Abstract
Panaeolus sphinctrinus, Panaeolus papilionaceus, and Coprinus friesii are described as producers of ligninolytic enzymes. P. papilionaceus and P. sphinctrinus both produced a laccase. In addition, P. sphinctrinus produced a manganese peroxidase. C. friesii secreted a laccase and two peroxidases similar to the peroxidase of Coprinus cinereus. The purified laccases and peroxidases were characterized by broad substrate specificities, significant enzyme activities at alkaline pH values, and remarkably high pH optima. The two peroxidases of C. friesii remained active at pH 7.0 and 60°C for up to 60 min of incubation. The peroxidases were inhibited by sodium azide and ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), whereas the laccases were inhibited by sodium azide and N,N-diethyldithiocarbamic acid. As determined by native polyacrylamide gel electrophoresis and isoelectric focusing, all three fungi produced laccase isoenzymes.
Laccases, peroxidases (including lignin peroxidases, manganese peroxidases, and manganese-independent peroxidases), and H2O2-generating oxidases are components of the lignin-degrading enzyme system (13). So far, wood-rotting fungi, such as white rot and soft rot fungi, are the only organisms known to be capable of extensively degrading lignin (42). There has been great interest in using fungal laccases and peroxidases for biotechnological processes due to their chemical and catalytic features (26, 37). Investigations of the use of wood-rotting fungi and their ligninolytic enzymes in biopulping processes in paper-making industries could lead to ways to reduce the energy and chemical requirements of those processes. A biobleaching process requires substantial enzyme activities at an alkaline pH and a high temperature. In addition, several other potential applications have been suggested for producers of ligninolytic enzymes; these applications have been reviewed by Heinzkill and Messner (16).
Laccases (EC 1.10.3.2; benzenediol:oxygen oxidoreductases) are for the most part extracellular copper-containing glycoproteins with molecular weights between 60,000 and 80,000 (40). Lignin peroxidases (EC 1.11.1.14; diarylpropane:oxygen, hydrogen peroxide oxidoreductases; molecular weights, 38,000 to 43,000) and manganese peroxidases [EC 1.11.1.13; Mn(II):H2O2 oxidoreductases; molecular weights, 43,000 to 49,000] are glycoproteins containing one protoporphyrin IX as a prosthetic group (12). The reactions catalyzed by laccases and peroxidases are very similar (17, 18, 20, 27). Both types of enzymes oxidize phenolic compounds and aromatic amines via one-electron oxidations, which creates radicals. Besides differences in the prosthetic groups, the laccases also differ from the peroxidases by generally having a lower oxidation potential (6, 14). Many producers of laccase (e.g., Ceriporiopsis subvermispora [34], Coriolus versicolor [29], and Panus tigrinus [25]) and producers of lignin peroxidases and manganese peroxidases (e.g., Phanerochaete chrysosporium [11] and Bjerkandera adusta [31]) secrete isoenzymes which differ in stability and catalytic features (7). The extracellular peroxidases isolated from Coprinus cinereus (28) and Arthromyces ramosus (22) differ from lignin peroxidases and manganese peroxidases by having broader substrate specificities and in the architecture of their active sites.
We recently discovered that the three basidiomycetes Panaeolus sphinctrinus, Panaeolus papilionaceus, and Coprinus friesii produce laccases and peroxidases, and the purpose of this study was to characterize these activities in more detail.
MATERIALS AND METHODS
Chemicals.
Sojamin 50 T was obtained from Lucas Meyer (Hamburg, Germany). 2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulfonic acid (ABTS) and EGTA were obtained from Sigma Chemical Co. (St. Louis, Mo.). 2,6-Dimethoxyphenol (2,6-DMP) and N,N-diethyldithiocarbamic acid (ammonium salt) (DEDTC) were purchased from Aldrich (Steinheim, Germany). All other chemicals were of reagent grade and were obtained from Merck (Darmstadt, Germany).
Organisms, media, and cultivation conditions.
Panaeolus sphinctrinus 82066 and Panaeolus papilionaceus CBS 630.95 were isolated from dung in France and Germany, respectively, while Coprinus friesii CBS 629.95 was isolated from a meadow in Denmark. The fungi were identified by using the methods of Moser (30) and Singer (38) and had all of the characteristics of the genera and species described previously (30, 38). All strains were positive for laccase activity when they were cultivated without shaking on a medium containing (per liter) 30 g of soy meal, 15 g of maltodextrin, and 5 g of Bacto Peptone (Difco Laboratories). In addition, Coprinus friesii was positive for peroxidase activity. Stocks of the strains were maintained on YMG agar (4 g of glucose per liter, 10 g of malt extract per liter, 4 g of yeast extract per liter, and 15 g of agar per liter in water adjusted to pH 5.5 before sterilization). Broth preparations used for purification of laccases and peroxidases were obtained by growing Panaeolus sphinctrinus, Panaeolus papilionaceus, and Coprinus friesii at 27°C in a Braun Biostat U apparatus containing 20 liters of soy meal medium (30 g of soy meal per liter, 15 g of maltose per liter, 15 g of Bacto Peptone per liter) and agitated at 120 to 150 rpm with aeration (4 liters/min). A 200-ml portion of a well-grown culture was used for inoculation in each case.
Ultrafiltration.
Fermentation broth preparations were initially centrifuged at 4°C for 20 min at 4,651 × g, and the resulting supernatants were filtered by using a 0.16-μm-pore-size tangential flow membrane filter with a 994-kDa cutoff and a Minisette ultrafiltration unit (Minisette omega series; Filtron, Karlstein, Germany). The filtrates were subsequently concentrated in the ultrafiltration unit by using a membrane filter with a 10-kDa cutoff and stored at −70°C until further purification. Before column chromatography the centrifuged, filtered, concentrated broth preparations were dialyzed against the equilibration buffer. Ultrafiltration with Amicon cells equipped with either a 10-kDa cutoff membrane filter or a 30-kDa cutoff membrane filter was used for buffer changes and concentration of pooled fractions.
Protein purification.
All protein purification procedures were carried out by using a BioLogic system (Bio-Rad, Hercules, Calif.) operated at 22°C. The columns and column materials used were Bio-Scale Q2 (Bio-Rad), Q-Sepharose FF (Sigma), DEAE-Sepharose FF (Sigma), Superdex 75 (Pharmacia, Uppsala, Sweden), and SP-Sepharose FF (Pharmacia). Purified Polyporus pinsitus laccase was obtained from Novo Nordisk.
Purification of a laccase from Panaeolus sphinctrinus.
Thirty milliliters of centrifuged, filtered, concentrated, dialyzed Panaeolus sphinctrinus culture broth was applied to a Q-Sepharose FF column (300 by 10 mm) equilibrated in 20 mM Tris-HCl–50 mM NaCl (pH 7.2). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted with a 200-ml linear gradient consisting of 50 mM to 0.5 M NaCl in 20 mM Tris-HCl (pH 7.2). The flow rate was 2 ml/min, and 8-ml fractions were collected and assayed for laccase activity. Laccase-containing fractions were pooled and concentrated, and the buffer was changed before application to a Bio-Scale Q2 column (52 by 7 mm) equilibrated in 20 mM Tris-HCl–50 mM NaCl (pH 7.2). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted with a 40-ml linear gradient consisting of 50 to 500 mM NaCl in 20 mM Tris-HCl (pH 7.2). The flow rate was 2 ml/min, and 5-ml fractions were collected and assayed for laccase activity. Laccase-containing fractions were pooled and concentrated, glycerol was added to a final concentration of 20%, and the preparation was stored at −70°C.
Purification of a manganese peroxidase from Panaeolus sphinctrinus.
One hundred milliliters of centrifuged, filtered, concentrated, dialyzed Panaeolus sphinctrinus culture broth was applied to a DEAE-Sepharose FF column (67 by 50 mm) equilibrated in 20 mM piperazine (pH 6.0). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted stepwise with 200-ml aliquots of equilibration buffer containing 50, 100, 200, 300, and 500 mM and 1 M NaCl. The flow rate was 6 ml/min, and 200-ml fractions were collected and assayed for manganese peroxidase activity. Manganese peroxidase-containing fractions were pooled and concentrated, and the buffer was changed before application to an SP-Sepharose FF column (100 by 26 mm) equilibrated in 20 mM sodium acetate–20 mM NaCl (pH 5.0). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted with a 400-ml linear gradient consisting of 20 to 400 mM NaCl in 20 mM sodium acetate (pH 5.0). The flow rate was 4 ml/min, and 8-ml fractions were collected and assayed for manganese peroxidase activity. Manganese peroxidase-containing fractions were pooled and concentrated, and the buffer was changed before application to a Superdex 75 column (300 by 10 mm) equilibrated in 20 mM sodium acetate–400 mM NaCl (pH 5.0). The flow rate was 0.6 ml/min, and 0.5-ml fractions were collected and assayed for manganese peroxidase activity. Manganese peroxidase-containing fractions were pooled and concentrated, glycerol was added to a final concentration of 20%, and the preparation was stored at −70°C.
Purification of a laccase from Panaeolus papilionaceus.
Twenty milliliters of centrifuged, filtered, concentrated, dialyzed Panaeolus papilionaceus culture broth was applied to a Q-Sepharose FF column (82 by 24 mm) equilibrated in 20 mM Tris-HCl–50 mM NaCl (pH 7.2). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted stepwise with 100-ml portions of equilibration buffer containing 50, 100, 200, 300, and 500 mM and 1 M NaCl. The flow rate was 5 ml/min, and 50-ml fractions were collected and assayed for laccase activity. The laccase eluted with 200 mM NaCl. The laccase-containing fraction was concentrated, and the buffer was changed before application to a DEAE-Sepharose FF column (82 by 24 mm) equilibrated in 20 mM Tris-HCl–50 mM NaCl (pH 7.2). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted with a 500-ml linear gradient consisting of 50 mM to 1 M NaCl in equilibration buffer. The flow rate was 2.5 ml/min, and 10-ml fractions were collected and assayed for laccase activity. Laccase-containing fractions were pooled and concentrated, glycerol was added to a final concentration of 20%, and the preparation was stored at −70°C.
Purification of one laccase and two peroxidases from Coprinus friesii.
Twenty milliliters of centrifuged, filtered, concentrated, dialyzed Coprinus friesii culture broth was applied to a Q-Sepharose FF column (300 by 10 mm) equilibrated in 20 mM Tris-HCl–50 mM NaCl (pH 7.2). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted with a 75-ml linear gradient consisting of 50 to 400 mM NaCl, followed by a 50-ml linear gradient consisting of 400 mM to 1 M NaCl in equilibration buffer. The flow rate was 2 ml/min, and 4-ml fractions were collected and assayed for laccase and peroxidase activities. The laccase eluted with 160 mM NaCl, while the peroxidase eluted with 180 mM NaCl. The laccase-containing fraction was concentrated, and the buffer was changed before application to a DEAE-Sepharose FF column (150 by 10 mm) equilibrated in 20 mM Tris-HCl–50 mM NaCl (pH 7.2). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted with NaCl in equilibration buffer as follows. The initial elution with a 15-ml linear gradient consisting of 50 to 150 mM NaCl was followed by a 30-ml isocratic wash with 150 mM NaCl, elution with a 15-ml linear gradient consisting of 150 to 200 mM NaCl, a 30-ml isocratic wash with 200 mM NaCl, elution with a 15-ml linear gradient consisting of 200 to 250 mM NaCl, and a 30-ml isocratic wash with 250 mM NaCl. The flow rate was 2 ml/min, and 4-ml fractions were collected and assayed for laccase activity. Laccase-containing fractions were pooled and concentrated, glycerol was added to a final concentration of 20%, and the preparation was stored at −70°C. The peroxidase-containing fractions were concentrated, and the buffer was changed before application to a DEAE-Sepharose FF column (150 by 10 mm) equilibrated in 20 mM Tris-HCl–50 mM NaCl (pH 7.2). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted with a 60-ml linear gradient consisting of 50 to 400 mM NaCl, followed by a 15-ml linear gradient consisting of 400 mM to 1 M NaCl in equilibration buffer. The flow rate was 1 ml/min, and 3-ml fractions were collected and assayed for peroxidase activity. Two pools of peroxidase activity were obtained. Peroxidase I eluted at an NaCl concentration of 125 mM, while peroxidase II eluted at an NaCl concentration of 220 nM. Following concentration and a buffer change both peroxidase pools were purified further by identical protocols. The peroxidase pools were applied to a DEAE-Sepharose FF column (150 by 10 mm) equilibrated in 20 mM Tris-HCl–50 mM NaCl (pH 7.2). The column was washed with equilibration buffer until the absorbance at 280 nm reached the baseline level. Bound protein was eluted with NaCl in equilibration buffer as follows. The initial elution with a 75-ml linear gradient consisting of 50 to 150 mM NaCl was followed by a 75-ml isocratic wash with 150 mM NaCl, elution with a 45-ml linear gradient consisting of 150 to 200 mM NaCl, a 45-ml isocratic wash with 200 mM NaCl, elution with a 30-ml linear gradient consisting of 200 to 250 mM NaCl, and a 30-ml isocratic wash with 250 mM NaCl. The flow rate was 2 ml/min, and 5-ml fractions were collected and assayed for peroxidase activity. Peroxidase-containing fractions were pooled and concentrated, glycerol was added to a final concentration of 20%, and the preparations were stored at −70°C.
Protein concentration determinations.
Protein concentrations were determined by the bicinchoninic acid protein reagent assay (Pierce, Rockford, Ill.).
PAGE and IEF.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of fractions from the purification procedures and of purified enzymes was performed as described by Sambrook et al. (36). Markers with molecular masses ranging from 14.4 to 97.4 kDa (Bio-Rad) were used as standards. Proteins were stained with Coomassie brilliant blue R-250 (36). Isoelectric focusing (IEF) of the purified enzymes was performed with a Multiphor II system (Pharmacia) at 4°C. Servalyt Precotes (pH 3 to 6; 300 μm; Serva, Heidelberg, Germany) and Ampholine PAG plates (pH 3.5 to 9.5; diameter, 1 mm; Pharmacia) were used. A standard calibration curve with different protein standards (low pI and broad pI calibration kit; Pharmacia) was used to determine the isoelectric points.
Determination of enzyme molecular masses.
The molecular masses were determined by SDS-PAGE and by gel permeation chromatography on a Superdex 75 column (see above). Protein standards (type MW-GF-200 kit; Sigma) were used to calculate the molecular masses of the enzymes.
N-terminal sequence analysis.
The N-terminal amino acid sequences of the purified laccases and peroxidases were determined following SDS-PAGE and electroblotting onto polyvinylidene difluoride membranes with an Applied Biosystems model 473A protein sequencer.
Enzyme assays and kinetics.
Laccase activity was determined at pH 5.0 and 7.0 by monitoring the oxidation of ABTS at 405 nm (ɛ405 = 36,000 M−1 cm−1) as follows (33, 46). A 100-μl sample was added to 100 μl of an ABTS solution in a 96-well microtiter plate, and the absorbance at 405 nm was determined for 5 min. The ABTS solutions used contained 3.6 mM ABTS in either 0.2 M sodium acetate (pH 5.0) or 0.2 M sodium phosphate (pH 7.0). Peroxidase activity was determined at pH 7.0 by monitoring the oxidation of ABTS at 405 nm. A 100-μl sample was added to 100 μl of an H2O2 solution in a 96-well microtiter plate. The enzymatic reaction was started by adding 100 μl of ABTS solution, and the absorbance at 405 nm was determined for 5 min. The H2O2 solution contained 0.5 mM H2O2 in 0.2 M sodium phosphate (pH 7.0), while the ABTS solution contained 3.6 mM ABTS in 0.2 M sodium phosphate (pH 7.0). The enzyme activities were expressed as units per milliliter, where 1 U was defined as 1 μmol of substrate oxidized per min. The peroxidase activity was always corrected for laccase activity. Manganese peroxidase activity and laccase activity were measured based on the oxidative dimerization of 2,6-DMP (ɛ469 = 49,000 M−1 cm−1) (4). To determine laccase activity, a 100-μl sample was added to 890 μl of a solution containing 50 mM sodium malonate and 1 mM 2,6-DMP (pH 4.5) in a 1-ml cuvette. The absorbance at 469 nm was determined for 2 min. To determine manganese peroxidase activity, a 100-μl sample was added to 873 μl of a solution containing 50 mM sodium malonate, 0.7 mM MnSO4, and 1 mM 2,6-DMP (pH 4.5) in a 1-ml cuvette. The enzymatic reaction was initiated by adding 10 μl of 10 mM H2O2. The absorbance at 469 nm was determined for 2 min. The enzyme activities were expressed as units per milliliter, where 1 U was defined as 1 μmol of substrate oxidized per min. The manganese peroxidase activity was always corrected for laccase activity.
The temperature optima of the isolated enzymes were determined with prewarmed substrate by using a thermostat-equipped cuvette and a UV-visible light spectrometer (model Lambda 16; Perkin-Elmer, Langen, Germany). For stability determinations the enzymes were incubated in a thermostatically controlled bath. After incubation the enzyme assays were performed at 22°C. To determine the pH optima, enzyme assays with the substrates ABTS and 2,6-DMP were performed at 22°C by using Britton-Robinson universal buffer. The Km and Ki values were calculated based on a Lineweaver-Burk plot (23).
RESULTS
Accumulation of laccase and peroxidase activities in the medium.
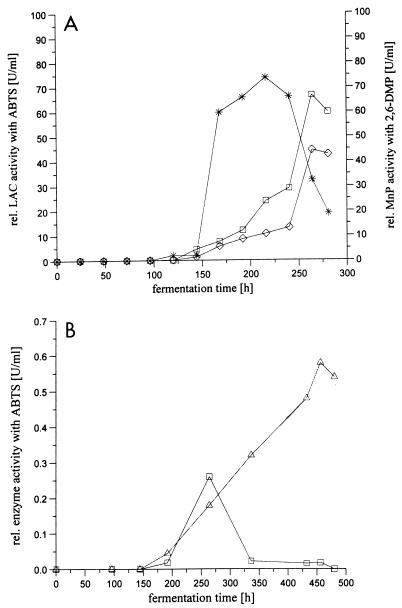
In an initial series of experiments it was established that the levels of laccase activity in Panaeolus sphinctrinus, Panaeolus papilionaceus, and Coprinus friesii cultures were higher when the organisms were cultivated in soy meal medium containing a surplus of nitrogen than when they were cultivated in the nitrogen-limited media which are normally used for inducing production of oxidoreductases in white rot fungi (12). As illustrated in Fig. 1A for Panaeolus sphinctrinus fermentation on the medium which provided the best yield, the laccase activity increased steadily over time after a short time lag. The same result was obtained for Panaeolus papilionaceus fermentation (data not shown). In addition, a manganese peroxidase was present in the Panaeolus sphinctrinus broth. Coprinus friesii fermentation resulted in a completely different time pattern for detection of laccase and peroxidase activities (Fig. 1B) as the activities were first detected after 170 h of fermentation. The occurrence of the enzyme activities was associated with a decrease in biomass. This could mean that the enzymes are released through cell lysis instead of being truly extracellular.
FIG. 1.
(A) Enzyme production by Panaeolus sphinctrinus in soybean medium. Symbols: □, laccase (LAC) activity at pH 5; ○, laccase activity at pH 7; ∗, manganese peroxidase activity at pH 4.5. (B) Enzyme production by Coprinus friesii in soybean medium. Symbols: □, laccase activity at pH 5; ▵, peroxidase activity at pH 7. rel., relative.
Purification of extracellular laccases and peroxidases.
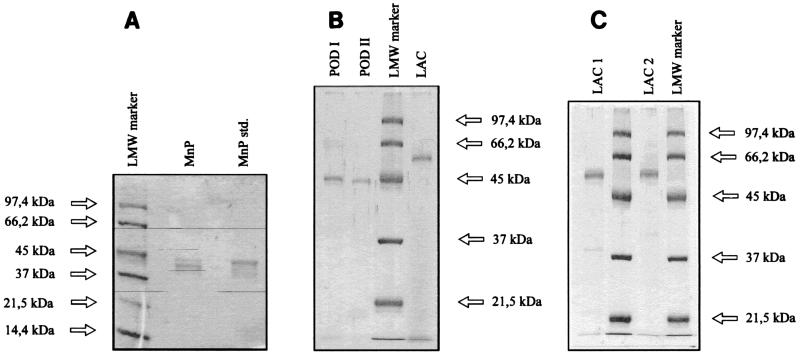
The procedures used for purification of the laccases and peroxidases from Panaeolus sphinctrinus, Panaeolus papilionaceus, and Coprinus friesii were carried out as described above. Figure 2 shows the results of the SDS-PAGE of the purified enzymes.
FIG. 2.
SDS-PAGE of purified enzymes. (A) Manganese peroxidases from Panaeolus sphinctrinus (lane MnP) and Phanerochaete crysosporium (lane MnP std.). (B) Peroxidases I (lane POD I) and II (lane POD II) and laccase (lane LAC) from Coprinus friesii. (C) Laccases from Panaeolus sphinctrinus (lane LAC1) and Panaeolus papilionaceus (lane LAC2). Lanes LMW marker contained molecular weight markers.
Molecular weights and isoelectric points.
The molecular weights of the laccases from Panaeolus sphinctrinus and Panaeolus papilionaceus were determined to be ca. 60,000 by gel filtration and SDS-PAGE. The molecular weight of the laccase isolated from Coprinus friesii was also estimated by SDS-PAGE to be ca. 60,000, whereas peroxidases I and II were found to have molecular weights of ca. 45,000. The manganese peroxidase from Panaeolus sphinctrinus was found to have a molecular weight of ca. 42,000. Tables 1 and 2 show the isoelectric points and molecular weights of the purified enzymes. All of the laccases were characterized by acidic isoelectric points around pH 3.5. On the basis of IEF results, as well as native PAGE results, it was evident that the purified laccases from the Panaeolus strains and from Coprinus friesii consist of three or four isoenzymes.
TABLE 1.
Characterization of purified laccases
| Source of laccase | Km for ABTS (μM) | Optimum pH with:
|
Remaining activity (%) at:
|
Ki (μM) for:
|
pI | Mol wt | |||
|---|---|---|---|---|---|---|---|---|---|
| ABTS | 2,6-DMP | 60°C | 90°C | NaN3 | DEDTC | ||||
| Panaeolus sphinctrinus | 32.4 | 3 | 7 | 0 (30)a | 0 (5) | 1.2 | 19.9 | <3.55 | ∼60,000 |
| Panaeolus papilionaceus | 50.6 | 3 | 8 | 0 (15) | 0 (5) | 1.3 | 11.2 | ≤3.6 | ∼60,000 |
| Coprinus friesii | 41.4 | 5 | 8 | 0 (20) | 0 (5) | NTb | NT | 3.5 | ∼60,000 |
| Polyporus pinsitus | 22.3 | 3 | 5 | 27 (60) | 0 (30) | 1.0 | 26 | ≤3.5 | 66,000 |
The numbers in parentheses are incubation times (in minutes).
NT, not tested.
TABLE 2.
Characterization of purified manganese peroxidases and peroxidases
| Species | Enzyme(s) |
Km (μM) for:
|
Saturation Mn2+ concn (μM) | Optimum H2O2 concn (μM) | Optimum pH with:
|
Remaining activity (%) at:
|
Ki (μM) for:
|
pI | Mol wt | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABTS | 2,6-DMP | ABTS | 2,6-DMP | 60°C | 90°C | NaN3 | EGTA | ||||||
| Panaeolus sphinctrinus | Manganese peroxidase | NTa | 11.7 | 100 | 50 | 4 | 5 | 0 (60)b | NT | 1.3 | 18.8 | 7.2 | ∼42,000 |
| Phanerochaete chrysosporium | Manganese peroxidase | NT | 8.2 | >50 | 50 | 4 | 4 | 0 (60) | NT | 1.1 | 17.8 | 3.9, 4.1 | 46,000 |
| Coprinus friesii | Peroxidase I | 344.8 | NT | 100 | 6 | 10 | 8 (60) | 0 (10) | 21.8 | 0.6 | 3.6 | ∼45,000 | |
| Peroxidase II | 71.4 | NT | 250 | 5 | 7 | 12 (60) | 0 (10) | 7.5 | 0.5 | 3.6 | ∼45,000 | ||
| Coprinus cinereus | Peroxidase | 52.6 | NT | 2,000 | 6 | 9 | 18 (60) | 0 (45) | 72.2 | 1.1 | ≤3.5 | 39,000 | |
NT, not tested.
The numbers in parentheses are incubation times (in minutes).
Kinetic studies of purified laccases and peroxidases.
Tables 1 and 2 summarize the results of kinetic studies of the purified laccases. All of the laccases and peroxidases follow typical Michaelis-Menten kinetics. Inhibition of laccase, peroxidase, and manganese peroxidase by sodium azide was determined to be competitive, whereas inhibition by DEDTC and EGTA was noncompetitive (1).
N-terminal amino acid sequences.
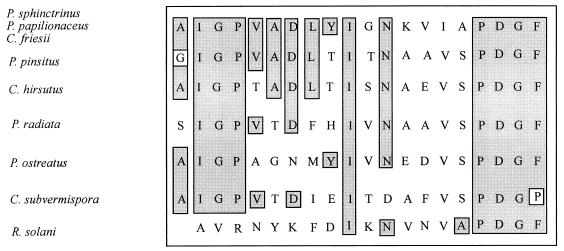
The N-terminal amino acid sequences of the three purified laccases were found to be identical and homologous to N-terminal amino acid sequences of other laccases, as shown in Fig. 3. The N-terminal amino acid sequence data for peroxidases I and II from Coprinus friesii and for the manganese peroxidase from Panaeolus sphinctrinus showed that the N-terminal amino group was blocked in these enzymes. Internal peptide sequence data obtained from the manganese peroxidase following proteolytic degradation revealed, however, possible homology to the manganese peroxidase from Phanerochaete chrysosporium.
FIG. 3.
Comparison of the N-terminal amino acid sequences of the purified laccases from Panaeolus sphinctrinus, Panaeolus papilionaceus, and Coprinus friesii with the laccase sequences of Polyporus pinsitus (47), Coriolus hirsutus (21), Phlebia radiata (35), Pleurotus ostreatus (32), Ceriporiopsis subvermispora (34), and Rhizoctonia solani (44).
DISCUSSION
For the first time two basidiomycetes belonging to the genus Panaeolus have been described as producers of laccases and peroxidases. As previously described, enzyme production is highly dependent on the cultivation conditions of an organism. Most white rot fungi, including Phanerochaete chrysosporium, start lignin degradation when nitrogen, carbon, or sulfur becomes limiting (12). Peroxidase production from Arthromyces ramosus is greatest when glucose and yeast extract or polypeptone are added at a ratio of 3:5 (41). The production of manganese peroxidase by the white rot fungus Lentinus edodes is suppressed by a high nitrogen concentration in the medium, whereas under these conditions laccase production reached its maximum level (3). However, the basidiomycetes Panaeolus sphinctrinus, Panaeolus papilionaceus, and Coprinus friesii produced both the highest level of laccase activity and the highest level of peroxidase-manganese peroxidase activity in soybean medium containing a surplus of nitrogen. Similar results were obtained by Youn et al. (48) during cultivation of Pleurotus ostreatus in a protein-rich medium. The different effects of nitrogen on enzyme production can be explained by the natural substrates of the coprophilic and saprophytic fungi used. Wood, which contains little nitrogen, is the substrate when the ligninolytic enzymes of most white rot fungi are produced. The coprophilic Panaeolus strains grow on nitrogen-rich dung. The alkaline pH of this natural substrate can also explain the high levels of laccase activity at pH values of ≥7.0.
All of the purified laccases are rather similar in affinity for the substrate ABTS, as indicated by the Km values (Table 2). The pH optima depend very much on the substrate. With ABTS the laccases exhibit optimum activity at rather acidic pH values (pH 3 to 5), like other fungal laccases (40). However, with 2,6-DMP as the substrate these new fungal laccases exhibit remarkably high levels of activity at pH values greater than 7.0, and the optimum pH is 7.0 to 8.0. The laccase isoenzymes isolated from Ceriporiopsis subvermispora are characterized by an optimum pH for ABTS of 2.0 to 3.0 (9). Other laccases with pH optima between 3.5 and 7.0 have been described (48). All purified laccases were inhibited by sodium azide, an inhibitor of metalloenzymes (39). The same results were obtained for the laccase of Ceroporiopsis subvermispora, which was inhibited by sodium azide and chelators such as thioglycol acid and DEDTC, whereas hydroxylamine and EDTA had no inhibitory effect (9).
The molecular weights of the purified laccases (ca. 60,000) are very similar to the molecular weights of most other fungal laccases, which have been found to be between 60,000 and 390,000 (40). Native PAGE (15) and IEF revealed that acid laccase isoforms were produced by the strains described in this study. The white rot fungi Coriolus versicolor (29) and Panus tigrinus (25) are also known producers of laccase isoforms. The N-terminal amino acid sequences of the purified laccases are clearly homologous to previously determined N-terminal amino acid sequences of fungal laccases, having between 35 and 65% identical residues.
The peroxidases isolated differ in affinity for ABTS. Compared to the peroxidase from Coprinus cinereus, peroxidases I and II from Coprinus friesii are much more sensitive to H2O2. The inhibition of peroxidase by sodium azide was determined to be competitive, and the inhibition of peroxidase by the iron chelator EGTA was determined to be noncompetitive. This observation was in contrast to the findings of De Pillis and Ortiz de Montellano (5), who discussed the inhibition of the peroxidase of Coprinus macrorhizus by sodium azide, which results in a mesoazidoheme.
The molecular weights and the pI values of peroxidases I and II are in the same ranges as the molecular weight and the pI of the Coprinus cinereus peroxidase (28). The N-terminal amino groups of the Coprinus friesii peroxidases were both inaccessible to sequencing, probably because of cyclization of a Gln residue, as found in the Coprinus cinereus peroxidase.
The manganese peroxidase isolated from Panaeolus sphinctrinus is similar to the previously described manganese peroxidase from Phanerochaete chrysosporium (10), except that the isoelectric point of the Panaeolus sphinctrinus enzyme (pI 7.2) is higher. Manganese peroxidase isoenzymes (pI 3.2 and 2.9) isolated from Coriolus versicolor have the greatest activity at low pH values (19). Six isoforms of manganese peroxidase and 21 isoenzymes of lignin peroxidase were detected in the culture broth of Phanerochaete chrysosporium (8). Ceriporiopsis subvermispora produces 11 manganese peroxidase isoenzymes with pI values between 3.2 and 4.58 (45) and three laccase isoenzymes with pI values of 3.71, 3.65, and 3.6 (35). The molecular size of the purified manganese peroxidase is similar to the molecular size of the manganese peroxidase from Phanerochaete chrysosporium (10). Urzúa et al. (43) isolated manganese peroxidase isoenzymes with a molecular weight of 52,500 from Ceriporiopsis subvermispora. Stationary cultures of the same fungus produced isoenzymes having a molecular weight of 62,500 (24). The N-terminal amino group of the manganese peroxidase was blocked, but an internal peptide sequence suggested that there was a relationship to the manganese peroxidase from Phanerochaete chrysosporium.
REFERENCES
- 1.Bisswanger H. Enzymkinetik. Theorie und Methoden. 2nd ed. Weinheim, Germany: VCH; 1994. [Google Scholar]
- 2.Bourbonnais R, Paice M G. Oxidation of nonphenolic substrates: an expanded role for laccase in lignin biodegradation. FEBS Lett. 1990;267:99–102. doi: 10.1016/0014-5793(90)80298-w. [DOI] [PubMed] [Google Scholar]
- 3.Buswell J A, Cai Y, Chang S T. Effect of nutrient and manganese on manganese peroxidase and laccase production by Lentinula (Lentinus) edodes. FEMS Microbiol Lett. 1995;128:81–88. [Google Scholar]
- 4.De Jong E. Physiological roles and metabolism of fungal aryl alcohols. Ph.D. dissertation. Wageningen, The Netherlands: Landbau Universität; 1993. [Google Scholar]
- 5.De Pillis G, Ortiz de Montellano P R. Substrate oxidation by the heme edge of fungal peroxidase. Reaction of Coprinus macrorhizus peroxidase with hydrazines and sodium azide. Biochemistry. 1988;28:7947–7952. doi: 10.1021/bi00445a059. [DOI] [PubMed] [Google Scholar]
- 6.Farhangrazi Z S, Copeland B R, Nakayama T, Amach T, Yamazaki I, Powers L S. Oxidation-reduction properties of compounds I and II of Arthromyces ramosus peroxidase. Biochemistry. 1994;33:5647–5652. doi: 10.1021/bi00184a038. [DOI] [PubMed] [Google Scholar]
- 7.Farrell R L, Murtagh K E, Tien M, Mozuch M D, Kirk T K. Physical and enzymatic properties of lignin peroxidase isoenzymes from Phanerochaete chrysosporium. Enzyme Microb Technol. 1989;11:322–328. [Google Scholar]
- 8.Fiechter A. Function and synthesis of enzymes involved in lignin degradation. J Biotechnol. 1993;30:49–55. [Google Scholar]
- 9.Fukushima Y, Kirk T K. Laccase component of the Ceriporiopsis subvermispora lignin-degrading system. Appl Environ Microbiol. 1995;61:872–876. doi: 10.1128/aem.61.3.872-876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenn J K, Gold M H. Purification and properties of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete Phanerochaete chrysosporium. Arch Biochem Biophys. 1985;242:329–341. doi: 10.1016/0003-9861(85)90217-6. [DOI] [PubMed] [Google Scholar]
- 11.Glumoff T. Lignin peroxidase from Phanerochaete chrysosporium. Eur J Biochem. 1990;187:515–520. doi: 10.1111/j.1432-1033.1990.tb15333.x. [DOI] [PubMed] [Google Scholar]
- 12.Gold M H, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatakka A. Lignin-modifying enzymes from selected white-rot fungi: production and role in lignin degradation. FEMS Microbiol Rev. 1994;13:125–135. [Google Scholar]
- 14.Hayashi Y, Yamazaki I. The oxidation-reduction potentials of compound I/compound II and compound II/ferric couples of horseradish peroxidases A2 and C. J Biol Chem. 1979;254:9101–9106. [PubMed] [Google Scholar]
- 15.Heinzkill M. Isolierung und Charakterisierung von Laccasen und Peroxidasen aus Basidiomyceten der Ordnung Agaricales. Ph.D. dissertation. Kaiserslautern, Germany: Universität Kaiserslautern; 1995. [Google Scholar]
- 16.Heinzkill M, Messner K. The ligninolytic system of fungi. In: Anke T, editor. Fungal biotechnology. Weinheim, Germany: Chapman & Hall; 1997. pp. 213–227. [Google Scholar]
- 17.Higuchi T. Mechanisms of lignin degradation by lignin peroxidase and laccase of white-rot fungi. Am Chem Soc Symp Ser. 1989;399:482–502. [Google Scholar]
- 18.Higuchi T. Lignin biochemistry: biosynthesis and biodegradation. Wood Sci Technol. 1990;24:23–63. [Google Scholar]
- 19.Johannson T, Nyman P O. Isoenzymes of lignin peroxidase and manganese(II) peroxidase from the white-rot basidiomycete Trametes versicolor. I. Isolation of enzyme forms and characterization of physical and catalytic properties. Arch Biochem Biophys. 1993;300:49–56. doi: 10.1006/abbi.1993.1007. [DOI] [PubMed] [Google Scholar]
- 20.Kersten P J, Kalyanaraman B, Hammel K E, Reinhammer B, Kirk T K. Comparison of lignin peroxidase, horseradish peroxidase and laccase in the oxidation of methoxybenzenes. Biochem J. 1990;268:475–480. doi: 10.1042/bj2680475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kojiama Y, Tsukuda Y, Kawai Y, Tsukamoto A, Suguiura J, Sakaino M, Kita Y. Cloning, sequence analysis and expression of ligninolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J Biol Chem. 1990;265:15224–15230. [PubMed] [Google Scholar]
- 22.Kunishima N, Fukuyama K, Matsubara H, Hatanaka H, Shibano Y, Amachi T. Crystal structure of the fungal peroxidase from Arthromyces ramosus at 1.9 Å resolution. J Mol Biol. 1994;235:331–344. doi: 10.1016/s0022-2836(05)80037-3. [DOI] [PubMed] [Google Scholar]
- 23.Lineweaver H, Burk D. The determination of enzyme dissociation constants. J Am Chem Soc. 1934;56:658–660. [Google Scholar]
- 24.Lobos S, Larrain J, Salas L, Cullen D, Vicuña R. Isozymes of manganese-dependent peroxidase and laccase produced by the lignin-degrading basidiomycete Ceriporiopsis subvermispora. Microbiology. 1994;140:2691–2698. doi: 10.1099/00221287-140-10-2691. [DOI] [PubMed] [Google Scholar]
- 25.Maltseva O V. Ligninolytic enzymes of the white-rot fungus Panus tigrinus. Biotechnol Appl Biochem. 1991;13:291–302. [Google Scholar]
- 26.Messner K, Srebotnik E. Biopulping: an overview of developments in an environmentally safe paper-making technology. FEMS Microbiol Rev. 1994;13:351–364. [Google Scholar]
- 27.Milstein O, Hüttermann A, Fründ R, Lüdemann H-D. Enzymatic co-polymerization of lignin with low molecular mass compounds. Appl Microbiol Biotechnol. 1994;40:760–767. [Google Scholar]
- 28.Morita Y, Yamashita H, Mikami B, Iwamoto H, Aibara S, Terada M, Minami J. Purification, crystallization and characterization of peroxidase from Coprinus cinereus. J Biochem. 1988;103:693–699. doi: 10.1093/oxfordjournals.jbchem.a122331. [DOI] [PubMed] [Google Scholar]
- 29.Moroshi N. Laccases from the ligninolytic fungus Coriolus versicolor. Am Chem Soc Symp Ser. 1991;460:204–207. [Google Scholar]
- 30.Moser M. Die Röhrlinge und Blätterpilze. Kleine Kryptogamenflora, Band IIb/2. Stuttgart, Germany: Gustav Fischer Verlag; 1983. [Google Scholar]
- 31.Muheim A, Leisola M S A, Schoemaker H E. Aryl-alcohol oxidase and lignin peroxidase from the white-rot fungus Bjerkandera adusta. J Biotechnol. 1990;13:159–168. [Google Scholar]
- 32.Palmieri G, Giardina P, Marzullu L, Desiderio B, Nitti G, Cannio R, Sannia G. Stability and activity of a phenol oxidase from the ligninolytic fungus Pleurotus ostreatus. Appl Microbiol Biotechnol. 1995;39:632–636. doi: 10.1007/BF00205066. [DOI] [PubMed] [Google Scholar]
- 33.Pütter J, Becker R. Peroxidases. In: Bergmeyer J, Gsaßl M, editors. Methods of enzymatic analysis. 3rd ed. III. Enzymes 1: oxidoreductases, transferases. Weinheim, Germany: Verlag Chemie GmbH; 1983. pp. 286–293. [Google Scholar]
- 34.Salas C, Lobos S, Larraín J, Salas L, Cullen D, Vicuña R. Properties of laccase isoenzymes produced by the basidiomycete Ceriporiopsis subvermispora. Biotechnol Appl Biochem. 1995;21:323–333. [PubMed] [Google Scholar]
- 35.Saloheimo M, Niku-Paavola M L, Knowles J K C. Isolation and structural analysis of the laccase gene from the lignin-degrading fungus Phlebia radiata. J Gen Microbiol. 1991;137:1537–1544. doi: 10.1099/00221287-137-7-1537. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sayadi S, Ellouz R. Roles of lignin peroxidase and manganese peroxidase from Phanerochaete chrysosporium in the decolorization of olive mill wastewaters. Appl Environ Microbiol. 1995;61:1098–1103. doi: 10.1128/aem.61.3.1098-1103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singer R. The Agaricales in modern taxonomy. Koenigstein, Germany: Koeltz Scientific Books; 1986. [Google Scholar]
- 39.Sugumaran M. A caution about the azide inhibition of enzymes associated with electrophilic metabolites. Biochem Biophys Res Commun. 1995;212:834–839. doi: 10.1006/bbrc.1995.2044. [DOI] [PubMed] [Google Scholar]
- 40.Thurston F. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- 41.Tsujimura H, Takaja M, Katano K, Matsumoto N, Park Y S, Okabe M. Peroxidase production by carbon and nitrogen sources feed-batch culture of Arthromyces ramosus. Biotechnol Lett. 1994;16:575–580. [Google Scholar]
- 42.Tuor U, Winterhalter K, Fiechter A. Enzymes of white-rot fungi involved in lignin degradation and ecological determinants for wood decay. J Biotechnol. 1995;41:1–17. [Google Scholar]
- 43.Urzúa U, Larrondo L, Lobos S, Larraín J, Vicuña R. Oxidation reactions catalyzed by manganese peroxidase isoenzymes from Ceriporiopsis subvermispora. FEBS Lett. 1995;371:132–136. doi: 10.1016/0014-5793(95)00874-9. [DOI] [PubMed] [Google Scholar]
- 44.Wahleithner J A, Xu F, Brown K M, Brown S H, Golightly E J, Halkier T, Kauppinen S, Pedersen A, Schneider P. The identification and characterization of 4 laccases from the plant-pathogenic fungus Rhizoctonia solani. Curr Genet. 1996;29:395–403. doi: 10.1007/BF02208621. [DOI] [PubMed] [Google Scholar]
- 45.Wolfenden B S, Willson R L. Radical cations as reference chromogens in kinetic studies of one-electron transfer reactions: pulse radiolysis studies of 2,2′-azinobis(-3-ethylbenz-thiazoline-6-sulphonate) J Chem Soc Perkin Trans II. 1982;1982:805–812. [Google Scholar]
- 46.Yaropolov A I, Skorobogat′ko O V, Vartanov S S, Varfolomeyev S D. Laccase: properties, catalytic mechanism and applicability. Appl Biochem Biotechnol. 1994;49:257–279. [Google Scholar]
- 47.Yaver D S, Xu F, Golightly E J, Brown K M, Brown S H, Rey M W, Schneider P, Halkier T, Mondorf K, Dalbøge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white-rot basidiomycete Trametes villosa. Appl Environ Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youn H-D, Kim K-J, Maeng J-S, Han Y-H, Jeong I-B, Jeong G, Kang S-O, Hah Y C. Single electron transfer by an extracellular laccase from the white-rot fungus Pleurotus ostreatus. Microbiology. 1995;141:393–398. doi: 10.1099/13500872-141-2-393. [DOI] [PubMed] [Google Scholar]