Abstract
PIK3CA pathogenic variants are responsible for a group of overgrowth syndromes, collectively known as PIK3CA-Related Overgrowth Spectrum (PROS). These gain-of-function variants arise postzygotically, and, according to time of onset, kind of embryonal tissue affected and regional body extension, give rise to heterogeneous phenotypes. PROS rarity and heterogeneity hamper the correct estimation of its epidemiology. Our work represents the first attempt to define the prevalence of PROS according to the established diagnostic criteria and molecular analysis and based on solid demographic data. We assessed the prevalence in Piedmont Region (Italy), including in the study all participants diagnosed with PROS born there from 1998 to 2021. The search identified 37 cases of PROS born across the 25-year period, providing a prevalence of 1:22,313 live births. Molecular analysis was positive in 81.0% of participants. Taking into account the cases with a detected variant in PIK3CA (n = 30), prevalence of molecularly positive PROS was 1:27,519.
Subject terms: Disease genetics, Genetic counselling
Introduction
PIK3CA-Related Overgrowth Spectrum (PROS) defines a heterogeneous group of rare congenital diseases, characterized by segmental/lateralized overgrowth of diverse body tissues and regions associated with malformations, having as a common genetic cause mosaic pathogenic gain-of-function variants in PIK3CA [1]. PROS groups a number of clinical entities with phenotypic overlap variously termed in the past depending on their clinical picture [1] including Fibroadipose Overgrowth (FAO) [2], Hemihyperplasia Multiple Lipomatosis (HHML), Klippel-Trenaunay syndrome (KTS) [3], Congenital Lipomatous Overgrowth, Vascular Malformations, Epidermal Nevi, Scoliosis/Skeletal and Spinal malformation syndrome (CLOVES), Type I macrodactyly [4], Muscle or Fibrous Hyperplasia (MH, FH) [2], Diffuse Capillary Malformation with Overgrowth (DCMO) [5], Facial Infiltrating Lipomatosis (FIL), Megalencephaly-Capillary Malformation (MCAP) [6], Dysplastic Megalencephaly (DMEG), Hemimegalencephaly (HMEG) [2], Focal Cortical Dysplasia (FCD) [1], Capillary malformation of the lower lip, Lymphatic malformation of the face and the neck, Asymmetry of the face and limbs and Partial or generalized Overgrowth (CLAPO) syndrome [2]. Some of such conditions are difficult to be classified in a specific syndrome, and present across the continuum of a clinical spectrum. Besides these phenotypes, a number of overgrowth/vascular-lymphatic malformation and conditions are increasingly recognized to be caused by PIK3CA pathogenic variants [7, 8]. To overcome their phenotypic heterogeneity, common diagnostic criteria were proposed in 2013, by Keppler-Noreuil et al. and currently employed in clinical practice [1].
The heterogeneity of these conditions reflects both the kind of tissue affected and the body extension of the overgrown area. These factors are connected with the timing of onset during fetal development of the somatic causative PIK3CA pathogenic variants reflecting the degree of mosaicism and the combination of tissues involved [9, 10]. Different gain-of-function variants in PIK3CA determine a variable degree of hyperactivation of the phosphatidylinositol 3 kinase (PI3K)/AKT/mTOR pathway [11], leading to a different severity in the abnormal proliferation of mesodermal and/or ectodermal tissues from embryogenesis onwards [10].
In spite of the advances in the knowledge of the PROS across the last decade, a consistent fraction of these phenotypes still remains orphan of a molecular diagnosis due to several factors: these include inappropriate tissue DNA sampling, very low-level mosaicism difficult to be detected, molecular mechanisms still to be unraveled. Some of the cases with PROS-like or overlapping phenotypes have recently shown to be caused by variants in genes of the same signal cascade (i.e., AKT1, AKT3, mTOR [9, 12–15]) or crosstalking molecular pathways (i.e., RAS/MAPK [16, 17], or vascular proliferation pathway [18, 19],). However, still nearly 30% of cases remain undiagnosed [9].
Given their only recently clarified molecular bases and classification, the epidemiologic data of the PROS are unknown and there are no incidence/prevalence studies available. The only data currently accessible are based on information for 5 disorders (KTS, CLOVES, MCAP, HHML and FAO) and estimated the prevalence of such conditions in 14 people per million (i.e., 1: 71,428 live birth) [20], with a male to female ratio of 1/1,3 [20]. Herein, we report the findings of a population study aimed at providing the first epidemiology data by locating all cases of PROS born in Piedmont, a large italian region.
Patients and methods
Study design and patients—Piedmont, a region of the North West Italy, counts 4,274,945 inhabitants (2021 data of the Italian National Institute for Statistics, www.istat.it). We included in the cohort all cases born in Piedmont between 1998 and 2021 and diagnosed, visited or followed at our Institution, both evaluated by the Clinical Genetic Unit and referred by other specialists. Participants ascertained through the search of the archives of the Clinical Genetic Unit of the Regina Margherita Children’s Hospital of Torino were cross-matched and implemented by additional sources: (1) the regional network of Clinical Genetics; (2) the Regional Section of the Italian Registry for Rare Diseases; (3) the archive of the Department of Pediatrics of the University of Torino; (4) the Italian Association of individuals with PROS (Associazione Italiana Macrodattilia e PROS—APS, https://www.associazione-nazionale-macrodattilia.org), and (5) contacting of the largest extra-regional laboratory in Italy offering specific molecular tests for PROS and PROS-like conditions (N.R.).
Year 1998 marks the start of the systematic collection of the cases evaluated at our Institution. All cases were reviewed at our Unit of Clinical Pediatric Genetics in Torino between 2020 and 2022 and re-assessed clinically according to 2013 criteria [1]. Clinical re-evaluation and molecular testing by high-depth NGS testing of somatic DNA have been offered since 2018 to all these participants and when not performed before.
Molecular studies—For all the participants, DNA was extracted either from peripheral venous blood, buccal swab and, whenever possible (n = 34), from fresh or frozen biopsies of affected body regions using the QIAamp Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions, and quantified on a Bio Spectrometer Plus (Eppendorf, Hamburg, Germany). Samples were submitted for genetic testing to the laboratory of the Department of Biomedical Sciences and Human Oncology, of the Medical Genetics of the University of Bari ‘Aldo Moro’ (NR). DNA samples were tested by Next Generation Sequencing techniques with two AmpliSeq Illumina Custom DNA Panels (2021 Illumina) including 21 genes in the PI3K/AKT/mTOR pathway (PIK3R1, PIK3R2, PIK3CA, PTEN, PDK1, PDK2, KRAS, AKT1, AKT2, AKT3, RICTOR, MAPKAP1, MLST8, MTOR, IRS1, GAB1, GAB2, THEM4, MAPK8I1, PTPN11, RAPTOR) [18]. Subjects with a wild-type (wt) PIK3CA allele at first test underwent a re-analysis by using a custom panel including RASA1, TEK, TSC2, GNAQ, TSC1, DEPDC5, CCND2, NPRL3, and GNA11. Both NGS panels were performed to obtain an average, unique on-target read depths >1000×. Sequencing runs and variant interpretation were carried out as already described [9].
Results
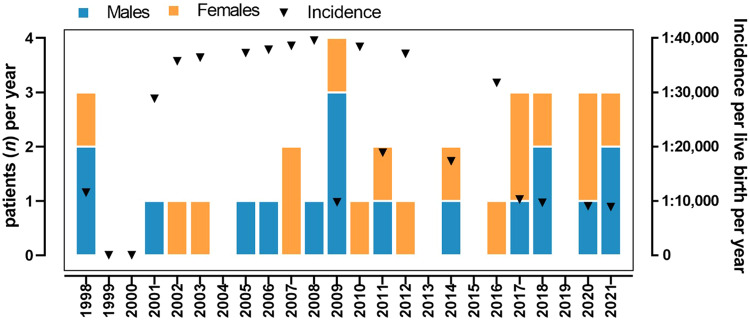
Births in Piedmont ranged from 34,658 in 1998 to 26,632 in 2021, with overall 825,593 births across this time period (mean 34,399 ± 4 040, median 35 685): 51.4% were boys (total 424,457, mean 17,686 ± 2213 per year), 48.6% were girls (total 401,136, mean 16,714 ± 1923 per year). Thirty-seven individuals affected by PROS born from 1998 and 2021 (18 males and 19 females) were found through our search. The incidence of PROS was therefore calculated as 1:22,313, corresponding to 4.48 per 100.000 live births (Fig. 1). This estimate was 1 in 23,580 live births in boys and 1 in 21,112 girls, with a male to female ratio of 1.17:1 and no statistically significant difference between the two sexes. All the individuals of the investigated cohort are still alive (two were lost at follow-up), so the incidence coincides with the prevalence rate in our case.
Fig. 1. Incidence per live birth per year.
Cases of PIKCA-related overgrowth spectrum (PROS) per year divided by sex (left axis) and incidence per year (right axis).
The phenotype classification based on those features is reported in Table 1, with respective variants in PIK3CA, found in 30 out of 37 participants (81.0%). Taking into account only the cases with a PIK3CA variant detected, the incidence of molecularly positive PROS was 1:27,519.
Table 1.
Patients’ classification in predominant phenotype according to clinical features and related PIK3CA variants.
| Phenotype | n | % | PIK3CA variant (n, VAF%) | Sample tested |
|---|---|---|---|---|
| KTS/DCMO | 11 | 29.8% |
c.323 G > A, p.Arg108His (n = 1, 6%) c.1136 G > A, p.Cys378Tyr (n = 1, 4%) c.1357 G > A, p.Glu453Lys (n = 1, 3%) c.1412 C > T, p.Pro471Leu (n = 1, 6%) c.1624A>G, p.Glu542Lys (n = 1, 4%) c.1636 C > A, p.Glu546Lys (n = 1, 15%) c.2176 G > A, p.Glu726Lys (n = 2, 13%, 6%) c.2740 A > G, p.Gly914Arg (n = 1, 8%) c.3127 A > G, p.Met1043Val (n = 1, 2%) c.3139 C > T, p.His194Tyr (n = 1, 15%) negative (n = 1) |
Skin/tissue biopsy (n = 11) |
| MCAP | 6 | 16.2% |
c.344 G > C, p.Arg115Pro (n = 1, 4.6%) c.1133 G > A, p.Cys378Tyr (n = 1, 37%) c.1356_1358AGA, p.Glu453del (n = 1, germline) c.2740 G > A, p.Gly914Arg (n = 1, 24%) c.3104 C > T, p.Ala135Val (n = 1, not availablea) c.3129 G > A, p.Met1043Ile (n = 1, 7%) |
Skin biopsy (n = 2) Buccal swab (n = 3) Blood (n = 2) |
| FIL | 4 | 10.8% |
c.3073 A > G, p.Thr1025Ala (n = 1, 24%) c.3140 A > G, p.His1047Arg (n = 2, 3%, 7.0%) negative (n = 1) |
Skin biopsy (n = 4) |
| Type I Macrodactyly | 4 | 10.8% |
c.3140 A > G, p.His1047Arg (n = 1, 9%) negative (n = 3) |
Skin biopsy (n = 4) |
| CLOVES | 3 | 8.1% |
c.1633G>A, p.Glu545Lys (n = 1, 3%) c.3073 A > G, p.Thr1025Ala (n = 1, 15%) negative (n = 1) |
Skin biopsy (n = 3) |
| FAO | 3 | 8.1% |
c.1132 T > C, p.Cys378Arg (n = 1, 7%) c.1636C>A, p.Glu546Lys (n = 1, 15%) c.3140 A > G, p.His1047Arg (n = 1, 15%) |
Skin biopsy (n = 3) |
| H/D-MEG | 2 | 5.4% |
c.3074 C > A, p.Thr1025Asn (n = 1, 6%) c.3140 A > G, p.His1047Arg (n = 1, germline) |
Skin biopsy (n = 2) |
| HHML | 2 | 5.4% |
c.1133 G > A, p.Cys378Tyr (n = 1, 26%) negative (n = 1) |
Skin biopsy (n = 2) |
| FH | 2 | 5.4% |
c.1133 G > A, p.Cys378Tyr (n = 1, 4%) c3139C>T p.His194Tyr (n = 1, 15%) |
Skin biopsy (n = 2) |
VAF Variant Allele Frequency, KTS Klippel-Trenaunay syndrome, DCMO Diffuse capillary malformation with overgrowth, MCAP Megalencephaly-Capillary Malformation, FIL Facial Infiltrating Lipomatosis, CLOVES Congenital Lipomatous Overgrowth, Vascular Malformations, Epidermal Nevi, Scoliosis/Skeletal and Spinal syndrome, FAO Fibroadipose Overgrowth, H/D-MEG Hemi- or Dysplastic Megalencephaly, HHML Hemihyperplasia Multiple Lipomatosis, FH Fibroadipose Hyperplasia.
aPresent in buccal swab-extracted DNA, absent in blood.
Discussion
This study represents the first attempt to provide epidemiologic data on PROS according to the established diagnostic criteria and specific molecular analysis. After a comprehensive literature review we just retrieved a previous estimate [20], based on available information for 5 disorders (KTS, CLOVES, MCAP, HHML and FAO) calculating a prevalence of 14 patients per million people (i.e., 1 in 71,428). Indeed, PROS rarity and heterogeneity likely hamper a correct estimation of its incidence and prevalence in the population. Also, the only recent discovery of its molecular bases and definition of clinical diagnostic criteria did not allow accumulating in medical literature historical estimates. The 2013 workshop that defined the spectrum had the goal of reconciling conflicting designations and overlapping phenotypes, bringing them together under the PROS acronym based on their molecular [1].
Our estimate relies on solid demographic data and represents the first attempt to provide epidemiologic definition of this group of disorders. This estimate—1 in 22,313—is much higher than the single one available previously. We acknowledge that our figure is a minimum estimate. Further cases could have been missed clinically or not being diagnosed for several reasons: a. the recent definition of the clinical criteria and molecular bases, b. the existence of mild phenotypes with little impact on health, c. the still poorly spread knowledge in general clinical practice of PROS as genetic diseases, d. the possibility that the most severe forms resulted in fetal demise or early childhood death before a diagnosis. Moreover, our data have the bias to be chiefly related to the syndromic forms of PROS and did not took into consideration systematically the non-syndromic PROS which are quite often not submitted to evaluation by a clinical geneticist. Actually, the syndromic cases, especially those presenting with tissue overgrowth or CNS involvement (i.e., MCAP, HMEG) are those submitted to evaluation in a pediatric clinical genetics setting. For these reasons, we believe that the real incidence of PROS is likely much higher than our estimate. As we registered no case of death over the study time period, the incidence rate results coincident with the prevalence data. On the other hand, in spite of the comprehensive NGS panel we employed to explore variants in genes connected to phenotypes overlapping PROS, some other genes, such as NPRL2, might contribute or cause the phenotype of our patients with negative molecular tests.
In conclusion, we report on the first estimate of syndromic PROS prevalence based on diagnostic criteria and molecular tests, providing an estimate much higher than previously gauged. More studies will be required to confirm these data on different populations. National and international registry data would be needed to determine the epidemiology and the clinical presentation of PROS worldwide.
Acknowledgements
We are grateful to the patients and their families who made this research possible by consenting to the use of data collected as part of their care. We thank the Italian Association of individuals with PROS (Associazione Italiana Macrodattilia e PROS—APS) who contributed to this study.
Author contributions
Conceptualization: AM, GR. Formal analysis and methodology: AM, GR. Investigation: AM, GR, SC, DC, ML, AG, PC, RLS, MP, RB, AT, CR. Resources: AM, SC, DC, ML, AG, PC, RLS, MP, RB, AT, CR, NR. Supervision: NR. Writing-original draft: AM, GR. Writing-review and editing: AM. All other authors approved this paper.
Funding
No specific grant for this research was received from any funding agency in the public, commercial or not-for-profit sectors.
Data availability
Depersonalized data that support the findings of this study are available within the paper and/or are available on request from the corresponding author AM.
Competing interests
The authors declare no competing interests.
Ethics approval
Given the retrospective nature of the study, approval by Ethic Committee was not required. Informed consent to genetic analysis on peripheral blood, tissue biopsy, or other samples was obtained by parents of participants and archived by authors according to the local ethic committee’s policy. Individual data included in this paper have been de-identified and presented in aggregate. The study was conducted in accordance with the ethical principles enshrined in the Helsinki Declaration.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Keppler-Noreuil KM, Rios JJ, Parker VER, Semple RK, Lindhurst MJ, Sapp JC, et al. PIK3CA-related overgrowth spectrum (PROS): Diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am J Med Genet Part A. 2015;167:287–95. doi: 10.1002/ajmg.a.36836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindhurst MJ, Parker VER, Payne F, Sapp JC, Rudge S, Harris J, et al. “Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat Genet. 2012;44:928–33. doi: 10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harnarayan P, Harnanan D. The Klippel-Trénaunay Syndrome in 2022: Unravelling Its Genetic and Molecular Profile and Its Link to the Limb Overgrowth Syndromes. Vasc Health Risk Manag. 2022;18:201–9. 10.2147/VHRM.S358849. [DOI] [PMC free article] [PubMed]
- 4.Rios JJ, Paria N, Burns DK, Israel BA, Cornelia R, Wise CA, et al. Somatic gain-of-function mutations in PIK3CA in patients with macrodactyly. Hum Mol Genet. 2013;22:444–51. doi: 10.1093/hmg/dds440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goss JA, Konczyk DJ, Smits P, Sudduth CL, Bischoff J, Liang MG, et al. Diffuse capillary malformation with overgrowth contains somatic PIK3CA variants. Clin Genet. 2020;97:736–40. doi: 10.1111/cge.13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarma K, Nayak MK, Mishra B, Gaikwad SB. Megalencephaly-Capillary Malformation-Polymicrogyria Syndrome (MCAP): A Rare Dynamic Genetic Disorder. Cureus. 2022;14:e25123. doi: 10.7759/cureus.25123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carli D, Kalantari S, Manicone R, Coppo P, Francia di Celle P, La Selva R, et al. Kaposiform hemangioendothelioma further broadens the phenotype of PIK3CA-related overgrowth spectrum. Clin Genet. 2021;100:624–7. doi: 10.1111/cge.14047. [DOI] [PubMed] [Google Scholar]
- 8.Weng J, Yang Y, Song D, Huo R, Li H, Chen Y, et al. Somatic MAP3K3 mutation defines a subclass of cerebral cavernous malformation. Am J Hum Genet. 2021;108:942–50. doi: 10.1016/j.ajhg.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mussa A, Leoni C, Iacoviello M, Carli D, Ranieri C, Pantaleo A et al. Genotypes and phenotypes heterogeneity in PIK3CA-related overgrowth spectrum and overlapping conditions: 150 novel patients and systematic review of 1007 patients with PIK3CA pathogenetic variants.” J Med Genet. 1–11:2022, 10.1136/jmedgenet-2021-108093. [DOI] [PubMed]
- 10.Madsen RR, Vanhaesebroeck B, Semple RK. Cancer-Associated PIK3CA Mutations in Overgrowth Disorders. Trends Mol Med. 2018;24:856–70. doi: 10.1016/j.molmed.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keppler-Noreuil KM, Sapp JC, Lindhurst MJ, Parker VER, Blumhorst C, Darling T, et al. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am J Med Genet Part A. 2014;164:1713–33. doi: 10.1002/ajmg.a.36552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang F, Liu L, Fang E, Zhang G, Chen T, Cao K, et al. Molecular Diagnosis of Mosaic Overgrowth Syndromes Using a Custom-Designed Next-Generation Sequencing Panel. J Mol Diagn. 2017;19:613–24. doi: 10.1016/j.jmoldx.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Mirzaa G, Timms AE, Conti V, Boyle EA, Girisha KM, Martin B et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight. 2016. 10.1172/jci.insight.87623. [DOI] [PMC free article] [PubMed]
- 14.Kuentz P, St-Onge J, Duffourd Y, Courcet JB, Carmignac V, Jouan T, et al. Molecular diagnosis of PIK3CA-related overgrowth spectrum (PROS) in 162 patients and recommendations for genetic testing. Genet Med J Am Coll Med Genet. 2017;19:989–97. doi: 10.1038/gim.2016.220. [DOI] [PubMed] [Google Scholar]
- 15.Carli D, Ferrero GB, Fusillo A, Coppo P, La Selva R, Zinali F, et al. A new case of Smith-Kingsmore syndrome with somatic MTOR pathogenic variant expands the phenotypic spectrum to lateralized overgrowth. Clin Genet. 2021;99:719–23. doi: 10.1111/cge.13931. [DOI] [PubMed] [Google Scholar]
- 16.Chang CA, Perrier R, Kurek KC, Estrada-Veras J, Lehman A, Yip S, et al. Novel findings and expansion of phenotype in a mosaic RASopathy caused by somatic KRAS variants. Am J Med Genet A. 2021;185:2829–45. doi: 10.1002/ajmg.a.62356. [DOI] [PubMed] [Google Scholar]
- 17.Mussa A, Carli D, Cardaropoli S, Ferrero GB, Resta N. Lateralized overgrowth with vascular malformation caused by a somatic PTPN11 pathogenic variant: Another piece added to the puzzle of mosaic RASopathies. Genes Chromosom Cancer. 2022;61:689–95. doi: 10.1002/gcc.23086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel DH, Cottrell CE, Streicher JL, Schilter KF, Basel DG, Baselga E, et al. Analyzing the Genetic Spectrum of Vascular Anomalies with Overgrowth via Cancer Genomics. J Investig Dermatol. 2018;138:957–67. doi: 10.1016/j.jid.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Mussa A, Carli D, Cardaropoli S, Ferrero GB, Resta N. Lateralized and Segmental Overgrowth in Children. Cancers (Basel). 2021;13. 10.3390/cancers13246166. [DOI] [PMC free article] [PubMed]
- 20.Data on file. Novartis Pharmaceuticals Corp; 2020. https://www.prosspectrum.com/about-pros/pros-overview/. (accessed Dec. 02, 2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Depersonalized data that support the findings of this study are available within the paper and/or are available on request from the corresponding author AM.