Abstract
Genomic studies of vertebrate chromosome evolution have long been hindered by the scarcity of chromosome-scale DNA sequences of some key taxa. One of those limiting taxa has been the elasmobranchs (sharks and rays), which harbor species often with numerous chromosomes and enlarged genomes. Here, we report the chromosome-scale genome assembly for the zebra shark Stegostoma tigrinum, an endangered species that has a relatively small genome among sharks (3.71 Gb), as well as for the whale shark Rhincodon typus. Our analysis using a male–female comparison identified an X Chromosome, the first genomically characterized shark sex chromosome. The X Chromosome harbors the Hox C cluster whose intact linkage has not been shown for an elasmobranch fish. The sequenced shark genomes show a gradualism of chromosome length with remarkable length-dependent characteristics—shorter chromosomes tend to have higher GC content, gene density, synonymous substitution rate, and simple tandem repeat content as well as smaller gene length and lower interspersed repeat content. We challenge the traditional binary classification of karyotypes as with and without so-called microchromosomes. Even without microchromosomes, the length-dependent characteristics persist widely in nonmammalian vertebrates. Our investigation of elasmobranch karyotypes underpins their unique characteristics and provides clues for understanding how vertebrate karyotypes accommodate intragenomic heterogeneity to realize a complex readout. It also paves the way to dissecting more genomes with variable sizes to be sequenced at high quality.
Genomes accommodate coexisting regions with differential characteristics, and these characteristics are manifested not only in DNA sequences (e.g., GC content) but also in intragenomic heterogeneity of nonsequence features such as chromatin openness, replication timing, and recombination frequency. These features are thought to be associated with how karyotypes of individual species are organized. For example, in the chicken genome, early replicating regions tend to be found in small chromosomes, known as “microchromosomes” (see below). It is still unknown how such intragenomic heterogeneity is accommodated by variable karyotypes as well as how it arose during evolution. To reconstruct the ancestor of all extant vertebrates and the evolutionary process thereafter, information from the evolutionary lineages that branched off in the early phase of vertebrate evolution is instrumental. Among those lineages, whole genome sequence information for cartilaginous fishes has been scarce. The importance of studying cartilaginous fishes is doubled when we consider their genomic trends. No additional whole genome duplication has been reported for cartilaginous fishes, whereas drastic lineage-specific genomic changes are being reported for the other extant nonosteichthyan taxon, cyclostomes (Nakatani et al. 2021).
Cartilaginous fishes (chondrichthyans) are divided into two groups, Holocephala (chimaera and ratfish) and Elasmobranchii (sharks and rays). Even long after whole genome sequences of a holocephalan species, Callorhinuchus milii, were made available (Venkatesh et al. 2014), those sequences have not been validated with any karyotyping report. This limitation stems mainly from the technical difficulty in reproducibly preparing chromosome spreads from a stable supply of metaphase cells. Only recently has our repeated sampling of fresh shark tissues (blood or embryos) (e.g., at aquariums) enabled karyotyping using culture cells for four orectolobiform shark species in Elasmobranchii (Uno et al. 2020). This has paved the way for rigid evaluation of whole genome sequences. Biological studies on cartilaginous fishes have been hindered by low accessibility to fresh material. Moreover, especially in studying elasmobranchs, genome analysis can encounter inherent difficulty incurred by their large genome sizes (for review, see Kuraku 2021). These factors have prevented previous efforts on cartilaginous fishes from obtaining a suite of genome sequences supported by karyotype and genome size estimate as well as transcriptome sequencing (Read et al. 2017; Marra et al. 2019; Weber et al. 2020; Zhang et al. 2020; Rhie et al. 2021; Tan et al. 2021).
Chromosome-level analysis is broadening our scope of comparative genomics (Deakin et al. 2019; Rhie et al. 2021). The difficulty of elasmobranch genome sequencing is manifested in the retrieval of the Hox C cluster, an array of homeobox-containing genes (for review, see Kuraku 2021). While their Hox A, B, and D gene clusters have been reliably assembled with few gaps (Mulley et al. 2009; Hara et al. 2018), assembling the Hox C cluster, which was initially reported as missing from elasmobranch genomes (King et al. 2011), suffered from high GC content and frequent repetitive elements, resulting in fragmentary sequences (Hara et al. 2018; for review, see Kuraku 2021). This difficulty is expected to be overcome by the application of a prevailing approach to scaffolding the genomic fragments up to the chromosome scale using chromatin contact data (Dudchenko et al. 2017; Yamaguchi et al. 2021) as well as long-read sequencing.
Another expectation from chromosome-level genome analysis is the identification of sex chromosomes, although sequencing and assembling sex chromosomes often suffer from difficulties caused by high repetitiveness or uneven sequence depth (Ma et al. 2021; Rhie et al. 2021). While sex determination mechanisms have been revealed by an increasing number of studies on osteichthyan vertebrates (Graves 2016; Pennell et al. 2018), no report is available for vertebrate species outside osteichthyans, namely cyclostomes and chondrichthyans. The quest for sex determination mechanisms can be initiated by the identification of sex chromosomes as already improvised in several vertebrate species (Franchini et al. 2018).
Some vertebrate karyotypes, including most bird karyotypes, consist of small-sized chromosomes, or microchromosomes (Fig. 1A) that were initially recognized as shorter than 1 μm in cytogenetic observations (Ohno et al. 1969; Ohno 1970). Microchromosomes are known to have higher GC content, higher gene density, and different chromatin states compared with the remaining macrochromosomes (Burt 2002; International Chicken Genome Sequencing Consortium 2004; Waters et al. 2021). Some genome sequence-based studies regard chromosomes smaller than 20 Mb as microchromosomes and investigated their possible common origin (International Chicken Genome Sequencing Consortium 2004; Nakatani et al. 2021), but they have not been unambiguously defined on a cross-species basis. Accumulating information from synteny-based analysis suggests that the last common jawed vertebrate ancestor already possessed microchromosomes (Nakatani et al. 2007, 2021; Braasch et al. 2016; Simakov et al. 2020; Meyer et al. 2021; Waters et al. 2021). However, this hypothesis needs to be examined by incorporating more diverse vertebrate taxa into the comparison, on a solid basis of experimentally validated karyotypic configurations of individual species.
Figure 1.
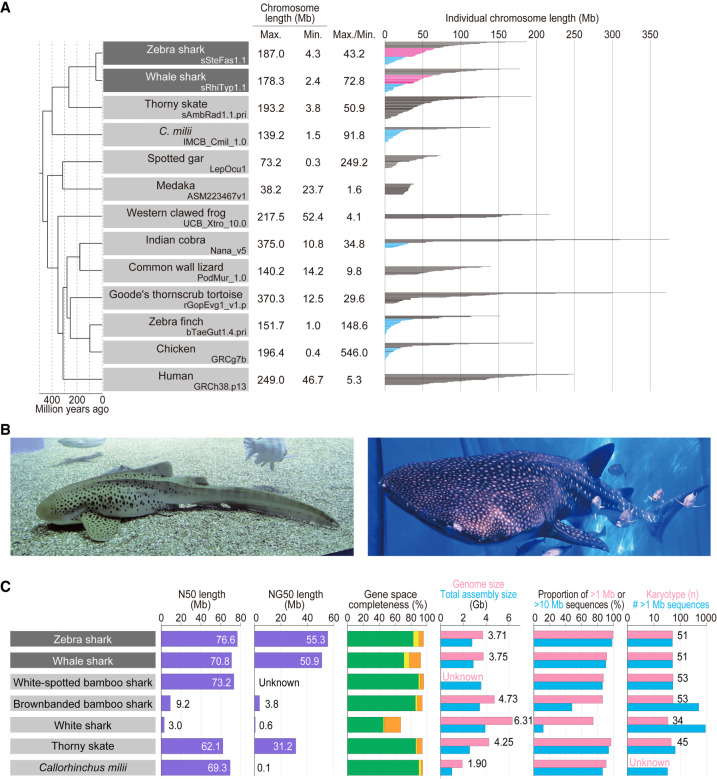
Shark species studied and comparative statistics of their genome assemblies. (A) The karyotypes of diverse vertebrate species are depicted with the length of individual chromosomal DNA sequences. The maximum and minimum chromosome lengths are based on the records in NCBI Genomes. Microchromosomes for osteichthyans are shown in light blue according to individual original reports (International Chicken Genome Sequencing Consortium 2004; Knief and Forstmeier 2016; Suryamohan et al. 2020; Nakatani et al. 2021), whereas the eMID and eMIC of the two shark species whose genomes have been sequenced in the present study (see Results) are shown in magenta and light blue, respectively. (B) Zebra shark (left) and whale shark (right). Photo credit: Shigehiro Kuraku (left), Rui Matsumoto (right). (C) Statistics of the genome assemblies. The identifiers of the genome assemblies, as well as statistical comparison of more metrics, are included in Supplemental Table 1. Gene space completeness shows the proportions of selected one-to-one protein-coding orthologs with “complete” (green), “duplicated” (yellow), and “fragmented” (orange) coverages retrieved in the sequences by the BUSCO pipeline (see Methods). The details of the genome sizes and karyotypes included are based on existing literature (Schwartz and Maddock 1986, 2002; Hardie and Hebert 2004; Uno et al. 2020).
In this study, we focused on the zebra shark Stegostoma tigrinum (or leopard shark; Fig. 1B) and report its whole-genome sequences for the first time. This species has the smallest genome size (3.71 Gb) among the elasmobranch species whose genomes have been sequenced to date (as of December 2022). Each of the resultant chromosome-scale genome assemblies of the zebra shark, as well as the whale shark (Fig. 1B), have been constructed using samples from a single individual (which was not achieved in earlier efforts: Read et al. 2017; Weber et al. 2020; Tan et al. 2021) and controlled by referring to their karyotypes. Using the obtained sequences, we performed comparative investigations to characterize the diversity of chromosomal organization.
Results
The smallest shark genome sequenced to date
We focused on the zebra shark (or leopard shark) Stegostoma tigrinum (formerly, S. fasciatum) with the haploid nuclear DNA content of 3.79 pg (3.71 Gb; Kadota et al. 2023) and the karyotype of 2n = 102 (Uno et al. 2020). Using genomic DNA extracted from blood cells of a female adult, the whole genome was sequenced and assembled with short reads. The resultant sequences were further scaffolded using Hi-C data obtained from blood cell nuclei of the same individual (see Methods). The number of output scaffold sequences longer than 1 Mbp (thereafter tentatively designated “chromosomes”) was 50, which closely approximates its chromosome number revealed by the cytogenetic observation and karyotypic organization previously characterized by us using primary cultured cells (Fig. 1C). The retrieval of the chromosomal scale was also guaranteed by the N50 scaffold length of 76.6 Mb (Fig. 1C).
We also performed de novo whole genome sequencing of an adult male whale shark, Rhincodon typus, using Linked-Read data and Hi-C scaffolding (see Methods). The number of sequences >1 Mbp matched the number of chromosomes in the karyotype (n = 51) (Fig. 1C), and the N50 scaffold length of the resultant assembly reached 70.8 Mb, significantly exceeding that of the assemblies previously published for this species (Fig. 1C). To our knowledge, to date, our product is the only chromosome-scale genome assembly for this species that was built consistently from a single individual.
While we recognize a significant gap between the estimated and retrieved total sequence lengths, genome assemblies for the zebra shark and whale shark showed high completeness of protein-coding gene space of more than 90% (Fig. 1C). Prediction of protein-coding genes on the zebra shark and whale shark genomes, performed by incorporating homolog sequences and transcriptome data, resulted in 33,222 and 35,334 genes, respectively (Supplemental Tables 1, 2), which allowed downstream molecular biological analysis.
Karyotypic trends in sharks
The obtained zebra shark genome assembly consisted of chromosome sequences of highly variable length with a gradual slope, in accordance with our previous cytogenetic observation (Uno et al. 2020), spanning from 187.0 Mb down to 4.3 Mb (Fig. 2A). This pattern does not resemble the length variation in the Callorhinchus milii or the chicken (Fig. 2A). The chicken especially shows a steep slope, marked by a number of chromosomes shorter than 20 Mb (conventionally called microchromosomes) (Figs. 1A, 2A). Gradualism in the chromosome length distribution is also observed in the whale shark (Supplemental Fig. 1), white-spotted bamboo shark (Zhang et al. 2020), and thorny skate (Rhie et al. 2021), and is assumed to be typical karyotypic organization of elasmobranchs (Supplemental Fig. 2). For simplicity, we tentatively designated (1) zebra shark Chromosome 1 to 14, longer than 70 Mb; (2) Chromosome 15 to 33, between 30 and 70 Mb; and (3) Chromosome 34 to 50, shorter than 30 Mb as elasmobranch macro-, middle-sized-, and micro-chromosomes (abbreviated into eMAC, eMID, and eMIC, respectively) that are differentially colored in Figures 1A and 2B. This categorization was also applied to the whale shark chromosomes (Fig. 1A; Supplemental Fig. 2).
Figure 2.
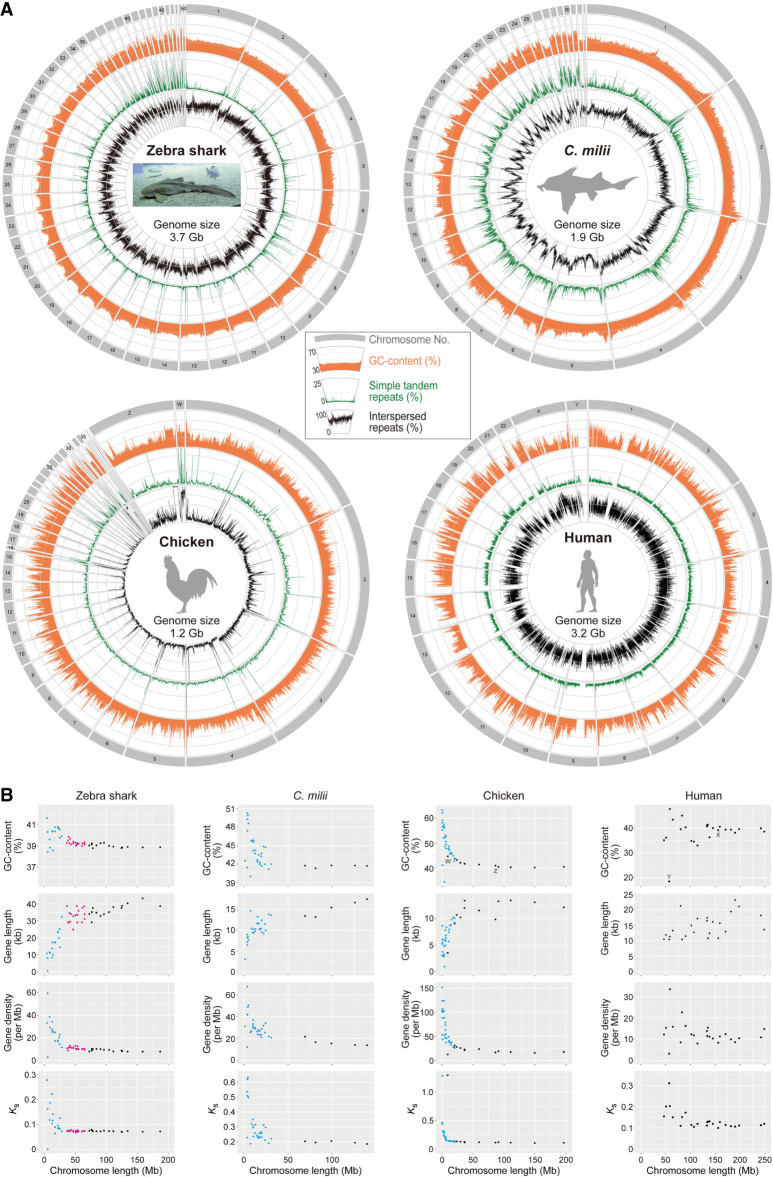
Chromosomal sequence compositions of the zebra shark and selected vertebrate species. (A) Sequence characterization of different chromosomal segments. The orange areas show GC content (30%–70%), whereas the green and black lines show content of simple tandem repeats (0%–25%) and interspersed repeats (0%–100%), respectively, in 100-kb-long nonoverlapping windows. (B) Two-dimensional plots of GC content and median values of gene length, gene density, and the median of synonymous substitutions per synonymous site (Ks) for protein-coding genes on individual chromosomes. See Methods for statistical tests for correlation of these features with chromosome length. Computation of Ks was performed for each of the four species in order by involving a pair species, namely, whale shark Rhincodon typus; small-eyed rabbitfish Hydrolagus affinis; helmeted guineafowl Numida meleagris; and common marmoset Callithrix jacchus. Zebra shark chromosomes are shown as three groups, eMAC, eMID, and eMIC (see text for details), in black, magenta, and cyan, respectively, whereas microchromosomes of the chicken and Callorhinchus milii are colored in cyan. At the moment, C. milii is the only holocephalan species with a chromosome-scale genome assembly.
The zebra shark genome shows consistent intrachromosomal GC content compared with other species, and its chromosome ends in general, as well as the shorter chromosomes, tend to have relatively higher GC content (Fig. 2A). Zebra shark DNA sequences show a uniformly high frequency of interspersed repeats throughout the genome (Fig. 2A). These characteristics are also observed in the whale shark genome (Supplemental Fig. 1A), but the co-occurrence of these two features (small intrachromosomal GC content variation and uniformly high interspersed repeat frequency) was not explicitly observed in the other species (Fig. 2A; Supplemental Fig. 1A).
Vertebrate-wide comparisons including elasmobranchs
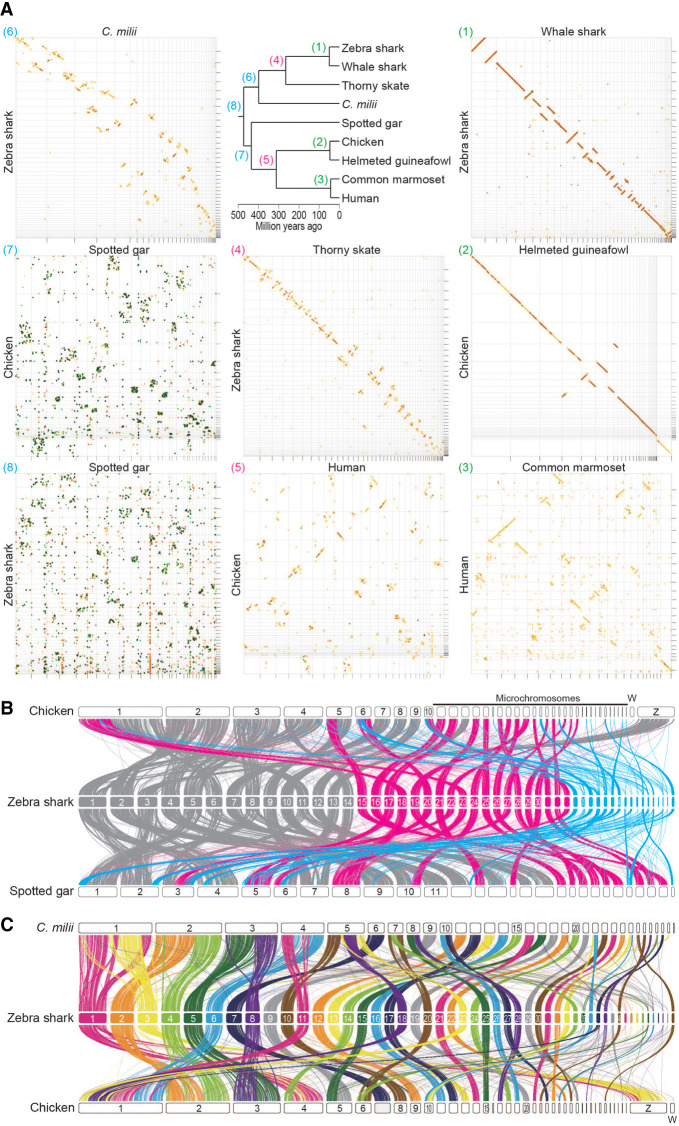
To analyze how the karyotypes of these shark species were derived, chromosomal nucleotide sequences were compared between species pairs with variable divergence times (Fig. 3A). The comparison between the zebra shark and the whale shark showed a high similarity in chromosomal organization with few intrachromosomal breaks (panel 1 in Fig. 3A). The high similarity of this orectolobiform shark species pair suggests high conservation of genomic sequences from around or earlier than 50 million yr ago (see Discussion), compared with osteichthyan species pairs in a similar divergence time range—the human-marmoset pair diverged about 43 million yr ago (panel 3 in Fig. 3A). The relatively high conservation of chondrichthyan chromosome organization is also supported by the comparisons between more distantly related species pairs. The similarity of the zebra shark genome sequences to the thorny skate (panel 4) and the C. milii (panel 6) exceeded those for the species pairs with about 300- and 400-million-yr divergences, respectively (panel 5 and panel 7 in Fig. 3A).
Figure 3.
Cross-species investigation of chromosomal homology. (A) Dot matrices showing genome sequence similarities for selected pairs of vertebrate species with variable divergence times. Sequences of high similarity are shown with diagonal lines by the program D-GENIES (Cabanettes and Klopp 2018) with the “Many repeats” mode. The numbers given to the individual panels (1) to (8), correspond to those at the nodes in the phylogenetic tree and are colored differentially to indicate comparable divergence times. Diagonal lines are colored according to the level of sequence divergence (dark green, 75%–100%; light green, 50%–75%; orange, 25%–50%; yellow, 0%–25%). (B) Chromosomal homology suggested by synteny conservation of one-to-one ortholog pairs. See Methods for details. The color of the ribbons connecting the synteny represents each of the three categories of the zebra shark chromosomes: eMAC (gray); eMID (magenta); and eMIC (cyan). (C) Chromosomal homology between the zebra shark and other vertebrates. Conserved synteny is visualized with the inter-specific correspondence of one-to-one orthologs (see Methods).
Previous studies sought to reconstruct the process of karyotypic evolution of vertebrates but often lacked elasmobranchs in the data set (Sacerdot et al. 2018; Nakatani et al. 2021). In the present study, we performed a gnathostome-wide comparison of the syntenic location of one-to-one orthologs, including the zebra shark (Fig. 3B; Supplemental Fig. 3). A considerable proportion of the one-to-one orthologs are shared between eMAC and large chromosomes of chicken and spotted gar (Fig. 3B). The majority of the genomic regions in the zebra shark eMIC were shown not to share one-to-one orthologs with the so-called microchromosomes of the chicken or spotted gar (Fig. 3B). Moreover, the smallest spotted gar chromosomes were frequently shown to be homologous to zebra shark eMID (chromosomes 15–33), and not to its eMIC (Chromosomes 34–50) (Fig. 3B). It was also shown that zebra shark Chromosomes 10, 25, 26, 27, and 28 have chicken homologs of similar size but their C. milii homologs have been fused into larger chromosomes, whereas zebra shark Chromosomes 2, 3, 4, 5, 6, 8, 11, 18, 20, 23, 33, 34, and 36 are likely to be products of fissions that occurred in the elasmobranch lineage (Fig. 3C; Supplemental Fig. 3). These results cast doubt on the common origins of microchromosomes among chicken, spotted gar, and zebra shark and indicate a more drastic reorganization of karyotypes at the base of jawed vertebrates than previously inferred from the comparison involving only the C. milii as a cartilaginous fish (Nakatani et al. 2021).
Length-dependent properties of chromosomes
Previous studies showed intragenomic heterogeneity of sequence features depending on chromosome lengths in birds and reptiles (Burt 2002; Kuraku et al. 2006; Matsubara et al. 2012; Srikulnath et al. 2021; Waters et al. 2021) To characterize shark chromosomes in depth, base compositions, gene length and density, and molecular evolutionary rates quantified with genic synonymous substitutions (Ks) were investigated (Fig. 2B). Our statistical tests supported a negative correlation for GC content, gene density, and Ks with chromosome length, as well as a positive correlation of gene length (Supplemental Table 3; see Methods). This chromosome length-dependent pattern was not supported in human, sea lamprey, and teleost fishes, but was supported for multiple features (among GC content, gene length and density, and Ks) in some other vertebrates with large chromosome length heterogeneity including other elasmobranch species (Fig. 2; Supplemental Fig. 2; Supplemental Table 3). The correlation of GC content with chromosome length was also observed for the western clawed frog Xenopus tropicalis which conventionally is thought to have no microchromosomes recognized cytogenetically (Uno et al. 2012) (Supplemental Fig. 2; Supplemental Table 3).
Although this chromosome length-dependent pattern is observed in phylogenetically diverse vertebrate species, some short chromosomes show exceptionally low GC content, such as Chromosome 26 of the C. milii, Chromosome 29 and 33 of the chicken, and Linkage Group 29 of the spotted gar (Fig. 2; Supplemental Figs. 1, 2). These exceptions evoke a caution for the generalization of common characteristics of short chromosomes. It needs to be carefully examined whether the relatively short sequences with exceptionally low GC content are fragments of large chromosomes that failed to be assembled to a chromosome scale. Such chromosomal scaffold sequences, with small length and relatively low GC content, may be the cause of the insufficient support for the chromosome length-dependent pattern in some species (Supplemental Table 3).
Do “macrochromosome” ends resemble microchromosomes?
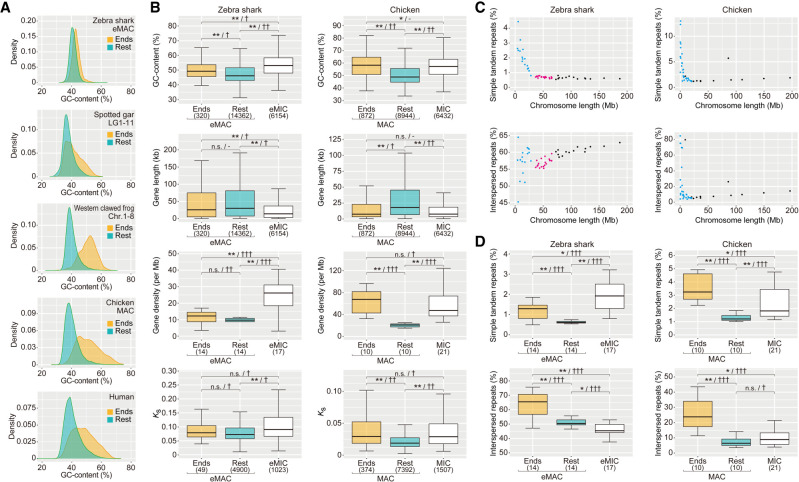
We analyzed regional variations within individual chromosomes of diverse vertebrates, including the zebra shark. To further characterize the distinct trend of chromosomal ends indicated in Figure 2, we separated the 1-Mb-long ends from relatively large chromosomes and analyzed the trends of genomic sequences in five vertebrate species (Fig. 4A). Intact chromosome ends are known to be occupied by telomeric or subtelomeric simple repeats. To eliminate the effect of those repeats, we focused on the regions harboring protein-coding genes, which recapitulated the higher GC content in the chromosome ends (Supplemental Fig. 4). To examine other characteristics of chromosome ends, we focused on zebra shark and chicken with a large chromosome length variation. Our further comparison consistently revealed an increase of the medians of GC content, gene density and synonymous substitution rate, as well as a decrease in gene length, in the ends of relatively large chromosomes, compared with their remainders (Fig. 4B). In both the zebra shark and chicken, the medians of these features for large chromosome ends were closer to those for small chromosomes (zebra shark eMIC and chicken MIC) than those for the remainders of large chromosomes (Fig. 4B). This pattern is less pronounced in the zebra shark than in chicken, according to the variable support levels from statistical tests (indicated with the number of the symbol “†” in Fig. 4B; Supplemental Table 4; also see Methods).
Figure 4.
Intrachromosomal heterogeneity of sequence characteristics. (A) Comparison of global GC content in 10-kb-long nonoverlapping windows between the 1-Mb-long ends and the remainders of relatively large chromosomes for diverse vertebrates. (B) Comparison of GC content of protein-coding regions, gene length, gene density, and synonymous substitution rate (Ks) among the 1-Mb-long chromosome ends, their remainders, and relatively small chromosomes (zebra shark eMIC and chicken microchromosomes [MIC]), for the zebra shark and chicken, respectively. (C) Two-dimensional plots of chromosome lengths and the coverage of interspersed repeats and simple tandem repeats. The coloring of the dots follows that in Figure 2B. (D) Differential distribution of simple tandem repeats and interspersed repeats. Proportions of the sequences identified as simple tandem repeats and interspersed repeats in 10-kb-long windows were compared between the 1-Mb-long ends of relatively large chromosomes (zebra shark eMAC and chicken macrochromosomes [MAC]), their remainders, and relatively small chromosomes (zebra shark eMIC and chicken MIC). In B and D, significance of difference is indicated as follows: (*) P-value < 0.05/number of tests, (**) P-value < 0.01/number of tests; (n.s.) not significant. The effect sizes in statistical tests are indicated with a hyphen for no effect, one dagger symbol “†” for small effect, two dagger symbols “††” for medium effect, and three dagger symbols “†††” for large effect. Numbers of the genomic regions sampled are included in parentheses. See Methods for more details about statistical tests.
Intragenomic repetitive element distribution
We also analyzed the distribution of repetitive elements on chromosomes of variable length. In fact, previous studies yielded equivocal observations. Some of those studies showed a higher abundance of repetitive elements on larger chromosomes (Koochekian et al. 2022), whereas others indicated localization biased toward smaller chromosomes (Hara et al. 2018). So far, no solid gnathostome-wide comparison has been made by taking the difference of repeat classes into account. In the newly obtained shark genome sequences, we separately quantified the sequence proportions identified as interspersed repeats (LINE, SINE, LTR, and DNA elements) and simple tandem repeats (simple repeats and low-complexity DNA sequences, including satellites). In the zebra shark, the interspersed repeat content is positively correlated with chromosome length, whereas the simple tandem repeat content shows a negative correlation (Fig. 4C; Supplemental Fig. 5; Supplemental Table 3). These patterns were also observed in the chicken and the C. milii but in a less pronounced manner, whereas they were not observed in human (Fig. 4C; Supplemental Fig. 5; Supplemental Table 3).
As examined above for other characteristics, we dissected the observed chromosome length-dependent trend of repeat distribution, again by isolating the 1-Mb-long ends versus the remainder of relatively large chromosomes (Fig. 4D; Supplemental Table 4). In this comparison, we observed a higher content of interspersed repeats in the ends of relatively large chromosomes, namely zebra shark eMAC and chicken MAC (Fig. 4D). The higher repeat content was commonly observed in relatively small chromosomes (zebra shark eMIC and chicken MIC), except that interspersed repeat content is reduced in zebra shark eMIC (Fig. 4D).
First genomic characterization of a shark sex chromosome
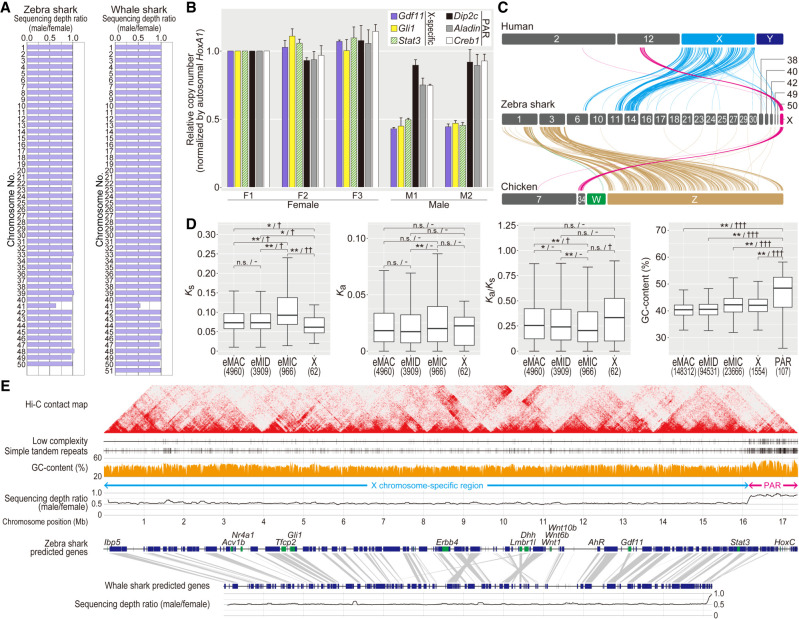
So far, there has been no intensive DNA sequence-based characterization of sex chromosomes for chondrichthyan species. Expecting that a sex chromosome will show a distinct male–female ratio of sequencing depth (Palmer et al. 2019), we performed short-read sequencing of the whole genomes for both sexes in zebra shark and whale shark. For these species, our previous cytogenetic analysis did not detect any heteromorphic sex chromosomes (Uno et al. 2020). Our comparison among different chromosomes detected a lower male-to-female sequencing depth ratio of close to 0.5 for Chromosome 41 of both these species (Fig. 5A). This suggests male as a heterogametic sex and the XY system for these species, which was validated for zebra shark by genomic quantitative polymerase chain reaction (PCR) (Fig. 5B). Next, we investigated the origin of the putative X Chromosome of these species—was it derived from the same ancestral chromosomes that were differentiated later into the sex chromosomes of other vertebrate lineages, particularly of mammals and birds that have long-standing sex chromosomes? Our comparison revealed chromosome-level homology of the putative X Chromosomes of the zebra shark to a part of human Chromosome 12 and chicken Chromosome 34 (Fig. 5C, magenta). Neither the human X, human Y, chicken Z, nor chicken W Chromosomes showed pronounced homology with the putative zebra shark Chromosome X. The putative shark X Chromosome identified in this study does harbor orthologs of a number of well-studied regulatory factors but does not harbor the orthologs of the master sex determination genes identified in other vertebrates including teleost fishes, that is, Dmrt1- or Sox3-related transcription factors as well as components of the TGFB signaling pathway, such as Amh, Amhr2, Bmpr1b, Gsdf, and Gdf6 (Bertho et al. 2021).
Figure 5.
Genomic identification of the zebra shark Chromosome X. (A) Male–female ratio of short-read sequencing depth in the shark chromosomes. (B) Male-female copy-number difference of zebra shark Chromosome X. Amplification levels of the genes on the PAR and the remainders of Chromosome X (scaffold 41) was quantified using real-time PCR controlled with amplification of autosomal genomic regions, by normalizing the PCR product abundance with that for the individual F1 (sSteFas1) (see Methods). (C) Cross-species synteny of sex chromosome-linked genes based on 1-to-1 orthologs. (D) Characteristic comparisons between the different chromosome categories in the zebra shark. Ks (synonymous substitution rate) and Ka (nonsynonymous substitution rate) values were calculated for 1-to-1 orthologs shared with the whale shark. Only for GC content, the PAR was shown separately, because it contained few genes. Significance of difference is indicated as follows: (*) P-value < 0.05/number of tests, (**) P-value < 0.01/number of tests; (n.s.) not significant. The effect sizes are indicated with a hyphen for no effect, one dagger symbol “†” for small effect, two dagger symbols “††” for medium effect, and three dagger symbols “†††” for large effect in statistical tests. Numbers of the genomic regions sampled are shown in parentheses. See Methods for more details about statistical tests. (E) Structural comparison between the zebra shark and whale shark X Chromosome sequences. We focused more on the zebra shark because we could not retrieve a part of the whale shark chromosome that is homologous to the putative zebra shark Chromosome X.
In the ∼17 Mb-long sequence of the putative zebra shark Chromosome X, one 1.5 Mb-long end showed a male–female sequencing depth ratio of nearly 1.0 (Fig. 5D). This region, with a sequence depth comparable to that of the autosomes, is deduced to be a pseudoautosomal region (PAR) that is likely shared between the heterogametic sex chromosomes X and Y (Smeds et al. 2014; Palmer et al. 2019). The identification of PAR was supported by a comparable level of genomic qPCR amplification for multiple genes in this region to that for autosomal regions (Fig. 5B). We characterized possible unique patterns of molecular evolution typical of sex chromosomes (Fig. 5D; Supplemental Table 5). In the X Chromosome, protein-coding genes showed a significant decrease of synonymous substitution rate (Ks) supported by a small effect and an increased median of nonsynonymous substitution rate (Ka), resulting in an increased Ka/Ks ratio. Our comparison also revealed higher frequency of low-complexity repeats as well as higher GC content in the PAR (Fig. 5D,E), which is a hallmark of PAR observed in other species resulting from accelerated recombination (Galtier et al. 2001; Galtier 2004; Smeds et al. 2014). The Hi-C contact map, derived from zebra shark blood, shows subproximal contacts suggesting intermittent chromatin compartmentalization within the PAR and the rest of Chromosome X (Fig. 5E).
Our comparison of the ortholog location between the putative X Chromosomes of the two species supported a high cross-species conservation of the chromosome structure (Fig. 5E). In the current whale shark assembly, Chromosome 41, which corresponds to the putative Chromosome X, may not cover the whole chromosome, possibly excluding one end of the PAR (Fig. 5E).
Identification of the Hox C cluster on the putative X Chromosome
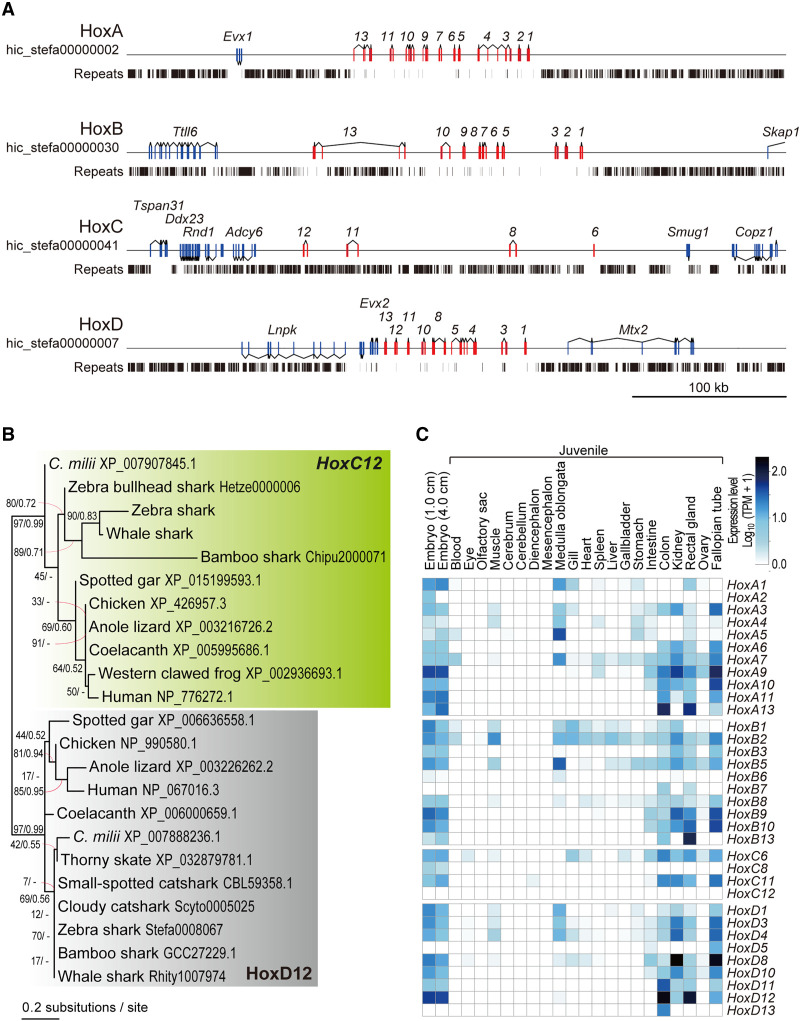
In the newly obtained sequence of the putative zebra shark X Chromosome, we identified an array of orthologs of the nonshark genes encoding homeobox proteins Hox C (Fig. 5E). In elasmobranchs, Hox C genes were long thought missing from the genome (King et al. 2011) but later identified in several shark species, as rogue open reading frames (ORFs) flanked by massively repetitive sequences (Hara et al. 2018; for review, see Kuraku 2021). Our genome-wide gene prediction for the zebra shark detected the ORF of Hoxc8, -c11, and -c12, whereas the partial ORF of the putative Hoxc6 ortholog was also identified by a manual search of the raw genomic sequence (Fig. 6A). These Hox C genes were located in a 180-kb-long genomic segment in the PAR of the putative Chromosome X (Fig. 6A), identified as a single cluster in an elasmobranch fish for the first time. Their orthologies were confirmed with molecular phylogenetic trees, which also indicated elevated molecular evolutionary rates with long branches for elasmobranch Hox C genes (Fig. 6B). Our RNA-seq data showed the transcription of these Hox C genes (except for Hoxc12) in embryos and juvenile tissues (Fig. 6C). The identified zebra shark Hox C cluster is massively invaded by repetitive elements unlike the other Hox gene clusters (A, B, and D clusters) of this and many other vertebrate species (Fig. 6A), as in the Hox C-containing genomic segments of other shark species (Hara et al. 2018). Our search for zebra shark orthologs of the protein-coding genes located near the human Hox C cluster (e.g., ATF7, CBX5) revealed poor conservation of the gene compositions. Some of the zebra shark orthologs were not identified in its entire genome sequence, suggesting a divergent nature of the genomic regions flanking the Hox C cluster.
Figure 6.
Zebra shark Hox C genes. (A) Genomic structure of the zebra shark Hox clusters and their neighboring regions. The exons of the Hox genes are shown in red boxes. (B) Molecular phylogenetic tree of Hox12 group of genes. The tree was inferred with the maximum-likelihood method as described in Methods. (C) Expression profiles of the zebra shark Hox genes in embryos and various tissues of a juvenile.
Discussion
In this study, we chose two orectolobiform shark species (zebra shark and whale shark) in Elasmobranchii and characterized their genomic organization with chromosome-scale DNA sequences. This study was achieved in support of epigenome and transcriptome data prepared using fresh tissue samples and previously obtained karyotype information. Our results suggest that their karyotypes are organized by chromosomes of gradual sizes marked with size-dependent sequence properties (Fig. 2B). The length gradualism is a remarkable feature of elasmobranch karyotypes, although we tentatively grouped those chromosomes into three length-dependent categories as proposed recently for other elasmobranch species (Marlétaz et al. 2023; Stanhope et al. 2023). The pattern in the shark chromosomal organization is unique in its diversity among vertebrates (Uno et al. 2020), which is characterized by abundant chromosomes (up to 106 for diploids) and variable chromosome sizes. The abundance and highly variable sizes of chromosomes are known for some avian species, but the shark genome organization is distinct from avian counterparts in that shark chromosomes generally have higher repeat content than those of the chicken (Fig. 2). In our comparison, the shark karyotype is also characterized by the ratio of the largest and smallest chromosome lengths of 40 to 100, compared with <10 for most vertebrates, except for species with microchromosomes (Fig. 1A). In fact, the length of the shortest chromosomal sequence in typical genome assemblies deposited currently in public databases is often dependent on sequence length cutoff established by the researcher. Especially for species with no solid karyotypic reference such as C. milii, the range of sequences considered as chromosomes needs to be carefully examined.
Our chromosome-scale genome sequencing and analysis were enabled by access to fresh tissue samples. Because of low accessibility, no previous efforts could provide a set of DNA sequences, karyotypic configuration, and reliable measure of nuclear DNA content for a single chondrichthyan species. Of these, the two latter elements serve as indispensable references to validate the output of sequencing. These requirements are satisfied for both zebra shark and whale shark in our study. Especially for the whale shark, no published studies used chromatin contact data for Hi-C scaffolding and transcriptome sequencing (Marra et al. 2019; Weber et al. 2020; Tan et al. 2021). In our study, the access to embryos and blood not only enabled chromosome-scale genome scaffolding but also provided transcriptional evidence of most shark Hox C genes that have been shown to exist in a cluster for the first time (Fig. 6).
Peculiar fractions of vertebrate karyotypes with small sizes and higher GC content have traditionally been designated as microchromosomes, which usually denote chromosomes shorter than 20 Mb (e.g., Nakatani et al. 2021) but have no uniform definition (see Introduction). Our investigation, focusing on various aspects of chromosomal DNA sequences, provided a novel view of vertebrate karyotypes that cannot be understood with a simple binary classification, namely with or without microchromosomes, or between macro- and microchromosomes. This view is supported by the common pattern of high intrachromosomal heterogeneity within individual macrochromosomes (Fig. 4; Supplemental Fig. 4) as well as interchromosomal heterogeneity among different microchromosomes (Fig. 2; Supplemental Figs. 1, 2). In particular, the heterogeneity within macrochromosomes, marked with high GC content, high gene density, and small gene length of their ends, may be shared widely among diverse vertebrates (Fig. 4A,B). Intragenomic heterogeneity of GC content was previously suggested to be caused by GC-biased gene conversion (Mugal et al. 2015). In addition, the uniform numbers of recombination per chromosome (Dumont and Payseur 2008) have been thought to explain the chromosome length-dependent GC content variation between different chromosomes. Our observations did not show pronounced length-dependent variation of GC content for chromosomes that were longer than 100 Mb (Fig. 2B; Supplemental Fig. 2). Taken together, we speculate that the peculiar nature of chromosome ends mainly account for the variation of GC content among different chromosomes (Fig. 4B). Importantly, “chromosome ends” in this context not only harbor telomeric or other simple repeats but also hold complex sequences including protein-coding genes in sequence stretches that are longer than 1 Mb (Fig. 4B; Supplemental Fig. 4). In the western clawed frog genome, such sequence stretches marked with elevated GC content span much longer ranges than in other genomes (Supplemental Figs. 1A, 2). The observed features of smaller chromosomes with larger proportions of such “ends” in length are more affected than those of longer chromosomes, which likely explains the length-dependent nature of chromosomes. The nature of macrochromosome ends (e.g., with higher GC content) is thought to be a remnant of the fusion of one or more microchromosome(s) to a macrochromosome (Waters et al. 2021). This hypothesis is not supported by our observation that even species possessing no explicit microchromosomes (e.g., western clawed frog) have chromosome ends with the peculiar nature of DNA sequences (Supplemental Figs. 1A, 4). No close relatives of the western clawed frog have been shown to possess microchromosomes; Tymowska 1991), and thus microchromosome fusions cannot account for the characteristics of their chromosome ends with higher GC content.
Our genome-wide sequencing depth investigation covering both sexes revealed the XY system for the two studied shark species and enabled the first sequence-based identification of shark sex chromosomes (namely, Chromosome X; Fig. 5A). The genes on the shark Chromosome X tend to show larger nonsynonymous substitution rates (Ka) and smaller synonymous substitution rates (Ks) than those on autosomes, resulting in a higher Ka/Ks ratio (Fig. 5D). This resembles the pattern known in other species including birds and insects, which is known as the “Faster X (or Faster Z) hypothesis” (Mank et al. 2007; Meisel and Connallon 2013; Charlesworth et al. 2018; Xu et al. 2019). The smaller Ks value is also indicative of the support for the male-driven evolution hypothesis (Miyata et al. 1987; Li 2002), which is to be examined by higher Ks values for genes on the presumptive Y chromosome that remains unidentified. Our comparison of protein-coding gene compositions showed that the zebra shark and the whale shark largely share Chromosome X that is homologous to each other (Fig. 5E). The Chromosome X harbors the Hox C cluster that was previously shown to be highly divergent and degenerative (Hara et al. 2018), but its localization in the PAR (Fig. 5E) suggests a balanced dosage of the Hox C genes and their expressions between males and females.
The whale shark is known as the largest extant “fish”, and one of its extant closest relatives is the zebra shark (Naylor et al. 2012). In elasmobranch evolution, the lineages leading to these two species diverged no later than 48.6 million yr ago (Long 1992). The high similarity of the chromosome-scale sequence organization (panel 1 in Fig. 3; Supplemental Fig. 3) as well as the gene compositions on the Chromosome X between these two species (Fig. 5E) indicates a lower rate of chromosomal rearrangement in these lineages compared with those of species pairs of similar divergence times in other vertebrate lineages (Fig. 3A). Among vertebrates, mammals and birds have relatively long-standing sex chromosomes (X/Y and Z/W, respectively) shared throughout these individual taxa that arose more than 48.6 million yr ago (Long 1992). Although still limited in number, sex chromosomes have been identified in some elasmobranch species by cytogenetic analyses, all of which have a male-heterogametic system (Uno et al. 2020). Our study with genome sequencing showed that it also holds for the zebra shark and whale shark. Sharks, or a phylogenetically wider subset of cartilaginous fishes, may possess even older sex chromosomes, depending on phylogenetic prevalence of their homologs in more distantly related shark and even ray species.
Our synteny analysis yielded novel insights into genome evolution encompassing the whole diversity of vertebrates. It showed homology of the shark Chromosome X to human Chromosome 12 and chicken Chromosome 34 (Fig. 5C), which suggests a difference in ancestral autosomes that were adopted as sex chromosomes between elasmobranchs and other vertebrates for which sex chromosomes have been characterized. Although sex chromosomes of the other chondrichthyans remain to be explored, these results suggest an independent origin of chondrichthyan sex chromosomes, which is in line with studies on other vertebrates showing repeated, independent recruitment of sex chromosomes from ancestral chordate chromosomes (Graves 2016). The synteny analysis involving sharks also provided clues to the origin of microchromosomes. It suggests that eMIC, the shark's small chromosomes, are homologous to several of the large chromosomes in both chicken and spotted gar and are not necessarily homologous to their microchromosomes (Fig. 3B). Also, microchromosomes of the chicken and gar were not shown to be homologous to eMICs (Fig. 3B). It is crucial for any effort for reconstructing the diversity of chromosome organization in vertebrates to incorporate diverse elasmobranchs into the comparisons.
Methods
Animals
Fresh blood from a female adult zebra shark (total length, 2.2 m); Individual ID, sSteFas1 (also called F1) and a male adult whale shark (total length, 8.8 m; Individual ID, sRhiTyp1) were sampled at Okinawa Churaumi Aquarium and used for the preparation of whole-genome shotgun DNA libraries, Hi-C libraries, and RNA-seq libraries as well as for measuring nuclear DNA content by flow cytometry. Likewise, fresh blood of a male zebra shark (total length, 2.1 m; Individual ID, sSteFas2 [also called M1]) and a female whale shark (total length, 8.0 m; Individual ID, sRhiTyp2) were sampled and used for quantifying the male/female ratios of individual chromosomal regions. Extraction of ultrahigh molecular weight DNA was performed by collecting blood cells by centrifugation, and the collected cells were embedded in agarose plugs (4.0 × 105 cells/plug). The agarose gel plugs were prepared and processed with the CHEF Mammalian Genomic DNA Plug Kit (Bio-Rad 1703591). Total RNAs used to construct RNA-seq libraries were extracted from various tissues of a female juvenile zebra shark (total length, 30 cm) born at Okinawa Churaumi Aquarium and a female juvenile whale shark (total length, 7.7 m; Individual ID, sRhiTyp3) (Supplemental Table 6). These animals were introduced into the aquarium in accordance with local regulations before those species were assessed as endangered. Animal handling and sample collections at the aquarium were conducted by veterinary staff without restraining the individuals under the experiment ID AT19002 approved by the Institutional Animal Care and Use Committee of the Okinawa Churashima Foundation in accordance with the Husbandry Guidelines approved by the Ethics and Welfare Committee of the Japanese Association of Zoos and Aquariums. All other experiments were conducted in accordance with the Guideline of the Institutional Animal Care and Use Committee (IACUC) of RIKEN Kobe Branch (Approval ID: H16-11).
Genome sequencing and scaffolding
For a female zebra shark, paired-end and mate-pair DNA libraries for de novo genome sequencing were prepared and sequenced as previously described (Hara et al. 2018; Yamaguchi et al. 2021). The amount of starting DNA and numbers of PCR cycles for the library preparation are included in Supplemental Table 6. The total sequencing coverage amounted to 95.8 times the genome size based on the reference measured previously by flow cytometry (3.71 Gb; Kadota et al., in prep.). Low-quality bases from paired-end reads were removed by TrimGalore v0.6.6 (https://github.com/FelixKrueger/TrimGalore, accessed 4 Jan 2019) with the options “‐‐stringency 2 ‐‐quality 20 ‐‐length 25 ‐‐paired ‐‐retain_unpaired”. As described previously (Hara et al. 2018), short-read assembly of the zebra shark, as well as scaffolding with mate-pair reads followed by gap closure, was performed using Platanus v1.2.4 (Kajitani et al. 2014).
Whole genome sequencing for a male whale shark used the 10x Genomics Chromium to produce Linked-Read data. A DNA library was prepared using 12 ng of gDNA extracted from blood cells according to the user guide of the Chromium Genome Library Kit v2 Chemistry using the Chromium Genome Library Kit & Gel Bead Kit v2 (10x Genomics 120258) and the Chromium Genome Chip Kit v2 (10x Genomics 120257). The library was sequenced on a HiSeq X (Illumina) platform to obtain 151 nt-long paired-end reads. Sequence assembly using the Linked-Read data of 46.4 times the genome size was performed with the program Supernova v2.0 (Weisenfeld et al. 2017). The resultant sequences were subjected to scaffolding with the program P_RNA_scaffolder (commit 7941e0f in GitHub) (Zhu et al. 2018) using the result of the alignment of transcriptome sequence reads (obtained as described below) performed with the program HISAT2 v2.1.0 onto those genome sequences (Kim et al. 2019).
Hi-C data production and chromosome-scale genome scaffolding
Hi-C libraries of the zebra shark and whale shark were constructed using restriction enzymes DpnII and HindIII, respectively, as previously reported (Kadota et al. 2020). Blood cells collected as described above were fixed in 1% formaldehyde solution. A fixed tissue containing 10 μg of DNA was used for the preparation of Hi-C DNA via in situ restriction digestion and ligation. The Hi-C library was prepared using 2 μg of the ligated DNA with five cycles of PCR amplification. Quality controls of the ligated DNA and the Hi-C libraries were performed as described previously (Kadota et al. 2020).
Each of the zebra shark and the whale shark genome assemblies was used for Hi-C read mapping with Juicer v1.5 (Durand et al. 2016a) and chromosome-scale scaffolding with the program 3d-dna (v180922) (Dudchenko et al. 2017). In the scaffolding, three different lengths were tested (5, 10, and 15 kb) for the option “-i” defining the input sequence length threshold. For each species, the three resulting scaffolding outputs, as well as the original assembly before Hi-C scaffolding, were assessed based on sequence length distribution and protein-coding gene completeness. Among all the scaffolding outputs compared, the output with the option “-i 10000” was judged to be optimal and was subjected to a “review” of the scaffolding results on Juicebox v1.11.08 (Durand et al. 2016b) to minimize inconsistent signals of chromatin contacts (Supplemental Fig. 6). The review was facilitated by referring to nucleotide sequence-level similarity between different scaffolding outputs visualized by SyMAP v5.0 (Soderlund et al. 2011). After the review, the sequences judged as contaminants from other organisms were removed, as previously reported (Hara et al. 2018).
Repeat identification
To obtain a species-specific repeat library, RepeatModeler v2.0.2a was run on the genome assembly of the individual species with default parameters (Smit and Hubley 2008). Detection of repeat elements in the genome was performed by RepeatMasker v4.1.2-p1 (Smit et al. 2013) with RMBlast v2.6.0+, using the species-specific repeat library obtained above. For quantification of the content of interspersed repeats and simple tandem repeats, RepeatMasker was run separately with the options “-nolow –norna” and “-noint –norna”, respectively.
Gene model construction
The program Braker v2.1.6 was used for gene prediction by inputting the results of RNA-seq read mapping to a genome assembly in which repetitive sequences are soft-masked by RepeatMasker with the options “-nolow –xsmall”, as well as amino acid sequences of closely related species as homolog hints (Smit et al. 2013; Brůna et al. 2021). To build the homolog hints based on the amino acid sequences, we used the previously reported amino acid sequence sets of the brownbanded bamboo shark and the whale shark.
Gene space completeness in Figure 1C was obtained by the BUSCO pipeline ver. 5 (Seppey et al. 2019) using the BUSCO's Vertebrata ortholog set.
Synonymous and nonsynonymous substitution quantification
To calculate the number of synonymous substitutions per synonymous site (Ks) and the number of nonsynonymous substitutions per nonsynonymous site (Ka), the 1-to-1 orthologs shared by the four elasmobranch species (zebra shark, whale shark, brownbanded bamboo shark, and cloudy catshark) were selected by SonicParanoid v1.3.4 (Cosentino and Iwasaki 2019) as follows. First, peptide sequences of the retrieved orthologs were aligned with MAFFT v7.475 with the option “-linsi” (Katoh and Standley 2013). The individual alignments were trimmed and back-translated into nucleotides with trimAl v1.4.rev15 with the options “-automated1 –backtrans” followed by the removal of gapped sites using trimAlL with the option “-nogaps” (Capella-Gutiérrez et al. 2009). Ortholog groups containing fewer than 100 aligned codons or a stop codon were discarded. For the selected ortholog groups, Ks and Ka were computed with yn00 in the PAML v4.9c88 (Yang 2007). Computed values larger than 0.01 and smaller than 99 were included in the results (Figs. 2B, 4B, 5D).
RNA-seq and transcriptome data processing
Total RNAs were extracted with TRIzol reagent (Thermo Fisher Scientific). Quality control of the RNA treated with DNase I was performed with Bioanalyzer 2100 (Agilent Technologies). Libraries were prepared with TruSeq RNA Sample Prep Kit (Illumina) or TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) as previously described (Hara et al. 2018). The amount of starting total RNA and numbers of PCR cycles are included in Supplemental Table 6. To remove adaptor sequences and low-quality bases, the obtained sequence reads were trimmed with Trim Galore! v0.6.6 as outlined above, and de novo transcriptome assembly was performed with the program Trinity v2.11.0 with the option “‐‐SS_lib_type RF” (Grabherr et al. 2011). The trimmed RNA-seq reads were aligned to the genome assembly using the program HISAT2 v2.1.1 (Kim et al. 2019), which was followed by gene expression quantification with StringTie v2.0.6 (Pertea et al. 2016).
Conserved synteny detection
Characterization of chromosomal homology among different species used predicted protein sequence data sets available at the NCBI RefSeq database. After alternative splicing variants were removed, one-to-one orthologs were selected by SonicParanoid v1.3.4 with the option “most-sensitive” (Cosentino and Iwasaki 2019). Conserved synteny between species was visualized based on single copy 1-to-1 orthologs using RIdeogram (Hao et al. 2020).
Statistical analysis
Relationships of chromosome lengths with chromosome sequence features were tested with the correlation coefficient through Pearson's correlation analysis (Figs. 2B, 4C; Supplemental Figs. 2, 5), and the details are included in Supplemental Table 3.
Statistical significance of between-group differences in the box plots in Figure 4B, 4D, and 5D was tested with the nonparametric Kruskal–Wallis test and the Mann–Whitney U test, and the detailed results are included in Supplemental Tables 4 and 5. The Kruskal–Wallis test was used to evaluate the significance of difference among groups, the Mann–Whitney U test was used to evaluate the significance of difference between two groups, and Rank-biserial correlation was calculated as effect size. Bonferroni correction was performed for multiple comparisons. A large number of samples resulted in a smaller standard error of the mean, which is more likely to be significant. Therefore, the degree of difference between two groups was evaluated not only by P-value, but also by effect size.
Molecular phylogeny inference
Amino acid sequences were retrieved from aLeaves (Kuraku et al. 2013). Multiple sequence alignment was performed with MAFFT with the option “-linsi”. The aligned sequence sets were processed using trimAl v1.4 rev15 with the option “-automated1” (Capella-Gutiérrez et al. 2009). This was followed by another trimAl run with the option “-nogaps”. Molecular phylogenetic trees were inferred by RAxML with the “-m PROTCATWAG -f a -# 1000” options unless stated otherwise (Stamatakis 2014). Tree inference in the Bayesian framework was performed with the program PhyloBayes v4.1c with the options “-cat -dgam 4 -wag -nchain 2 1000 0.3 50” unless stated otherwise. This was followed by an execution of bpcomp in the PhyloBayes v4.1c package with the option “-x 100” (Lartillot et al. 2009). The support values at the nodes of molecular phylogenetic trees included are, in order, bootstrap values and Bayesian posterior probabilities. The latter was shown only when the relationship at the node in the visualized tree was supported by the Bayesian inference.
Identification of the shark X Chromosome
To identify a chromosome-scale scaffold with a distinct male–female sequencing depth ratio in the zebra shark, the same number of trimmed genomic shotgun reads (293,686,584) was prepared for both sexes. The reads were mapped with BWA-MEM (v2.2.1) onto a genome assembly (Li and Durbin 2009). Mapped reads were counted for male and female using the bamtobed program in the package BEDTools v2.29.2, for each scaffold in 10 kb nonoverlapping windows, and male–female ratios were calculated (Quinlan and Hall 2010). Sequencing depth of male and female reads on the identified Chromosome X was also calculated for 10 kb nonoverlapping windows. Windows with the proportion of ambiguous bases of more than 50% were excluded from computation. This procedure was also applied to the whale shark, using 220,785,436 trimmed reads.
The sequencing-based identification of the putative zebra shark Chromosome X was validated with multiple male and female individuals by real-time quantitative PCR (qPCR). The PCR was performed with the Luna Universal qPCR Master Mix (New England Biolabs) according to the manufacturer's instruction using the DNA template of 68.4 ng and oligonucleotide primers designed to avoid intronic regions (Supplemental Table 7). The reaction was performed in triplicate on a CFX96 Real-time PCR System (Bio-Rad) with preheating at 95°C for 1 min and two-step cycling (40 cycles) of denaturing at 95°C for 15 sec and annealing/extension at 60°C for 30 sec, followed by a postamplification step for dissociation curve analysis. Male-female difference was quantified with the 2−ΔΔCt method (Rao et al. 2013) as shown conventionally in validating sex difference (e.g., Sheffer et al. 2022).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Genome (https://www.ncbi.nlm.nih.gov/genome/) under accession numbers JAHMAH000000000 and JAFIRC000000000 and the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA703743.
Supplementary Material
Acknowledgments
We thank Tomoyuki Furuyashiki and Masayuki Taniguchi for their cooperation in obtaining Linked-Read sequence data, Chiharu Tanegashima and Kaori Tatsumi for their support in sequence data acquisition, Hatsune Makino-Itou for qPCR experiments, animal caretakers including Yano Nagisa at Okinawa Churaumi Aquarium, Keisuke Yonehara, and Akane Kawaguchi for their assistance, and Yuichiro Hara, Yukiko Imai, and Taiki Niwa for valuable discussion. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics. This study was funded by intramural budgets granted by RIKEN and the National Institute of Genetics, as well as JSPS KAKENHI Grants No. 20H03269 and No. 22K15088.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276840.122.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Bertho S, Herpin A, Schartl M, Guiguen Y. 2021. Lessons from an unusual vertebrate sex-determining gene. Philos Trans R Soc Lond B Biol Sci 376: 20200092. 10.1098/rstb.2020.0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, et al. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48: 427–437. 10.1038/ng.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brůna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M. 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform 3: lqaa108. 10.1093/nargab/lqaa108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt DW. 2002. Origin and evolution of avian microchromosomes. Cytogenet Genome Res 96: 97–112. 10.1159/000063018 [DOI] [PubMed] [Google Scholar]
- Cabanettes F, Klopp C. 2018. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6: e4958. 10.7717/peerj.4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Campos JL, Jackson BC. 2018. Faster-X evolution: Theory and evidence from Drosophila. Mol Ecol 27: 3753–3771. 10.1111/mec.14534 [DOI] [PubMed] [Google Scholar]
- Cosentino S, Iwasaki W. 2019. SonicParanoid: fast, accurate and easy orthology inference. Bioinformatics 35: 149–151. 10.1093/bioinformatics/bty631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Potter S, O'Neill R, Ruiz-Herrera A, Cioffi MB, Eldridge MDB, Fukui K, Marshall Graves JA, Griffin D, Grutzner F, et al. 2019. Chromosomics: bridging the gap between genomes and chromosomes. Genes (Basel) 10: 627. 10.3390/genes10080627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356: 92–95. 10.1126/science.aal3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. 2008. Evolution of the genomic rate of recombination in mammals. Evolution (N Y) 62: 276–294. 10.1111/j.1558-5646.2007.00278.x [DOI] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL. 2016a. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. 2016b. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3: 99–101. 10.1016/j.cels.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini P, Jones JC, Xiong P, Kneitz S, Gompert Z, Warren WC, Walter RB, Meyer A, Schartl M. 2018. Long-term experimental hybridisation results in the evolution of a new sex chromosome in swordtail fish. Nat Commun 9: 5136. 10.1038/s41467-018-07648-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N. 2004. Recombination, GC content and the human pseudoautosomal boundary paradox. Trends Genet 20: 347–349. 10.1016/j.tig.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. 2001. GC content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics 159: 907–911. 10.1093/genetics/159.2.907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. 2016. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet 17: 33–46. 10.1038/nrg.2015.2 [DOI] [PubMed] [Google Scholar]
- Hao Z, Lv D, Ge Y, Shi J, Weijers D, Yu G, Chen J. 2020. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput Sci 6: e251. 10.7717/peerj-cs.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Yamaguchi K, Onimaru K, Kadota M, Koyanagi M, Keeley SD, Tatsumi K, Tanaka K, Motone F, Kageyama Y, et al. 2018. Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat Ecol Evol 2: 1761–1771. 10.1038/s41559-018-0673-5 [DOI] [PubMed] [Google Scholar]
- Hardie DC, Hebert PDN. 2004. Genome-size evolution in fishes. Can J Fish Aquat Sci 61: 1636–1646. 10.1139/f04-106 [DOI] [Google Scholar]
- International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Kadota M, Nishimura O, Miura H, Tanaka K, Hiratani I, Kuraku S. 2020. Multifaceted Hi-C benchmarking: what makes a difference in chromosome-scale genome scaffolding? GigaScience 9: giz158. 10.1093/gigascience/giz158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota M, Tatsumi K, Yamaguchi K, Uno Y, Kuraku S. 2023. Shark and ray genome size estimation: methodological optimization for inclusive and controllable biodiversity genomics. bioRxiv 10.1101/2023.02.23.529029 [DOI]
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 24: 1384–1395. 10.1101/gr.170720.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BL, Gillis JA, Carlisle HR, Dahn RD. 2011. A natural deletion of the HoxC cluster in elasmobranch fishes. Science 334: 1517. 10.1126/science.1210912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief U, Forstmeier W. 2016. Mapping centromeres of microchromosomes in the zebra finch (Taeniopygia guttata) using half-tetrad analysis. Chromosoma 125: 757–768. 10.1007/s00412-015-0560-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koochekian N, Ascanio A, Farleigh K, Card DC, Schield DR, Castoe TA, Jezkova T. 2022. A chromosome-level genome assembly and annotation of the desert horned lizard, Phrynosoma platyrhinos, provides insight into chromosomal rearrangements among reptiles. GigaScience 11: giab098. 10.1093/gigascience/giab098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S. 2021. Shark and ray genomics for disentangling their morphological diversity and vertebrate evolution. Dev Biol 477: 262–272. 10.1016/j.ydbio.2021.06.001 [DOI] [PubMed] [Google Scholar]
- Kuraku S, Ishijima J, Nishida-Umehara C, Agata K, Kuratani S, Matsuda Y. 2006. cDNA-based gene mapping and GC3 profiling in the soft-shelled turtle suggest a chromosomal size-dependent GC bias shared by sauropsids. Chromosome Res 14: 187–202. 10.1007/s10577-006-1035-8 [DOI] [PubMed] [Google Scholar]
- Kuraku S, Zmasek CM, Nishimura O, Katoh K. 2013. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res 41: W22–W28. 10.1093/nar/gkt389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288. 10.1093/bioinformatics/btp368 [DOI] [PubMed] [Google Scholar]
- Li W-H. 2002. Male-driven evolution. Curr Opin Genet Dev 12: 650–656. 10.1016/S0959-437X(02)00354-4 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DJ. 1992. Sharks from the La Meseta formation (Eocene), Seymour Island, Antarctic Peninsula. J Vertebr Paleontol 12: 11–32. 10.1080/02724634.1992.10011428 [DOI] [Google Scholar]
- Ma W, Xu L, Hua H, Chen M, Guo M, He K, Zhao J, Li F. 2021. Chromosomal-level genomes of three rice planthoppers provide new insights into sex chromosome evolution. Mol Ecol Resour 21: 226–237. 10.1111/1755-0998.13242 [DOI] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H. 2007. Fast-X on the Z: Rapid evolution of sex-linked genes in birds. Genome Res 17: 618–624. 10.1101/gr.6031907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlétaz F, de la Calle-Mustienes E, Acemel RD, Paliou C, Naranjo S, Martínez-García PM, Cases I, Sleight VA, Hirschberger C, Marcet-Houben M, et al. 2023. The little skate genome and the evolutionary emergence of wing-like fins. Nature 616: 495–503. 10.1038/s41586-023-05868-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra NJ, Stanhope MJ, Jue NK, Wang M, Sun Q, Bitar PP, Richards VP, Komissarov A, Rayko M, Kliver S, et al. 2019. White shark genome reveals ancient elasmobranch adaptations associated with wound healing and the maintenance of genome stability. Proc Natl Acad Sci 116: 4446–4455. 10.1073/pnas.1819778116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Kuraku S, Tarui H, Nishimura O, Nishida C, Agata K, Kumazawa Y, Matsuda Y. 2012. Intra-genomic GC heterogeneity in sauropsids: evolutionary insights from cDNA mapping and GC3 profiling in snake. BMC Genomics 13: 604. 10.1186/1471-2164-13-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T. 2013. The faster-X effect: integrating theory and data. Trends Genet 29: 537–544. 10.1016/j.tig.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Schloissnig S, Franchini P, Du K, Woltering JM, Irisarri I, Wong WY, Nowoshilow S, Kneitz S, Kawaguchi A, et al. 2021. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 590: 284–289. 10.1038/s41586-021-03198-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. 1987. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol 52: 863–867. 10.1101/SQB.1987.052.01.094 [DOI] [PubMed] [Google Scholar]
- Mugal CF, Weber CC, Ellegren H. 2015. GC-biased gene conversion links the recombination landscape and demography to genomic base composition. Bioessays 37: 1317–1326. 10.1002/bies.201500058 [DOI] [PubMed] [Google Scholar]
- Mulley JF, Zhong Y-F, Holland PW. 2009. Comparative genomics of chondrichthyan Hoxa clusters. BMC Evol Biol 9: 218. 10.1186/1471-2148-9-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S. 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res 17: 1254–1265. 10.1101/gr.6316407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Shingate P, Ravi V, Pillai NE, Prasad A, McLysaght A, Venkatesh B. 2021. Reconstruction of proto-vertebrate, protocyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nat Commun 12: 4489. 10.1038/s41467-021-24573-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor G, Caira K, Jensen K, White W, Last P. 2012. A DNA sequence–based approach to the identification of shark and ray species and its implications for global elasmobranch diversity and parasitology. Bull Am Mus Nat Hist 367: 1–262. 10.1206/754.1 [DOI] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer Science & Business Media, Berlin. [Google Scholar]
- Ohno S, Muramoto J, Stenius C, Christian L, Kittrell WA, Atkin NB. 1969. Microchromosomes in holocephalian, chondrostean and holostean fishes. Chromosoma 26: 35–40. 10.1007/BF00319498 [DOI] [PubMed] [Google Scholar]
- Palmer DH, Rogers TF, Dean R, Wright AE. 2019. How to identify sex chromosomes and their turnover. Mol Ecol 28: 4709–4724. 10.1111/mec.15245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell MW, Mank JE, Peichel CL. 2018. Transitions in sex determination and sex chromosomes across vertebrate species. Mol Ecol 27: 3950–3963. 10.1111/mec.14540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, stringTie and ballgown. Nat Protoc 11: 1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Huang X, Zhou Z, Lin X. 2013. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 3: 71–85. [PMC free article] [PubMed] [Google Scholar]
- Read TD, Petit RA, Joseph SJ, Alam T, Weil MR, Ahmad M, Bhimani R, Vuong JS, Haase CP, Webb DH, et al. 2017. Draft sequencing and assembly of the genome of the world's largest fish, the whale shark: Rhincodon typus smith 1828. BMC Genomics 18: 755. 10.1186/s12864-017-4138-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592: 737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C, Louis A, Bon C, Berthelot C, Roest Crollius H. 2018. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol 19: 166. 10.1186/s13059-018-1559-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz FJ, Maddock MB. 1986. Comparisons of karyotypes and cellular DNA contents within and between major lines of elasmobranchs. In Indo-Pacific fish biology, pp. 148–157. Ichthyological Soc. of Japan, Tokyo. [Google Scholar]
- Schwartz FJ, Maddock MB. 2002. Cytogenetics of the elasmobranchs: genome evolution and phylogenetic implications. Mar Freshw Res 53: 491–502. 10.1071/MF01139 [DOI] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. In Gene prediction methods in molecular biology (ed. Kollmar M), pp. 227–245. Humana Press, New York. [DOI] [PubMed] [Google Scholar]
- Sheffer MM, Cordellier M, Forman M, Grewoldt M, Hoffmann K, Jensen C, Kotz M, Král J, Kuss AW, Líznarová E, et al. 2022. Identification of sex chromosomes using genomic and cytogenetic methods in a range-expanding spider, Argiope bruennichi (Araneae: Araneidae). Biological J Linn Soc 136: 405–416. 10.1093/biolinnean/blac039 [DOI] [Google Scholar]
- Simakov O, Marlétaz F, Yue JX, O'Connell B, Jenkins J, Brandt A, Calef R, Tung CH, Huang TK, Schmutz J, et al. 2020. Deeply conserved synteny resolves early events in vertebrate evolution. Nat Ecol Evol 4: 820–830. 10.1038/s41559-020-1156-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, Kawakami T, Burri R, Bolivar P, Husby A, Qvarnström A, Uebbing S, Ellegren H. 2014. Genomic identification and characterization of the pseudoautosomal region in highly differentiated avian sex chromosomes. Nat Commun 5: 5448. 10.1038/ncomms6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R. 2008. RepeatModeler Open-1.0. http://www.repeatmasker.org.
- Smit AFA, Hubley R, Green P. 2013. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Soderlund C, Bomhoff M, Nelson WM. 2011. SyMAP v3.4: A turnkey synteny system with application to plant genomes. Nucleic Acids Res 39: e68–e68. 10.1093/nar/gkr123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K, Ahmad SF, Singchat W, Panthum T. 2021. Why do some vertebrates have microchromosomes? Cells 10: 2182. 10.3390/cells10092182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhope MJ, Ceres KM, Sun Q, Wang M, Zehr JD, Marra NJ, Wilder AP, Zou C, Bernard AM, Pavinski-Bitar P, et al. 2023. Genomes of endangered great hammerhead and shortfin mako sharks reveal historic population declines and high levels of inbreeding in great hammerhead. iScience 26: 105815. 10.1016/j.isci.2022.105815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryamohan K, Krishnankutty SP, Guillory J, Jevit M, Schröder MS, Wu M, Kuriakose B, Mathew OK, Perumal RC, Koludarov I, et al. 2020. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat Genet 52: 106–117. 10.1038/s41588-019-0559-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Redmond AK, Dooley H, Nozu R, Sato K, Kuraku S, Koren S, Phillippy AM, Dove ADM, Read TD. 2021. The whale shark genome reveals patterns of vertebrate gene family evolution. eLife 10: e65394. 10.7554/eLife.65394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymowska J. 1991. Polyploidy and cytogenetic variation in frogs of the genus Xenopus. In Amphibian cytogenetics and evolution (ed. Green DM, Sessions SK), pp. 259–297. Academic Press, Cambridge. [Google Scholar]
- Uno Y, Nishida C, Tarui H, Ishishita S, Takagi C, Nishimura O, Ishijima J, Ota H, Kosaka A, Matsubara K, et al. 2012. Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS One 7: e53027. 10.1371/journal.pone.0053027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Nozu R, Kiyatake I, Higashiguchi N, Sodeyama S, Murakumo K, Sato K, Kuraku S. 2020. Cell culture-based karyotyping of orectolobiform sharks for chromosome-scale genome analysis. Commun Biol 3: 652. 10.1038/s42003-020-01373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, et al. 2014. Corrigendum: Elephant shark genome provides unique insights into gnathostome evolution. Nature 513: 574. 10.1038/nature13699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PD, Patel HR, Ruiz-Herrera A, Álvarez-González L, Lister NC, Simakov O, Ezaz T, Kaur P, Frere C, Grutzner F, et al. 2021. Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc Natl Acad Sci 118: e2112494118. 10.1073/pnas.2112494118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA, Park SG, Luria V, Jeon S, Kim HM, Jeon Y, Bhak Y, Jun JH, Kim SW, Hong WH, et al. 2020. The whale shark genome reveals how genomic and physiological properties scale with body size. Proc Natl Acad Sci 117: 20662–20671. 10.1073/pnas.1922576117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. 2017. Direct determination of diploid genome sequences. Genome Res 27: 757–767. 10.1101/gr.214874.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wa Sin SY, Grayson P, Edwards SV, Sackton TB. 2019. Evolutionary dynamics of sex chromosomes of paleognathous birds. Genome Biol Evol 11: 2376–2390. 10.1093/gbe/evz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Kadota M, Nishimura O, Ohishi Y, Naito Y, Kuraku S. 2021. Technical considerations in Hi-C scaffolding and evaluation of chromosome-scale genome assemblies. Mol Ecol 30: 5923–5934. 10.1111/mec.16146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao H, Li H, Guo J, Ouyang B, Wang M, Xu Q, Wang J, Lv M, Guo X, et al. 2020. The white-spotted bamboo shark genome reveals chromosome rearrangements and fast-evolving immune genes of cartilaginous fish. iScience 23: 101754. 10.1016/j.isci.2020.101754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B-H, Xiao J, Xue W, Xu G-C, Sun M-Y, Li J-T. 2018. P_RNA_scaffolder: A fast and accurate genome scaffolder using paired-end RNA-sequencing reads. BMC Genomics 19: 175. 10.1186/s12864-018-4567-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.