Abstract
Early detection of breast cancer is crucial for a better prognosis. Various studies have been conducted where tumor lesions are detected and localized on images. This is a narrative review where the studies reviewed are related to five different image modalities: histopathological, mammogram, magnetic resonance imaging (MRI), ultrasound, and computed tomography (CT) images, making it different from other review studies where fewer image modalities are reviewed. The goal is to have the necessary information, such as pre-processing techniques and CNN-based diagnosis techniques for the five modalities, readily available in one place for future studies. Each modality has pros and cons, such as mammograms might give a high false positive rate for radiographically dense breasts, while ultrasounds with low soft tissue contrast result in early-stage false detection, and MRI provides a three-dimensional volumetric image, but it is expensive and cannot be used as a routine test. Various studies were manually reviewed using particular inclusion and exclusion criteria; as a result, 91 recent studies that classify and detect tumor lesions on breast cancer images from 2017 to 2022 related to the five image modalities were included. For histopathological images, the maximum accuracy achieved was around 99 , and the maximum sensitivity achieved was 97.29 by using DenseNet, ResNet34, and ResNet50 architecture. For mammogram images, the maximum accuracy achieved was 96.52 using a customized CNN architecture. For MRI, the maximum accuracy achieved was 98.33 using customized CNN architecture. For ultrasound, the maximum accuracy achieved was around 99 by using DarkNet-53, ResNet-50, G-CNN, and VGG. For CT, the maximum sensitivity achieved was 96 by using Xception architecture. Histopathological and ultrasound images achieved higher accuracy of around 99 by using ResNet34, ResNet50, DarkNet-53, G-CNN, and VGG compared to other modalities for either of the following reasons: use of pre-trained architectures with pre-processing techniques, use of modified architectures with pre-processing techniques, use of two-stage CNN, and higher number of studies available for Artificial Intelligence (AI)/machine learning (ML) researchers to reference. One of the gaps we found is that only a single image modality is used for CNN-based diagnosis; in the future, a multiple image modality approach can be used to design a CNN architecture with higher accuracy.
Keywords: Breast cancer diagnosis, Convolutional Neural Network (CNN), Medical image analysis, Deep learning, Machine learning
Introduction
Breast cancer is the most common cancer among women; it had the highest incident rate of 126.9 (per 100,000) for the year 2014–2018 in the USA, compared to other cancers in women [1]. According to the American Cancer Society, there will be 290,560 new cases of breast cancer in both sexes which is the second-highest of all cancer cases [1]. There are various ways of diagnosing and localizing breast cancer, such as histopathological images, mammograms, magnetic resonance imaging (MRI), ultrasound images, and computed tomography (CT) scans. Histopathological images or biopsies are some of the initial screening methods that help to diagnose cancer. These are used for those at risk of having breast cancer. Biopsies are usually used to confirm a suspected site for cancer. MRI, CT scans, and ultrasounds are used at different stages of breast cancer. Experienced specialists such as pathologists or radiologists study these image modalities to detect cancer. However, due to the vast amount of details in the images, there can be cases where the specialist might miss diagnosing a tumor lesion on the image. According to a study done with histopathological images, 10.2 of cases that were diagnosed showed disagreement among the different pathologists [2]. Therefore, using a Convolutional Neural Network (CNN) to automatically detect and segment tumor lesions is very helpful in avoiding such misdiagnosis.
As an interdisciplinary research group for breast cancer detection, we have developed tools and used the electro-mechanical properties, viscoelastic parameters of formalin-fixed paraffin-embedded human skin tissue, and electrothermomechanical properties to detect the presence of cancer [3–7]. In addition, we have also investigated breast cancer diagnosis with multiple machine learning techniques such as support vector machine (SVM) and faster region-based CNN (RCNN) on the biopsied human breast tissue samples [8, 9]. This study is a narrative overview [10] that reviews studies on CNN-based detection and segmentation of breast cancer in five modalities: histopathological, CT, mammogram, ultrasound, and MRI images. The studies included in this paper are state-of-the-art research trends related to different image modalities. To the best of our knowledge, this review paper is the first to include five of the most common image modalities used for breast cancer diagnosis. The motivation behind this study is to have a comparative analysis of the recent research trends related to five commonly used image modalities for breast cancer diagnoses in one place. The study focuses on determining some of the best pre-processing methods and architectures combinations for each of the five modalities and whether some standard techniques and architectures can be used effectively for all mentioned image modalities. The comparison includes challenges related to these image types, the pre-processing techniques, and computational diagnosis using CNN. This makes all the necessary information, from accessing the data to developing a model for the five image modalities, readily available for future studies. The study reflects that though a similar structure has been adopted for the development and intelligent detection of tumors in the image modalities, each modality has supremacy based on the cancer formation stage (Table 7). Summarizing the state-of-the-art CNNs for breast cancer diagnosis in medical images commonly used in clinics will promote interdisciplinary approaches and collaborations among researchers in medicine, computer science, and engineering for more accurate computer-aided decision-making processes in breast cancer diagnosis. The contributions of this study are (1) a summary of challenges of the five image modalities; (2) a review of state-of-the-art CNN-based studies for breast cancer diagnosis with a summary of data sources, pre-processing techniques, and CNN architectures; and (3) envisioning future direction for breast cancer diagnosis in the image modalities.
Table 7.
Image modalities summarized
| Image modality | Supremacy | Popular CNN-based architectures |
|---|---|---|
| Histopathological | 1. Customized existing architectures for improved efficiency | Customized CNN, ALexNet, GoogleNet, ResNet, DenseNet 121 |
| 2. Patch sampling strategy using CNN and K-means [31] | ||
| 3. Fine-tuned pre-trained architectures | ||
| Mammogram | 1. Customized existing architectures for improved efficiency | Customized CNN, AlexNet, VGG, ResNet101, Inception v3 |
| 2. Use of block-based CNN [52], Class activation Map [62] and fused feature set [56] | ||
| 3. Classification of both ROI patches and whole images | ||
| MRI | 1. Used to detect ALN metastasis [75], predict breast cancer molecular sub-type [70, 82] and tumor lesions | Customized CNN, SegNet, UNet, VGG19, ResNet18 |
| 2. Machine learning classifier used in conjunction with CNN [72] | ||
| 3. Use of high dimensional feature map for increased efficiency [83] | ||
| Ultrasound | 1.US is radiation-free, non-invasive, real-time, well-tolerated by women [100]. | ImageNet, AlexNet, VGGNet, CaffeNet, MobileNet, ZFNet, ResNet, DarkNet-53, Inception-v3, InceptionResNet, Xception, DenseNet. |
| 2. Imaging can be performed with Different angular orientations [136]. | ||
| 3. For dense breasts where mammography shows decreased diagnostic accuracy, US is considered as a more accurate imaging modality for detecting breast cancer [137]. | ||
| CT | 1. PET/CT can detect lymph nodes and distant metastases, axial, and internal mammary nodes [138]. | VGGNet, U-Net, MobileNet, ResNet, DarkNet-53, Inception-v3, InceptionResNet |
| 2. When traditional imaging methods are equivocal, PET/CT has been recommended in asymptomatic breast cancer patients because it has high sensitivity and specificity to detect loco-regional recurrence [139]. | ||
| 3. Breast CT acquires images views over 360 for each breast in one scan, thus reducing the radiation dose [140]. |
Challenges to Overcome
There are many challenges related to breast cancer diagnosis in the image modalities. Though expert radiologists can detect tumors with some accuracy using their experience, their expertise is perishable with time. There is also variability in radiologists’ observations that can impede accurate breast cancer diagnosis [11–14]. Therefore, computer-assisted intelligent techniques are necessary for better diagnosis, though applying AI technology in medical applications such as cancer detection has limitations to overcome. However, once a successful AI framework is developed, it can diagnose multiple types of diseases in less time with decent efficiency, providing additional assistance to radiologists. Therefore, in the case of cancer diagnosis, opportunities in CNN-based approaches are considered highly promising.
Functional CNN models deal with a large number of learnable parameters, requiring extensive data sets to train for an optimum outcome. But there are different challenges to data acquisition and processing of the various available image modalities. Depending on patient physiology, many false positive data points may occur in diagnosis. In radiographically dense breasts, mammography gives a high false positive rate of breast cancer [15]. On the other hand, ultrasound using a non-invasive acoustic pressure, with low soft tissue contrast, results in early-stage false detection [16]. MRI can provide a three-dimensional volumetric image but is time-consuming, expensive, and unsuitable for routine tests. PET/CT has high sensitivity but has some limitations for low proliferative tumors.
There is a series of steps that have been adopted in previous studies to diagnose breast cancer in different types of image modalities. First, the images are acquired from an open-source or retrospective study. Next, data pre-processing techniques are used on the images for enhancement purposes and to reduce reluctant falsifications, preparing the data for efficient tumor detection, localization, or segmentation [17]. These techniques include operations such as geometric transformations [18, 19], pixel brightness transformations [20], image filtering [21], and Fourier transform and image restoration [22]. Most of the time, data augmentation techniques are applied to expand the training data, this contributes to the efficiency of the trained model. Data augmentation involves creating a copy of the image and using rotation, flipping, scaling, padding, translation, affine transformation, or cropping on the same [23]. Once the images are pre-processed and augmented, the images are fed to the designed CNN or a pre-trained CNN in the case of transfer learning. Transfer learning is used when the data set is not large enough to train the model well; therefore, knowledge is extracted from one or more pre-trained models for detection or segmentation purposes. In the next section, we have reviewed the pre-processing techniques, data augmentation techniques, CNN models used, and application of transfer learning for histopathological, mammogram, MRI, ultrasound, and computed tomography images.
Methods
The databases used for the search of studies were Google Scholar and ScienceDirect Elsevier. The databases were searched using the terms ‘breast cancer diagnosis using CNN’, ‘breast cancer diagnosis’, ‘cancer diagnosis using CNN’, ‘breast cancer diagnosis in histopathological images’, ‘breast cancer diagnosis in histopathological images using CNN’, ‘breast cancer diagnosis in mammogram images’, ‘breast cancer diagnosis in mammogram images using CNN’, ‘breast cancer diagnosis in MRI’, ‘breast cancer diagnosis in MRI using CNN’, ‘breast cancer diagnosis in Ultrasound images’, ‘breast cancer diagnosis in Ultrasound images using CNN’, ‘breast cancer diagnosis in CT images’, and ‘breast cancer diagnosis in CT images using CNN’ from 2021 through April 2022. We used some search phrases without the term ‘CNN’ to avoid excluding papers that do not have the exact term but might refer to the CNN architecture with different names. The criteria for inclusion were that they should be published in English between the period starting from 2017 to 2022 (April), should be related to CNN-based breast cancer diagnosis in histopathological, mammogram, MRI, ultrasound, or CT images, and include quantitative evaluation of the CNN architecture as we wanted to have quantitative comparison based on the model performance. Studies that belonged to any of the following categories were excluded: published in a different language, related to a different type of cancer other than breast cancer, did not include CNN as a cancer detection technique, lacked quantitative evaluation, related to image modalities other than histopathological, mammogram, MRI, ultrasound, and CT, and published before 2017 as we wanted to include only recent studies. The studies were manually reviewed using the inclusion and exclusion criteria mentioned above. After the screening, 91 studies were included for review (Fig. 1). The studies are grouped into different image modalities, each image modality section is categorized into pre-processing techniques used, studies related to CNN architectures built from scratch, studies associated with CNN architectures using transfer learning, and studies that compare the performance of different CNN architectures. Each image modality consists of a summary table where we have included most of the studies with higher accuracies and mentioned important information related to them, such as the dataset used, pre-processing techniques, image size, comparison models, the novel technique used, CNN architecture, and performance.
Fig. 1.
PRISMA flow diagram of review strategy
Histopathological Images
Histopathological slides are obtained from biopsy procedures and are one of the most comprehensive image modalities to diagnose cancer [24]. However, due to the number of details in histopathological images, there can be cases when tumor lesions are not correctly diagnosed. According to one study, approximately 10.2 of cases that were diagnosed showed disagreement [2]. There can be several reasons for pathologists’ observation variability, such as experience and human error. Therefore, plenty of automation studies related to cancer diagnosis in histopathological images have been conducted to assist pathologists and reduce misdiagnosis.
Pre-Processing and Data Augmentation Techniques
The most common stain used to stain histopathological images is the hematoxylin and eosin (H &E) stain; the stain binds differently with different biological structures of the tissue. Hematoxylin is bluish-purple and binds strongly with the nuclei, and eosin, a red-pink dye, binds with the proteins in cytoplasm [25]. Because of the difference in stains, a lot of studies perform stain normalization on the images [9, 26–32]. There are various methods used for stain normalization, such as [25, 33–36]. Another pre-processing technique is based on the lightness component called Contrast Limited Adaptive Histogram Equalization (CLAHE); this is not a common technique to be used for histopathological images, but Wang et al. used it as a pre-processing technique for their study [32]. Togacar et al. used random brightness contrast in their study [37]. All these techniques help CNN to learn the features better. Another technique that helps CNN identify the features is reducing the size of the images and creating patches. This technique increases the number of images and helps the CNN train faster and reduce overfitting. There are several studies such as [9, 26, 27, 30–32, 38–42] which have used this technique. In our previous work, full-size images were stain normalized and divided into patches, and the effect of these techniques on the performance of the model was compared [9]. Senan et al. used bilinear interpolation to resize images to 224224 from the original 460700 size [27]. Gupta and Chawla rescaled the images by using Python/TensorFlow-Keras ImageDataGenerator class [43] and converted the scale of pixels from [0, 255] to [0, 1] [39]. Li et al. extracted patches using the sliding window mechanism. Some patches were non-overlapping of size 128128 to contain cell-related characteristics, and others were of size 512512 with 50 overlap to contain continuous morphological information [31]. Wahab et al. divided the images into patches of size 512512 with an overlap of 80 pixels to avoid the exclusion of mitotic nuclei [41]. Vesal et al. created patches of size 512512 with 50 overlap similar to [32, 34] to avoid exclusion of class information related to the image [42]. Data augmentation techniques help to generate more data from the existing datasets. Different studies such as [26, 27, 29, 37, 41, 42] used a combination of rotation by 90, 180, 270, horizontal flipping, and vertical flipping.
Customized CNN and Comparison Studies
Some studies created CNN models from scratch, some fine-tuned existing ones, and others used machine learning models in conjunction with CNN. Some studies are direct applications without any significant change in the data input; others compare different models and use various data augmentation techniques to get a better result. Li et al. extracted features at the cell- and tissue-level from smaller and larger size patches to classify them using a CNN architecture. The dataset used was from the bioimaging 2015 breast histology classification challenge with images having a magnification of 200. The images were stain-normalized, and a patch sampling strategy was proposed to get patches with discriminative features that contain cell- and tissue-level features; this method was based on CNN and K-means algorithm. The classification framework extracts features from patches and computes features for the whole image so that it can be classified from the classifier. The accuracy of the overall method was 88.89 [31]. In one of the studies, co-FCN and UNet were used to segment the tumor lesions separately, and their results were compared [44]. In another study, the cGan model is used to segment tumor lesions in different organs, and the input fed into the model is a synthetic histopathological image dataset [45]. A dictionary of different size nuclei was created and used to generate the synthetic annotated dataset. The synthetic data with the original dataset is used as training data to train the cGan model for nuclei segmentation. In another study, Wang et al. enhanced the basic Efficientnet model to obtain a boosted Efficientnet model [46]. The boosted efficient net model is then compared with other models such as ResNet-50, DenseNet-121, and basic EfficientNet. Boosted EfficientNet performed the best among all other CNNs. The authors also introduced a novel data augmentation technique Random Center Cropping. This technique was used in combination with the Reduced Downsampling Scale (RDS), Feature Fusion, and Attention feature; all these techniques improved the result of the boosted Efficientnet model.
In one study, the authors used random forest, SVN, eXtreme Gradient Boost, and multi-layer perceptron to classify the histopathological images based on some textured features [47]. The authors used the phylogenetic diversity index to extract characteristics from the images. The index gives information related to the gray levels of the image, the number of pixels in these different gray levels, and the number of edges between these different gray levels. The indices are used to describe each image, and the classifier uses these parameters to classify the image into one of the four categories. Different experiments were run to distinguish between normal and abnormal, benign and malignant, invasive and in situ carcinoma. SVM performed the best in all experiments with 92.5 to 100 accuracy. In another study, different models of CNNs with different classification layers such as the Logistic Regression layer, K-NN layer, and SVM layer were used [48]. The CNN model with SVM as the classification layer performed the best with 99.84 accuracy. In the study conducted by Gupta et al., the residual neural network was modified to detect breast cancer and classify it into benign and malignant. The images were pre-processed using resizing, zooming, rotation, random flip, and vertical flip. And the network was trained using cyclic learning rates, test time augmentation, stochastic gradient descent, and discriminative layer learning. Even though the model was trained with 40 magnification images, it performed well for 100, 200, and 400. The model was compared with other existing studies and resulted in the best performance with average classification accuracy obtained was 99.75 , the precision of 99.18 , and recall of 99.37 [49].
Transfer Learning
Many studies use transfer learning to use existing weights and solve the problem of fewer data. Transfer learning is a technique in which a previously trained CNN on a big dataset is used for a new problem. Wahab et al. used a pre-trained CNN on the ImageNet dataset to perform segmentation and output segmented patches. These patches were then fed as input to the hybrid CNN to classify mitotic and non-mitotic nuclei. By using the pre-trained model, the class imbalance problem was reduced from 1:61 to 1:12. The proposed measure resulted in an F-Measure of 0.71 compared to the other existing models with 0.68 as F-Measure [41]. Senan et al. proposed a method to extract deep features and diagnose breast cancer as benign or malignant based on AlexNet architecture. Experiments were performed based on different image magnification values (Fig. 2) [27]. The pre-trained AlexNet initially trained on ImageNet was fine-tuned so that the last layer of the model can classify only two classes, malignant and benign. The accuracy of the fine-tuned model was 95 .
Fig. 2.
Magnification factors (a) 40, (b) 100, (c) 200, and (d) 400. Reprinted with permissions from [27]
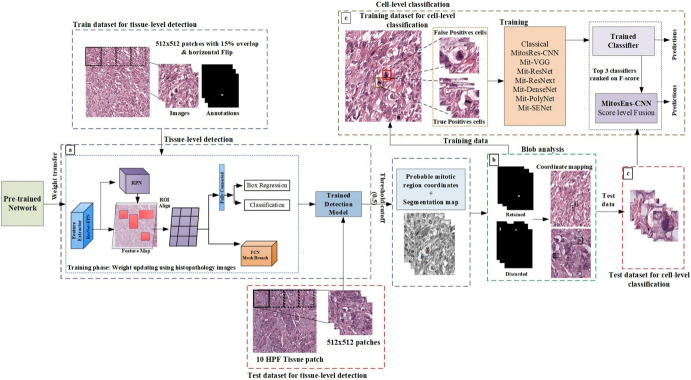
Sohail et al. proposed a framework for multi-phase mitosis detection named MP-MitDet to identify the mitotic nuclei [28]. There were different phases related to the framework; the first was to refine the weak annotated labels. This pre-trained Mask Region-based CNN (RCNN) trained on MITOS12 was fine-tuned to label the images at the pixel level. Next, Mask RCNN’s multi-objective loss function was used for region selection at the tissue level. The predictions made by Mask RCNN were filtered based on a threshold cut-off of 0.5. The selected mitotic nuclei were considered as blobs, and the blobs with an area of more than 600 pixels were retained. Mitos-Res-CNN was proposed to reduce false positives and enhance mitosis detection at the cell level. The full method is displayed in Fig. 3. The proposed system performed the best with an F-Score of 0.75, a precision of 0.734, and a recall of 0.768. These studies are summarized in Table 1.
Fig. 3.
An example for multi-phase mitosis detection: (a) Mitotic region selection at tissue-level using multi-objective deep instance-based detection and segmentation model. (b) Blob analysis. (c) Enhancement using cell-level classification. Reprinted with permission from [28]
Table 1.
Histopathological studies summarized
| Pre-processing | Size of images | Model used | Novel technique | Performance | Dataset used | Ref., year |
|---|---|---|---|---|---|---|
| Reinhard et al. method RGB channel processing [50] | 128 128, 512 512 | CNN from scratch | Patch sampling strategy using CNN and K-means | Accuracy (acc) 88.89 overall | Bioimaging 2015 breast histology classification challenge | [31], 2019 |
| 2040 1536 | SVM with Xception, ResNet, NasNet, VGG19 architectures | Feature extraction using phylogenetic diverse indexes, and classification of normal, abnormal, benign, malignant, in-situ | Acc (NasNet for classifying normal vs abnormal) 75 , precision 56 , acc (VGG19 for classifying benign vs malignant) 72 , precision 51 | BACH 2018 | [47], 2020 | |
| Image resizing, zooming, rotation, random flip, vertical flip | 230 230, 460 460 | ResNet34, ResNet50 | Modified ResNet for classification | Acc 99.75 , precision 99.18 , recall 99.37 | BreakHis | [49], 2021 |
| Cropping, rotation, flip | 512 512, 80 80 | TL-Mit-Seg | Hybrid-CNN with transferred weights for classification of mitosis | Precision 0.772, recall 0.663, F-measure 0.713 | TUPAC16, MITOS12, MITOS14 | [41], 2019 |
| Translation, scaling, rotation, flipping, bilinear interpolation (image resized), stain normalization | 224 224 | AlexNet | Customized CNN to extract deep features for benign and malignant classification | Acc 95 , SN 97 , SP 90 , AUC 99.36 | BreakHis | [27]. 2021 |
| Image normalization, rotation, reflection | 64 64 | CNN + LSTM | Combination of CNN and LSTM for classification and nuclear atypic grading | Acc 86.67 , SP 0.9278, F-Score 0.8663 | MITOS-ATYPIA-14 | [26], 2021 |
| Image normalization [33] | 512 512 | AlexNet | Custom CNN classifier for label refiner, tissue-level mitotic region selection, blob analysis and cell level refinement | F-Score 0.75, recall 0.76, precision 0.71 | TUPAC16, MITOS12, MITOS14 | [28], 2021 |
| Python/keras. preprocessing library | 144 96 | ResNet50 | Pre-trained ResNet50 to classify benign and malignant tissue | Acc 99.10 | BreakHis | [51], 2020 |
| Patches, image normalization, rotation, flip | 224 224 | Pre-trained DenseNet | finetuned DenseNet-121 to classify lymph node metastasis | Acc 97.96 , AUC 99.68 , SN 97.29 , SP 97.65 | BreakHis | [46], 2021 |
| Flip, rotation, contrast enhancement | 224 224, 227 227 | BreastNet | BreasNet using hypercolumn technique, convolutional, pooling, residual and dense blocks for classification | Acc 98.80 , F-Score 98.59 | BreakHis | [37], 2020 |
Mammogram Images
Mammograms are used as an early source of breast cancer diagnosis. There are different types of mammogram images. Digital mammogram images are created by taking a single image, whereas digitized images are generated to get a 3D view of the breast through 3D mammography. Mammogram images are grayscale images (single-channel images), unlike histopathological images with three channels (i.e., RGB).
Pre-Processing and Data Augmentation Techniques
Mammogram images have mostly high-intensity or low-intensity visual information. Some of the reasons that make mass detection difficult for CNNs are that mammogram images lack high quality, they have irregular shapes of masses, the mass size varies a lot, and sometimes it is difficult to distinguish between the dense region and the mass region [52]. Variation of mammogram density is one of the reasons for misdiagnosis [53]; therefore, image enhancement before feeding the images to CNN models is essential. Studies have used various pre-processing and data augmentation techniques with CNNs to classify and segment the tumor lesions on mammogram images.
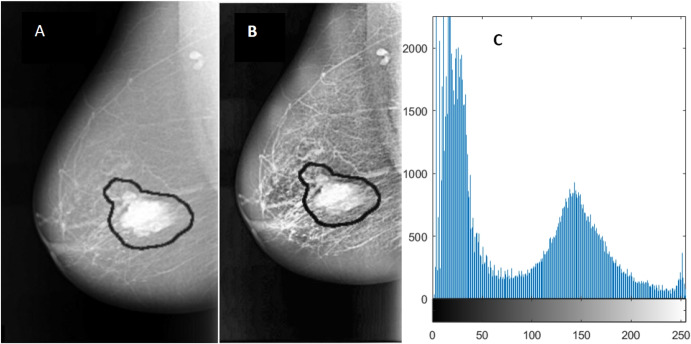
Due to variations in intensity, the small region of tumor masses, and the presence of pectoral muscle, it is usually difficult to use the whole image as an input to CNNs for tumor detection. Before feeding the mammogram images to CNN, it is important that the images are pre-processed, and data augmentation techniques are applied to them. Also, generating more data using data augmentation techniques helps solve the problem of scarce data. In one study, Feature Wise Data Augmentation technique is used on images of size 1024 1024. The region of interest (ROI) is extracted and labeled. Image patches are created and then rotated clockwise to angles 90, 180, 270, and 360. After each patch is rotated, it is flipped vertically. The data augmentation results in 8 patches, resulting in more data and smaller image size. The final size of the image obtained is 128 128 [54]. There is another study on Contrast-Limited Adaptive Histogram Equalization (CLAHE) technique for image enhancement. This technique enhances the image without enhancing the noise in the image. It does not result in noise enhancement because it uses a clip level for each pixel which restricts the contrast enhancement (see Fig. 4) [55]. In one study, Gaussian filter was applied to remove image noise, and the image was downscaled to 16, resulting in an image size of 64 64. Each of downscaled images was flipped horizontally (reflection operation), and both downscaled and reflected images were rotated by different angles such as 90, 180, and 270, giving a total of 2576 images [56]. In another study, sliding window approach is used to scan the whole breast and extract possible patches while controlling the minimum overlap between the two consecutive patches. All patches are classified based on the annotations provided with the dataset. If the central pixel of the patch lies inside the mass, it is considered positive. All positive patches are extracted for CBIS-DDSM data, as no negative images are present. Then, an equal number of negative patches are randomly selected to balance the data. For INbreast data, positive patches are extracted only from positive images, and an equal number of negative patches from the normal images [57]. Houby et al. used MIAS and INbreast for their study. The two datasets have different formats of images; therefore, all images were converted to png format. A median filter was used to remove noise and preserve image edges. Images were then converted to binary images using the Otsu thresholding method to remove labels. Images were then sharpened to remove edges. CLAHE was used as an image enhancement method. ROI was cropped in the form of bounding boxes by using different methods for the two databases. For data augmentation, images were rotated and flipped, and finally, all images were resized to 208 208 [58]. Tavakoli et al. used Otsu’s thresholding method [59] and modified the method to get the final thresholding to convert the images into binary format. A mask was generated, and the breast region was obtained by multiplying the mask with the original image. Finally, the breast region was extracted. As pectoral muscle has similar intensity as an abnormal mass, it is preferred to remove those. The authors used Jen and Yu’s method [60] to remove the pectoral muscle. The orientation of the breast was determined, image contrast was enhanced using gamma correction equalization, a binary image was obtained, and the candidate component at the corner of the image was eliminated. In the third step, a mask was created to distinguish between background pixels, healthy tissue pixels, and abnormal pixels with black, white, and gray colors, respectively. Finally, the image was enhanced by using CLAHE [52]. Samala et al. used Chan et al.’s background correction method to normalize the grayscale background of the images [61]. Xi et al. used image patches from the database CBIS-DDSM and tested them on full mammogram images to locate tumor masses. Rotation and random X and Y reflection were applied to the training data as data augmentation techniques [62]. Jadoon et al. used rotation at angles 90, 180, and 270 and flipping transformations as data augmentation techniques. CLAHE was used for image enhancement. Two dimensional, Discrete Wavelet Transform, Discrete curvelet, and Dense Scale Invariant Feature Transform were used for clear edges and image decomposition into different components [63]. Platania et al. removed the black area of the images by flipping right view images and trimming the black area from left to right for all images. Images were then rotated five times at random angles, and reflection across Y was performed. The image was then normalized, and rectangular boxes were created around the ROIs [64].
Fig. 4.
Image enhancement using CLAHE. A Original malignant mass, B enhanced mass using CLAHE, and C image histogram. Reprinted with permission from [55] (DOI: 10.7717/peerj.6201/fig-3)
Comparison Studies
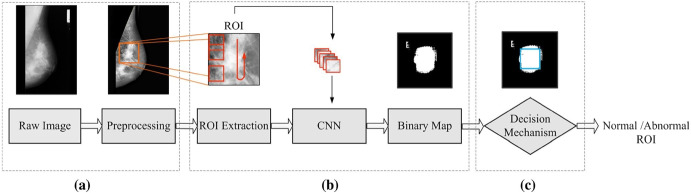
Studies have been conducted on whole mammogram images or ROI patches to locate and classify mass. In a study done by Houby and Yassin, a simple CNN was developed with small-size filters to classify breast lesions as malignant and non-malignant. The dataset used was from MIAS, DDSM, and Inbreast datasets. To solve the issue of class imbalance, more data was generated by data augmentation. Some of the techniques used were format unification, noise removal from the image, ROI extraction, augmentation, and image resizing. The performance of the developed CNN was compared with some reviewed studies. The novel CNN developed was the best among all, with a sensitivity of 96.55 , specificity of 96.49 , accuracy of 96.52 , and AUC of 98 [58]. In another study, Ragab et al. finetuned DCNN with AlexNet architecture to classify benign and mass tumors in the mammogram images. The last layer of the DCNN was replaced by SVM to get better results. Two segmentation techniques were used: the first was the global thresholding method, and the second was region-based segmentation. The dataset used was DDSM and CBIS DDSM with an image size of 227 227. The accuracy, AUC, sensitivity, specificity, precision, and F1 score for the DDSM dataset were 80.5 , 77.4 , 84.2 , 86 , and 81.5 , respectively, while the same measures for CBIS-DDSM dataset were 87.2 , 94 , 86.2 , 87.7 , 88 , and 87.1 , respectively [55]. Ting et al. developed CNNI-BCC for the classification of breast cancer using CNN for classification and an interactive detection-based lesion locator (IDBLL). The images are from the MIAS dataset, whose ground truth has been examined by a medical doctor. The large image is decomposed into small patches of size 128 128 using feature-wise data augmentation while disregarding the non-significant patches. Each patch is then rotated clockwise to 90, 180, 270, and 360 and then flipped vertically. The method’s performance is compared with previously reviewed studies. It performed the best with sensitivity of 89.47 , accuracy of 90.5 , and specificity of 90.71 [54]. Wang et al. proposed a mass detection method based on sub-domain CNN deep features and US-ELM clustering. US-ELM is a semi-supervised learning algorithm, as this can give better results with a small amount of data. First, the images were processed with noise reduction and contrast enhancement. Next, the ROI was extracted by mass segmentation so that features can be extracted properly for later steps. Deep, morphological, texture, and density features were extracted using CNN. The ROI was divided into non-overlapping sub-regions using a sliding window of size 48 48, and deep features were extracted from the ROI patches. The deep features were classified using US-ELM clustering into two categories: suspicious and non-suspicious mass areas using the fused features. The proposed method performed best with an accuracy of 86.5 [56]. Tavakoli et al. used CNN to classify breast tissue into normal and abnormal. First, some pre-processing methods were applied, such as breast region extraction, suppression of pectoral muscle, breast mask creation, and contrast enhancement. ROIs were not rescaled to maintain quality; instead, a block around each pixel was created and fed to CNN (see Fig. 5). The CNN was trained with 50 of normal blocks and 50 abnormal blocks to avoid class imbalance. Next binary map of ROI was generated, which was then used for the decision mechanism. Two methods were compared for the decision mechanism: the first thresholding technique and the second another CNN. The method with the thresholding technique performed the best with a thresholding value of 0.6, a block size of 32 32, and 94.68 accuracy [52]. Xi et al. used two different CNN models, one for localization of mass and the other for classification. When a full-size image was fed into the patch classifier, a Class Activation Map (CAM) [65] was generated that resulted in a heatmap that highlights one class. The resNet architecture was used for generating CAM. The classification CNN was trained on cropped ROI image patches and tested on the whole image [62]. Sun et al. developed semi-supervised deep CNN, which could use a large amount of unlabeled data. The dimension reduction method that performed best was MDS with an accuracy of 82.43 [66]. Jadoon et al. proposed a three-class classification model which could classify normal, benign, and malignant masses. The authors used two methods: CNN-DW (discrete wavelet transform) and CNN-CT (curvelet transform). As a part of pre-processing, images were divided into patches and treated with CLAHE contrast enhancement method. During the CNN-DW method, enhanced images were decomposed into four sub-bands by means of a two-dimensional discrete wavelet transform (2D-DWT). In the case of CNN-CT, discrete curvelet transform was used for decomposing into sub-bands. The input data matrix containing sub-band features was created as a result of the two methods, which was fed as input to the CNN. The performance of the SoftMax layer and SVM layer as classification layer were compared. SVM layer as classification performed the best with a CNN-DW accuracy of 81.83 and a CNN-CT accuracy of 83.74 [63].
Fig. 5.
Tavakoli et al.’s proposed method: (a) Pre-processing, (b) CNN pixel classification, and (c) label assignment. Reprinted by permission from Springer Nature Customer Service Centre GmbH: Springer Nature Journal of Ambient Intelligence and Humanized Computing [52]
Transfer Learning
Some studies have used transfer learning as part of the proposed framework or as the base model. Agarwal et al. analyzed the performance of VGG16, ResNet-50, and Inception V3 for mass and non-mass classification and used the best architecture to show domain adaption between the CBIS-DDSM digitized dataset and INbreast digital dataset using different CNN architectures. Inception V3 performed the best, with a True Positive Rate of 0.98 in classifying mass and non-mass breast regions. This was compared with domain adaption natural images database ImageNet. A sliding window approach was used to scan the whole breast image, and ROI patches were extracted. Patches were then classified based on the annotation of the central pixel. For domain adaption from ImageNet to INbreast, CNNs were trained with 224 224 3, where three is for RGB channels; therefore, each grayscale mammogram patch (224 224 1) was replicated onto RGB channel [57]. Tsochatzidis et al. compared to the state-of-the-arts pre-trained CNN to classify benign or malignant ROIs. Two scenarios were considered: first where pre-trained weights were used for classification and second where random weights were initialized. Transfer learning scenario and ResNet-101 performed the best for the DDSM-400 database with an accuracy of 0.785, and ResNet-152 for CBIS-DDSM with an accuracy of 0.755 [67]. Platania et al. proposed a framework for automatic detection and diagnosis named BC-DROID. BCDROID was first pre-trained based on physician-defined regions of interest in mammogram images. Both detection and classification were done in a single step, and the whole mammogram image was used as an input for training. The authors adapted You Only Look Once (YOLO) [68] to identify and label ROI. The adapted YOLO gave several outputs such as the width and height of ROI, coordinates of the center of ROI, and class label vector with the probability of benign and cancerous class. These predicted values were then fitted to rue values using a second CNN. Pre-trained weights are extracted from the first CNN and used as initialization values for the second CNN in the main training process. The resulting CNN network was able to detect and classify ROI as benign or cancerous with a detection accuracy of 90 and classification accuracy of 93.5 [64]. These studies are summarized in Table 2.
Table 2.
Mammogram studies summarized
| Pre-processing technique | Size of images | Compared with | Model used | Novel technique | Performance | Dataset used | Ref., year |
|---|---|---|---|---|---|---|---|
| Conversion to same image format, Otsu thresholding [59], noise removal, image sharpening, CLAHE for image enhancement, ROI cropping, image resizing, rotation, flip | 208208 | Previous studies | Customized CNN | Customized CNN to classify both ROI patches and whole images | Acc 96.52 , SN 96.55 , SP 96.49 , AUC 0.98 | MIAS, DDSM, INbreast | [58], 2021 |
| Image enhancement using CLAHE, rotation | 227227 | AlexNet + SVM | Pre-trained AlexNet customized to classify 2 classes | Acc 87.2 , AUC 0.94 | DDSM, CBIS DDSM | [55], 2019 | |
| Feature-wise data augmentation, patches, rotation, flip | 128128 | Previous studies | Customized CNN | CNNI-BCC: feature wise pre-processing + CNN classification + interactive lesion detector | SN 89.47, SP 90.71, acc 90.5, AUC 0.90 | MIAS | [54], 2019 |
| Noise reduction, contrast enhancement | 4848 | CNN + ELM | Fused feature set using CNN + fused feature set used by ELM for classification | SN 85.1, SP 88.02, acc (benign) 88.5, acc (malignant) 84.5, AUC 0.923 | [56], 2019 | ||
| Breast region extraction, pectoral muscle suppression, breast mask creation, contrast enhancement, block creation, | Previous studies | Customized CNN | Block-based CNN and decision mechanism | AUC 0.95, acc 94.68 , SN 93.33 , SP 95.31 | MIAS | [52], 2019 | |
| ROI patches, rotation, random X and Y flip | AlexNet, GoogleNet, ResNet, VGGNet | ResNet + VGGNet | ResNet for Class Activation Maps, VGGNet for classification | Overall acc 92.53 | CBIS-DDSM | [62], 2018 | |
| Rotation, reflection, CLAHE, 2-D DWT, Discrete Curvelet Transform, Dense Scale Invariant Feature Transform for clear edges | 128128 | Previous studies | CNN-DW (discrete wavelet), CNN-DT (curvelet transform) | CNN-DW/CNN-DT + SVM classifier | CNN-DW: acc 81.83 , SN 87.6 , SP 81.9 ; CNN-CT: acc 83.74 , SN 88.8 , SP 80.1 | IRMA | [63], 2017 |
| Patch extraction (sliding window), | 224224 | VGG16, ResNet50, InceptionV3 | InceptionV3 | Customized Inception V3 for classification and detection | Acc 88.86 | CBIS-DDSM, INbreast | [57], 2019 |
| Image format conversion, ROI extraction | 224224 | AlexNet, VGG-16, VGG-19, ResNet 50, ResNet 101,ResNet152, GoogLeNet, Inception-BN (v2) | ResNet101 | Modified pre-trained model | Acc 85.9 | DDSM-400, CBIS-DDSM | [67], 2019 |
Magnetic Resonance Imaging (MRI)
MRI is usually performed when the patient is at high risk of getting breast cancer to reduce the chance of a misdiagnosis. Another reason is to detect the proper location of the detected mass.
Pre-Processing and Data Augmentation Techniques
To get an efficient result in detecting tumors through MRI images, the image noise must be reduced, and the images must be pre-processed using different techniques. These techniques increase the quality of the images and increase the size of data, which is important as, most of the time, there is not enough data for the CNN to learn features. Adoui et al. used a bias field correction filter and annotated the tumors on the breast images. For data augmentation, techniques such as translations, rotations, scale, and flip were used [69]. Ha et al. used random affine transformation to alter mass so that the same looks different to the network, random rotation by 30, 90 across X-, Y-, and Z-axes. The techniques were only applied to 50 of the data to reduce the network’s bias towards augmented data. The noise was also introduced by using a random Gaussian noise matrix, random contrast jittering, and random brightness so that the network could learn marginalized noise [70]. Yurttakal et al. pre-processed the images by cropping tumor regions and then resizing the image to 50 50. Then, the images were normalized, and the noise was reduced using the denoise deep neural network DnCNN. For data augmentation, images were randomly translated by 3 pixels and then rotated to an angle of 20 [71]. Lesions were automatically segmented, and segmented lesions were used to obtain the ROI regions, which were used as an input to the classification process to enable ROI construction [72]. Haarburger et al. used reflection and rotation around Z-axis by 15 [73]. The tumor was segmented using fuzzy c-means clustering algorithm by subtracting the pre-contrast image from DCE postcontrast image. Square ROI was placed on the highest intensity region, and the tumor within the region was enhanced using the unsharp filter. Fuzzy c-mean was applied on all voxels belonging to the tumor in the mask. The segmented mask results were verified by experienced radiologists [74]. Ren et al. used ITK-SNAP to segment the axillary lymph nodes. A uniform 32 32 pixel bounding box was created around the center of mass. For data augmentation, flipping, rotating, and shearing were used [75]. Zhou et al. used Frangi et al.’s [76] method on 2D slices of MRI to obtain boundaries for the breast area, pectoral muscle, and breast glands. 2D binary masks were obtained by a threshold of the filtered slice, connected component analysis, and hole filling. 2D masks were stacked to obtain a 3D mask, and then the resultant mask was smoothed using a Gaussian filter. After the bounding box was obtained, the image was normalized [77]. Dalmics et al. obtained subtraction volume by using the motion-corrected postcontrast image; additionally, the relative enhancement volume was obtained by normalizing the image intensities [78]. Rasti et al. first reduced the image background using first post-contrast subtraction, enhanced the image contrast, and performed breast region cropping that helped reduce the other structures. Otsu thresholding [59] and morphological top-hat filtering [79] were also used to remove the non-lesion structure. The next step was to apply radius-based filtering to select regions within a certain range. Localized active contour (LAC) segmentation was applied using the Chan-Vese active contour model [80] to recover tumor pixels that were removed by previous operations. Compact-based filter was applied to reduce the false positives and to select the ROI based on the compactness score [81].
Direct Application of CNN and Comparison Studies
Studies have used various architectures and frameworks to segment tumor regions on images such as UNet, Seg-Net, and Mask RCNN. Some studies considered ground truth from PET scores and histopathological images.
In one study by Adoui et al., two CNN approaches were proposed based on Seg-Net and UNet architectures for tumor segmentation. The study used a dataset of 86 MRI images of 43 patients from the collaborator institute Jules Bordet Institute-Brussels, Belgium. For pre-processing operations of translation, rotation, flip and scale were applied randomly to images to obtain a greater number of images at each epoch. UNet and Seg-Net were used as the encoder-decoder architectures and tuned to create tumor masks and compared for segmentation performance. UNet had an accuracy of 76.14 and Seg-Net of 68.88 . Therefore, UNet architecture was considered the best among the two [69]. Ren et al. use CNN to detect axillary lymph node metastasis on MRI images. The dataset used is from Stony Brook Hospital patient data and consists of 66 abnormal nodes. The data is from peak contrast dynamic images taken by 1.5 Tesla MRI scanners at the pre-neoadjuvant chemotherapy stage. The ground truth was taken from the PET score and was determined by experienced readers. The model achieved an accuracy of 84.8 , ROC of 0.91, specificity of 79.3 , and sensitivity of 92.1 [75].
Yurttakal et al. designed a multi-layer CNN using one-pixel information with online data augmentation to classify lesions as malignant or benign. The dataset was from Haseki Training and Research Hospital in Istanbul, Turkey. MRI dataset consisted of 200 tumorous regions. The ground truth was created by two radiologists using the BI-RADS lesion characteristics. The MRI images were cropped to get only the tumor region and were resized to 50 50. Pixels were normalized, and then DnCNN was used to reduce the noise in the images. The resulting images were then processed randomly with operations such as translating vertically and horizontally and rotating with an angle of 20. The images were used as input for the proposed CNN model. The proposed model had an accuracy of 98.33 , a sensitivity of 1, a specificity of 0.9688, and a precision of 0.9655 [71]. Another study by Haarburger et al. emphasized that criteria other than the lesions, such as background enhancement and location within the breast, are important for diagnosis. These are difficult to capture from object detection models. To solve this problem, a 3D CNN with the architecture of ResNet-18 and a multi-scale curriculum learning strategy to classify malignancy globally on an MRI is proposed. The data consisted of 408 DCE MR images; out of those, 305 were malignant and 103 benign. Images were obtained from a local institution with 512 512 32 resolution. The proposed method is compared with Mask RCNN and Retina U-Net and has obtained an AUROC of 0.89 and an accuracy of 0.81 [73]. Ha et al. performed a study in which a CNN model with 14 layers was developed to predict the molecular subtype of breast cancer: luminal A, luminal B, and HER2+. The data of 216 patients were obtained from a conducted study that consisted of MRIs and immunohistochemical staining pathology data. The ground truth was recorded by a breast imaging radiologist. Data augmentation included random rotation by 30 and 90, and augmentation was performed only on 50 of the data to avoid bias towards augmented data. Also, random noise was introduced, and the size of inputs was 32 32 pixel bounding boxes consisting of the size normalized lesions. The customized CNN was tested with other models such as ResNet-52, Inception v4, and DenseNet. The accuracy of the customized model was 70 , class normalized ROC was 0.853, non-normalized AUC was 0.871, average sensitivity was 0.603, and average specificity was 0.958 [70].
Transfer Learning
Zhang et al. used CNN and CLSTM and re-tuned them after transfer learning to classify the three breast cancer molecular subtypes on MRI images. The three molecular subtypes classified were (HR+/HER2-), HER2+, and triple-negative (TN). The data used was from a hospital study of 244 patients. The dataset was divided into training, testing 1, and testing 2. The models were used on both testing data. For transfer learning, the pre-trained model used on training data was used as the base, and then the resulted model was trained on testing data 1 and tested on testing data 2. The accuracy of CNN after transfer learning improved from 47 to 78 , and for CLSTM, it improved from 39 to 74 [82]. Hu et al. developed a CNN model to diagnose breast cancer with the help of transfer learning and multi-parametric MRI. The dataset consisted of DCE-MRI and T2-weighted MRI for each study. A pre-trained CNN was used to extract features for these images, and SVM was used to classify the region as benign or malignant.
The CNN model was trained on the images separately as a single sequence method. For multi-parametric sequence, the two types of images, DCE-MRI and T2-weighted MRI, were integrated with image fusion, feature fusion, and classifier fusion. The feature fusion method outperformed all other single sequence and other multi-parametric sequence methods [72]. In another study, Lu et al. developed a framework to classify and segment breast cancer with data obtained by merging four imaging modes using a refined UNet architecture. The four imaging modes were T1-weighted (T1W), T2-weighted (T2W), diffusion-weighted and eTHRIVE sequences (DW1), and DCE-MRI images. The four modes of images help solve the variation in the images that lead to an inconsistent diagnosis. The data comprised 67 breast examinations which consisted of 8132 images. Out of these, 6000 images were used for classification and 1800 for segmentation. The augmentation techniques used were creating patches, random-size cropping, and flipping the images horizontally. In this study, a pre-trained CNN was used in addition to the trained CNN. A higher dimensional feature map was used, which was obtained using feature maps from the four images. For classification, CNN was compared with VGG16, ResNet-50, Inception V3, and DenseNet. For segmentation, UNet was refined and initialized using the network used for classification. The accuracy achieved was 0.942 [83]. These studies are summarized in Table 3.
Table 3.
MRI studies summarized
| Pre-processing technique | Size of images | Compared with | Model used | Novel technique | Performance | Dataset used | Ref., year |
|---|---|---|---|---|---|---|---|
| Bias field correction filter, selection of breast with tumor, translations, rotations, scale, flip | 32 32 | SegNet, UNet | UNet | Tuned UNet and SegNet for tumor segmentation | Acc (IoU) 76.14 | Private dataset, Jules Bordet institute-Brussels, Belgium | [69], 2019 |
| ITK-SNAP to segment the axillary lymph nodes, flip, rotation, shear | 32 32 | Previous Studies | Customized CNN | Detected ALN metastasis using CNN | Acc 84.8 , ROC 0.91, SN 92.1, SP 79.3 | Retrospective study, Stony Brook Hospital | [75], 2020 |
| Cropping tumor regions, resizing, image normalization, noise reduction using DnCNN, random translation, rotation | 50 50 | Other ML methods such as SVM, KNN | Customized CNN | Customized multi-layer CNN for classification of benign and malignant masses | Acc 98.33, SN 1, SP 0.9688, precision 0.9655 | Training and Research Hospital in Istanbul, Turkey | [71], 2020 |
| Reflection, rotation around z axis | 256 256 32 voxels (x, y, z) | Mask R-CNN, Retina U-Net | ResNet18 | 3D ResNet18 CNN o generate feature maps with classification head | AUROC 0.89, acc 81 | Dataset from local institution | [73], 2019 |
| Random transformation, random rotation to 50 of data, noise introduction (Gaussian noise matrix), random contrast jittering, random brightness | 32 32 | ResNet52, Inception v4, DenseNet | Customized CNN | CNN to predict breast cancer molecular subtype | ROC 0.853, AUC 0.871, SN 0.603, SP 0.958 | Retrospective study | [70], 2019 |
| Contrast enhancement maps, lesion bounding box | 32 32 | Customized CNN | CNN and CLSTM used to classify breast cancer molecular subtype with transfer learning for retuning | CNN acc 0.91 , CLSTM acc 0.83 | Private dataset from hospital | [82], 2021 | |
| Automatic segmentation of lesions and single sequence method, multiparametric methods (image fusion, feature fusion, classifier fusion) | VGG19 | Pre-trained VGG19 for feature extraction with SVM classifier | AUC (feature fusion) 0.87 | Retrospective study dataset | [72], 2020 | ||
| Cropping, flipping | VGG16, ResNet-50, Inception V3, DenseNet | UNet | Refined UNet used to classify breast cancer by using high dimensional feature map | Acc 0.942 | Data from Tianjin Center Obstetrics and Gynecology Hospital | [83], 2019 |
Ultrasound (US) Images
US image modality is most commonly used in breast cancer diagnosis. The recent development of deep learning techniques for image classification, segmentation, image synthesis, and density-varied object detection shows promising results. Eventually, it was realized that, for breast US images, the use of deep learning prediction has significant diagnostic value. Many recent studies have shown that different improved and transfer CNN applied in US images results in high accuracy of breast cancer diagnosis, comparable to experienced radiologist’s diagnosis outcomes [84–86].
For investigating Artificial Intelligence (AI) on breast cancer diagnosis accuracy, recent trends in research have followed specific steps to improve the processes. In particular, for the application of deep-learning in US image modalities, some basic approaches are to collect authentic US image data and define the dataset, apply pre-processing techniques for preparing the images as the readable inputs for the CNN model, define transfer learning CNN models, propose novel or modified CNN architecture, comparing the result with the existing AI algorithm, or to the radiologist evaluations; these are some practicing research trends so far.
Dataset
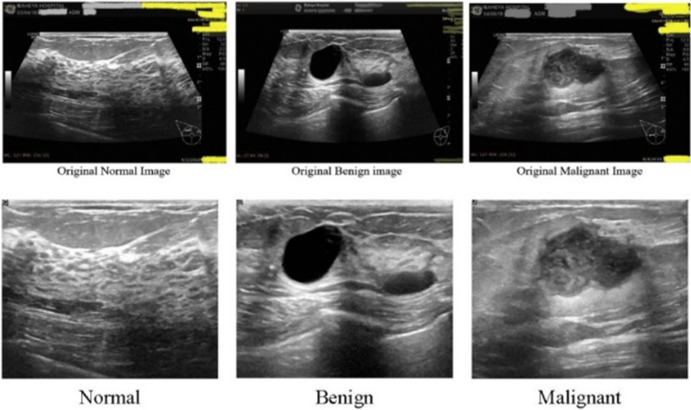
Different studies have used different sets of data to train, test, and evaluate the developed CNN architecture in deep learning research of US image modality. Some of these datasets are publicly available for use. Mendeley, Breast Ultrasound Images (BUSI), and BreakHis are some popular open-source US image datasets that have been used in many studies. Other studies have prepared unique datasets from certain investigative collection methods and clinical trials. Wilding et al. [88] used two datasets, namely, Dataset BUSI and Dataset B. This dataset B was collected in 2012 from the UDIAT Diagnostic Centre, Spain [89]. Karthik et al. [90] used the ‘Dataset of breast ultrasound images,’ collected from Baheya Hospital, Cairo, Egypt [87]. A US image of normal, benign, and malignant breast is shown in Fig. 6 from dataset of breast ultrasound images [87]. Yu et al. [91] collected data from a random remote location. Jiang et al. [92] collected data from ‘Breast Imaging Database’ at Wuhan Tongji Hospital. In another study, Lee et al. [93] collected US image data of 153 women (age range from 24 to 91 years, mean: 52.37 ± 12.26 years) from the National Taiwan University Hospital (NTUH), and Zhang et al. [94] obtained original US breast image dataset from a database of Harbin Medical University Cancer Hospital (a total of 17,226 images from 2542 patients). Wang et al. [95], Zhou et al. [96], and Huang et al. [97] have also used certain private clinical datasets for the US image-based deep learning diagnosis approach. A summary of the open-source data set is included in Table 4.
Fig. 6.
US images of normal, benign, and malignant breast. Reprinted with permission from [87]
Table 4.
Available open source data set
| Dataset name | Location | Used in |
|---|---|---|
| Mendeley | https://data.mendeley.com/datasets | [98–100] |
| Breast Ultrasound Images (BUSI) | https://scholar.cu.edu.eg/?q=afahmy/pages/dataset | [88, 101–104] |
| kaggle BUSI | https://www.kaggle.com/aryashah2k/breast-ultrasound-images-dataset | [105] |
| MT-Small-Dataset (BUSI) | https://www.kaggle.com/mohammedtgadallah/mt-small-dataset. | [98, 106] |
| BreaKHis | https://web.inf.ufpr.br/vri/databases /breast-cancer-histopathological-database-breakhis/ | [107, 108] |
Pre-Processing Techniques
When working with US images, image pre-processing techniques (such as cropping, rotation, flipping, segmentation, resampling, noise removal, and image separation) are needed to obtain an effective deep-learning analysis. Karthik et al. [90] applied basic geometric augmentation, and Zhou et al. [96] used Keras ImageDataGenerator in Python for data augmentation, which was employed for population increase and balancing of the initial dataset. Ayana et al. [98] used open-source OpenCV and scikit-image in Python for image pre-processing as binary and noise-free images. Among other studies, Wang et al. [95] used multiview CNN without any manual pre-processing step and extracted features directly. Misra et al. [109] conducted a performance comparison by testing with and without an image cropping dataset as the input. Yu et al. [91] proposed a 5GB remote e-health where the cloud center performs ROI extraction, image resolution adjustment, data normalization, and data augmentation on US image. Boumaraf et al. [110] pre-processed the dataset for subsequent stages by normalizing the stain-color of images, which method was initially proposed by Vahadane et al. [111].
For data pre-processing, some studies have used a deep CNN model and fuzzy threshold level segmentation and noise removal [106, 112]. In a deep learning study, Huang et al. [97] applied the ROI-CNN model and the G-CNN model for data augmentation.
Transfer Learning and Convolutional Neural Network
In a deep learning study, a CNN architecture can be trained from scratch when a large dataset is available, which is known as De-Novo [113]. Pre-trained CNNs trained on large datasets can also be used in some cases. In the case of US breast images, the availability of a large dataset is quite challenging. In this context, most recent studies used the transfer learning approach and a few De-Novo to develop an intelligent breast cancer diagnosis system using US images.
Transfer learning (TL) technique in US image-based breast cancer diagnosis shows promising results in many recent studies. Many popular pre-trained CNN architectures, such as DarkNet-53 [101], ResNet-50 [88, 98, 106, 108], ResNet-101 [88, 113], Inception-V3 [91, 95, 98], EfficientNetB2 [98], VGG-16 [108, 114, 115], VGG1-19 [107, 108, 114], Xception[94], AlexNet, and DenseNet [116], have been used with modification and robust optimization to develop a novel transfer learning (TL) method intending to increase the automatic diagnostic value. In a recent study, Jabeen et al. [101] used a transfer learning for breast cancer classification by applying reformed deferential evolution (RED) reformed gray wolf (RGW) optimization, which delivered a promising outcome. This TL method can be applied in multistage [98] or in a certain stage and can be optimized and fine-tuned with the augmented datasets. It is observed that diverse studies developed transfer learning (TL)-based CNNs from a single pre-trained architecture [102] or combined more than one pre-trained CNN architecture [106] to infer the best outcome.
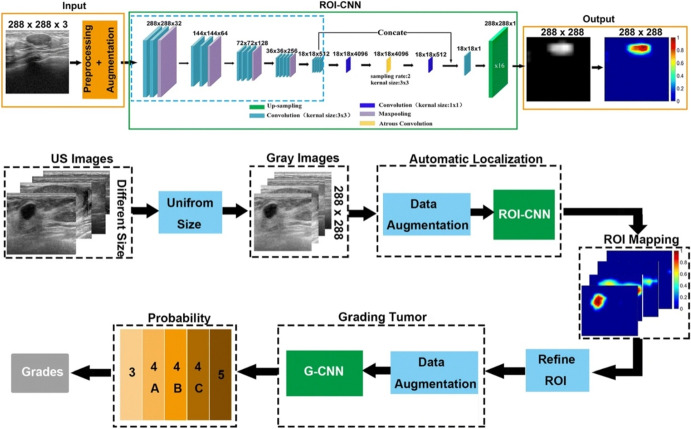
Some studies also developed new CNN architecture, mostly when they were affluent with a satisfactory large dataset or when the intention was to construct an alternative and evaluate a De-Novo CNN. Like, Karthik et al. [90] proposed a Gaussian dropout-based novel configuration of a Stacking Ensemble CNN, which was capable of classifying US images of breast tumors efficiently. Furthermore, Jiang et al. [92] train a ResNet-50 DCNN model from scratch and evaluate the performance. Also, Huang et al. [97] proposed a two-stage grading system, Breast Imaging Reporting and Data System (BI-RADS), to evaluate breast tumors from US images automatically. In Fig. 7, Huang et al.’s proposed model’s steps are shown.
Fig. 7.
Huang et al.’s proposed method of two-stage CNNs for computerized BI-RADS categorization in breast ultrasound images. Reprinted with permission from [97]
Evaluation Criteria and Comparison Study
Individual studies have utilized various evaluation metrics to evaluate the performance of machine learning models, such as accuracy, sensitivity, specificity, precision, recall, F1-score, and AUC. Furthermore, by considering these evaluating metrics, different studies utilized various approaches of comparison study to measure the impact of the proposed ML model.
Here, TP = true positive, TN = true negative, FP = false positive, and FN = false negative.
These US image-based deep learning studies usually compared the outcome to the other study’s evaluation metrics or to radiologist diagnosis outcomes to assess the significance of the studies. Other studies compare its performances based on other parallel techniques. Many recent studies developed based on deep learning utilized US breast cancer images to show equal or better diagnostic performance compared to radiologists’ diagnoses [117]. By using the Mendeley dataset, Ayana et al. [98] developed a transfer learning for US breast cancer image classification and compared it with the other models based on the Mendeley dataset, which shows its supremacy with the test accuracy of 99 ± 0.612 . In another study, Hijab et al. [115] show that the US image-based fine-tuned detector outperforms the mammogram-based fine-tuned detector. Some studies tested identical datasets with different classifiers and compared the outcomes to determine the best-fit result [106, 110]. The US studies are summarized in Table 5.
Table 5.
US studies summarized
| Pre-processing technique | Pre-trained model used | Novel technique | Performance | Ref., year |
|---|---|---|---|---|
| Data augmentation | DarkNet-53 | CNN Optimized CNN (RDE & RGW optimizer) | Acc 99.1 | [101], 2022 |
| Oversampling, data augmentation | ResNet-50; ResNet-101 | Dynamic U-Net with a ResNet-50 encoder backbone. | Acc: classifier A (normal vs abnormal) 96 & classifier B (benign vs abnormal) 85 | [88], 2022 |
| Image processing in OpenCV & scikit-image (Python) | EficientNetB2, Inception-V3, ResNet-50 | Multistage transfer learning (MSTL). optimizers: Adam, Adagrad, and stochastic gradient descent (SGD) | Acc: Mendeley dataset 99 & MT-Small-Dataset 98.7 | [98], 2022 |
| Basic geometric augmentation | – | Gaussian Dropout Based Stacked Ensemble CNN model with meta-learners | Acc 92.15%, F1-score 92.21 , precision 92.26 , recall 92.17 . | [90], 2021 |
| ROI extraction, image resolution adjustment, data normalization, data augmentation. | Inception-V3 | Fine-tuned Inception-v3 architecture | Acc 98.19 | [91], 2021 |
| – | – | Shape-Adaptive Convolutional (SAC) Operator with K-NN & Self-attention coefficient U-Net with VGG-16 & ResNet-101. | ResNet-101 mean IoU 82.15 (multi-object segmentation) and IoU 77.9 & 72.12 (Public BUSI). | [104], 2021 |
| Image resized & normalized | ResNet-101 pre-trained on RGB images. | Novel transfer learning technique based on deep representation scaling (DRS) layers. | AUC: 0.955, acc: 0.915 | [118], 2021 |
| Focal loss strategy, data augmentation, rotation, horizontal or vertical flipping, random cropping, random channel shifts. | − | ResNet-50 | Test cohort A’s acc: 80.07 to 97.02, AUC: 0.87, PPV: 93.29, MCC: 0.59. Test cohort B’s acc: 87.94 to 98.83, AUC: 0.83, PPV: 88.21, MCC: 0.79. | [92], 2021 |
| − | ImageNet-based pre-trained weights. | DNN; deepest layers were fine-tuned by minimizing the focal loss. | AUC: 0.70 ((95 CI,0.630.77; on 354 TMA samples) & 0.67 (95 CI,0.620.71; on 712 whole-slide image). | [114], 2021 |
| Enhanced by fuzzy preprocessing. | FCN-AlexNet, UNet, SegNet-VGG16, SegNetVGG19, DeepLabV3 (ResNet-18, ResNet-50, MobileNet-V2, Xception). | A scheme based on combining fuzzy logic (FL) and deep learning (DL). | Global acc: 95.45, mean IoU: 78.70, mean Boundary F1: 68.08 . | [106], 2021 |
| DL-based data augmentation & online augmentation. | Pre-trained AlexNet & ResNet | Fine-tuned ensemble CNN | Acc: 90 | [109], 2021 |
| Coordinate marking, image cutting, mark removal. | Pretrained Xception CNN | Optimized deep learning model (DLM) | For DLM, acc: 89.7 , SN: 91.3 , SP: 86.9 , AUC: 0.96. For DLM in BI-RADS, acc: 92.86 . false negative rate 10.4. | [94], 2021 |
| Normalizing image stain-color | Pre-trained VGG-19 | Block-wise fine-tuned VGG-19 model with softmax classifier on top. | Acc: 94.05 to 98.13 | [110], 2021 |
| Augmentation: flipping, rotation, gaussian blur, scalar multiplication. | Pre-trained on the MS COCO dataset | Deep learning-based computer-aided prediction (CAP) system. Mask R-CNN, DenseNet-121 | Acc: 81.05, SN: 81.36, SP: 80.85, AUC: 0.8054. | [93], 2021 |
| Data augmentation, rotation | Inception-v3 | Modified Inception-v3 | AUC: 0.9468, SN: 0.886, Specificity: 0.876. | [95], 2020 |
| Data augmentation: random rotation, random shear, random zoom. | − | DenseNet | Raining/testing cohorts AUCs: 0.957/0.912 (combined region), 0.944/0.775 (peritumoral region), (0.937/0.748 (intratumoral region). | [100], 2020 |
| Data augmentation: random geometric image transformations, flipping, rotation, scaling, shifting, resizing | – | Inception-V3, Inception-ResNet-V2, ResNet-101 | Inception-V3’s AUC: 0.89 (95 CI: 0.83, 0.95), SN: 85 (35 of 41 images; 95 CI: 70, 94), SP: 73(29 of 40 images; 95 CI: 56, 85) | [96], 2020 |
| Data augmentation: flipping, translation, scaling, and rotation technique. | VGG16, VGG19, ResNet-50 | Finetuned CNN | VGG16 with linear SVM’s patch-based accuracies: (93.97 for 40-, 92.92 for 100-, 91.23 for 200-, 91.79 for 400-); patient-based accuracies: (93.25 for 40-, 91.87 for 100-, 91.5 for 200-,92.31 for 400-). | [108], 2020 |
| .jpeg conversion, trimmed, resized | − | GoogLeNet CNN | SN: 0.958, SP: 0.875, acc: 0.925, AUC: 0.913. | [117], 2019 |
| Data augmentation: used ROI-CNN & G-CNN model | − | Two-stage grading. ROI-CNN, G-CNN | Acc = 0.998 | [97], 2019 |
| Data augmentation | VGG16 CNN | Fine-tuned deep learning parameters | Acc: 0.973, AUC: 0.98 | [115], 2019 |
Computed Tomography (CT) Images
CT image analysis is not very popular in practice in diagnosing breast cancer. However, many recent studies have shown promising results in detecting breast cancer at different stages of malignancy by using CT-guided deep-learning techniques.
Despite the variety in image modalities, the basic steps for developing a deep learning study are identical, such as preparing datasets, pre-processing images, constructing novel CNN or using pre-trained CNN, and defining the evaluation criteria. However, for the breast CT imaging modality studies, studies are still in the initial stages, and fewer studies exist. Transfer learning has also been applied to this modality with some optimistic results.
Dataset
The trends of using CT imaging modality in automatic breast cancer detection are quite new; hence, there are no widely used publicly available datasets to train and test the developed neural network. Most of the CT image breast cancer study datasets are prepared individually and kept private. In many current studies, investigators collected CT image data from associated medical centers or reproduced existing clinical datasets due to limited access to reliable open sources [120, 121]. Like, Koh et al. [122] collect chest CT images obtained after the diagnosis of breast cancer from one tertiary medical center (Severance Hospital) and includes 1070 chest CT scans for the experiment. Takahashi et al. [123] and Li et al. [124] also collect data of female patients who underwent F-FDG-PET/CT at a hospital. Moreau et al. [125] collect baseline and follow-up PET/CT clinical data at two sites and prepare two datasets for training and test assessment. In another study, Wang et al. [126] collected mammography breast data from the CBIS-DDSM image library [127] to train a CNN architecture, for mainly constructing the breast cancer CT image detection model.
Pre-Processing Techniques
In breast CT-based deep learning studies have adopted different techniques for data pre-processing. Among some studies, data is processed with the help of expert radiologists, manually segmenting the ROIs [128, 129], and some studies use data augmentation (shifting, flipping, rotation, mirroring, shearing, etc.) with the assistance of software [120, 121, 130]. Other studies used computational techniques such as fuzzy clustering [126], CNN architecture, or different coding platform packages (like Python imaging library of Pillow 3.3.1) [123] for data augmentation and preparation. In a breast CT imaging deep learning study, Caballo et al. [129] applied manual segmentation, software-aided data augmentation, and Generative Adversarial Network (GAN) for data augmentation simultaneously. Although some deep learning studies trained the model with raw CT data without pre-processing [122, 124, 131]. In most cases, different processing techniques are applied to enhance the dataset size and quality and ensure better performance of the proposed deep learning models.
Transfer Learning and Novel CNNs
When ambiguity remains with the other image modalities diagnosis, PET/CT imaging assessments are recommended. This framework can realize the significance of CT image-based deep learning approaches for breast cancer diagnosis. Recent studies have observed both De-Novo and the transfer learning approaches in breast CT image analysis.
An earlier study found that both the whole body and organ-wise manual quantification of the metabolic tumor volume (MTV) were significant prognosticators of overall survival in advanced breast cancer [119]. It refers that the neural network with regard to lesion detection, anatomical position determination, and total tumor volume quantification shows a certain level of accuracy to examine breast cancer, even though the neural network involved in PARS (PET-Assisted Reporting System) was not trained on breast cancer F-FDG PET/CT image data [119]. A breast axial FDG PET/CT image is shown in Fig. 8. Other pre-trained transfer learning studies also show a decent level of accuracy, like Liu et al. [121] proposed a deformable attention VCC19 (DA-VGG19) CNN model, which used a pre-trained VGG16 architecture trained on ImageNet. The DA-VGG19 model was trained with the axillary lymph nodes (ALN) datasets, keeping the first four convolutional layers stable and fine-tuning the other layers; the final model performed with higher accuracy (0.9088). Yang et al. [132] also proposed a convolutional neural network fast (CNN-F) model which was pre-trained on the ILSVRC-2012 dataset. This model evaluated the human epidermal growth factor receptor 2 (HER2) status in patients with breast cancer using multidetector computed tomography (MDCT); the full method is displayed in Fig. 9. For PET/CT image classification, Takahashi et al. [123] proposed a 2DL model based on Xecption architecture, which had 36 convolutional layers. This CNN was pre-trained on ImageNet datasets and fine-tuned to extract the best performance.
Fig. 8.
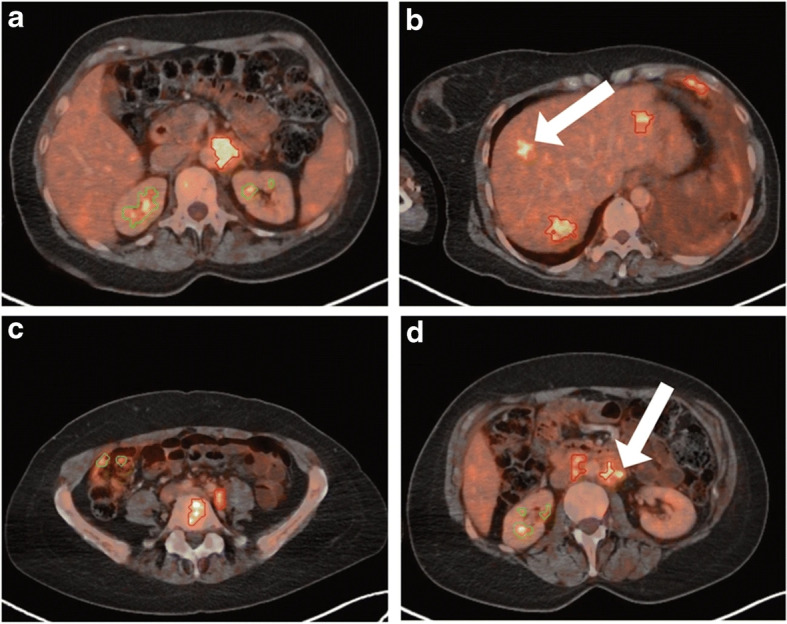
Breast axial FDG PET/CT image. Reprinted with permission from [119]
Fig. 9.
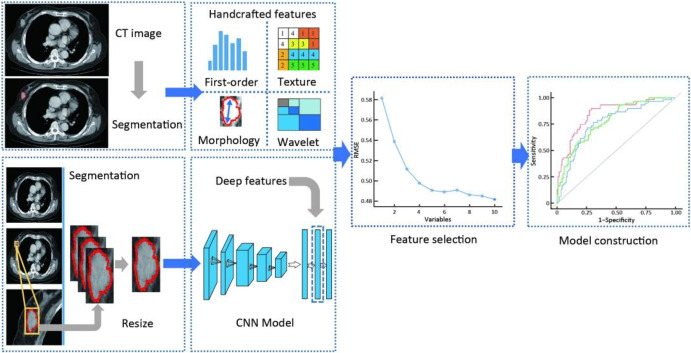
Flowcharts of CT with deep learning and handcrafted radiomics features. The handcrafted radiomics features were extracted from the manually segmented ROIs. A rectangular cross-section of the maximum tumor area was cropped and resized to a fixed size. Deep features were extracted from a generated RGB image after inputting into a pre-trained CNN model. Reprinted with permission from [132]
Other studies proposed some CNN models trained from scratch using clinically obtained CT images. U-Net and U-Net networks’ architectures are proposed for automatic segmentation of metastatic breast cancer by Moreau et al. [125]. Four imaging bio-markers, SUL, Total Lesion Glycolysis (TLG), PET Bone Index (PBI), and PET Liver Index (PLI), were computed and evaluated. Ma et al. [120] proposed a deep learning neural architecture search CNN which was trained and tested with contrast-enhanced breast cone-beam CT images.
Evaluation Criteria and Comparison Study
Classical evaluation metrics (accuracy, sensitivity, specificity, precision, recall, F1-score, AUC, etc.) are used for CT image-based deep learning studies. Some breast CT imaging deep learning studies sets have used imaging biomarkers such as SUL and SUV as evaluation metrics [125, 133].
The outcomes of CT image-based deep learning studies are often being compared with the outcomes of other popular image modalities [126]. For testing the reliability of a proposed deep learning breast CT image model, the outcomes are compared with the radiologist’s diagnosis outcome [123]. A comparative analysis is performed by comparing the proposed model with pre-existing robust models to get a comprehensive idea about the performance indicator of the proposed model. Ma et al. [120] compared their proposed neural architecture search (NAS)-generated CNN with the pre-trained ResNet models and found that with an AUC of 0.727, the proposed NAS-CNN surpassed the performance of well-performed best-known ResNet-50. This way, transfer-learned CNN models are frequently compared with the previously pre-trained models’ performance. As so, Liu et al. [121] meticulously designed the deformable attention VGG19 (DA-VGG19) CNN model from a pre-trained VGG16 model and achieved higher accuracy in comparison. The CT studies are summarized in Table 6.
Table 6.
CT studies summarized
| Pre-processing technique | Pre-trained model used | Novel technique | Performance | Ref., year |
|---|---|---|---|---|
| – | – | Deep learning algorithm based on RetinaNet | For internal test set- per-image SN: 91.9, per-lesion SN: 96.5 , precision: 18.2, FPs/case: 13.5. for external test set- per-image SN: 90.3, per-lesion SN: 96.1 , precision: 34.2, FPs/case: 15.6. | [122], 2022 |
| Processed and augmented with code written in Python 3.7.0 and Python imaging library of Pillow 3.3.1 | ImageNet pretraining model | Xpection CNN. Applied fine-tuned and Stochastic gradient descent optimizer | 4-degree model’s SN: 96, SP: 80, AUC = 0.936 (CI: 95, 0.8900.982 ). 0-degree model’s SN: 82, SP: 88, AUC = 0.918 (CI: 95, 0.8590.968, p = 0.078). | [123], 2022 |
| Dual-phase contrast-enhanced (reconstructed by blending factor of 0.5) DECT scan of the thorax. | – | Univariate analysis, logistic regression, XGBoost, SGD, LDA, AdaBoost, RF, decision tree, and SVM-based model. | On training dataset, AUROC: 0.880.99, SN: 0.850.98, SP: 0.921.0, F1 score: 0.870.98. On testing dataset, AUROC: 0.830.96, SN: 0.720.92, SP: 0.761.0, F1 score: 0.750.91. | [134], 2022 |
| Data augmentation: 90 rotation, grayscale-value reversing, 90 rotation of generated images. | – | Primal-dual hybrid gradient (PDHG) methods based algorithm. FI-Net to replace the computation. | Structural similarity measure (SSIM): 0.94, root mean squared error (RMSE): 0.1. | [130], 2022 |
| Data augmentation: horizontal and vertical shifting, flipping. | Compared with pre-trained ResNet models | Neural architecture search (NAS)-generated CNN | AUC: 0.727, SN: 80 (95 CI), SP: 60 (95 CI). | [135], 2021 |
| Data augmentation: flipped the training set images horizontally and vertically. labeled samples preparation. | Pre-trained VGG16; pre-trained in ImageNet | Deformable attention, DA-VGG19 is proposed | AUC: 0.9696, acc: 0.9088, PPV: 0.8786, NPV: 0.9469, SN: 0.9500, SP: 0.8675. | [121], 2021 |
| – | – | 3D residual CNN equipped with an attention mechanism. | SN: 68.6 & 64.2 | [124], 2021 |
| Data augmentation: elastic deformations, random scaling, random rotation, gamma augmentation. | – | U-Net & U-Net network’s architectures | For SUL biomarker; AUC: 0.89, SN: 87 , SP: 87 , optimal cutoff value: -32 , value: 0.001. | [125], 2021 |
| Contrast-enhanced. image reconstruction-ordered subset expectation maximization algorithm. | Pre-trained on lymphoma and lung cancer F-FDG PET/CT data. | PET-Assisted Reporting System (PARS) prototype that uses a neural network. | SN: 92, SP: 98, acc: 98, region:88 | [119], 2021 |
| Image optimization with fuzzy C-means clustering algorithm (FCM). Gray-gradient two-dimensional histogram generated. | – | Convolution and deconvolution neural network (CDNN) through the CNN. | SN: 80 (FP rate: 0,1). Detection acc: 78.4 (CI: 0.95) | [126], 2021 |
| Cropping, resizing, manual segmentation of ROI. | CNN-F pre-trained in ILSVRC-2012 dataset | CNN-F- consisted of five convolutional layers and three fully connected layers. | Combined model brier score: 0.159 (primary cohort) & 0.211 (validation cohort). | [128], 2020 |
| Manual segmentation, rotation (1–20, 10–30,20–40), mirroring, shearing. Generative Adversarial Network (GAN) for data augmentation. | - | U-Net based CNN architecture | Average DICE: 0.93 ± 0.03, SN: 0.92 ± 0.03, precision: 0.93 ± 0.05, conformity: 0.85 ± 0.06. | [129], 2020 |
| PET/CT fusion images attenuation-corrected by radiologists, Define spherical ROI with a radius of 2.4 cm., data augmentation. | – | Novel 3D CNN | Predicted SUV associated with real SUV ( estimate = 0.83, p !‘ 0.0001) and with FDG avidity (p !‘ 0.0001), ROC AUC: 0.85. | [133], 2019 |
| No adjustment | – | AlexNet trained from scratch with 3D CT case. | Classify breast density correctly 72 (training samples) & 76 (testing samples). | [131], 2017 |
Discussion and Conclusion
Mammogram and biopsy (histopathological images) are two of the initial breast cancer screening methods used. These are used in patients with breast cancer risk, whereas MRI is usually used to locate the tumor. CT is used to detect cancer at different stages of malignancy. Unlike other image modalities, CT research is still at the initial stages, and fewer studies exist related to the automated detection of breast cancer. Image datasets for histopathological, mammogram, MRI, US, and CT images are either obtained through public datasets or retrospective studies and clinical trials. There has always been a shortage, no matter where the data is obtained from. Therefore, image data augmentation techniques such as reflection, rotation, cropping, and scaling are applied to the data. For the CNN models to function efficiently, it is crucial to improve the quality of the images, and therefore, pre-processing techniques such as noise reduction, contrast enhancement, and image normalization are used. One of the significant differences between histopathological images and the other four image modalities is that histopathological has three image channels (RGB format), whereas others are only single channel as they are grayscale; therefore, some of the pre-processing techniques are different based on the number of image channels. Contrast enhancement and noise reduction techniques are most popular in grayscale images as a significant area might be out of the ROI. Most of the time, the pre-processing methods are accomplished manually by python libraries or other tools. Still, few studies have used CNN and other computation techniques to pre-process the data [71, 74, 97, 106, 112, 126, 129]. After pre-processing of data, neural networks or a combination of machine learning models and neural networks are used for tumor detection. Some neural networks used for detection and segmentation are UNet, ResNet50, VGG16, VGG19, Mask RCNN, GAN, Faster RCNN, AlexNet, ImageNet, and DenseNet. Few studies use a combination of SVM, K-means, and CNN for classification and segmentation. For histopathological image studies mentioned, some of the most common CNN architectures achieved an accuracy reaching up to 99 [48]. One of the most common architectures used for mammogram images is AlexNet or a customized CNN. For the mammogram studies, the highest accuracy reached is around 96 [58]. For MRI images, ResNet or customized CNN are the most common architectures, and the highest accuracy achieved is around 98 [71]. For ultrasound images, the highest accuracy reached was 99.8 with ROI-CNN and G-CNN [97]. For CT images, the highest sensitivity achieved was around 96 with Xpection CNN [123]. MRI and CT image modalities are still developing in the research area and therefore have fewer studies available compared to other image modalities. The supremacies of the five image modalities are summarized in Table 7
In conclusion, state-of-the-art neural network systems can perform the task of detection and segmentation with an accuracy ranging from 96 to 99 . Pre-trained models can be used and customized to save training time and have more efficient results. Also, it is crucial to use the proper pre-processing techniques based on the type of image to provide better input to the models so that the tumor detection tasks can be performed more efficiently.
Future Scope
Collecting extensive labeled data sets from a single image modality can be cumbersome; one way to solve this is by using collaborative learning from multi-modal image data. Our subsequent work focuses on feature fusion from multiple image modalities for more efficient tumor localization and segmentation. Imbalanced data is another problem related to each imaging modality, as samples exhibiting tissue abnormalities are less than the normal samples. A large number of normal samples dominates the training data, so the classifier favors classes with a more significant number of labeled samples. Data augmentation is one of the most common methods used to solve the issue of imbalanced class data, but it increases the computational cost. Therefore, future studies can utilize a surrogate evolutionary algorithm to optimize the computationally expensive models [141]. Another way of improving the efficiency of the computational model is by using the correct combination of pre-processing techniques. One of our future works involves comparing the performance of different CNN architectures by applying a combination of patching and stain normalization techniques. A different approach can be designing models that can differentiate noise in the input images. Many studies eliminate noise or other structures, such as pectoral muscle, from the images. Models can be created to train with some amount of noise, similar to the study by Ha et al. [70], so that noise in target images may not reduce the efficiency of tumor detection. Such models can be developed using a pre-trained model with machine learning methods such as SVM.
Author Contributions
Study conception and design were performed by Kihan Park and Pratibha Harrison. Material preparation, data collection, and analysis were performed by Pratibha Harrison and Rakib Hasan. The first draft of the manuscript was written by Pratibha Harrison and Rakib Hasan, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This research was based upon work supported by the National Science Foundation (NSF) under Grant No. 2153430.
Availability of Data and Materials
Not applicable
Code Availability
Not applicable
Declarations
Ethics Approval
Not applicable
Consent to Participate
Not applicable
Consent for Publication
Not applicable
Conflict of Interest
The authors declare no conflict of interest. The funder had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Cancer facts & figures 2022. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2022.html. Accessed 22 Mar 2022
- 2.Andrion A, Magnani C, Betta P, Donna A, Mollo F, Scelsi M, Bernardi P, Botta M, Terracini B. Malignant mesothelioma of the pleura: interobserver variability. J Clin Pathol. 1995;48(9):856–860. doi: 10.1136/jcp.48.9.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandya HJ, Park K, Desai JP. Design and fabrication of a flexible MEMS-based electro-mechanical sensor array for breast cancer diagnosis. Journal of Micromechanics and Microengineering. 2015;25(7):075025. doi: 10.1088/0960-1317/25/7/075025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park K, Harrison P, Desai JP (2020) Micro–scale viscoelastic characterization of human skin tissues as a biomarker for melanoma. In: 2020 International conference on manipulation, automation and robotics at small scales (MARSS). IEEE pp 1–6
- 5.Pandya HJ, Park K, Chen W, Goodell LA, Foran DJ, Desai JP. Toward a portable cancer diagnostic tool using a disposable MEMS-based biochip. IEEE Trans Biomed Eng. 2016;63(7):1347–1353. doi: 10.1109/TBME.2016.2535364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandya HJ, Park K, Chen W, Chekmareva MA, Foran DJ, Desai JP. Simultaneous MEMS-based electro-mechanical phenotyping of breast cancer. Lab on a Chip. 2015;15(18):3695–3706. doi: 10.1039/C5LC00491H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park K, Chen W, Chekmareva MA, Foran DJ, Desai JP. Electromechanical coupling factor of breast tissue as a biomarker for breast cancer. IEEE Trans Biomed Eng. 2017;65(1):96–103. doi: 10.1109/TBME.2017.2695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park K, Desai JP (2017) Machine learning approach for breast cancer localization. 2017 International Conference on Manipulation. Automation and Robotics at Small Scales (MARSS), IEEE, pp 1–6
- 9.Harrison P, Park K (2021) Tumor detection in breast histopathological images using faster r–cnn. In: 2021 International symposium on medical robotics (ISMR). IEEE pp 1–7
- 10.Green BN, Johnson CD, Adams A. Writing narrative literature reviews for peer-reviewed journals: secrets of the trade. J Chiropr Med. 2006;5(3):101–117. doi: 10.1016/S0899-3467(07)60142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witt JK, Warden AC, Dodd MD, Edney EE. Visual bias could impede diagnostic accuracy of breast cancer calcifications. J Med Imaging. 2022;9(3):035503. doi: 10.1117/1.JMI.9.3.035503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beam CA, Layde PM, Sullivan DC. Variability in the interpretation of screening mammograms by us radiologists: findings from a national sample. Arch Intern Med. 1996;156(2):209–213. doi: 10.1001/archinte.1996.00440020119016. [DOI] [PubMed] [Google Scholar]
- 13.Elmore JG, Jackson SL, Abraham L, Miglioretti DL, Carney PA, Geller BM, Yankaskas BC, Kerlikowske K, Onega T, Rosenberg RD, et al. Variability in interpretive performance at screening mammography and radiologists’ characteristics associated with accuracy. Radiology. 2009;253(3):641. doi: 10.1148/radiol.2533082308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunyé TT, Mercan E, Weaver DL, Elmore JG. Accuracy is in the eyes of the pathologist: the visual interpretive process and diagnostic accuracy with digital whole slide images. J Biomed Inform. 2017;66:171–179. doi: 10.1016/j.jbi.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le MT, Mothersill CE, Seymour CB, McNeill FE. Is the false-positive rate in mammography in North America too high? The British journal of radiology. 2016;89(1065):20160045. doi: 10.1259/bjr.20160045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert FJ, Pinker-Domenig K. Diagnosis and staging of breast cancer: when and how to use mammography, tomosynthesis, ultrasound, contrast-enhanced mammography, and magnetic resonance imaging. Dis Chest, Breast, Heart and Vessels. 2019;2019–2022:155–166. [PubMed] [Google Scholar]
- 17.Chaki J, Dey N. A beginner’s guide to image preprocessing techniques. Boca Raton, FL, USA: CRC Press; 2018. [Google Scholar]
- 18.de la Rosa FL, Gómez–Sirvent JL, Sánchez–Reolid R, Morales R, Fernández–Caballero A (2022) Geometric transformation–based data augmentation on defect classification of segmented images of semiconductor materials using a ResNet50 convolutional neural network. Expert Systems with Applications, 117731
- 19.Jasti V, Zamani AS, Arumugam K, Naved M, Pallathadka H, Sammy F, Raghuvanshi A, Kaliyaperumal K (2022) Computational technique based on machine learning and image processing for medical image analysis of breast cancer diagnosis. Security and Communication Networks 2022
- 20.Yelmanov S, Romanyshyn Y (2022) Image enhancement using gain–limited contrast stretching technique. In: 2022 IEEE 16th International conference on advanced trends in radioelectronics, telecommunications and computer engineering (TCSET). IEEE pp 447–450
- 21.Stepanyuk O, Kozarev K, Nedal M (2022) Multi–scale image preprocessing and feature tracking for remote CME characterization. arXiv preprint arXiv:2205.15088
- 22.Mudeng V, Kim M, Choe S-w. Prospects of structural similarity index for medical image analysis. Appl Sci. 2022;12(8):3754. doi: 10.3390/app12083754. [DOI] [Google Scholar]
- 23.Sznitman R, Yuan Y, Zhu D, Engelhardt S, Huang SX, Xue Y, Oksuz I, Nguyen H, Heller N, Mukhopadhyay A. Deep generative models, and data augmentation, labelling, and imperfections: first workshop, DGM4MICCAI 2021, and first workshop, DALI 2021, held in conjunction with MICCAI 2021, Strasbourg. Springer, New York, NY, USA: France. O; 2021. [Google Scholar]
- 24.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M, Steiger K, Schlitter AM, Esposito I, Navab N. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging. 2016;35(8):1962–1971. doi: 10.1109/TMI.2016.2529665. [DOI] [PubMed] [Google Scholar]
- 26.Karimi Jafarbigloo S, Danyali H. Nuclear atypia grading in breast cancer histopathological images based on CNN feature extraction and LSTM classification. CAAI Trans Intell Technol. 2021;6(4):426–439. doi: 10.1049/cit2.12061. [DOI] [Google Scholar]
- 27.Senan EM, Alsaade FW, Al-Mashhadani MIA, Theyazn H, Al-Adhaileh MH, et al. Classification of histopathological images for early detection of breast cancer using deep learning. J Appl Sci Eng. 2021;24(3):323–329. [Google Scholar]
- 28.Sohail A, Khan A, Wahab N, Zameer A, Khan S. A multiphase deep CNN based mitosis detection framework for breast cancer histopathological images. Scientific Reports. 2021;11(1):1–18. doi: 10.1038/s41598-021-85652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Shen X, Zhou Y, Wang X, Li T-Q. Classification of breast cancer histopathological images using interleaved DenseNet with SENet (IDSNet) PloS one. 2020;15(5):0232127. doi: 10.1371/journal.pone.0232127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu L, Daigle BJ., Jr Prognostic analysis of histopathological images using pre-trained convolutional neural networks: application to hepatocellular carcinoma. PeerJ. 2020;8:8668. doi: 10.7717/peerj.8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Wu J, Wu Q. Classification of breast cancer histology images using multi-size and discriminative patches based on deep learning. IEEE Access. 2019;7:21400–21408. doi: 10.1109/ACCESS.2019.2898044. [DOI] [Google Scholar]
- 32.Wang Z, Dong N, Dai W, Rosario SD, Xing EP (2018) Classification of breast cancer histopathological images using convolutional neural networks with hierarchical loss and global pooling. In: International conference image analysis and recognition. Springer pp 745–753
- 33.Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Guan X, Schmitt C, Thomas NE (2009) A method for normalizing histology slides for quantitative analysis. In: 2009 IEEE International symposium on biomedical imaging: from nano to macro. IEEE pp 1107–1110
- 34.Araùjo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, Polónia A, Campilho A. Classification of breast cancer histology images using convolutional neural networks. PloS one. 2017;12(6):0177544. doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rakhlin A, Shvets A, Iglovikov V, Kalinin AA (2018) Deep convolutional neural networks for breast cancer histology image analysis. In: International Conference Image Analysis and Recognition, Springer pp 737–744
- 36.Khan AM, Rajpoot N, Treanor D, Magee, (2014) A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans Biomed Eng 61(6):1729–1738 [DOI] [PubMed]
- 37.Toǧacar M, Özkurt KB, Ergen B, Cömert Z. BreastNet: a novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Phys A Stat Mech Appl. 2020;545:123592. doi: 10.1016/j.physa.2019.123592. [DOI] [Google Scholar]
- 38.Wang P, Wang J, Li Y, Li P, Li L, Jiang M. Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing. Biomed Signal Process Control. 2021;65:102341. doi: 10.1016/j.bspc.2020.102341. [DOI] [Google Scholar]
- 39.Gupta K, Chawla N (2020) Analysis of histopathological images for prediction of breast cancer using traditional classifiers with pre–trained CNN. Procedia Computer Science 167–878–889
- 40.Das K, Conjeti S, Chatterjee J, Sheet D. Detection of breast cancer from whole slide histopathological images using deep multiple instance CNN. IEEE Access. 2020;8:213502–213511. doi: 10.1109/ACCESS.2020.3040106. [DOI] [Google Scholar]
- 41.Wahab N, Khan A, Lee YS. Transfer learning based deep CNN for segmentation and detection of mitoses in breast cancer histopathological images. Microscopy. 2019;68(3):216–233. doi: 10.1093/jmicro/dfz002. [DOI] [PubMed] [Google Scholar]
- 42.Vesal S, Ravikumar N, Davari A, Ellmann S, Maier A (2018) Classification of breast cancer histology images using transfer learning. In: International conference image analysis and recognition. Springer pp 812–819
- 43.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Mané D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Viégas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, Zheng X (2015) TensorFlow: large–scale machine learning on heterogeneous systems. Software available from tensorflow.org. https://www.tensorflow.org/
- 44.Gerard G, Piastra M (2021) Conditional deep convolutional neural networks for improving the automated screening of histopathological images. arXiv preprint arXiv:2105.14338
- 45.Mahmood F, Borders D, Chen RJ, McKay GN, Salimian KJ, Baras A, Durr NJ. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans Med Imaging. 2019;39(11):3257–3267. doi: 10.1109/TMI.2019.2927182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Liu Q, Xie H, Yang Z, Zhou H. Boosted efficientnet: detection of lymph node metastases in breast cancer using convolutional neural networks. Cancers. 2021;13(4):661. doi: 10.3390/cancers13040661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho ED, Antonio Filho O, Silva RR, Araujo FH, Diniz JO, Silva AC, Paiva AC, Gattass M. Breast cancer diagnosis from histopathological images using textural features and CBIR. Artif Intell Med. 2020;105:101845. doi: 10.1016/j.artmed.2020.101845. [DOI] [PubMed] [Google Scholar]
- 48.Kiambe K, Kiambe K. Breast histopathological image feature extraction with convolutional neural networks for classification. ICSES Trans. Image Process. Pattern Recognit. (ITIPPR) 2018;4:4–12. [Google Scholar]
- 49.Gupta V, Vasudev M, Doegar A, Sambyal N. Breast cancer detection from histopathology images using modified residual neural networks. Biocybernetics and Biomedical Engineering. 2021;41(4):1272–1287. doi: 10.1016/j.bbe.2021.08.011. [DOI] [Google Scholar]
- 50.Reinhard E, Adhikhmin M, Gooch B, Shirley P. Color transfer between images. IEEE Comput Graph Appl. 2001;21(5):34–41. doi: 10.1109/38.946629. [DOI] [Google Scholar]
- 51.Al–Haija QA, Adebanjo A (2020) Breast cancer diagnosis in histopathological images using ResNet-50 convolutional neural network. In: 2020 IEEE International IOT, electronics and mechatronics conference (IEMTRONICS). IEEE pp 1–7
- 52.Tavakoli N, Karimi M, Norouzi A, Karimi N, Samavi S, Soroushmehr S (2019) Detection of abnormalities in mammograms using deep features. J Ambient Intell Humanized Comput 1–13
- 53.Byng JW, Yaffe MJ, Jong RA, Shumak RS, Lockwood GA, Tritchler DL, Boyd NF. Analysis of mammographic density and breast cancer risk from digitized mammograms. Radiographics. 1998;18(6):1587–1598. doi: 10.1148/radiographics.18.6.9821201. [DOI] [PubMed] [Google Scholar]
- 54.Ting FF, Tan YJ, Sim KS. Convolutional neural network improvement for breast cancer classification. Expert Syst Appl. 2019;120:103–115. doi: 10.1016/j.eswa.2018.11.008. [DOI] [Google Scholar]
- 55.Ragab DA, Sharkas M, Marshall S, Ren J. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ. 2019;7:6201. doi: 10.7717/peerj.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Li M, Wang H, Jiang H, Yao Y, Zhang H, Xin J. Breast cancer detection using extreme learning machine based on feature fusion with CNN deep features. IEEE Access. 2019;7:105146–105158. doi: 10.1109/ACCESS.2019.2892795. [DOI] [Google Scholar]
- 57.Agarwal R, Diaz O, Lladó X, Yap MH, Martí R. Automatic mass detection in mammograms using deep convolutional neural networks. J Med Imaging. 2019;6(3):031409. doi: 10.1117/1.JMI.6.3.031409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El Houby EM, Yassin NI. Malignant and nonmalignant classification of breast lesions in mammograms using convolutional neural networks. Biomed Signal Process Control. 2021;70:102954. doi: 10.1016/j.bspc.2021.102954. [DOI] [Google Scholar]
- 59.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- 60.Jen C-C, Yu S-S. Automatic detection of abnormal mammograms in mammographic images. Expert Syst Appl. 2015;42(6):3048–3055. doi: 10.1016/j.eswa.2014.11.061. [DOI] [Google Scholar]
- 61.Samala RK, Chan H-P, Hadjiiski LM, Helvie MA, Cha KH, Richter CD. Multi-task transfer learning deep convolutional neural network: application to computer-aided diagnosis of breast cancer on mammograms. Phys Med & Biol. 2017;62(23):8894. doi: 10.1088/1361-6560/aa93d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xi P, Shu C, Goubran R (2018) Abnormality detection in mammography using deep convolutional neural networks. In: 2018 IEEE International symposium on medical measurements and applications (MeMeA). IEEE pp 1–6
- 63.Jadoon MM, Zhang Q, Haq IU, Butt S, Jadoon A (2017) Three–class mammogram classification based on descriptive CNN features. BioMed research international 2017 [DOI] [PMC free article] [PubMed]
- 64.Platania R, Shams S, Yang S, Zhang J, Lee K, Park S–J (2017) Automated breast cancer diagnosis using deep learning and region of interest detection (bc–droid). In: Proceedings of the 8th ACM international conference on bioinformatics, computational biology, and health informatics, pp 536–543
- 65.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning deep features for discriminative localization. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2921–2929
- 66.Sun W, Tseng T-LB, Zhang J, Qian, (2017) Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput Med Imaging Graph 57:4–9 [DOI] [PubMed]
- 67.Tsochatzidis L, Costaridou L, Pratikakis I. Deep learning for breast cancer diagnosis from mammograms-a comparative study. J Imaging. 2019;5(3):37. doi: 10.3390/jimaging5030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Redmon J, Divvala S, Girshick R, Farhadi A (2016) You only look once: unified, real–time object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 779–788
- 69.El Adoui M, Mahmoudi SA, Larhmam MA, Benjelloun M. MRI breast tumor segmentation using different encoder and decoder CNN architectures. Computers. 2019;8(3):52. doi: 10.3390/computers8030052. [DOI] [Google Scholar]
- 70.Ha R, Mutasa S, Karcich J, Gupta N, Pascual Van Sant E, Nemer J, Sun M, Chang P, Liu MZ, Jambawalikar S. Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. Journal of Digital Imaging. 2019;32(2):276–282. doi: 10.1007/s10278-019-00179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yurttakal AH, Erbay H, İkizceli T, Karaçavuş S. Detection of breast cancer via deep convolution neural networks using MRI images. Multimedia Tools and Applications. 2020;79(21):15555–15573. doi: 10.1007/s11042-019-7479-6. [DOI] [Google Scholar]
- 72.Hu Q, Whitney HM, Giger ML. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-67441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haarburger C, Baumgartner M, Truhn D, Broeckmann M, Schneider H, Schrading S, Kuhl C, Merhof D (2019) Multi scale curriculum CNN for context–aware breast MRI malignancy classification. In: International conference on medical image computing and computer–assisted intervention. Springer pp 495–503
- 74.Zhang Y, Chan S, Park VY, Chang K–T, Mehta S, Kim MJ, Combs FJ, Chang P, Chow D, Parajuli R, et al (2020) Automatic detection and segmentation of breast cancer on MRI using mask R–CNN trained on non–fat–sat images and tested on fat–sat images. Acad Radiol [DOI] [PMC free article] [PubMed]
- 75.Ren T, Cattell R, Duanmu H, Huang P, Li H, Vanguri R, Liu MZ, Jambawalikar S, Ha R, Wang F, et al. Convolutional neural network detection of axillary lymph node metastasis using standard clinical breast MRI. Clinical breast cancer. 2020;20(3):301–308. doi: 10.1016/j.clbc.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 76.Frangi AF, Niessen WJ, Vincken KL, Viergever MA (1998) Multiscale vessel enhancement filtering. In: International conference on medical image computing and computer–assisted intervention. Springer pp 130–137
- 77.Zhou J, Luo L-Y, Dou Q, Chen H, Chen C, Li G-J, Jiang Z-F, Heng P-A. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J Magn Reson Imaging. 2019;50(4):1144–1151. doi: 10.1002/jmri.26721. [DOI] [PubMed] [Google Scholar]
- 78.Dalmış MU, Vreemann S, Kooi T, Mann RM, Karssemeijer N, Gubern-Mérida A. Fully automated detection of breast cancer in screening MRI using convolutional neural networks. Journal of Medical Imaging. 2018;5(1):014502. doi: 10.1117/1.JMI.5.1.014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dougherty ER, Lotufo RA (2003) Hands–on morphological image processing, vol 59. SPIE press, Bellingham, Washington
- 80.Chan TF, Vese LA. Active contours without edges. IEEE Trans Image Process. 2001;10(2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 81.Rasti R, Teshnehlab M, Phung SL. Breast cancer diagnosis in DCE-MRI using mixture ensemble of convolutional neural networks. Pattern Recogn. 2017;72:381–390. doi: 10.1016/j.patcog.2017.08.004. [DOI] [Google Scholar]
- 82.Zhang Y, Chen J-H, Lin Y, Chan S, Zhou J, Chow D, Chang P, Kwong T, Yeh D-C, Wang X, et al. Prediction of breast cancer molecular subtypes on DCE-MRI using convolutional neural network with transfer learning between two centers. Eur Radiol. 2021;31(4):2559–2567. doi: 10.1007/s00330-020-07274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu W, Wang Z, He Y, Yu H, Xiong N, Wei J (2019) Breast cancer detection based on merging four modes MRI using convolutional neural networks. In: ICASSP 2019–2019 IEEE International conference on acoustics, speech and signal processing (ICASSP). IEEE pp 1035–1039
- 84.Agarwal P, Yadav A, Mathur P (2022) Breast cancer prediction on breakhis dataset using deep cnn and transfer learning model. In: Data Engineering for Smart Systems. Springer, ??? pp 77–88
- 85.Xiao T, Liu L, Li K, Qin W, Yu S, Li Z (2018) Comparison of transferred deep neural networks in ultrasonic breast masses discrimination. BioMed research international 2018 [DOI] [PMC free article] [PubMed]
- 86.Kim K, Song MK, Kim E-K, Yoon JH. Clinical application of S-Detect to breast masses on ultrasonography: a study evaluating the diagnostic performance and agreement with a dedicated breast radiologist. Ultrasonography. 2017;36(1):3. doi: 10.14366/usg.16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data in brief. 2020;28:104863. doi: 10.1016/j.dib.2019.104863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilding R, Sheraton VM, Soto L, Chotai N, Tan EY (2022) Deep learning applied to breast imaging classification and segmentation with human expert intervention. Journal of Ultrasound, 1–8 [DOI] [PMC free article] [PubMed]
- 89.Yap MH, Pons G, Martí J, Ganau S, Sentís M, Zwiggelaar R, Davison AK, Marti R. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Health Inform. 2017;22(4):1218–1226. doi: 10.1109/JBHI.2017.2731873. [DOI] [PubMed] [Google Scholar]
- 90.Karthik R, Menaka R, Kathiresan G, Anirudh M, Nagharjun M (2021) Gaussian dropout based stacked ensemble CNN for classification of breast tumor in ultrasound images. IRBM
- 91.Yu K, Tan L, Lin L, Cheng X, Yi Z, Sato T. Deep-learning-empowered breast cancer auxiliary diagnosis for 5GB remote E-health. IEEE Wirel Commun. 2021;28(3):54–61. doi: 10.1109/MWC.001.2000374. [DOI] [Google Scholar]
- 92.Jiang M, Zhang D, Tang S-C, Luo X-M, Chuan Z-R, Lv W-Z, Jiang F, Ni X-J, Cui X-W, Dietrich CF. Deep learning with convolutional neural network in the assessment of breast cancer molecular subtypes based on us images: a multicenter retrospective study. Eur Radiol. 2021;31(6):3673–3682. doi: 10.1007/s00330-020-07544-8. [DOI] [PubMed] [Google Scholar]
- 93.Lee Y-W, Huang C-S, Shih C-C, Chang R-F. Axillary lymph node metastasis status prediction of early-stage breast cancer using convolutional neural networks. Comput Biol Med. 2021;130:104206. doi: 10.1016/j.compbiomed.2020.104206. [DOI] [PubMed] [Google Scholar]
- 94.Zhang X, Li H, Wang C, Cheng W, Zhu Y, Li D, Jing H, Li S, Hou J, Li J, et al. Evaluating the accuracy of breast cancer and molecular subtype diagnosis by ultrasound image deep learning model. Frontiers Oncol. 2021;11:606. doi: 10.3389/fonc.2021.623506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y, Choi EJ, Choi Y, Zhang H, Jin GY, Ko S-B. Breast cancer classification in automated breast ultrasound using multiview convolutional neural network with transfer learning. Ultrasound Med & Biol. 2020;46(5):1119–1132. doi: 10.1016/j.ultrasmedbio.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 96.Zhou L-Q, Wu X-L, Huang S-Y, Wu G-G, Ye H-R, Wei Q, Bao L-Y, Deng-B Li X-R, Cui X-W, et al. Lymph node metastasis prediction from primary breast cancer US images using deep learning. Radiology. 2020;294(1):19–28. doi: 10.1148/radiol.2019190372. [DOI] [PubMed] [Google Scholar]
- 97.Huang Y, Han L, Dou H, Luo H, Yuan Z, Liu Q, Zhang J, Yin G. Two-stage CNNs for computerized BI-RADS categorization in breast ultrasound images. Biomedical engineering online. 2019;18(1):1–18. doi: 10.1186/s12938-019-0626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ayana G, Park J, Jeong J-W, Choe S-w. A novel multistage transfer learning for ultrasound breast cancer image classification. Diagnostics. 2022;12(1):135. doi: 10.3390/diagnostics12010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Latif G, Butt MO, Al Anezi FY, Alghazo J (2020) Ultrasound image despeckling and detection of breast cancer using deep CNN. In: 2020 RIVF International conference on computing and communication technologies (RIVF). IEEE pp 1–5
- 100.Sun Q, Lin X, Zhao Y, Li L, Yan K, Liang D, Sun D, Li Z–C (2020) Deep learning vs. radiomics for predicting axillary lymph node metastasis of breast cancer using ultrasound images: don’t forget the peritumoral region. Frontiers Oncol 10:53 [DOI] [PMC free article] [PubMed]
- 101.Jabeen K, Khan MA, Alhaisoni M, Tariq U, Zhang Y-D, Hamza A, Mickus A, Damaševičius R. Breast cancer classification from ultrasound images using probability-based optimal deep learning feature fusion. Sensors. 2022;22:807. doi: 10.3390/s22030807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Byra M. Breast mass classification with transfer learning based on scaling of deep representations. Biomed Signal Process Control. 2021;69:102828. doi: 10.1016/j.bspc.2021.102828. [DOI] [Google Scholar]
- 103.Mishra AK, Roy P, Bandyopadhyay S, Das SK. Breast ultrasound tumour classification: a machine learning-radiomics based approach. Expert Syst. 2021;38(7):12713. doi: 10.1111/exsy.12713. [DOI] [Google Scholar]
- 104.Huang K, Zhang Y, Cheng H, Xing P (2021) Shape–adaptive convolutional operator for breast ultrasound image segmentation. In: 2021 IEEE International conference on multimedia and expo (ICME). IEEE pp 1–6
- 105.Pourasad Y, Zarouri E, Salemizadeh Parizi M, Salih Mohammed A (2021) Presentation of novel architecture for diagnosis and identifying breast cancer location based on ultrasound images using machine learning. Diagnostics 11(10):1870 [DOI] [PMC free article] [PubMed]
- 106.Badawy SM, Mohamed AE-NA, Hefnawy AA, Zidan HE, GadAllah MT, El-Banby GM. Automatic semantic segmentation of breast tumors in ultrasound images based on combining fuzzy logic and deep learning-a feasibility study. PloS one. 2021;16(5):0251899. doi: 10.1371/journal.pone.0251899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boumaraf S, Liu X, Wan Y, Zheng Z, Ferkous C, Ma X, Li Z, Bardou D. Conventional machine learning versus deep learning for magnification dependent histopathological breast cancer image classification: a comparative study with visual explanation. Diagnostics. 2021;11(3):528. doi: 10.3390/diagnostics11030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sharma S, Mehra R. Conventional machine learning and deep learning approach for multi-classification of breast cancer histopathology images-a comparative insight. J Digital Imaging. 2020;33(3):632–654. doi: 10.1007/s10278-019-00307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Misra S, Jeon S, Managuli R, Lee S, Kim G, Yoon C, Lee S, Barr RG, Kim C. Bi-modal transfer learning for classifying breast cancers via combined b-mode and ultrasound strain imaging. IEEE Transactions on ultrasonics, ferroelectrics, and frequency control. 2021;69(1):222–232. doi: 10.1109/TUFFC.2021.3119251. [DOI] [PubMed] [Google Scholar]
- 110.Boumaraf S, Liu X, Wan Y, Zheng Z, Ferkous C, Ma X, Li Z, Bardou D. Conventional machine learning versus deep learning for magnification dependent histopathological breast cancer image classification: a comparative study with visual explanation. Diagnostics. 2021;11(3):528. doi: 10.3390/diagnostics11030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M, Steiger K, Schlitter AM, Esposito I, Navab N. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging. 2016;35(8):1962–1971. doi: 10.1109/TMI.2016.2529665. [DOI] [PubMed] [Google Scholar]
- 112.Sharif M, Hussain A, Jaffar MA, Choi T-S. Fuzzy similarity based non local means filter for rician noise removal. Multimedia tools and applications. 2015;74(15):5533–5556. doi: 10.1007/s11042-014-1867-8. [DOI] [Google Scholar]
- 113.Murtaza G, Shuib L, Wahab AWA, Mujtaba G, Raza G. Ensembled deep convolution neural network-based breast cancer classification with misclassification reduction algorithms. Multimed Tools Appl. 2020;79(25):18447–18479. doi: 10.1007/s11042-020-08692-1. [DOI] [Google Scholar]
- 114.Bychkov D, Linder N, Tiulpin A, Kücükel H, Lundin M, Nordling S, Sihto H, Isola J, Lehtimäki T, Kellokumpu-Lehtinen P-L, et al. Deep learning identifies morphological features in breast cancer predictive of cancer ERBB2 status and trastuzumab treatment efficacy. Sci Reports. 2021;11(1):1–10. doi: 10.1038/s41598-021-83102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hijab A, Rushdi MA, Gomaa MM, Eldeib A (2019) Breast cancer classification in ultrasound images using transfer learning. In: 2019 Fifth International Conference on Advances in Biomedical Engineering (ICABME). IEEE pp 1–4
- 116.Cao Z, Duan L, Yang G, Yue T, Chen Q. An experimental study on breast lesion detection and classification from ultrasound images using deep learning architectures. BMC Med Imaging. 2019;19(1):1–9. doi: 10.1186/s12880-019-0349-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fujioka T, Kubota K, Mori M, Kikuchi Y, Katsuta L, Kasahara M, Oda G, Ishiba T, Nakagawa T, Tateishi U. Distinction between benign and malignant breast masses at breast ultrasound using deep learning method with convolutional neural network. Japan J Radiol. 2019;37(6):466–472. doi: 10.1007/s11604-019-00831-5. [DOI] [PubMed] [Google Scholar]
- 118.Byra M. Breast mass classification with transfer learning based on scaling of deep representations. Biomed Signal Process Control. 2021;69:102828. doi: 10.1016/j.bspc.2021.102828. [DOI] [Google Scholar]
- 119.Weber M, Kersting D, Umutlu L, Schafers M, Rischpler C, Fendler WP, Buvat I, Herrmann K, Seifert R (2021) Just another “Clever Hans”? Neural networks and FDG PET–CT to predict the outcome of patients with breast cancer. European journal of nuclear medicine and molecular imaging, 1–10 [DOI] [PMC free article] [PubMed]
- 120.Ma J, He N, Yoon JH, Ha R, Li J, Ma W, Meng T, Lu L, Schwartz LH, Wu Y, et al. Distinguishing benign and malignant lesions on contrast-enhanced breast cone-beam ct with deep learning neural architecture search. Eur J Radiol. 2021;142:109878. doi: 10.1016/j.ejrad.2021.109878. [DOI] [PubMed] [Google Scholar]
- 121.Liu Z, Ni S, Yang C, Sun W, Huang D, Su H, Shu J, Qin N. Axillary lymph node metastasis prediction by contrast-enhanced computed tomography images for breast cancer patients based on deep earning. Comput Biol Med. 2021;136:104715. doi: 10.1016/j.compbiomed.2021.104715. [DOI] [PubMed] [Google Scholar]
- 122.Koh J, Yoon Y, Kim S, Han K, Kim E-K. Deep learning for the detection of breast cancers on chest computed tomography. Clin Breast Cancer. 2022;22(1):26–31. doi: 10.1016/j.clbc.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 123.Takahashi K, Fujioka T, Oyama J, Mori M, Yamaga E, Yashima Y, Imokawa T, Hayashi A, Kujiraoka Y, Tsuchiya J, et al (2022) Deep learning using multiple degrees of maximum–intensity projection for PET/CT image classification in breast cancer.Tomography 8(1):131–141 [DOI] [PMC free article] [PubMed]
- 124.Li Z, Kitajima K, Hirata K, Togo R, Takenaka J, Miyoshi Y, Kudo K, Ogawa T, Haseyama M. Preliminary study of AI-assisted diagnosis using FDG-PET/CT for axillary lymph node metastasis in patients with breast cancer. EJNMMI Research. 2021;11(1):1–10. doi: 10.1186/s13550-021-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moreau N, Rousseau C, Fourcade C, Santini G, Brennan A, Ferrer L, Lacombe M, Guillerminet C, Colombié M, Jézéquel P, et al. Automatic segmentation of metastatic breast cancer lesions on 18F-FDG PET/CT longitudinal acquisitions for treatment response assessment. Cancers. 2021;14(1):101. doi: 10.3390/cancers14010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Y, Yang F, Zhang J, Wang H, Yue X, Liu S. Application of artificial intelligence based on deep learning in breast cancer screening and imaging diagnosis. Neural Comput Appl. 2021;33(15):9637–9647. doi: 10.1007/s00521-021-05728-x. [DOI] [Google Scholar]
- 127.Lee RS, Gimenez F, Hoogi A, Miyake KK, Gorovoy M, Rubin DL. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci Data. 2017;4(1):1–9. doi: 10.1038/sdata.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang X, Wu L, Zhao K, Ye W, Liu W, Wang Y, Li J, Li H, Huang X, Zhang W, et al. Evaluation of human epidermal growth factor receptor 2 status of breast cancer using preoperative multidetector computed tomography with deep learning and handcrafted radiomics features. Chin J Cancer Res. 2020;32(2):175. doi: 10.21147/j.issn.1000-9604.2020.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Caballo M, Pangallo DR, Mann RM, Sechopoulos I. Deep learning-based segmentation of breast masses in dedicated breast CT imaging: radiomic feature stability between radiologists and artificial intelligence. Comput Biol Med. 2020;118:103629. doi: 10.1016/j.compbiomed.2020.103629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fan Y, Wang H, Gemmeke H, Hopp T, Hesser J. Model-data-driven image reconstruction with neural networks for ultrasound computed tomography breast imaging. Neurocomputing. 2022;467:10–21. doi: 10.1016/j.neucom.2021.09.035. [DOI] [Google Scholar]
- 131.Zhou X, Kano T, Koyasu H, Li S, Zhou X, Hara T, Matsuo M, Fujita H (2017) Automated assessment of breast tissue density in non-contrast 3D CT images without image segmentation based on a deep CNN. In: Medical imaging 2017: computer-aided diagnosis. Spie vol 10134, pp 704–709
- 132.Yang X, Wu L, Zhao K, Ye W, Liu W, Wang Y, Li J, Li H, Huang X, Zhang W, et al. Evaluation of human epidermal growth factor receptor 2 status of breast cancer using preoperative multidetector computed tomography with deep learning and handcrafted radiomics features. Chin J Cancer Res. 2020;32(2):175. doi: 10.21147/j.issn.1000-9604.2020.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shaish H, Mutasa S, Makkar J, Chang P, Schwartz L, Ahmed F. Prediction of lymph node maximum standardized uptake value in patients with cancer using a 3D convolutional neural network: a proof-of-concept study. Am J Roentgenol. 2019;212(2):238–244. doi: 10.2214/AJR.18.20094. [DOI] [PubMed] [Google Scholar]
- 134.Lan X, Wang X, Qi J, Chen H, Zeng X, Shi J, Liu D, Shen H, Zhang J. Application of machine learning with multiparametric dual-energy computed tomography of the breast to differentiate between benign and malignant lesions. Quantitative Imaging in Medicine and Surgery. 2022;12(1):810. doi: 10.21037/qims-21-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ma J, He N, Yoon JH, Ha R, Li J, Ma W, Meng T, Lu L, Schwartz LH, Wu Y, et al. Distinguishing benign and malignant lesions on contrast-enhanced breast cone-beam CT with deep learning neural architecture search. Eur J Radiol. 2021;142:109878. doi: 10.1016/j.ejrad.2021.109878. [DOI] [PubMed] [Google Scholar]
- 136.Lai KK-L, Lee TT-Y, Lee MK-S, Hui JC-H, Zheng Y-P. Validation of scolioscan air-portable radiation-free three-dimensional ultrasound imaging assessment system for scoliosis. Sensors. 2021;21(8):2858. doi: 10.3390/s21082858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang Z, Wang W, Wang X, Yu X, Zhu Y, Zhan H, Chen Z, Li B, Huang J. Breast-specific gamma imaging or ultrasonography as adjunct imaging diagnostics in women with mammographically dense breasts. European Radiology. 2020;30(11):6062–6071. doi: 10.1007/s00330-020-06950-2. [DOI] [PubMed] [Google Scholar]
- 138.Lin X, Li Y, Wang S, Zhang Y, Chen X, Wei M, Zhu H, Wu A, Yang Z, Wang X (2022) 68Ga–FAPI improves tumor staging in patients with colorectal cancer: comparing to 18F–FDG PET/CT
- 139.Piva R, Ticconi F, Ceriani V, Scalorbi F, Fiz F, Capitanio S, Bauckneht M, Cittadini G, Sambuceti G, Morbelli S. Comparative diagnostic accuracy of 18F-FDG PET/CT for breast cancer recurrence. Breast Cancer: Targets and Ther. 2017;9:461. doi: 10.2147/BCTT.S111098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cong W, Shan H, Zhang X, Liu S, Ning R, Wang G (2019) Deep–learning–based breast CT for radiation dose reduction. In: Developments in X–ray tomography XII. International Society for Optics and Photonics vol. 11113, pp 111131
- 141.Gonçalves CB, Souza JR, Fernandes H (2022) CNN optimization using surrogate evolutionary algorithm for breast cancer detection using infrared images. In: 2022 IEEE 35th international symposium on computer-based medical systems (CBMS). IEEE pp 84–89
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable
Not applicable