Abstract
Citrate metabolism in the lactic acid bacterium Leuconostoc mesenteroides generates an electrochemical proton gradient across the membrane by a secondary mechanism (C. Marty-Teysset, C. Posthuma, J. S. Lolkema, P. Schmitt, C. Divies, and W. N. Konings, J. Bacteriol. 178:2178–2185, 1996). Reports on the energetics of citrate metabolism in the related organism Lactococcus lactis are contradictory, and this study was performed to clarify this issue. Cloning of the membrane potential-generating citrate transporter (CitP) of Leuconostoc mesenteroides revealed an amino acid sequence that is almost identical to the known sequence of the CitP of Lactococcus lactis. The cloned gene was expressed in a Lactococcus lactis Cit− strain, and the gene product was functionally characterized in membrane vesicles. Uptake of citrate was counteracted by the membrane potential, and the transporter efficiently catalyzed heterologous citrate-lactate exchange. These properties are essential for generation of a membrane potential under physiological conditions and show that the Leuconostoc CitP retains its properties when it is embedded in the cytoplasmic membrane of Lactococcus lactis. Furthermore, using the same criteria and experimental approach, we demonstrated that the endogenous CitP of Lactococcus lactis has the same properties, showing that the few differences in the amino acid sequences of the CitPs of members of the two genera do not result in different catalytic mechanisms. The results strongly suggest that the energetics of citrate degradation in Lactococcus lactis and Leuconostoc mesenteroides are the same; i.e., citrate metabolism in Lactococcus lactis is a proton motive force-generating process.
Only a few strains of lactic acid bacteria are able to ferment citrate. The ability to metabolize citrate is invariably linked to endogenous plasmids that contain the gene encoding the transporter that is responsible for uptake of citrate from the medium. Citrate transporters (CitPs) have been found in strains belonging to the genera Lactococcus and Leuconostoc. In the dairy industry, these Lactococcus and Leuconostoc strains are used in mixed cultures, and metabolism of citrate is important in many fermentation processes because of the formation of carbon dioxide, diacetyl, acetoin, and butanediol, compounds that contribute to the organoleptic properties of the fermentation products (for a review see reference 11).
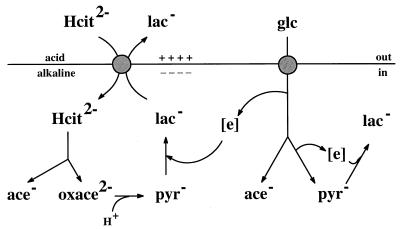
In recent studies we have described in detail the energetics of citrate metabolism in Leuconostoc mesenteroides (26, 27). Citrate fermentation results in the formation of an electrochemical proton gradient across the cell membrane (proton motive force) by a secondary mechanism (16, 21) in which the CitP plays a crucial role. The transporter catalyzes exchange of divalent anionic citrate and monovalent lactate, which results in the generation of a membrane potential with physiological polarity (i.e., the inside is negative) (Fig. 1). Lactate is a product of citrate fermentation during cometabolism with a carbohydrate (citrolactic fermentation). Upon entering the cell, citrate is cleaved by citrate lyase, which yields acetate and oxaloacetate. Decarboxylation of the latter compound yields carbon dioxide and pyruvate. Pyruvate functions as a sink for the reducing equivalents produced in the heterofermentative carbohydrate degradation pathway and is converted to the end product lactate (3), which leaves the cell in exchange for citrate (precursor-product exchange). The decarboxylation of oxaloacetate plays an additional role in metabolic energy generation since it consumes a cytoplasmic proton, which results in the generation of a transmembrane pH gradient with physiological polarity (i.e., the inside is alkaline). Together, the charge translocation catalyzed by the transporter and the proton consumption in the decarboxylation step are equivalent to the pumping of a proton out of the cell (20, 21).
FIG. 1.
Schematic diagram of the mechanism of proton motive force generation by citrolactic fermentation in Leuconostoc mesenteroides. Citrate metabolism and glucose metabolism are shown on the left and right, respectively. Electrogenic exchange of citrate and lactate by CitP results in the generation of a membrane potential. Scalar proton consumption in the decarboxylation of oxaloacetate results in the generation of a transmembrane pH gradient. During cometabolism, redox equivalents are shunted from glucose metabolism to citrate metabolism. Hcit, citrate; lac, lactate; ace, acetate; oxace, oxaloacetate; pyr, pyruvate; glc, glucose.
In citrate-fermenting Lactococcus species the initial steps of citrate breakdown proceed through the same intermediates as those observed in Leuconostoc species, and the homofermentative carbohydrate metabolism in lactococci is likely to produce enough lactate for the CitP to function as a citrate-lactate exchanger (35). Moreover, the genes coding for the citrate carriers of Lactococcus lactis (citPll) and Leuconostoc lactis (citPlcl) have been cloned and sequenced and have been found to be almost identical (4, 41). All of this strongly suggests that the energetics of the citrate metabolic pathway described above for Leuconostoc mesenteroides is also valid for lactococci. However, reports on the energy-coupling mechanism of the Lactococcus lactis transporter are contradictory. Uptake studies performed with membrane vesicles prepared from Escherichia coli expressing the transporter of Lactococcus lactis indicated that citrate transport was driven by the proton motive force. Consequently, it was concluded that uptake of citrate in Lactococcus lactis costs metabolic energy (4). In contrast, measurements of energetic parameters in whole cells of Lactococcus lactis indicated that metabolic energy was conserved during citrate metabolism, possibly via electrogenic exchange of divalent citrate with monovalent acetate or pyruvate (11). The results of yet another study, which involved citrate uptake measurements in whole cells of Lactococcus lactis, again seemed to indicate that proton motive force-driven uptake of citrate occurs (25).
Different energetics in the citrate degradation pathways of Lactococcus lactis and Leuconostoc mesenteroides would mean that the energy-coupling mechanisms of the CitPs must be different. The transporter of Leuconostoc mesenteroides has not been cloned and sequenced and could be a different protein. Alternatively, small differences in the amino acid sequences of the transporters could be responsible for different mechanisms, or the different lipid environments in the two types of lactic acid bacteria could change the energy-coupling mechanism, as was recently described for the glutamate transporter (GltT) of the thermophilic bacterium Bacillus stearothermophilus (38). The present study demonstrates that the energetics of the CitPs of Lactococcus and Leuconostoc species are the same. This conclusion strongly suggests that citrate metabolism in Lactococcus lactis results in the generation of a proton motive force.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Leuconostoc mesenteroides subsp. mesenteroides 19D was obtained from the collection of the Institute National de la Recherche Agronomique (Jouy en Josas, France) and was grown at 30°C in MRS medium supplemented with 0.5% glucose with or without 10 mM citrate (5). Lactococcus lactis subsp. lactis biovar diacetylactis NCDO176, which was obtained from the Dutch Institute of Dairy Research (Ede, The Netherlands), and Lactococcus lactis MG1363 (9) harboring plasmid pMBcitP were grown at 30°C in M17 medium supplemented with 0.5% lactose with or without 10 mM citrate. Plasmid pMBcitP is an E. coli-Lactococcus lactis shuttle vector that contains the citP gene of Leuconostoc mesenteroides under control of the citP promoter of Lactococcus lactis and has been described in more detail elsewhere (1). E. coli JM101 and JM109(DE3) (32) were routinely grown in Luria-Bertani medium at 37°C with vigorous shaking, and when appropriate, the medium was supplemented with carbenicilin or ampicillin at a concentration of 50 μg/ml. Citrate utilization by Leuconostoc mesenteroides was detected by using the medium of Kempler and McKay (14). Citrate utilization by E. coli was examined by using Simmons agar (33) and Christensen agar (2).
Genetic manipulations.
Unless otherwise indicated, all genetic engineering was performed by standard procedures (32). Endogenous plasmids of Leuconostoc mesenteroides were isolated on a small scale as described previously (30) and on a large scale by alkaline lysis, with the following modifications. Harvested cells were washed twice in TES buffer (30 mM Tris-HCl, 5 mM EDTA, 50 mM NaCl; pH 8.0) (18), and the CsCl-ethidium bromide gradient was replaced by ethanol precipitation (17). Lactococcus lactis MG1363 was transformed by electroporation (10), and the E. coli strains were transformed either by electroporation or by the standard CaCl2 treatment procedure. Southern blot hybridizations (34) were performed by using a VacuGene XL vacuum blotting system (Pharmacia) and a 0.45-μm-pore-size nylon membrane filter (Pharmacia). Labelling of DNA probes, hybridization, and detection were performed by using the DIG labelling and detection kit protocols (Boehringer, Mannheim, Germany). DNA hybridization and stringent washes in 150 mM NaCl–15 mM sodium citrate were carried out at 68°C.
Membrane preparations.
Cells of Lactococcus lactis MG1363 harboring plasmid pMBcitP and Lactococcus lactis NCDO176 were harvested at an optical density at 660 nm (OD660) of 1, washed once, resuspended in 50 mM potassium phosphate (pH 7.0) to an OD660 of 500, and then rapidly frozen and stored in liquid nitrogen until they were used. Right-side-out membrane vesicles were prepared by the osmotic shock lysis procedure essentially as described previously (31). Membrane vesicles were rapidly frozen and stored in liquid nitrogen until they were used. The protein concentration was determined by the method of Lowry et al. (23). The membranes were fused to proteoliposomes containing beef heart cytochrome c oxidase or to liposomes essentially as described previously (6). l-α-Phosphatidylethanolamine was purified from 1 g of E. coli extract (Avanti polar lipids) by successive washing with acetone and diethyl ether, after which the concentration was determined (7). Cytochrome c oxidase isolated from beef heart mitochondria was reconstituted into liposomes by detergent dialysis and then was reconstituted in a mixture containing the purified E. coli lipids and egg phosphatidylcholine at a ratio 3:1. Proteoliposomes containing cytochrome c oxidase were fused with the membrane vesicles of Lactococcus lactis at a ratio of 1 mg of protein per 10 mg of lipids by freezing the preparation in liquid nitrogen and then slowly thawing it at room temperature (6). The resulting hybrid membranes were made unilamelar by sonication (eight cycles; 15 s on and 45 s off; amplitude, 4 to 6 μm) or by extrusion by using successively 400- and 200-nm-pore-size polycarbonate filters (29). Right-side-out membrane vesicles of E. coli JM101 were prepared by the osmotic lysis procedure (13).
Transport assays. (i) Proton motive force-driven uptake in vesicles.
The experiments to determine proton motive force-driven uptake in vesicles were performed in 50 mM potassium phosphate (pH 6) under a flow of water-saturated air at 30°C. The membranes were incubated for 10 min in the presence or in the absence of valinomycin and nigericin at concentrations of 1 and 0.5 μM, respectively. Membrane vesicles of Lactococcus lactis fused to proteoliposomes containing cytochrome c oxidase were energized by adding 200 μM N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD), 20 μM cytochrome c (from horse heart; Sigma), and 10 mM potassium ascorbate. Membrane vesicles of E. coli were energized by adding 200 μM phenazine methosulfate (PMS) and 10 mM potassium ascorbate. After incubation for 1 min, [1,5-14C]citrate was added at a concentration of 4.5 μM. Samples (100 μl) were removed at different times, placed in 2 ml of ice-cold 0.1 mM LiCl to stop the reaction, and filtered through 0.45-μm-pore-size cellulose nitrate filters (Schleicher & Schuell GmbH). The filters were rinsed with 2 ml of ice-cold 0.1 M LiCl and transferred to scintillation vials.
(ii) Exchange measurements.
Hybrid membranes obtained by fusion of membrane vesicles of Lactococcus lactis with liposomes lacking cytochrome c oxidase were concentrated by centrifugation (250,000 × g, 45 min, 4°C) and incubated with 5 mM [1,5-14C]citrate in 50 mM potassium phosphate (pH 6) for 30 min at 20°C. After incubation with valinomycin and nigericin, the hybrid membranes were diluted 100-fold with the same buffer with or without 5 mM unlabelled substrates. The samples were processed as described above.
Nucleotide sequence accession number.
The nucleotide sequence of the insert in pNA1008 determined in this study has been deposited in the GenBank database under accession no. L29572.
RESULTS
Cloning and sequencing of the citP gene of Leuconostoc mesenteroides 19D.
Cells of Leuconostoc mesenteroides 19D Cit+ produce dark blue colonies on Kempler-McKay indicator plates. The Cit+ phenotype is easily and spontaneously lost when citrate is omitted from the growth medium, as shown by the appearance of white colonies on the same plates. Prolonged growth of cells with the Cit− phenotype in medium containing citrate does not result in revertants, which is consistent with the notion that the Cit+ phenotype is plasmid encoded (18). Total plasmid DNAs from Leuconostoc mesenteroides Cit+ and Cit− cultures were isolated, and a size analysis revealed that a 22-kb plasmid was not present in the Cit− derivatives. The citP gene of Lactococcus lactis NCDO176 hybridized strongly and exclusively with the 22-kb plasmid from the Leuconostoc mesenteroides Cit+ cells and not with any of the plasmid DNAs of the Cit− derivative. No hybridization was observed with chromosomal DNA isolated from either the parental strain or the Cit− strain (data not shown). These results strongly suggest that the citP gene of Leuconostoc mesenteroides 19D is located exclusively on the 22-kb plasmid.
Total plasmid DNA from Leuconostoc mesenteroides 19D was partially restricted with Sau3A. Fragments ranging in size from 4 to 9 kb were cloned into the compatible BamHI site of plasmid pUC19. About 400 recombinant plasmids were screened by hybridization by using the lactococcal citP gene as a probe. Strong hybridization was obtained with plasmid pNA1006, which contained a 5.5-kb insert. Further subcloning was achieved by deleting a HindIII fragment; this resulted in plasmid pNA1008, which contained a 3.6-kb insert. Plasmid pNA1008 was transformed into E. coli JM101, and the transformant was grown on citrate indicator plates. E. coli cannot metabolize citrate because it lacks a transporter for citrate. The presence of the complete citP gene of Leuconostoc mesenteroides on plasmid pNA1008 was demonstrated by the clear Cit+ phenotype of the transformed E. coli cells on Christensen medium, which contained glucose as an additional energy and carbon source. Growth of the cells on minimal citrate medium (Simmons citrate agar) was poor.
A restriction map of pNA1008 was constructed (data not shown), and fragments were cloned into the phagemids pBluescript KS(+) and pBluescript KS(−). Single-stranded DNA was isolated from both types of plasmids by infection with a helper phage, which allowed us to determine the nucleotide sequence of the inserts in both directions. Reconstruction of the sequence of the insert in pNA1008 revealed a 1,329-bp open reading frame. Two ATG initiation codons were preceded by a putative ribosomal binding site sequence (GGAGA). No terminator of transcription was detected behind the stop codon.
Sequence analysis.
The Leuconostoc mesenteroides 19D citP gene (citPlcm) is almost identical to the citP genes of Lactococcus lactis NCDO176 (citPll) and Leuconostoc lactis NZ6070 (citPlcl). The Leuconostoc mesenteroides gene is one and two codons longer than the Lactococcus lactis and Leuconostoc lactis, genes, respectively. In the translated amino acid sequences of the proteins the differences in length cluster in the region around position 260, and in the Leuconostoc mesenteroides sequence one and two alanine residues are inserted compared to the other two sequences (Fig. 2). Just preceding this position differences in the base sequences between the two Leuconostoc genes and the Lactococcus gene result in the mutations Asp-256-Lys and Ala-257-Ser. This region in the sequences is hydrophilic, and the structure of the region was predicted to be an interhelical loop; interhelical loops are often found to be quite variable in homologous membrane proteins (19, 40). Finally, conservative substitutions in the Leuconostoc and Lactococcus genes occur at positions 33 and 103, where Val is substituted for Ile and Met is substituted for Ile, respectively. The CitP proteins are homologous to the membrane potential-generating malate transporter of Lactococcus lactis involved in malolactic fermentation and the Na+-dependent citrate transporter CitS of Klebsiella pneumoniae. Together, these transporters form the family of 2-hydroxycarboxylate transporters (1).
FIG. 2.
Amino acid sequences of CitPs of lactic acid bacteria. Differences between the sequences are indicated by x. The CitP sequences of Leuconostoc mesenteroides 19D (CitPlcm), Leuconostoc lactis (CitPlcl), and Lactococcus lactis (CitPll) are shown.
Characterization of CitPlcm of Leuconostoc mesenteroides expressed in Lactococcus lactis.
Transport catalyzed by the cloned CitPlcm transporter expressed in Lactococcus lactis was characterized with respect to energy coupling and heterologous exchange. The citPlcm gene of Leuconostoc mesenteroides was cloned in expression vector pMB, yielding pMBcitP, in which expression is under control of the citP promoter of Lactococcus lactis (1, 22). The plasmid was transformed into Lactococcus lactis MG1363, a plasmid-free strain that is not able to ferment citrate.
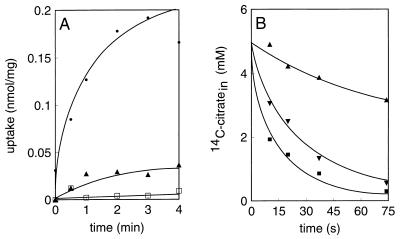
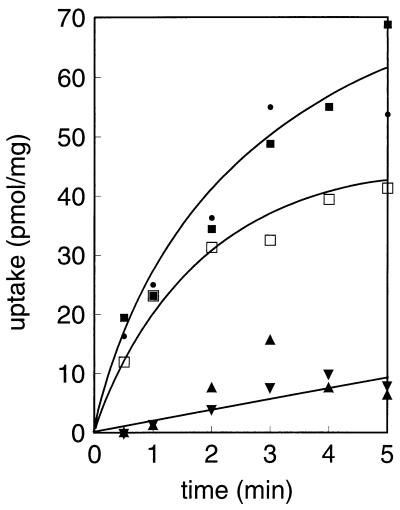
Cytoplasmic membranes with a right-side-out orientation were prepared from Lactococcus lactis harboring plasmid pMBcitP, and the membranes were fused to proteoliposomes reconstituted with purified beef heart cytochrome c oxidase. This enzyme catalyzes electron transfer from reduced cytochrome c to molecular oxygen while it conserves the free energy of the reaction in the form of an electrochemical proton gradient across the membrane by pumping out protons. Generation of an electrochemical proton gradient or proton motive force across the membranes resulted in a low level of citrate uptake; however, the level of citrate uptake was significantly higher than the level of uptake observed in the absence of the electron donor (Fig. 3A). No uptake was observed with membranes of Lactococcus lactis MG1363 without plasmid pMBcitP. The proton motive force is composed of a pH gradient and the electrical membrane potential, which can be selectively dissipated by the ionophores nigericin (a K+-H+ antiporter) and valinomycin (a K+ pore), respectively. Dissipation of the pH gradient completely prevented citrate uptake, while dissipation of the membrane potential resulted in marked stimulation of citrate uptake. In conclusion, uptake of citrate in membrane vesicles is driven by the pH gradient and is counteracted by the membrane potential.
FIG. 3.
Citrate transport catalyzed by CitP of Leuconostoc mesenteroides expressed in Lactococcus lactis. (A) Membrane vesicles isolated from Lactococcus lactis MG1363 expressing CitPlcm were fused to liposomes containing cytochrome c oxidase. Uptake of 4.5 μM [1,5-14C]citrate was measured in the presence of no ionophores (▴), valinomycin (•), and nigericin (□). The fused membranes were energized with the cytochrome c-TMPD-ascorbate electron donor system. (B) The same membrane vesicles were fused to liposomes, loaded with 5 mM [1,5-14C]citrate, and subsequently diluted 100-fold with buffer containing no further additions (▴), 5 mM citrate (▪), or 5 mM d-lactate (▾). In both experiments valinomycin and nigericin were present at concentrations of 1 and 0.5 μM, respectively. Uptake is expressed as nanomoles per milligram of membrane protein.
Membrane vesicles prepared from Lactococcus lactis MG1363 expressing CitPlcm were fused to liposomes to improve the tightness of the membranes and were loaded with 5 mM [14C]citrate. When the membranes were diluted with buffer without citrate at room temperature, a slow efflux of labelled citrate occurred in the first 75 s (Fig. 3B). On the other hand, dilution with a buffer containing unlabelled citrate resulted in a rapid release of label from the membranes by homologous citrate-citrate exchange. Similarly, dilution with buffer containing an equivalent concentration of d-lactate demonstrated that the transporter catalyzes heterologous citrate–d-lactate exchange. The results show that the catalytic properties of cloned CitPlcm of Leuconostoc mesenteroides expressed in Lactococcus lactis are similar to the properties of the transporter in its native environment, the Leuconostoc mesenteroides membrane (26).
Characterization of citrate carrier CitPll of Lactococcus lactis NCDO176.
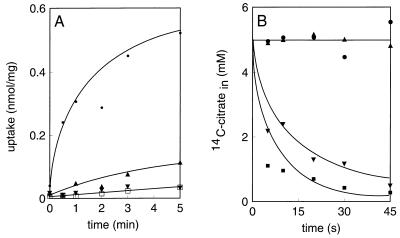
CitPll of Lactococcus lactis NCDO176 was characterized by using the same experimental approach as that described above for the transporter of Leuconostoc mesenteroides. Thus, right-side-out membranes of Lactococcus lactis NCDO176 were fused to proteoliposomes containing cytochrome c oxidase and to liposomes and were assayed for proton motive force-driven uptake and heterologous exchange, respectively. The results are presented in Fig. 4 and are virtually the same as the results observed with CitPlcm of Leuconostoc mesenteroides. The uptake activity was clearly stimulated by dissipation of the membrane potential, and no uptake was observed when the pH gradient was dissipated (Fig. 4A). The transporter catalyzed homologous exchange at a high rate when no efflux was observed and d-lactate was a substrate of the carrier in heterologous exchange (Fig. 4B). Acetate is an end product of citrate metabolism in lactic acid bacteria and has been implicated in the catalysis of CitPll of Lactococcus lactis (12). However, the data presented in Fig. 4B clearly shows that acetate is not a substrate of the transporter. In conclusion, the citrate carriers of Lactococcus lactis and Leuconostoc mesenteroides are mechanistically similar on the basis of these criteria.
FIG. 4.
Citrate transport in membrane vesicles derived from Lactococcus lactis NCDO176. The conditions were the same as the conditions described in the legend to Fig. 3. (A) Uptake of 4.5 μM [1,5-14C]citrate was measured in the presence of no ionophores (▴), valinomycin (•), and nigericin (□) or in the absence of the cytochrome c-TMPD-ascorbate electron donor system (▾). (B) Exchange. Symbols: ▴, no further additions, ▪, 5 mM citrate; ▾, 5 mM d-lactate; •, 5 mM acetate.
Expression and characterization of CitPlcm in E. coli.
Apparently, the data obtained with CitPll of Lactococcus lactis NCDO176 are at variance with the data in a previous report which described the characteristics of the cloned CitPll expressed in E. coli. An experiment similar to the experiment shown in Fig. 4A did not reveal stimulation of uptake when the membrane potential was dissipated by adding valinomycin (4). We repeated these experiments with Leuconostoc mesenteroides CitPlcm to complete the series of experiments performed with the lactococcal and Leuconostoc CitPs.
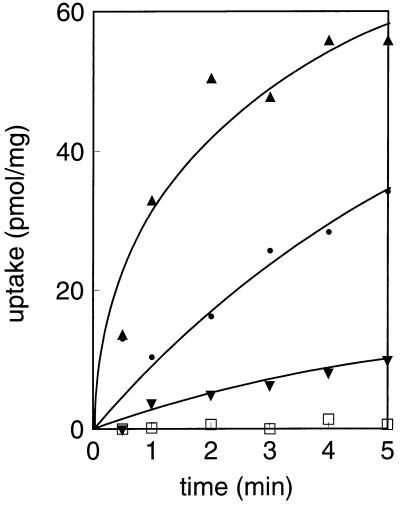
From the initial stages of the cloning of the gene coding for CitPlcm it was clear that functional expression of the protein in E. coli was poor. In fact, the clone was detected by hybridization with Lactococcus lactis citPll DNA rather than by functional selection of plasmids containing the Leuconostoc mesenteroides DNA fragments on citrate indicator plates. We constructed a series of plasmids containing the citPlcm gene under control of different promoters and transformed them into different strains of E. coli that were grown in minimal and rich media with and without induction, but the functional expression of CitPlcm was invariably low. Attempts to increase the expression of CitP in the membrane resulted in inhibition of growth of the cells, a phenomenon often observed when a membrane protein is overexpressed. In the experiment shown in Fig. 5 the citP gene was cloned behind the T7 promoter and transformed into E. coli JM109(DE3) that contains a chromosomal copy of the T7 polymerase under control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (36). In the absence of inducer, a small amount of the polymerase was synthesized due to leakage of the promoter, and a low but significant level of uptake of citrate into the cells was observed. Addition of increasing concentrations of the inducer IPTG resulted, at first, in an increase in the expression of CitP, but unfortunately, at higher IPTG concentrations the uptake decreased dramatically. The decreased uptake activity was paralleled by a decreased growth rate of the cells, showing the toxicity of the CitP protein in E. coli.
FIG. 5.
Functional expression of CitPlcm in E. coli. Cells of E. coli JM109(DE3) harboring plasmid pSKcitP#3 (▾, ▴, and •), which encodes CitPlcm under control of the T7 polymerase, or control plasmid pBlueScript SK with no insert (□) were grown in the presence of no IPTG (•), 20 μM IPTG (▴), and 50 μM IPTG (▾). No IPTG was added to the control cells. The cells were resuspended to an OD660 of 5, and [1,5-14C]citrate was added to a concentration of 4.5 μM. Uptake is expressed as picomoles per milligram of cell protein.
The low uptake activity observed in whole cells might be a consequence of the mechanism by which CitPlcm catalyzes citrate transport. Resting cells of E. coli maintain a high membrane potential that is expected to counteract transport. Valinomycin cannot be used to selectively dissipate the membrane potential since added ionophore is scavenged by the outer membrane. Therefore, cytoplasmic membrane vesicles were prepared from cells expressing CitPlcm, which allow manipulation of the composition of the proton motive force. Nevertheless, Fig. 6 shows that the uptake activity was at least 2 orders of magnitude lower than the uptake activity observed with vesicles prepared from cells expressing, for instance, the Na+-dependent CitS of K. pneumoniae, a transporter that is homologous to the CitPs of lactic acid bacteria (39). Surprisingly, but consistent with the results obtained previously with Lactococcus lactis CitPll expressed in E. coli, no stimulation of uptake was observed when the membrane potential was selectively dissipated with valinomycin. At higher concentrations, valinomycin was slightly inhibitory. Dissipation of the pH gradient completely prevented any uptake of citrate above background levels. Together, the results of these experiments suggest that citrate uptake by CitPlcm expressed in E. coli is catalyzed by an electroneutral proton symport mechanism.
FIG. 6.
Energy coupling to citrate transport catalyzed by CitPlcm of Leuconostoc mesenteroides expressed in E. coli. Right-side-out membrane vesicles were prepared from E. coli JM101 cells harboring plasmid pSKcitP#56, which encodes CitPlcm under control of the Lac promoter. The membranes were resuspended at a concentration of 0.8 mg of protein per ml and were not energized (▴) or energized (•, ▾, ▪, and □) by adding 10 mM potassium ascorbate and 0.2 mM PMS. Uptake of [1,5-14C]citrate at a concentration of 4.5 μM was measured with no further additions (•) and in the presence of 0.5 μM nigericin (▾), 0.125 μM valinomycin (▪), and 1 μM valinomycin (□). Uptake is expressed as picomoles per milligram of membrane protein.
DISCUSSION
In lactic acid bacteria the ability to ferment citrate is associated with endogenous plasmids that contain the gene that codes for the system responsible for uptake of citrate into the cells. Genes coding for CitPs have been described for strains of Lactococcus lactis and Leuconostoc lactis (4, 41). The cloned lactococcal CitP is encoded on a 7.9-kb plasmid which appears to be present in all citrate-fermenting Lactococcus strains analyzed so far (15). In Leuconostoc strains the plasmids that are indispensable for citrate metabolism are larger and more variable in size in the different strains (41). Consistent with this observation, in this study we found that a citP gene is present on a 22.4-kb endogenous plasmid of Leuconostoc mesenteroides 19D. A strong signal was obtained when plasmid DNA isolated from Leuconostoc mesenteroides was hybridized with a probe derived from the Lactococcus lactis citP gene, and subsequent cloning and sequencing revealed an open reading frame whose sequence was almost identical to the other two known citP nucleotide sequences. Clearly, although the transporters are almost identical, the genetic context of the citP genes is different in Lactococcus and Leuconostoc strains, and it has been shown that the mechanisms that control expression of the genes are different (22, 24, 28).
Citrate metabolism in Leuconostoc mesenteroides results in the generation of a proton motive force by a secondary mechanism in which the CitP is responsible for generation of the membrane potential (26, 27). Evidence that the citP gene of Leuconostoc mesenteroides described in this report codes for the membrane potential-generating transporter comes from the following observations: (i) loss of the Cit+ phenotype correlates with loss of the 22-kb plasmid; (ii) loss of the 22-kb plasmid correlates with loss of citrate uptake in membrane vesicles (26); and (iii) the characteristics of citrate transport in membrane vesicles derived from Lactococcus lactis MG1363 expressing citPlcm are similar to the characteristics observed with membrane vesicles derived from Leuconostoc mesenteroides (Fig. 3).
The physiological mode of transport of CitP of Leuconostoc mesenteroides is precursor-product exchange. Divalent citrate is exchanged for monovalent lactate which is the product of citrate metabolism during glucose-citrate cometabolism (27). In membrane vesicles in which the citrate metabolic enzymes are absent and no lactate is formed, CitP functions as a proton symporter that translocates divalent citrate with a single proton (26). Both modes of transport are counteracted by the membrane potential, and together they are diagnostic for secondary transporters that function in a secondary proton motive force-generating pathway. The unidirectional symport and the exchange reaction show that the transporters function as membrane potential generators and precursor-product exchangers, respectively. When these criteria were used, the citP gene product described in this paper could be identified as the CitP that is operative in citrate metabolism in Leuconostoc mesenteroides (Fig. 3). Moreover, by using the same criteria, it was shown that the transporter of Lactococcus lactis, CitPll, catalyzes citrate transport via the same mechanism (Fig. 4). The results show that neither the few differences in the amino acid sequences of CitPlcm and CitPll nor the differences in the cytoplasmic membrane compositions of Leuconostoc mesenteroides and Lactococcus lactis affect the mechanism of the two transporters.
The results of previous experiments performed with the cloned lactococcal CitPll expressed in E. coli suggested that there is a proton motive force-driven transport mechanism, which was considered evidence that energy-dependent citrate uptake by CitPll occurs in Lactococcus lactis (4). The initial rate of citrate uptake in membrane vesicles derived from E. coli expressing CitPll was reduced rather than stimulated when the membrane potential was dissipated, indicating that the membrane potential does not counteract transport. In this study we confirmed these observations by using cloned CitPlcm of Leuconostoc mesenteroides. Under the conditions of the experiments performed in buffer at pH 6, the ascorbate-PMS electron donor system has been shown to generate a significant membrane potential in E. coli membrane vesicles (37). Nevertheless, dissipation of the membrane potential did not result in stimulation of the uptake of citrate (Fig. 6). Apparently, the results obtained with cloned CitPll and CitPlcl expressed in E. coli are at variance with the results obtained with the same transporters in their native environment (Fig. 3 and 4). A similar observation has been made with GltT of the thermophilic bacterium B. stearothermophilus. Studies performed with membrane vesicles derived from B. stearothermophilus showed that GltT translocates glutamate in symport with two cations, a proton, and an Na+ ion. Surprisingly, expression of GltT in E. coli resulted in a complete loss of the dependence on Na+ (8, 38). These observations were explained by suggesting that the lipid environment plays a role in the cation specificity of the transporter.
In a secondary metabolic energy-generating pathway, the two components of the proton motive force are generated in separate steps; the membrane potential is generated in the transport step (electrogenic citrate-lactate exchange), and the pH gradient is a consequence of the consumption of a cytoplasmic proton in the decarboxylation step (oxaloacetate decarboxylation). Nevertheless, it can be demonstrated that the two events are coupled indirectly (20). Thus, membrane potential generation by citrate uptake via CitPll of Lactococcus lactis necessarily results in generation of a pH gradient across the cytoplasmic membrane when a proton is consumed in the oxaloacetate decarboxylation step. Therefore, the identical mechanisms of the CitPs of Lactococcus lactis and Leuconostoc mesenteroides demonstrated in this study strongly suggest that citrate metabolism in Lactococcus lactis is a secondary metabolic energy-generating process.
REFERENCES
- 1.Bandell M, Ansanay V, Rachidi N, Dequin S, Lolkema J S. Membrane potential generating malate (MleP) and citrate (CitP) transporters of lactic acid bacteria are homologous proteins. Substrate specificity of the 2-hydroxy-carboxylate transporter family. J Biol Chem. 1997;272:18140–18146. doi: 10.1074/jbc.272.29.18140. [DOI] [PubMed] [Google Scholar]
- 2.Christensen W B. Hydrogen sulfide production and citrate utilization in the differentiation of enteric pathogens and coliform bacteria. Res Bull Weld County Health Dep Greeley Colo. 1949;1:3–16. [Google Scholar]
- 3.Cogan T M. Co-metabolism of citrate and glucose by Leuconostoc spp.: effects on growth, substrates and products. J Appl Bacteriol. 1987;63:551–558. [Google Scholar]
- 4.David S, Van Der Rest M E, Driessen A J M, Simons G, De Vos W M. Nucleotide sequence and expression in Escherichia coli of the Lactococcus lactis citrate permease gene. J Bacteriol. 1990;172:5789–5794. doi: 10.1128/jb.172.10.5789-5794.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Man J C, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 6.Driessen A J M, de Vrij W, Konings W N. Incorporation of beef heart cytochrome c oxidase as a proton motive force generating mechanism in bacterial membrane vesicles. Proc Natl Acad Sci USA. 1985;82:7555–7559. doi: 10.1073/pnas.82.22.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driessen A J M, Brundage L, Hendrick J P, Schiebel E, Wickner W. Preprotein translocase of Escherichia coli: solubilization, purification and reconstitution of the integral membrane subunits SecY/E. Methods Cell Biol. 1991;34:147–165. doi: 10.1016/s0091-679x(08)61679-9. [DOI] [PubMed] [Google Scholar]
- 8.Gaillard I, Slotboom D-J, Knol J, Lolkema J S, Konings W N. Purification and reconstitution of the glutamate carrier GltT of the thermophilic bacterium Bacillus stearothermophilus. Biochemistry. 1995;35:6150–6156. doi: 10.1021/bi953005v. [DOI] [PubMed] [Google Scholar]
- 9.Gasson M. Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holo H, Ness I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugenholtz J. Citrate metabolism in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:165–178. [Google Scholar]
- 12.Hugenholtz J, Perdon L, Abee T. Growth and energy generation by Lactococcus lactis subsp. lactis biovar diacetylactis during citrate metabolism. Appl Environ Microbiol. 1993;59:4216–4222. doi: 10.1128/aem.59.12.4216-4222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaback H R. Bacterial membranes. Methods Enzymol. 1971;22:99–120. [Google Scholar]
- 14.Kempler G M, McKay L L. Improved medium for detection of Streptococcus lactis subsp. diacetylactis. Appl Environ Microbiol. 1980;39:926–927. doi: 10.1128/aem.39.4.926-927.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kempler G M, McKay L L. Biochemistry and genetics of citrate utilization in Streptococcus lactis subsp. diacetylactis. J Dairy Sci. 1981;64:1527–1531. [Google Scholar]
- 16.Konings W N, Lolkema J S, Poolman B. The generation of metabolic energy by solute transport. Arch Microbiol. 1995;164:235–242. [Google Scholar]
- 17.Lamoureux M, Prevost H, Cavin J F, Diviès C. Recognition of Leuconostoc oenos strains by the use of DNA restriction profiles. Appl Microbiol Biotechnol. 1993;39:547–552. doi: 10.1007/BF00205049. [DOI] [PubMed] [Google Scholar]
- 18.Lin J, Schmitt P, Divies C. Characterization of a citrate-negative mutant of Leuconostoc mesenteroides subsp. mesenteroides: metabolic and plasmidic properties. Appl Microbiol Biotechnol. 1991;34:628–631. doi: 10.1007/BF00172737. [DOI] [PubMed] [Google Scholar]
- 19.Lolkema J S, Speelmans G, Konings W N. Na+-coupled versus H+-coupled energy transduction in bacteria. Biochim Biophys Acta. 1994;1187:211–215. doi: 10.1016/0005-2728(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 20.Lolkema J S, Poolman B, Konings W N. Role of scalar protons in metabolic energy generation in lactic acid bacteria. J Bioenerg Biomembr. 1995;27:467–473. doi: 10.1007/BF02110009. [DOI] [PubMed] [Google Scholar]
- 21.Lolkema J S, Poolman B, Konings W N. Secondary transporters and metabolic energy generation in bacteria. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 229–260. [Google Scholar]
- 22.López de Felipe F, Magni C, de Mendoza D, López P. Citrate utilization gene cluster of the Lactococcus lactis biovar diacetylactis: organization and regulation of expression. Mol Gen Genet. 1995;246:590–599. doi: 10.1007/BF00298965. [DOI] [PubMed] [Google Scholar]
- 23.Lowry O H, Rosebrough N J, Farr A J, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Magni C, López de Felipe F, Sesma F, López P, de Mendoza D. Citrate transport in Lactococcus lactis subsp. lactis biovar diacetylactis. Expression of the citrate permease P. FEMS Microbiol Lett. 1994;118:75–82. [Google Scholar]
- 25.Magni C, López P, de Mendoza D. The properties of citrate transport catalyzed by CitP of Lactococcus lactis subsp. lactis biovar diacetylactis. FEMS Microbiol Lett. 1996;142:265–269. doi: 10.1111/j.1574-6968.1996.tb08063.x. [DOI] [PubMed] [Google Scholar]
- 26.Marty-Teysset C, Lolkema J S, Schmitt P, Divies C, Konings W N. Membrane potential generating transport of citrate and malate catalysed by CitP of Leuconostoc mesenteroides. J Biol Chem. 1995;270:25370–25376. doi: 10.1074/jbc.270.43.25370. [DOI] [PubMed] [Google Scholar]
- 27.Marty-Teysset C, Posthuma C, Lolkema J S, Schmitt P, Divies C, Konings W N. Proton motive force generation by citrolactic fermentation in Leuconostoc mesenteroides. J Bacteriol. 1996;178:2178–2185. doi: 10.1128/jb.178.8.2178-2185.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marty-Teysset C, Lolkema J S, Schmitt P, Divies C, Konings W N. The citrate metabolic pathway in Leuconostoc mesenteroides. Expression, amino acid synthesis, and α-ketocarboxylate transport. J Bacteriol. 1996;178:6209–6215. doi: 10.1128/jb.178.21.6209-6215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer D L, Hope M J, Cullis P R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim Biophys Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 30.O’Sullivan D J, Klaenhammer T R. Rapid mini-preparation of high-quality DNA from Lactococcus and Lactococcus spp. Appl Environ Microbiol. 1993;59:2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otto R, Lageveen R G, Veldkamp H, Konings W N. Lactate efflux-induced electrical potential in membrane vesicles of Streptococcus cremoris. J Bacteriol. 1982;149:733–738. doi: 10.1128/jb.149.2.733-738.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Simmons J S. A culture medium for differentiating organisms of typhoidcodon aerogenes and for isolating of certain fungi. J Infect Dis. 1926;39:209–214. [Google Scholar]
- 34.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 35.Starrenburg M, Hugenholtz J. Citrate fermentation by Lactococcus and Leuconostoc spp. Appl Environ Microbiol. 1991;57:3535–3540. doi: 10.1128/aem.57.12.3535-3540.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 37.Ten Brink B, Konings W N. Generation of an electrochemical proton gradient by lactate efflux in Escherichia coli membrane vesicles. Eur J Biochem. 1980;111:59–66. doi: 10.1111/j.1432-1033.1980.tb06074.x. [DOI] [PubMed] [Google Scholar]
- 38.Tolner B, Ubbink-Kok T, Poolman B, Konings W N. Cation-selectivity of the l-glutamate transporters of Escherichia coli, Bacillus stearothermophilus and Bacillus caldotenax. Mol Microbiol. 1995;18:123–133. doi: 10.1111/j.1365-2958.1995.mmi_18010123.x. [DOI] [PubMed] [Google Scholar]
- 39.Van der Rest M E, Siewe R M, Abee T, Schartz E, Oesterhelt D. Nucleotide sequence and functional properties of a sodium-dependent citrate transport system from Klebsiella pneumoniae. J Biol Chem. 1992;267:8971–8976. [PubMed] [Google Scholar]
- 40.Van Geest M, Lolkema J S. Membrane topology of the Na+-dependent citrate transporter of Klebsiella pneumoniae. Evidence for a new structural class of secondary transporters. J Biol Chem. 1996;271:25582–25589. doi: 10.1074/jbc.271.41.25582. [DOI] [PubMed] [Google Scholar]
- 41.Vaughan E E, David S, Harrington A, Daly C, Fitzgerald G F, De Vos W M. Characterization of plasmid-encoded citrate permease (citP) genes from Leuconostoc species reveals high sequence homology conservation with the Lactococcus lactis citP gene. Appl Environ Microbiol. 1996;61:3172–3176. doi: 10.1128/aem.61.8.3172-3176.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]