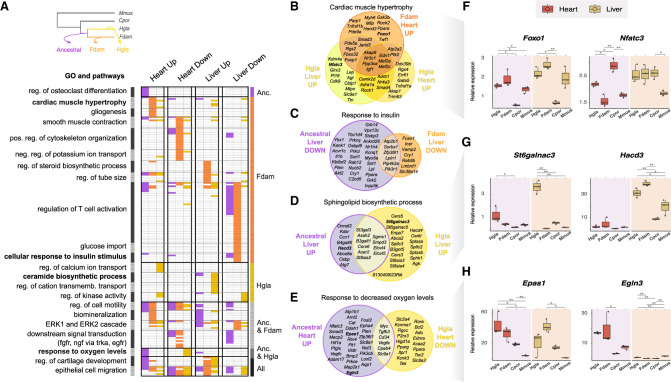
Comparison of GOs and pathways enriched in enhancers with an activity shift in ancestral and single-species mole-rat branches. (A) GO terms significantly associated to enhancers with activity shifts in mole rats across different branches (top left inset) and tissues. Similar GO terms and pathways were grouped in clusters (left axis; Methods). Each row in the heatmap is a GO term or pathway within a cluster, with its presence (colored) or absence (white) indicated in each enhancer set. Columns correspond to the different tissues and shift directions; colors, to the branches (top left inset). Clusters are ordered to highlight terms enriched in particular branches (right axis). The complete list of enriched terms and cluster membership is available in Supplemental Table S10. (B–E) Selected examples of GO terms enriched across enhancers of different branches or tissues. Venn diagrams show the overlap between genes associated with shifted enhancers of each set (Methods). (F–H) Relative gene expression levels for loci associated with cardiac hypertrophy (F), sphingolipid biosynthesis (G), and response to hypoxia (H). Representation as in Figure 3F. See also Supplemental Figures S7 and S9 and Supplemental Table S10.
First, this comparison revealed temporal associations of GOs and pathways along the three branches. We observe most GO enrichments were driven by one branch, such as cardiac muscle hypertrophy or response to insulin in the Damaraland mole-rat branch and such as ceramide biosynthesis in the naked mole-rat branch. However, several processes appear to harbor regulatory rewiring on both the ancestral and single-species branches, possibly in response to continued evolutionary pressure. We further explored some of these pathways by analyzing whether enhancer shifts across branches are proximal to the same genes (Fig. 4B–E).
Genes associated with cardiac muscle hypertrophy accumulate Up enhancers in both single-species branches: in the heart for the Damaraland mole-rat and in both the heart and liver for the naked mole-rat (Fig. 4B). Cardiomyocyte hypertrophy is a complex process mediated by signals arising from multiple cell types (McLellan et al. 2020), and a study comparing mice and naked mole-rats reported reduced aging-associated hypertrophy in naked mole-rat hearts compared with those of mice (Can et al. 2022). Indeed, we found individual hypertrophy-related genes in each branch/tissue often had corresponding tissue-specific expression levels (Supplemental Fig. S9). Moreover, we measured mRNA levels of several hypertrophy loci across the mole-rat and rodent samples (Fig. 4F; Supplemental Fig. S7), and observed increased expression of Foxo1 and Foxp1 in mole-rat tissues and Nfatc3 in the mole-rat liver. Our results thus suggest mole-rats have recently and convergently modified their response to cardiac hypertrophy, primarily in the heart but potentially also at liver loci.
Genes in the insulin response pathway are known to be differentially expressed in mole-rats (Fang et al. 2014), with individual genes in this response being either induced or repressed. Down enhancers are the predominant gene regulatory change in this response, in both the ancestral and Damaraland mole-rat branches (Fig. 4C). Although we detect a lower number of associated loci in the single-species branch, these include both upstream (Insr) and downstream (Foxo1) response genes. Nevertheless, locus-specific gene expression measurements (Supplemental Fig. S7) suggest these regulatory changes can be buffered by other enhancers and may not associate with decreased gene expression in mole-rat liver. Similarly, we found loci associated with ceramide biosynthesis in Up liver enhancers for both the ancestral and naked mole-rat branches (Fig. 4D). There is evidence ceramides can cross the blood–brain barrier (Zimmermann et al. 2001; Eguchi et al. 2020); thus, our observation may inform reported high levels of short-chain ceramides in naked mole-rat brain lipids (Frankel et al. 2020). We verified increased expression of several genes involved in sphingolipid and ceramide biosynthesis in the mole-rat liver (Fig. 4G; Supplemental Fig. S7), which may result in differences in liver lipid secretion between mole-rats and other rodents.
Last, we detected genomic loci associated with the response to hypoxia among heart enhancer shifts in both ancestral and naked mole-rat branches (Fig. 4A,E). Protein levels of HIF1A are thought to be increased in mole-rats (Kim et al. 2011), and we detected Up enhancers in the ancestral mole-rat branch for both core regulators of hypoxic gene expression (Hif1a, Epas1, and Arnt2) and bona fide HIF target genes (Vegfa, Egln3, and Aqp1). For Epas1 and Egln3, we also observed increased gene expression in the mole-rat heart (Fig. 4H). However, we also found a number of hypoxia response gene loci harbor Down enhancers in the naked mole-rat branch. These results suggest that, in addition of an up-regulation in the response to hypoxia in mole-rats, the naked mole-rat may also tune down gene regulatory landscapes for a subset of hypoxia-inducible genes. In support of this hypothesis, we tested whether regulatory changes in the ancestral and mole-rat branches affect distinct sets of genes in the hypoxia response pathway (Fig. 4E) and confirmed that loci associated with enhancer shifts in each branch overlap more rarely than expected at random (permutation test with 100,000 random resampling iterations: P-value = 0.009).
Nonalignable mole-rat enhancers associate with SINE repetitive elements and rewire metabolic transcription factor networks
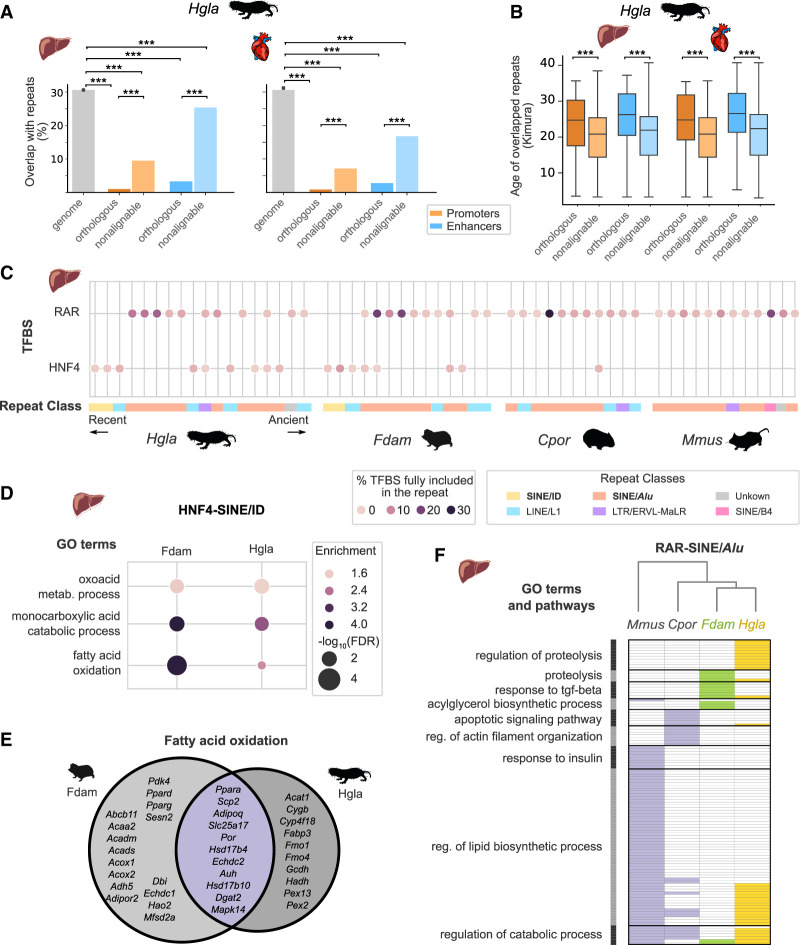
The phylogenetic modeling approach we developed here focuses on alignable segments within regulatory regions across our four study species. Nevertheless, we noticed some of the genomic loci previously linked to mole-rat traits are flanked by regulatory elements that cannot be aligned to the mouse genome, which we term nonalignable. Repetitive genomic regions (which are difficult to align across species) have been previously linked to rewiring of regulatory networks (Lynch et al. 2011; Trizzino et al. 2017; Roller et al. 2021). We thus investigated the genomic properties of nonalignable promoters and enhancers in our study and their potential contribution to mole-rat-specific gene regulation.
As expected, we observed that nonalignable regulatory elements in mole-rats consistently enrich for repetitive element sequences estimated to be of young age (Fig. 5A,B). Moreover, nonalignable enhancers significantly overlap with several repetitive element families (Supplemental Fig. S10). Among these, we identified enriched transcription factor binding motifs suggestive of regulatory co-option (Fig. 5C; Supplemental Table S11). First, we found a consistent enrichment of RAR binding motifs across nonalignable liver enhancers in all four species, as well as in sequences overlapping SINE/Alu repeats, which suggests rodent SINE/Alu elements contribute to RAR-mediated gene regulation, as previously proposed in primates (Polak and Domany 2006). In contrast, enrichment of HNF4A binding motifs in SINE/ID repeats was specific to mole-rat liver enhancers in both species. In both cases, a substantial fraction of these repeats was bound by the corresponding TF in mole-rats (Supplemental Table S11). Moreover, HNF4-SINE/ID sequences enrich for monocarboxylic and fatty acid catabolism GOs (Fig. 5D), which include transcription factor loci with key roles in lipid metabolism, such as Ppar genes (Fig. 5E; Supplemental Fig. S7). For RAR-SINE/Alu sequences enriched in nonalignable liver enhancers, we clustered GO terms associated to their flanking genes across the four species (Fig. 5F). We found, in agreement with the known roles of RAR/RXR in fatty acid oxidation and lipid metabolism (Li et al. 2021), terms related to lipid biosynthesis and catabolism consistently across all species. However, for mole-rats RAR-SINE/Alu sequences also associated with TGFB signaling and proteolysis, suggesting mole-rat-specific repeats may rewire protein degradation and immunomodulation via the RAR network.
Figure 5.
Nonalignable mole-rat enhancers associate with specific repetitive elements and transcription factor binding sites. (A) Overlap with repetitive elements for naked mole-rat orthologous and nonalignable promoters and enhancers. Nonalignable enhancers and promoters overlap significantly more with repeats than orthologous elements, whereas all regulatory element sets are significantly depleted in repeats compared with the genomic background: (***) Benjamini–Hochberg corrected permutation-based P-values < 0.001 (Methods). (B) Nonalignable naked mole-rat promoters and enhancers overlap significantly younger repeats than orthologous promoters and enhancers: Mann–Whitney U tests, with Benjamini–Hochberg correction for multiple testing, (***) corrected P-values < 0.001. Repeats ages were estimated with RepeatModeller (Methods) and are expressed as Kimura distances. (C) Transcription factor binding motifs for RAR and HNF4 in liver nonalignable enhancers significantly associate with specific repetitive elements. Circles indicate significant association between a specific repeat and a motif, with color intensity proportional to the fraction of total TF motifs in nonalignable liver enhancers included in the repeat. Repeats are ordered by age and colored by repeat class. RAR motifs are significantly associated with SINE/Alu across all four species, whereas the HNF4–ID SINE/ID enrichment is specific to mole-rats. (D) GO terms significantly associated with nonalignable enhancers presenting a HNF4-SINE/ID instances in naked mole-rat and Damaraland mole-rat liver enhancers (GREAT enrichment tests; Methods). (E) Venn diagram of genes associated to nonalignable mole-rat liver enhancers with HNF4-SINE/ID instances and annotated with the “fatty acid oxidation” GO term. (F) GO terms associated to RAR-SINE/Alu expansions across the four species. Representation as in Figure 4A: GO terms were grouped in clusters (shown on the left), and the absence (white) and presence (colored) of GO terms in each species are indicated. The complete list of enriched terms and cluster membership is available in Supplemental Table S11. For results across both the heart and liver, see also Supplemental Figure S10 and Supplemental Table S11.
In conclusion, our results are consistent with a body of evidence on the importance of repetitive, transposon-derived elements providing a source of regulatory potential that can associate with functional innovation in mammals (Lynch et al. 2011; Trizzino et al. 2017; Deniz et al. 2019; Roller et al. 2021).
Discussion
Evolutionary differences between mammals are expected to be largely driven by alterations in gene regulation rather than changes in protein sequences. Previous comparative studies (Vierstra et al. 2014; Villar et al. 2015; Young et al. 2015) have identified a continuum of regulatory regions from highly conserved to lineage specific (Fong and Capra 2022), with the former being associated with pleiotropy and core tissue functions. However, the extent to which lineage-specific changes in regulatory activity contribute to tissue-level phenotypic adaptations is debated (Cooper and Brown 2008; Kellis et al. 2014), with the exception of a few well-characterized examples (Gerbault et al. 2011; Lynch et al. 2011; Chuong et al. 2016; Kvon et al. 2016).
To date, most comparative analyses of regulatory landscapes have been based on presence or absence of orthologous promoters and enhancers and thus have a number of methodological limitations (Dunn et al. 2018; Price et al. 2022). These approaches do not normalize or quantitatively compare epigenomic levels of promoter and enhancer activity across species and typically do not account for varying phylogenetic distances between species. To extend and improve these analyses, we applied and validated a phylogenetic modeling approach to promoter and enhancer epigenomic signals in two tissues from a four-species phylogeny: comparing two mole-rat species with the guinea pig and mouse as outgroup rodents. This strategy allowed us to quantitatively identify lineage-specific shifts in promoters and enhancers across mole-rat branches, investigate the associated biological processes, and assess the potential contributions of mole-rat-specific changes in gene regulation to their characteristic evolutionary traits. The method we describe here improves on previously known issues with comparative analyses of functional genomics data and could be applied to similar data sets across species, tissues, and developmental stages. Our work adds to ongoing efforts to extend statistically sound, quantitative evolutionary models to the analysis of functional genomics data (Trizzino et al. 2017; Dunn et al. 2018; Yang et al. 2018; Dukler et al. 2020).
Our results reveal potential contributions of mole-rat gene regulation to tissue-specific processes. First, we find a consistent up-regulation of enhancers associated with Ppar/Pppargc1a loci, especially in the heart but also in the liver. These mole-rat-specific changes may fine-tune the role of PPARGC1A-PPARs in lipid metabolism and preservation of heart function (Zhu et al. 2019; Pergande et al. 2021). Moreover, we found lineage-specific liver enhancers in mole-rats enrich for SINE repeat families and associate with fatty acid oxidation processes, which is in agreement with the reported increase in fatty acid utilization in the mole-rat liver (Heinze et al. 2018). In the heart, some of the strongest up-regulated enhancers associate with myocardial conduction processes and include genes such as Nebl, Cacna1c, and Myh7. These changes in heart-specific gene regulation in mole-rats could contribute to the morphological and conduction properties of the mole-rat myocardium (Grimes et al. 2017; Can et al. 2022).
Comparing across ancestral, naked mole-rat, and Damaraland mole-rat branches, our results identified recurrent regulatory changes associated with specific pathways. We found up-regulated enhancers associated with cardiac hypertrophy in both the Damaraland mole-rat and naked mole-rat branch. These included Foxo1 and Foxp1 transcriptional repressor loci in the heart and the Nfatc3 locus in the liver, corresponding to transcription factors known to regulate hypertrophy (Ni et al. 2006; Bai and Kerppola 2011). This observation suggests a complex landscape of mole-rat changes in this pathway that may inform reduced levels of hypertrophy in the mole-rat versus mouse myocardium (Faulkes et al. 2019; Can et al. 2022). In contrast, for enhancers associated with the insulin response, we found a consistent down-regulation in the mole-rat liver, in both the ancestral and Damaraland mole-rat branches. Previous work documented both up-regulated and down-regulated expression of insulin response genes in mole-rats (Fang et al. 2014), and our findings suggest epigenomic enhancer down-regulation is significant in this response. Last, for loci involved in the response to hypoxia, we found evidence of both epigenomic up-regulation in the ancestral branch and down-regulation in the naked mole-rat branch (in heart). Changes in the HIF1A protein sequence in mole-rats are thought to result in reduced proteasomal degradation (Kim et al. 2011), albeit protein levels were reported to be unaltered in the naked mole-rat heart compared with the mouse heart (Xiao et al. 2017). Nevertheless, we observed increased gene expression of both Epas1 and Egln3 in the mole-rat heart, suggesting an EPAS1-mediated response. Additionally, the epigenomic enhancer down-regulation we observed in the naked mole-rat branch suggests a second wave of rewiring in this response that may fine-tune hypoxic regulation at a subset of loci. Although entirely speculative, a possible interpretation is down-regulated naked mole-rat enhancers in this response may enrich for HIF target genes, as suggested by their overlap with hypoxia-inducible genes in humans (70%) (Ward et al. 2021) and lower affinity in HIF binding sites compared with the mouse (Supplemental Fig. S8). Nevertheless, further experimental work is required to substantiate this hypothesis.
There are a number of limitations to our approach. First, our ability to compare epigenomic signals across species is partly dependent on reference genome assemblies, alignments, and annotations, which were of variable quality and completeness across our study phylogeny. The use of improved genomic resources, such as those recently reported by the Zoonomia consortium (Christmas et al. 2023), holds promise for enhanced resolution in future comparative studies.
Second, we identified promoter and enhancer shifts based on epigenomic enrichment of H3K27ac, which strongly associates with canonical promoter and enhancer elements across the genome. H3K27ac levels display strong phylogenetic signal across species, thus representing a good quantitative proxy of regulatory evolution. Moreover, enhancers harboring this histone mark have been experimentally validated in developmental models, and >60% drive expected expression patterns (Cotney et al. 2013; Nord et al. 2013). Nevertheless, noncanonical enhancers are increasingly recognized as an additional source of regulatory potential (Pradeepa et al. 2016). Such enhancers are often not associated with H3K27ac levels and are thus invisible to our approach, potentially limiting the completeness of the lineage-specific shifts we identified here.
Third, our phylogenetic modeling approach is affected by the size of the study phylogeny, which is constrained by available genomic resources in this clade, and sample accessibility limitations for wild mole-rat species. Thus, our data represent a strategic compromise between phylogenetic coverage and epigenomic completeness. Nevertheless, it is likely a denser or deeper phylogenetic sampling would impact performance of this approach by improving sensitivity and reducing false-discovery estimates. Similarly, our analyses of nonalignable promoters and enhancers in mole-rats rely on our ability to identify lineage-specific sequences in the study phylogeny, which would be enhanced by a denser phylogenetic sampling in this clade.
In summary, this study presents a quantitative phylogenetic framework with which to investigate the long-standing question of how lineage-specific changes in gene regulation may contribute to phenotypic traits. By modeling epigenomic activities of promoters and enhancers within the mole-rat clade, we show the utility of this approach to identify promoter and enhancer shifts across ancestral and single-species branches, as well as connect these lineage-specific innovations with candidate tissue-specific processes rewired in mole-rats.
Methods
ChIP and high-throughput sequencing
We performed ChIP experiments followed by high throughput sequencing (ChIP-seq) using liver and heart tissue samples isolated from the naked mole-rat, Damaraland mole-rat, guinea pig, and mouse. The origin, number of replicates, sex, and age for each species’ samples are detailed in Supplemental Table S1. Chromatin from 50–200 mg of dounced tissue was used for each ChIP experiment using antibodies against H3K4me3 (Millipore 05-1339), H3K27ac (Abcam ab4729), H3K4me1 (Abcam ab8895), TBX5 (Insight bio sc-515536), RXRA (sc-553), FOXA1 (ab70382), HNF4A (ARP31946), CEBPA (sc-9314), and ONECUT1 (sc-13050).
Cross-mapping of regulatory regions across four rodent species
Sequencing reads were aligned to the appropriate reference genome with BWA v.0.7.17 (Li and Durbin 2009) (Supplemental Table S1) and regions of enrichment determined with MACS2 (Zhang et al. 2008). Regions enriched in two to four biological replicates and overlapping by a minimum of 50% of their length were merged and categorized into promoters, enhancers, and primed enhancers independently for each species and tissue. We defined a set of “high-confidence orthologous regions” as four-way orthologous regions for which we required robust liftOver (Kuhn et al. 2013) mapping of regulatory elements across the four genomes. In this set, we homogenized the assignation of each region to a regulatory element type using a majority rule, and excluded graylisted regions with unusually elevated signal in input ChIP experiments (GreyListChIP R package) (R Core Team 2021).
Sequence conservation of orthologous and nonalignable regulatory elements
We downloaded sequence conservation scores (“phastCons”) for the mouse genome from the UCSC (http://hgdownload.cse.ucsc.edu/goldenpath/mm10/phastCons60way/), which were computed from a multiple alignment of 60 vertebrate genomes. We used the UCSC tool bigWigAverageOverBed (version 357) to extract average phastCons scores over each regulatory region included in the “nonalignable,” “orthologous,” and “genome” regulatory element sets. The “random” element sets were built from 100 permutations of the “nonalignable” element sets on the mouse genome, excluding exons.
Phylogenetic modeling of shifts in regulatory activity on mole-rat branches
Read density normalization for phylogenetic modeling
We first normalize the H3K27ac FPKM signal for each library with the corresponding input control, retaining the log2 fold change of signal FPKM over input FPKM. Specifically, we extracted H3K27ac read density (FPKM) at orthologous enhancers and promoters for each replicate and its corresponding input control experiment with the BAMscale cov utility (Pongor et al. 2020), retaining only confidently mapped reads (-q 13). We next used quantile normalization to normalize fold changes across species and replicates and verified that samples group according to the species phylogeny after normalization (Supplemental Fig. S3). This normalized data serve as the basic input for phylogenetic modeling of regulatory activity across species.
Phylogenetic modeling and detection of regulatory activity shifts
We modelled the evolution of regulatory activity along the study phylogeny using the EVE model, an improved Ornstein–Ulhenbeck model for continuous trait evolution under selection and drift (Rohlfs and Nielsen 2015). Specifically, the EVE's null model has four parameters that govern how traits evolve: the sigma parameter (strength of drift), the alpha parameter (strength of selection), the theta parameter (optimal trait value), and the beta parameter (ratio of intra- to interspecies variation). To detect significant shift in regulatory activity of enhancers and promoters on specific branches of the study phylogeny, we took advantage of EVE's branch shift test. The branch shift test conducts likelihood ratio tests (LRTs) comparing two nested models: the four-parameters null model to a five-parameters model with a branch-specific shift in the optimal trait value. In the branch model, foreground branches have a distinct optimum (theta1) from background branches (theta0). By leveraging replicates and explicitly accounting for within-species variation (with the parameter beta), the EVE model significantly improves upon classical OU models, as it reduces false-positive inferences of stabilizing selection and optimum shifts (Rohlfs and Nielsen 2015; Price et al. 2022). A challenge to applying EVE's branch shift test to ChIP-seq data is its relatively low statistical power, typically requiring a large amount of data (number of replicates and size of the phylogeny) that is unusual even in RNA-seq experiments (Rohlfs et al. 2014). In addition, on small phylogenies, values of the test statistic (LRT) can depart from the chi-square distribution. Therefore, we performed computational simulations to (1) determine the distribution of LRT in our data and compute accurate P-values and (2) evaluate the true-positive rate and select an appropriate alpha threshold for the branch shift test.
We ran EVE's branch shift test over normalized ChIP-seq reads using the evemodel R package (Gillard et al. 2021) to detect regulatory activity shifts on the following branches of the four-species phylogeny: the ancestral mole-rat branch (ancestral), the naked-mole rat branch (Hgla), and the Damaraland mole-rat branch (Fdam). Branch lengths in substitutions per site were extracted from Ensembl Compara v99 (specifically, from the species tree computed from pairwise whole-genome alignments, available at https://github.com/Ensembl/ensembl-compara/blob/release/99/conf/vertebrates/species_tree.branch_len.nw) (Cunningham et al. 2022). For each of the four orthologous regulatory elements sets (enhancer heart, enhancer liver, promoter heart, and promoter liver), we first estimated model parameters under the null model (selection without shift), in order to obtain realistic parameter values for simulations. We then performed n = 1000 simulations under the null model for each of the four sets, using the mean value of the estimated model parameters (alpha, beta, theta0, and sigma). We next tested whether the study phylogeny was sufficiently large for LRTs to be distributed as a chi-square distribution: we computed LRT comparing likelihoods of the simulated data under the null model and under a model with a shift in the ancestral mole-rats branch and found that LRT did not follow a chi-square distribution. We thus used these simulations under the null model to compute empirical P-values (Supplemental Fig. S3), as recommended previously (Rohlfs and Nielsen 2015).
Because the branch shift test has relatively low statistical power, we performed additional simulations to evaluate the false-positive and true-positive rates under a variety of simulation settings in order to select suitable thresholds on (1) the alpha significance level and (2) the absolute shift value abs(theta1–theta0) estimated by EVE. We again performed n = 1000 simulations for each of the four sets (enhancer heart, enhancer liver, promoter heart, and promoter liver) and with varying proportions of simulations under the null and shift models (from 5% to 17.5% of simulations with a shift). We performed simulations under the null and shift models using the average parameter values estimated from the data (parameters alpha, beta, theta0, and sigma), whereas the additional shift parameter value for the simulations with shift was drawn from a beta distribution beta(alpha = 8, beta = 2) × 3, selected to resemble empirically estimated shift values. On average and across the four sets, testing for shift on the ancestral branch at alpha = 0.05 and without filtering on the value of the estimated shift yielded a true-positive rate of 0.46. We next selected thresholds on the alpha significance and estimated shift value in two successive steps, with the objective to increase the true-positive rate while maintaining the false-positive rate around 0.05–0.1. First, we selected the optimal alpha threshold using the commonly used Youden method (i.e., the alpha that maximizes the Youden index = sensitivity + specifically – 1). This method identified an optimal alpha significance threshold of around 0.2 for most sets (Supplemental Table S2A). Second, we selected the threshold on the absolute shift value in order to recover at most 0.05–0.1 false-positive rates on most sets. We found that selecting alpha = 0.20 and filtering on absolute shift values >1.5 yielded an acceptable false-positive rate (0.08), while increasing the true-positive rate to 0.77 (Supplemental Table S2). Similar results were obtained with the corresponding simulations for shifts in each of the naked mole-rat (Hgla) and Damaraland mole-rat (Fdam) branches (Supplemental Table S2A,B).
Based on the above, for the four orthologous regulatory elements sets (enhancer heart, enhancer liver, promoter heart, and promoter liver) and each of the three tested branches, we obtained P-values for the branch shift test and estimated shift values for each regulatory region. We then retained as differentially active elements all elements with empirical P-value < 0.2 and absolute shift value >1.5. Read density heatmaps and profile plots were drawn with deepTools version 3.5.0 (Ramírez et al. 2016).
We compared these results with regulatory activity shifts using parsimony (binary presence or absence of peaks) and differential binding (DiffBind R package) (Ross-Innes et al. 2012).
Chromatin 3D contacts of Up and Down elements in the mouse
We used previously published 5-kb resolution Hi-C data in mouse liver and heart (Chapski et al. 2018) to test for enrichment/depletion of 3D contacts for Up and Down enhancers. We started from the list of significant 3D interactions between 5-kb windows on the mouse genome to count the number of Up/Down elements falling in windows with predicted 3D contacts. In the two tissues, Up enhancers have significantly fewer 3D contacts in the mouse than do Down elements. Specifically, 43% of heart Up enhancers display 3D contacts as opposed to 49% for Down enhancers (P-value < 10−5, Fisher's exact test), and 37% of liver Up enhancers display 3D contacts as opposed to 40% for Down enhancers (P-value < 0.05, Fisher's exact test). The limited correspondence between H3K27ac levels and 3D contacts density for Up and Down enhancers could be owing to the different resolution of the two experimental approaches or to potentially compensatory 3D contacts between other regulatory elements in the vicinity of Up/Down enhancers.
4C-seq of chromatin 3D contacts at candidate loci
We used 4C-seq (Krijger et al. 2020) to identify physical contacts between bait gene promoters and associated enhancer shifts, using restriction enzymes and primers indicated in Supplemental Table S12. Sequencing data were processed with the pipe4C pipeline (Krijger et al. 2020) to generate normalized 4C coverage tracks. Coverage was normalized to sum up to 10,000 reads over the captured loci scaffold and plotted as a running average across 21 fragment ends, clipped at the bait to maximize the range of nonbait signal. We called peaks using peakC (Geeven et al. 2018) with default parameters to identify regions in significant contact with the promoter.
RNA isolation and RT-QPCR
Total RNA was extracted with RNA universal mini kit (Qiagen) and treated with DNase (Ambion am1907) to remove contaminating genomic DNA. RNA (1 μg) was retrotranscribed using a high-capacity cDNA synthesis kit (Thermo Fisher Scientific), and cDNA was diluted 1/10 for qPCRs using KAPA SYBR FAST (Sigma-Aldrich KK4610). RT–qPCR was performed on a Roche LC480 for 40 cycles, using primers indicated in Supplemental Table S12.
GOs, pathways, and gene enrichment tests
We downloaded mouse GO data from the MGI database on April 20, 2022, as well as C2 pathway annotations from MgDB at https://bioinf.wehi.edu.au/software/MSigDB/. We filtered GO data to retain only GO of the Biological Process (BP) domain, as well as C2 pathways to retain only 1397 pathways (mostly REACTOME, KEGG, and BIOCARTA pathways). We transferred mouse GO and pathway annotations to the naked mole-rat and Damaraland mole-rat using gene orthologies from Ensembl compara version 102, extracted with Ensembl BioMart (Cunningham et al. 2022), and reimplemented the region-based GO tests implemented in GREAT (McLean et al. 2010). We used GREAT's default gene-to-region association rule and implemented its three association tests, using similar rules for GO propagation and filters as in GREAT. To increase statistical power and alleviate redundancy, we filter GO to only test for the most specific GO terms among all GO associated to the exact same foreground genes. For GO and pathways enrichment tests, we used GREAT recommended tests a and b (i.e., tests against the whole genome as background), retained terms found enriched with both tests, ranked them according to BH adjusted P-value < 0.05 of test a, and selected the top 100 GO terms and top 20 C2 pathways.
Transcription-factor binding motif enrichment analysis
We used the HOMER software suite version 4.11 (Heinz et al. 2010) to identify enriched TF motifs in sets of regulatory regions (findMotifsGenome.pl script with default parameters, with “-h” to conduct hypergeometric tests and “-size given” to only search for motifs within regulatory regions). We used the Damaraland mole-rat genome as a background to search for enriched motifs in the regulatory elements with an activity shift on the ancestral branch. We retain the top 10 enriched TF for each element set (all BH-corrected P-values < 0.05).
We tested for significant TF motif–GO term association within each element set using the shuffling procedure implemented in the RemapEnrich R package (Hammal et al. 2022). We identified significant association among the top 10 TF motifs and top 100 GO enriched in each set (Supplemental Fig. S8A) and retained pairs with corrected P-values < 0.05 (tests were conducted for each TF separately and corrected for multiple testing using the Benjamini–Yekutieli procedure to account for a possible dependency structure in the P-values distribution). We used the same approach to test for significant associations between GO terms and TF binding sites (Supplemental Fig. S8B).
Clustering of GO terms for visualization of enrichment across branches and tissues
To compare functional enrichments across sets of shifted regulatory elements in different tissues and branches of the phylogeny, we first removed redundancies by filtering out from each set the elements found shifted in two branches (i.e., “ambiguous” elements, found shifted in the ancestral and in one mole rat branch). We next used the GREAT approach to identify enriched GO terms and pathways in each filtered element set, retaining the top 100 GO (all corrected P-values < 0.05) and 20 C2 (all corrected P-values < 0.1) pathways (for implementation details, see section GOs, Pathways, and Gene Enrichment Tests). For visualization, we grouped together similar GO terms and pathways found within or across sets, based on the overlap of associated genes. Specifically, two GO (pathways) were grouped in the same cluster if the Jaccard index of the overlap was >0.35. We next relax the Jaccard index threshold to 0.2 to tentatively connect remaining singleton GOs to existing clusters. We retained all clusters of a size greater than three GO terms (and/or pathways) for visualization (Fig. 4A).
Overlaps between regulatory elements and repeats
We constructed de novo repeat libraries for the naked mole-rat and Damaraland mole-rat using RepeatModeler version 2.0.2a (Flynn et al. 2020) and used precomputed de novo repeat libraries from the Dfam database (Storer et al. 2021) for the guinea pig and mouse. We next annotated the location of repeats in mole-rat, guinea pig, and mouse genomes with RepeatMasker version 4.1.2-p1. We used BEDTools version v2.29.2 (Quinlan and Hall 2010) to compute overlaps between elements and repeats and used BEDTools Jaccard to obtain the corresponding Jaccard index (ratio of the intersection to the union of the data sets, in number of bases). “Genome” sets were constructed from n = 10,000 random permutations of the corresponding “orthologous” or “nonalignable” set on the whole genome. We tested for significant differences in overlaps with repeats across sets and used the Jaccard test to define significantly enriched repeat families.
Enrichment of TF binding motifs in repeats
We identified enriched TF motifs in the set of nonalignable enhancers of each mole-rat using the HOMER software (see section Transcription-Factor Binding Motif Enrichment Analysis). We again used a permutation of intervals approach to identify motifs significantly found fully included in a repeat more often than expected by chance. The null distribution was obtained from n = 100 random permutations of motifs within nonalignable enhancers. In mole-rats, we tested for significant association between enriched repeats and the top 10 enriched motifs. In the outgroups, we tested only for significant association between HNF4 and RAR TF motifs and enriched repeats.
Finally, we used GREAT (for details, see section GOs, Pathways, and Gene Enrichment Tests) to test for functional enrichment for enhancers with the co-occurring RAR motifs and SINE/Alu repeats and HNF4 motifs and Sine/ID repeats.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE222972. Aligned BAM files have been deposited in Zenodo (https://doi.org/10.5281/zenodo.8272788). All original code and data sets generated in this study have been deposited in Zenodo (https://doi.org/10.5281/zenodo.7442105) and uploaded as Supplemental Material.
Supplementary Material
Acknowledgments
This project has been funded by the British Heart Foundation (fellowship FS/18/39/33684 to D.V.), the Royal Society (RGS\R2\202330 to D.V.), the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement no. 851360 to C.B.), Cancer Research UK (travel grants C46232/A15517 and 22761 to D.V.), and the European Molecular Biology Organisation (short-term fellowship to D.V.). E.P. is supported by a Newton International Fellowship from the Royal Society (NIF\R1\222125); D.F.A., by a Postdoctoral Fellowship from Alfonso Martin Escudero Foundation. We thank the QMUL and CRUK-CI Genomics cores, David Gaynor and Katy Goddard (Kalahari Research Trust), and Nigel Bennet (University of Pretoria) for technical assistance; Tim Clutton-Brock (Kalahari Research Trust) for the permission to collect tissues from Damaraland mole-rats; Ferdinand Marletaz (UCL, London), Radu Zabet (QMUL, London), Juan J. Tena, and Silvia Naranjo (CABD, Seville) for technical advice; Pierre Vincens (IBENS, Paris), Bronwen L. Aken (European Bioinformatics Institute), and Queen Mary's Apocrita HPC facility, supported by QMUL Research-IT, for the coordination of computing resources; and Alexandre de Mendoza (QMUL) and Masa Roller (European Bioinformatics Institute) for critical comments on the manuscript.
Author contributions: E.P., D.V., and C.B. designed experiments; D.V., D.F.A., S.F., and A.U. performed experiments; E.P., D.V., and C.B. analysed the data; T.J.P., M.Z., and E.J.S. provided tissue samples; and D.V., E.P., and C.B. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277715.123.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Andersson R, Sandelin A. 2020. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet 21: 71–87. 10.1038/s41576-019-0173-8 [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. 2014. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet 46: 685–692. 10.1038/ng.3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S, St-Pierre J. 2012. PGC1α and mitochondrial metabolism: emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci 125: 4963–4971. 10.1242/jcs.113662 [DOI] [PubMed] [Google Scholar]
- Bai S, Kerppola TK. 2011. Opposing roles of FoxP1 and Nfat3 in transcriptional control of cardiomyocyte hypertrophy. Mol Cell Biol 31: 3068–3080. 10.1128/MCB.00925-10 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bennett NC, Ganswindt A, Ganswindt SB, Jarvis JUM, Zottl M, Faulkes CG. 2018. Evidence for contrasting roles for prolactin in eusocial naked mole-rats, Heterocephalus glaber and Damaraland mole-rats, Fukomys damarensis. Biol Lett 14: 20180150. 10.1098/rsbl.2018.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot C, Villar D, Horvath JE, Odom DT, Flicek P. 2018. Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat Ecol Evol 2: 152–163. 10.1038/s41559-017-0377-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R, Amoroso V, Andziak B, Avdieiev S, Azpurua J, Barker AJ, Bennett NC, Brieño-Enríquez MA, Bronner GN, Coen C, et al. 2022. The naked truth: a comprehensive clarification and classification of current ‘myths’ in naked mole-rat biology. Biol Rev Camb Philos Soc 97: 115–140. 10.1111/brv.12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can E, Smith M, Boukens BJ, Coronel R, Buffenstein R, Riegler J. 2022. Naked mole-rats maintain cardiac function and body composition well into their fourth decade of life. Geroscience 44: 731–746. 10.1007/s11357-022-00522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelijns B, Baak ML, Geeven G, Vermunt MW, Wiggers CRM, Timpanaro IS, Kondova I, de Laat W, Creyghton MP. 2020. Recently evolved enhancers emerge with high interindividual variability and less frequently associate with disease. Cell Rep 31: 107799. 10.1016/j.celrep.2020.107799 [DOI] [PubMed] [Google Scholar]
- Chapski DJ, Rosa-Garrido M, Hua N, Alber F, Vondriska TM. 2018. Spatial principles of chromatin architecture associated with organ-specific gene regulation. Front Cardiovasc Med 5: 186. 10.3389/fcvm.2018.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin YA, Dhillon R, Dowell J, Denu JM, Pamenter ME. 2022. Metabolomic analysis of carbohydrate and amino acid changes induced by hypoxia in naked mole-rat brain and liver. Metabolites 12: 56. 10.3390/metabo12010056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmas MJ, Kaplow IM, Genereux DP, Dong MX, Hughes GM, Li X, Sullivan PF, Hindle AG, Andrews G, Armstrong JC, et al. 2023. Evolutionary constraint and innovation across hundreds of placental mammals. Science 380: eabn3943. 10.1126/science.abn3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Brown CD. 2008. Qualifying the relationship between sequence conservation and molecular function. Genome Res 18: 201–205. 10.1101/gr.7205808 [DOI] [PubMed] [Google Scholar]
- Cotney J, Leng J, Yin J, Reilly SK, DeMare LE, Emera D, Ayoub AE, Rakic P, Noonan JP. 2013. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 154: 185–196. 10.1016/j.cell.2013.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R, et al. 2022. Ensembl 2022. Nucleic Acids Res 50: D988–D995. 10.1093/nar/gkab1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KT, Bennett NC, Tsagkogeorga G, Rossiter SJ, Faulkes CG. 2015. Family wide molecular adaptations to underground life in African mole-rats revealed by phylogenomic analysis. Mol Biol Evol 32: 3089–3107. 10.1093/molbev/msv175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz O, Frost JM, Branco MR. 2019. Regulation of transposable elements by DNA modifications. Nat Rev Genet 20: 417–431. 10.1038/s41576-019-0106-6 [DOI] [PubMed] [Google Scholar]
- Dukler N, Huang YF, Siepel A. 2020. Phylogenetic modeling of regulatory element turnover based on epigenomic data. Mol Biol Evol 37: 2137–2152. 10.1093/molbev/msaa073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Zapata F, Munro C, Siebert S, Hejnol A. 2018. Pairwise comparisons across species are problematic when analyzing functional genomic data. Proc Natl Acad Sci 115: E409–E417. 10.1073/pnas.1719369115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi K, Mikami D, Sun H, Tsumita T, Takahashi K, Mukai K, Yuyama K, Igarashi Y. 2020. Blood-brain barrier permeability analysis of plant ceramides. PLoS One 15: e0241640. 10.1371/journal.pone.0241640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, Yin J, Reilly SK, Gockley J, Noonan JP. 2016. Origin and evolution of developmental enhancers in the mammalian neocortex. Proc Natl Acad Sci 113: E2617–E2626. 10.1073/pnas.1603718113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmrich S, Trapp A, Tolibzoda Zakusilo F, Straight ME, Ying AK, Tyshkovskiy A, Mariotti M, Gray S, Zhang Z, Drage MG, et al. 2022. Characterization of naked mole-rat hematopoiesis reveals unique stem and progenitor cell patterns and neotenic traits. EMBO J 41: e109694. 10.15252/embj.2021109694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Seim I, Huang Z, Gerashchenko MV, Xiong Z, Turanov AA, Zhu Y, Lobanov AV, Fan D, Yim SH, et al. 2014. Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep 8: 1354–1364. 10.1016/j.celrep.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat E, Devereaux MEM, Pamenter ME, Weber JM. 2020. Naked mole-rats suppress energy metabolism and modulate membrane cholesterol in chronic hypoxia. Am J Physiol Regul Integr Comp Physiol 319: R148–R155. 10.1152/ajpregu.00057.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkes CG, Eykyn TR, Aksentijevic D. 2019. Cardiac metabolomic profile of the naked mole-rat: glycogen to the rescue. Biol Lett 15: 20190710. 10.1098/rsbl.2019.0710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci 117: 9451–9457. 10.1073/pnas.1921046117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong SL, Capra JA. 2022. Function and constraint in enhancer sequences with multiple evolutionary origins. Genome Biol Evol 14: evac159. 10.1093/gbe/evac159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel D, Davies M, Bhushan B, Kulaberoglu Y, Urriola-Munoz P, Bertrand-Michel J, Pergande MR, Smith AA, Preet S, Park TJ, et al. 2020. Cholesterol-rich naked mole-rat brain lipid membranes are susceptible to amyloid β-induced damage in vitro. Aging (Albany NY) 12: 22266–22290. 10.18632/aging.202138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeven G, Teunissen H, de Laat W, de Wit E. 2018. peakC: a flexible, non-parametric peak calling package for 4C and capture-C data. Nucleic Acids Res 46: e91. 10.1093/nar/gky443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbault P, Liebert A, Itan Y, Powell A, Currat M, Burger J, Swallow DM, Thomas MG. 2011. Evolution of lactase persistence: an example of human niche construction. Philos Trans R Soc Lond B Biol Sci 366: 863–877. 10.1098/rstb.2010.0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard GB, Grønvold L, Røsaeg LL, Holen MM, Monsen O, Koop BF, Rondeau EB, Gundappa MK, Mendoza J, Macqueen DJ, et al. 2021. Comparative regulomics supports pervasive selection on gene dosage following whole genome duplication. Genome Biol 22: 103. 10.1186/s13059-021-02323-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes KM, Barefield DY, Kumar M, McNamara JW, Weintraub ST, de Tombe PP, Sadayappan S, Buffenstein R. 2017. The naked mole-rat exhibits an unusual cardiac myofilament protein profile providing new insights into heart function of this naturally subterranean rodent. Pflugers Arch 469: 1603–1613. 10.1007/s00424-017-2046-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammal F, de Langen P, Bergon A, Lopez F, Ballester B. 2022. Remap 2022: a database of human, mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res 50: D316–D325. 10.1093/nar/gkab996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze I, Bens M, Calzia E, Holtze S, Dakhovnik O, Sahm A, Kirkpatrick JM, Szafranski K, Romanov N, Sama SN, et al. 2018. Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging. BMC Biol 16: 82. 10.1186/s12915-018-0547-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, et al. 2014. Defining functional DNA elements in the human genome. Proc Natl Acad Sci 111: 6131–6138. 10.1073/pnas.1318948111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, et al. 2011. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479: 223–227. 10.1038/nature10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger PHL, Geeven G, Bianchi V, Hilvering CRE, de Laat W. 2020. 4C-seq from beginning to end: a detailed protocol for sample preparation and data analysis. Methods 170: 17–32. 10.1016/j.ymeth.2019.07.014 [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, Kent WJ. 2013. The UCSC genome browser and associated tools. Brief Bioinform 14: 144–161. 10.1093/bib/bbs038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissières V, Pickle CS, Plajzer-Frick I, Lee EA, et al. 2016. Progressive loss of function in a limb enhancer during snake evolution. Cell 167: 633–642.e11. 10.1016/j.cell.2016.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J, Park TJ. 2009. Extreme hypoxia tolerance of naked mole-rat brain. Neuroreport 20: 1634–1637. 10.1097/WNR.0b013e32833370cf [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Cai SY, Boyer JL. 2021. The role of the retinoid receptor, RAR/RXR heterodimer, in liver physiology. Biochim Biophys Acta Mol Basis Dis 1867: 166085. 10.1016/j.bbadis.2021.166085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP. 2011. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet 43: 1154–1159. 10.1038/ng.917 [DOI] [PubMed] [Google Scholar]
- Ma S, Yim SH, Lee SG, Kim EB, Lee SR, Chang KT, Buffenstein R, Lewis KN, Park TJ, Miller RA, et al. 2015. Organization of the mammalian metabolome according to organ function, lineage specialization, and longevity. Cell Metab 22: 332–343. 10.1016/j.cmet.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan MA, Skelly DA, Dona MSI, Squiers GT, Farrugia GE, Gaynor TL, Cohen CD, Pandey R, Diep H, Vinh A, et al. 2020. High-resolution transcriptomic profiling of the heart during chronic stress reveals cellular drivers of cardiac fibrosis and hypertrophy. Circulation 142: 1448–1463. 10.1161/CIRCULATIONAHA.119.045115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy SD, Davidson S, Shchuka VM, Singh G, Malek-Gilani N, Langroudi L, Martchenko A, So V, Macpherson NN, Mitchell JA. 2017. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res 27: 246–258. 10.1101/gr.210930.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni YG, Berenji K, Wang N, Oh M, Sachan N, Dey A, Cheng J, Lu G, Morris DJ, Castrillon DH, et al. 2006. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation 114: 1159–1168. 10.1161/CIRCULATIONAHA.106.637124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. 2013. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155: 1521–1531. 10.1016/j.cell.2013.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka SI, Sabry AD, Cawley KM, Warren JS. 2020. Multiple levels of PGC-1α dysregulation in heart failure. Front Cardiovasc Med 7: 2. 10.3389/fcvm.2020.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Reznick J. 2023. Extreme physiology extreme tolerance to hypoxia, hypercapnia, and pain in the naked mole-rat. J Muscle Res Cell Motil 44: 61–72 10.1007/s10974-022-09623-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Reznick J, Peterson BL, Blass G, Omerbašić D, Bennett NC, Kuich P, Zasada C, Browe BM, Hamann W, et al. 2017. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science 356: 307–311. 10.1126/science.aab3896 [DOI] [PubMed] [Google Scholar]
- Park TJ, Smith ESJ, Reznick J, Bennett NC, Applegate DT, Larson J, Lewin GR. 2021. African naked mole-rats demonstrate extreme tolerance to hypoxia and hypercapnia. Adv Exp Med Biol 1319: 255–269. 10.1007/978-3-030-65943-1_9 [DOI] [PubMed] [Google Scholar]
- Partha R, Chauhan BK, Ferreira Z, Robinson JD, Lathrop K, Nischal KK, Chikina M, Clark NL. 2017. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife 6: e25884. 10.7554/eLife.25884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergande MR, Amoroso VG, Nguyen TTA, Li W, Vice E, Park TJ, Cologna SM. 2021. PPARα and PPARγ signaling is enhanced in the brain of the naked mole-rat, a mammal that shows intrinsic neuroprotection from oxygen deprivation. J Proteome Res 20: 4258–4271. 10.1021/acs.jproteome.1c00131 [DOI] [PubMed] [Google Scholar]
- Polak P, Domany E. 2006. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics 7: 133. 10.1186/1471-2164-7-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongor LS, Gross JM, Vera Alvarez R, Murai J, Jang SM, Zhang H, Redon C, Fu H, Huang SY, Thakur B, et al. 2020. BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks. Epigenetics Chromatin 13: 21. 10.1186/s13072-020-00343-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeepa MM, Grimes GR, Kumar Y, Olley G, Taylor GC, Schneider R, Bickmore WA. 2016. Histone H3 globular domain acetylation identifies a new class of enhancers. Nat Genet 48: 681–686. 10.1038/ng.3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PD, Palmer Droguett DH, Taylor JA, Kim DW, Place ES, Rogers TF, Mank JE, Cooney CR, Wright AE. 2022. Detecting signatures of selection on gene expression. Nat Ecol Evol 6: 1035–1045. 10.1038/s41559-022-01761-8 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. 2015. Evolutionary genomics: evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347: 1155–1159. 10.1126/science.1260943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs RV, Nielsen R. 2015. Phylogenetic ANOVA: the expression variance and evolution model for quantitative trait evolution. Syst Biol 64: 695–708. 10.1093/sysbio/syv042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs RV, Harrigan P, Nielsen R. 2014. Modeling gene expression evolution with an extended Ornstein–Uhlenbeck process accounting for within-species variation. Mol Biol Evol 31: 201–211. 10.1093/molbev/mst190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller M, Stamper E, Villar D, Izuogu O, Martin F, Redmond AM, Ramachanderan R, Harewood L, Odom DT, Flicek P. 2021. LINE retrotransposons characterize mammalian tissue-specific and evolutionarily dynamic regulatory regions. Genome Biol 22: 62. 10.1186/s13059-021-02260-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm A, Bens M, Szafranski K, Holtze S, Groth M, Görlach M, Calkhoven C, Müller C, Schwab M, Kraus J, et al. 2018. Long-lived rodents reveal signatures of positive selection in genes associated with lifespan. PLoS Genet 14: e1007272. 10.1371/journal.pgen.1007272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ES, Omerbašić D, Lechner SG, Anirudhan G, Lapatsina L, Lewin GR. 2011. The molecular basis of acid insensitivity in the African naked mole-rat. Science 334: 1557–1560. 10.1126/science.1213760 [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Storer J, Hubley R, Rosen J, Wheeler TJ, Smit AF. 2021. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA 12: 2. 10.1186/s13100-020-00230-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res 27: 1623–1633. 10.1101/gr.218149.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOudenhove J, Yankee TN, Wilderman A, Cotney J. 2020. Epigenomic and transcriptomic dynamics during human heart organogenesis. Circ Res 127: e184–e209. 10.1161/CIRCRESAHA.120.316704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346: 1007–1012. 10.1126/science.1246426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160: 554–566. 10.1016/j.cell.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MC, Banovich NE, Sarkar A, Stephens M, Gilad Y. 2021. Dynamic effects of genetic variation on gene expression revealed following hypoxic stress in cardiomyocytes. eLife 10: e57345. 10.7554/eLife.57345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Zheng D, Tan SZ, Bower NL, Garside V, Vanwalleghem G, Gaiti F, Scott E, Hogan BM, Kikuchi K, et al. 2020. Deep conservation of the enhancer regulatory code in animals. Science 370: eaax8137. 10.1126/science.aax813 [DOI] [PubMed] [Google Scholar]
- Wong HS, Freeman DA, Zhang Y. 2022. Not just a cousin of the naked mole-rat: Damaraland mole-rats offer unique insights into biomedicine. Comp Biochem Physiol B Biochem Mol Biol 262: 110772. 10.1016/j.cbpb.2022.110772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Wang S, Yang G, Sun X, Zhao S, Lin L, Cheng J, Yang W, Cong W, Sun W, et al. 2017. HIF-1α contributes to hypoxia adaptation of the naked mole rat. Oncotarget 8: 109941–109951. 10.18632/oncotarget.22767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gu Q, Zhang Y, Sasaki T, Crivello J, O’Neill RJ, Gilbert DM, Ma J. 2018. Continuous-trait probabilistic model for comparing multi-species functional genomic data. Cell Syst 7: 208–218.e11. 10.1016/j.cels.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KN, Wong HS, Ramanathan C, Rodriguez-Wagner CA, Roberts MD, Freeman DA, Buffenstein R, Zhang Y. 2022. Naked mole-rat and Damaraland mole-rat exhibit lower respiration in mitochondria, cellular and organismal levels. Biochim Biophys Acta Bioenerg 1863: 148582. 10.1016/j.bbabio.2022.148582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RS, Hayashizaki Y, Andersson R, Sandelin A, Kawaji H, Itoh M, Lassmann T, Carninci P, Consortium F, Bickmore WA, et al. 2015. The frequent evolutionary birth and death of functional promoters in mouse and human. Genome Res 25: 1546–1557. 10.1101/gr.190546.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Shen W, Yao K, Wang H, Liu B, Li T, Song L, Diao D, Mao G, Huang P, et al. 2019. Fine-tuning of PGC1α expression regulates cardiac function and longevity. Circ Res 125: 707–719. 10.1161/CIRCRESAHA.119.315529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C, Ginis I, Furuya K, Klimanis D, Ruetzler C, Spatz M, Hallenbeck JM. 2001. Lipopolysaccharide-induced ischemic tolerance is associated with increased levels of ceramide in brain and in plasma. Brain Res 895: 59–65. 10.1016/S0006-8993(01)02028-5 [DOI] [PubMed] [Google Scholar]
- Zöttl M, Vullioud P, Mendonça R, Torrents Ticó M, Gaynor D, Mitchell A, Clutton-Brock T. 2016. Differences in cooperative behavior among Damaraland mole rats are consequences of an age-related polyethism. Proc Natl Acad Sci 113: 10382–10387. 10.1073/pnas.1607885113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Gong N, Cui Y, Wang X, Cui A, Chen Q, Jiao T, Dong X, Yang H, Zhang S, et al. 2015. Forkhead box P1 (FOXP1) transcription factor regulates hepatic glucose homeostasis. J Biol Chem 290: 30607–30615. 10.1074/jbc.M115.681627 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.