Abstract
The gutD gene of Clostridium beijerinckii NCIMB 8052 encoding glucitol 6-phosphate dehydrogenase was cloned on a 5.7-kbp chromosomal DNA fragment by complementing an Escherichia coli gutD mutant strain and selecting for growth on glucitol. Five open reading frames (ORFs) in the order gutA1 gutA2 orfX gutB gutD were identified in a 4.0-kbp region of the cloned DNA. The deduced products of four of these ORFs were homologous to components of the glucitol phosphotransferase system (PTS) and glucitol 6-phosphate dehydrogenase from E. coli, while the remaining ORF (orfX) encoded an enzyme which had similarities to members of a family of transaldolases. A strain in which gutD was inactivated by targeted integration lacked glucitol 6-phosphate dehydrogenase activity. The gutA1 and gutA2 genes encoded two polypeptides forming enzyme IIBC of the glucitol PTS comprising three domains in the order CBC. Domain IIA of the glucitol PTS was encoded by gutB. Glucitol phosphorylation assays in which soluble and membrane fractions of cells grown on glucose (which did not synthesize the glucitol PTS) or cells grown on glucitol were used confirmed that there is a separate, soluble, glucitol-specific PTS component, which is the product of the gutB gene. The gut genes were regulated at the level of transcription and were induced in the presence of glucitol. Cells grown in the presence of glucose and glucitol utilized glucose preferentially. Following depletion of glucose, the glucitol PTS and glucitol 6-phosphate dehydrogenase were synthesized, and glucitol was removed from the culture medium. RNA analysis showed that the gut genes were not expressed until glucose was depleted.
The clostridia are a diverse group of heterotrophic anaerobes, many of which are able to metabolize a wide range of carbohydrate substrates. The solventogenic clostridia have attracted interest principally as a result of their ability to produce organic acids and alcohols by fermentation (9, 20). In the industrial-scale acetone-butanol fermentation process which was used successfully earlier this century, Clostridium acetobutylicum was used to ferment starch or molasses (7, 8), but this process is currently uneconomic. Revival of this process and development of related processes will depend among other things on a thorough understanding of the biochemistry, physiology, and genetics of the organisms, which should allow optimization of conversion of the substrate to the desired end product. Despite the fact that accumulation of substrate is a potentially important metabolic control point, few detailed studies have been carried out on this aspect of the physiology of clostridia (14, 15).
The synthesis of membrane-bound transport systems and associated catabolic enzymes often depends on growth conditions. A variety of mechanisms that control gene expression have been identified (22), but in each case a set of genes are expressed efficiently only when the gene products are required. It is apparent that like other bacteria, the clostridia specifically regulate synthesis of enzymes in response to their metabolic needs, and a number of cases of preferential use of sugars in a mixture by clostridial strains have been reported (3, 14, 15). For example, Clostridium thermosaccharolyticum exhibits classical diauxic growth in the presence of the sugars glucose and xylose; glucose is the preferred sugar, and when it is present, the synthesis of xylose isomerase, xylulokinase, and the xylose transport system is repressed (1). Regulation of this kind in response to substrate availability has profound implications for industrial fermentations performed with feedstocks which contain several fermentable sources of carbon. We are now involved in a study of carbohydrate utilization by Clostridium beijerinckii (formerly C. acetobutylicum) NCIMB 8052, a strain which has received considerable attention due to the fact that it can be manipulated genetically with relative ease (11). This strain shows a marked preference for glucose, which by as-yet-undefined mechanisms regulates the metabolism of alternative substrates, such as glucitol, galactose, and the disaccharides cellobiose, lactose, maltose, and sucrose (2, 13, 15). In cells not induced for metabolism of these substrates, utilization of each of them is strongly inhibited by glucose. Following induction, however, different patterns of utilization in the presence of glucose have been observed, indicating that it is likely that there are distinct mechanisms of regulation directed at the synthesis and activity of catabolic enzymes. We are currently performing molecular studies of some of the regulated gene systems in an effort to better understand the mechanisms involved. In this paper, we report the cloning, sequencing, and analysis of a gene system concerned with glucitol metabolism in C. beijerinckii and demonstrate that the expression of this system is subject to catabolite repression by glucose.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
C. beijerinckii NCIMB 8052 was maintained as a spore suspension at 4°C. Spores were heat shocked at 80°C for 10 min and inoculated into 20 ml of reinforced clostridial medium (RCM) (Oxoid). Starter cultures that were grown overnight at 37°C in an anaerobic cabinet (Forma Scientific, Marietta, Ohio) under an N2-H2-CO2 (80:10:10) atmosphere were subcultured in a defined medium (supplemented with the appropriate carbon source) and incubated anaerobically at 37°C to provide working cultures. Vessels containing up to 1 liter were incubated in the anaerobic cabinet. For experiments performed with 10-liter cultures, a 1-liter culture was grown overnight, and this culture was subsequently used as the inoculum for a larger vessel, which was incubated at 37°C and was sparged with nitrogen. The defined medium contained (per liter) 2.2 g of ammonium acetate, 0.5 g of KH2PO4, 0.45 g of K2HPO4, 0.2 g of MgSO4 · 7H2O, 0.01 g of MnSO4 · 4H2O, 0.01 g of NaCl, 0.01 g of FeSO4 · 7H2O, 0.01 g of p-aminobenzoic acid, and 0.001 g of d-biotin. Carbon sources were sterilized separately and added to the medium after cooling. C. beijerinckii AA219 (31) was grown in synthetic RCM (17) containing 10 μg of erythromycin per ml and 0.5% glucitol as the only fermentable carbon source. Escherichia coli TG1 (26), JM83 (30), and WM4 were grown aerobically at 37°C in nutrient broth (Oxoid) or L broth, supplemented when necessary with ampicillin (100 μg ml−1) or tetracycline (10 μg ml−1). WM4 was constructed by P1 transduction of JM83 with a lysate prepared from E. coli JC10279 (srl301::Tn10 srlD50 gutC300) (5) by the method described by Silhavy et al. (27) and selection for tetracycline resistance and a glucitol-negative phenotype on MacConkey agar (Oxoid). Screening for the C. beijerinckii gutD gene was carried out by plating transformed E. coli cells onto M9 minimal medium containing 0.2% glucitol as the sole carbon and energy source.
DNA manipulations.
A C. beijerinckii NCIMB 8052 gene library in plasmid pAT153 (29) was constructed by procedures which have been described previously (12). E. coli was transformed by electroporation with a Bio-Rad Gene Pulser by using the protocol described in the manufacturer’s manual. Plasmid DNA from chloramphenicol-amplified cultures was purified as described previously (12). Restriction endonuclease and DNA ligase were purchased from Bethesda Research Laboratories and were used under the conditions recommended by the supplier. Nucleotide sequencing was performed by the chain termination method (Sequenase version 2.0 kit; U.S. Biochemicals, Cleveland, Ohio). A 2.2-kbp HindIII fragment of pSORB1 was ligated into pMTL23, reisolated, and then circularized and sonicated as described previously (4). The random sequences obtained were sorted into a contiguous sequence with DNASTAR software, and sequencing of both strands was completed by using site-specific primers and pSORB1 as the template. Additional smaller restriction fragments from pSORB1 were cloned in opposite orientations into M13 mp18 and mp19 (34) and were sequenced by walking along the insert by using site-specific primers to initiate reactions. The contiguous nature of these fragments and the HindIII fragment was confirmed by performing sequencing reactions with pSORB1 as the template and appropriate primers to ensure that the sequence obtained included the restriction sites at the junctions.
Assay to determine sugar concentrations in culture supernatants.
Culture samples (1 ml) were removed and centrifuged at intervals. The glucose concentrations in supernatants were determined with a Sigma assay kit (catalog no. 510). Glucitol concentrations were estimated with a d-sorbitol–xylitol assay kit (Boehringer).
Preparation of cell extracts.
Cell extracts were prepared essentially by the method of Mitchell and Booth (16). Cultures were harvested and washed in Tris-HCl buffer (20 mM Tris-HCl [pH 7.6], 5 mM MgCl2, 1 mM dithiothreitol). Washed cells were routinely resuspended in buffer at a ratio of 4 ml per g (wet weight) and were ruptured by passage through a French press. For 10-liter cultures, 0.5- or 1-liter aliquots (depending on the cell density) were removed at intervals, and the washed cell pellets were resuspended at a ratio of 8 ml per g (wet weight). Extracts were fractionated as described previously (16). Protein concentrations in cell extracts were determined by the microbiuret assay, as described by Zamenhof (35), by using bovine serum albumin as the standard.
Sugar phosphorylation assays performed with cell extracts.
Sugar phosphorylation assays were carried out by the method of Gachelin (6), as described by Tangney et al. (28). Routinely, 0.4-ml aliquots of extract were diluted into buffer which, when appropriate, contained 1 mM phosphoenolpyruvate (PEP) in a total assay volume of 1 ml. Each mixture was equilibrated at 37°C for 3 min prior to the assay. Radiolabelled sugar (9.5 mM; 1.05 Ci mol−1) was added to a concentration of 0.21 mM, and 0.15-ml samples were removed at appropriate intervals to estimate sugar phosphate contents; the assay time course varied between 10 min and 1 h so that accurate values for rates of activity could be obtained. Samples were added to 2 ml of 1% (wt/vol) barium bromide in 80% (vol/vol) ethanol. The resulting phosphate precipitates which formed were removed by filtration on glass fiber discs (Whatman type GF/F) and were washed with 5 ml of 80% ethanol. The discs were dried under a heat lamp, and the radioactive counts on each disc were determined in 4 ml of scintillation cocktail O (BDH Scintran).
Glucitol 6-phosphate dehydrogenase activity in cell extracts.
The glucitol 6-phosphate dehydrogenase activities in C. beijerinckii cell extracts were determined quantitatively as described previously (21), except that the pH was 9.3. The activity was proportional to the extract concentration in the range studied. A qualitative assay was also performed with extracts of both C. beijerinckii and E. coli strains. This assay was based on the production of dark-colored formazan from the reduction of a light-sensitive tetrazolium salt. Each assay mixture (total volume, 1 ml) contained 50 mM Tris-HCl (pH 9.3), 1 mM NAD+, 0.075 mM phenazine methosulfate, 0.5 mM 3-(4,5-dimethylthiazolyl-2)-2,5 diphenyltetrazolium bromide, and up to 100 μl of cell extract. The mixture was incubated at room temperature in the dark for 5 min, and the reaction was then started by adding glucitol 6-phosphate to a final concentration of 1 mM. The reaction was allowed to proceed in the dark for up to 10 min, after which a dark colloidal suspension was observed in tubes which contained active glucitol 6-phosphate dehydrogenase.
Preparation of RNA and labelling of probes.
RNA was isolated from cells with an RNeasy Total RNA kit from Qiagen. The protocol was slightly modified for use with C. beijerinckii by increasing the concentration of lysozyme in the cell lysis solution from 1 to 10 mg ml−1. DNA probes (internal fragments of each open reading frame [ORF]) were prepared by PCR and labelled with digoxigenin-dUTP, which was obtained from Boehringer. The following primers were used for PCR (the positions are nucleotide positions [see Fig. 2], and the reverse primers had sequences complementary to the regions indicated): gutA1, positions 195 to 215 and 673 to 653; gutA2, positions 720 to 740 and 1698 to 1678; orfX, positions 1843 to 1863 and 2501 to 2481; gutB, positions 2579 to 2599 and 2925 to 2905; and gutD, positions 3037 to 3057 and 3823 to 3803.
FIG. 2.
Nucleotide sequence of the DNA fragment encoding genes associated with glucitol metabolism in C. beijerinckii. The deduced amino acid sequences of the five ORFs are shown below the coding sequence. Putative ribosome binding sites are underlined.
Slot blot hybridization for detection of mRNA.
Slot blots were prepared with approximately 1 μg of total RNA immobilized on nitrocellulose membrane filters by using a Minifold II system (Schleicher & Schuell). The filters were hybridized with digoxigenin-labelled DNA probes at 50°C overnight. Then the filters were washed under low-stringency conditions (2× SSC containing 0.1% [wt/vol] sodium dodecyl sulfate at room temperature [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) and high-stringency conditions (0.5× SSC containing 0.1% [wt/vol] sodium dodecyl sulfate at 62°C) conditions, and bound RNA-DNA hybrids were detected with a Boehringer Mannheim detection kit.
Nucleotide sequence accession number.
The DNA sequence data described in this paper have been deposited in the EMBL nucleotide sequence database under accession no. AJ002527.
RESULTS
Glucitol metabolism by C. beijerinckii.
C. beijerinckii NCIMB 8052 can grow on glucitol as a sole fermentable carbon source. It has been shown that this substrate is accumulated via a PEP-dependent phosphotransferase system (PTS) whose synthesis is induced by glucitol (13). The result of PTS-mediated uptake is intracellular accumulation of glucitol 6-phosphate. Extracts prepared from cells grown on glucitol exhibited an NAD-specific glucitol 6-phosphate dehydrogenase activity which was detected in both the qualitative assay and the quantitative assay, and as observed for the PTS, this activity was absent from cells grown on other carbon sources. The mechanism of glucitol metabolism in C. beijerinckii, therefore, appeared to be the same as the mechanism of glucitol metabolism in Clostridium pasteurianum and E. coli, i.e., accumulation and phosphorylation via a PTS, followed by oxidation of the resulting glucitol 6-phosphate to form fructose 6-phosphate (21, 33). The synthesis of these activities was apparently coordinately controlled. Since the PTS is a complex, multiprotein system, the most straightforward approach to obtain the gut genes of C. beijerinckii was to clone the gutD gene encoding the glucitol 6-phosphate dehydrogenase by complementing a gutD mutant strain of E. coli.
Construction of E. coli WM4 and cloning of the C. beijerinckii gutD gene.
To facilitate cloning of the C. beijerinckii gutD gene, an E. coli host strain carrying a gutD mutation in a defined genetic background was constructed. A P1 transducing lysate was prepared from strain JC10279 and used to infect strain JM83. Thirteen colonies were isolated on MacConkey agar containing glucitol and tetracycline, and all of these colonies had a negative fermentation phenotype. One transductant (WM4) was selected for further analysis. This strain was not able to grow on M9 minimal medium containing glucitol as the only source of carbon and exhibited no glucitol 6-phosphate dehydrogenase activity in the qualitative assay. Therefore, this strain was used as a host for cloning of the C. beijerinckii gutD gene.
A C. beijerinckii NCIMB 8052 gene library was constructed in pAT153 as described above and was used to transform E. coli WM4. Transformants were plated onto M9 minimal medium containing 0.2% glucitol, and only transformants which carried an intact copy of the gutD gene should have been able to grow on this medium. A plasmid isolated from one of the transformant colonies was retransformed into strain WM4, which gained the ability to grow on glucitol and exhibited glucitol 6-phosphate dehydrogenase activity. This plasmid, pSORB1, was found to carry a 5.7-kbp DNA insert which was shown by Southern hybridization to be derived from C. beijerinckii (data not shown).
Nucleotide sequence analysis and identification of the gut genes.
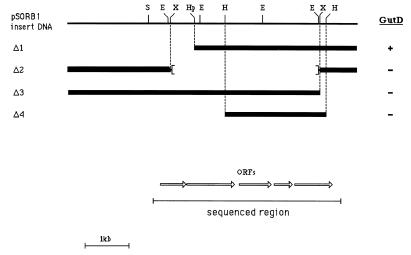
After the positions of restriction enzyme digestion sites were determined, deletions of pSORB1 were constructed, and the abilities of the deleted plasmids to complement E. coli WM4 for growth on glucitol were investigated. The results (Fig. 1) indicated that deletion of an internal 3.3-kbp XbaI fragment or of a terminal 0.66-kbp region of the insert resulted in loss of gutD function. A 2.2-kbp HindIII fragment, which was also unable to complement E. coli WM4, and flanking regions subcloned as discrete restriction fragments were sequenced, and the sequence revealed that there were five ORFs (Fig. 1 and 2), each of which was preceded by a putative ribosome binding site with a spacing of 7 to 12 nucleotides. The deduced amino acid sequences of four of the ORFs exhibited significant homology to proteins encoded by the gut operon of E. coli (Fig. 3A). The most distal ORF, gutD (nucleotides 3029 to 3841), apparently encodes glucitol 6-phosphate dehydrogenase. The gene product is homologous throughout its length (25% identity) to the glucitol 6-phosphate dehydrogenase from E. coli (Fig. 3B). It is also homologous to a number of other dehydrogenases and is most closely related (58.7% identity) to the glucitol 6-phosphate dehydrogenase of Klebsiella aerogenes. A mutant strain of C. beijerinckii, AA219, has been constructed in which the gutD gene is inactivated due to integration of a nonreplicative plasmid (31). We examined this strain, which is unable to grow on glucitol, and found that extracts prepared following growth on synthetic RCM containing glucitol as the only fermentable carbon source lacked glucitol 6-phosphate dehydrogenase activity. Therefore, we concluded that the gutD gene does encode glucitol 6-phosphate dehydrogenase.
FIG. 1.
Restriction map of C. beijerinckii DNA insert in pSORB1. The abilities of various deletion derivatives of the 5.7-kbp insert to complement E. coli WM4 for growth on glucitol are indicated on the right. The sequence data revealing five ORFs were obtained by sequencing (i) the internal HindIII fragment (Δ4) (ii) subcloning and sequencing HindIII-HpaI, HpaI-EcoRI, and EcoRI-ScaI fragments upstream, and (iii) subcloning the entire downstream region as a HindIII-SalI fragment (SalI site within the vector) and sequencing to a position beyond the final ORF (see text). S, ScaI; E, EcoRI; X, XbaI; Hp, HpaI; H, HindIII.
FIG. 3.
Comparison of the cloned C. beijerinckii sequence with the Gut system of E. coli. (A) Schematic diagram showing the arrangement of the ORFs of the cloned C. beijerinckii DNA and the gut operon of E. coli. The arrows indicate the directions of transcription. (B) Alignment of the C. beijerinckii sequences with the Gut proteins of E. coli. The deduced amino acid sequences of clostridial ORFs A1, A2, B, and D are aligned with the E. coli proteins GutA, GutB, and GutD (32). The clostridial sequences are preceded by the letter C. The numbers indicate the positions in the protein sequences. Dashes in the sequences indicate gaps that resulted in optimal alignment, while dots indicate identical residues. The protein sequence for A1 is aligned with the N-terminal portion of the E. coli GutA protein. The final amino acid of A1 (at position 182) is underlined. The A2 sequence is aligned with the remainder of the E. coli GutA protein. The first amino acid of the deduced A2 sequence is underlined and marked by the number 1.
The glucitol operon of E. coli comprises three structural genes encoding the membrane-bound (gutA; enzyme IICBCgut) and soluble (gutB; enzyme IIAgut) components of the glucitol PTS, as well as glucitol 6-phosphate dehydrogenase (18, 32, 33) (Fig. 3A). Three of the ORFs in the gut operon of C. beijerinckii encode components of the glucitol-specific PTS. The first two gene products are homologous to enzyme IICBCgut of E. coli, while the third is homologous to enzyme IIAgut (Fig. 3). A notable difference between the C. beijerinckii and E. coli systems is that the GutA (enzyme IICBCgut) protein of C. beijerinckii is apparently synthesized as two polypeptide chains, GutA1 (182 amino acids; predicted Mr, 20,187) and GutA2 (336 amino acids; predicted Mr, 35,179). The sequence data were supported by the results of in vitro transcription-translation studies, which showed that the largest protein band expressed uniquely from pSORB1 had an estimated Mr of approximately 35,000 (data not shown), which corresponds to the Mr of the putative GutA2 protein, which is the largest product encoded by the predicted ORFs in the gut system. This molecule is considerably smaller than the entire GutA protein, which has a predicted Mr of more than 55,000. The ORFs gutA1 (nucleotides 140 to 685) and gutA2 (nucleotides 706 to 1713), which encode the N- and C-terminal portions of enzyme II, respectively, were separated by 20 nucleotides. The two segments of enzyme II were homologous to the corresponding regions of E. coli enzyme IICBCgut, and alignment of the sequences indicated that there was no gap in either protein (Fig. 3B).
The product of the ORF gutB (nucleotides 2578 to 2943) is homologous throughout its length (33.6% identity) to enzyme IIAgut of E. coli (Fig. 3B). Therefore, the gene order in E. coli, in which gutB precedes gutD, is conserved in C. beijerinckii. The putative protein comprises 122 amino acids and has a predicted Mr of 13,245. An alternative start codon was identified at nucleotides 2566 to 2568, but this start codon is not favored due to its proximity to the putative ribosome binding site.
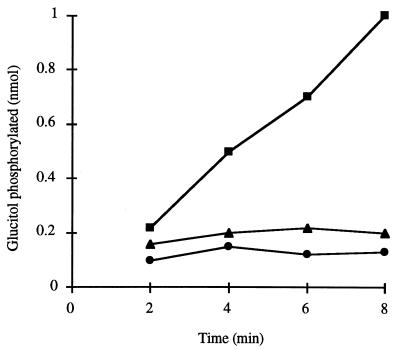
The existence of distinct gutA and gutB genes encoding enzymes IICBC and IIA indicates that the glucitol PTS comprises a specific protein in both the membrane and the cytoplasm. All phosphotransferases are dependent on a substrate-specific, membrane-bound permease and on the general, soluble proteins enzyme I and HPr, and activity in extracts therefore requires both membrane and cytoplasmic fractions. The presence of a substrate-specific soluble protein can, however, be demonstrated by combining fractions from different extracts which are induced or uninduced for the system and assaying for phosphorylation of the test substrate. Glucose-grown cells of C. beijerinckii have been shown to lack glucitol PTS activity (13). The ability of a membrane or soluble fraction of an extract prepared from these cells to complement the other fraction from glucitol-grown cells for glucitol phosphorylation was therefore examined, and the results are shown in Fig. 4. Although the combined fractions from glucitol-grown cell extracts were clearly active, glucose-grown cell membranes, as expected, were not able to function in the presence of the glucitol-grown cell soluble fraction due to a lack of membrane-bound enzyme II. In addition, the glucose-grown cell soluble fraction could not complement glucitol-grown cell membranes, despite the fact that since glucose is a PTS substrate, both enzyme I and HPr must have been present (17). These results show that a glucitol-specific, soluble protein is not present in the glucose-grown cell extract, confirming that, as in E. coli, the enzyme IIA function (i.e., the gutB gene product) is synthesized as a separate protein.
FIG. 4.
Glucitol phosphorylation in reconstituted cell extracts of C. beijerinckii. Extracts were prepared from cultures grown on either glucose or glucitol as the sole carbon source. Extracts were fractionated into membrane and cytosol components. Extract preparation and fractionation were performed as described in the text. Combined purified membranes and cytosols were assayed for glucitol PTS activity in phosphorylation assays, as described in the text. The assay mixtures contained the following combinations of membrane and cytosol: glucitol-grown cell membranes and glucose-grown cell cytosol (•); glucitol-grown cell cytosol and glucose-grown cell membranes (▴); and glucitol-grown cell membranes and glucitol-grown cell cytosol (▪).
The fifth ORF, located between gutA2 and gutB at nucleotides 1833 to 2510, encodes a putative 226-amino-acid protein (predicted Mr, 25,137). There is no equivalent gene in the E. coli gut operon. The coding region of this ORF starts with the codon GTG. The protein product is not homologous to any known PTS component or to enzymes directly involved in metabolism of glucitol or glucitol 6-phosphate. It is most closely related (37.4% identity) to the talC gene product of E. coli, which has recently been shown to be a member of the transaldolase protein family (19). This suggests that OrfX may be an enzyme with transaldolase-like activity. The possible significance of such a function is discussed below.
Expression of the gut genes.
Cells of C. beijerinckii grown on glucitol possess glucitol PTS and glucitol 6-phosphate dehydrogenase activities, which are absent from cells grown on other carbon sources, such as glucose and xylose. In addition, glucose has been shown to prevent utilization of glucitol in cultures containing both carbon sources (13). Therefore, expression of the gut genes was studied both by monitoring enzyme activities and by detecting mRNA as described above. Induction of the glucitol system at the level of transcription was observed after glucitol was added to cells which had been grown on xylose (data not shown). To examine the effect of glucose on expression of the gut system, a 10-liter culture was prepared, and samples were taken periodically and used to prepare extracts, isolate and analyze mRNA, and monitor sugar utilization in the culture. As shown in Fig. 5A, the cells initially grew rapidly on glucose; the glucose was completely depleted after 8 h, at which point the growth rate slowed markedly. Glucitol utilization was first detected at around 6 h and continued until the end of the experiment at 28 h. Enzyme assays revealed that neither glucitol PTS activity nor glucitol 6-phosphate dehydrogenase activity could be detected during the first phase of growth (Table 1), but these activities were induced later. On the other hand, glucose PTS activity was present throughout the culture period and in fact increased four- to fivefold during the glucitol utilization phase. RNA produced from the gut genes was detected in slot blots by using DNA probes directed against internal regions of each ORF. As shown in Fig. 5B for expression of gutA2, gut-specific RNA was not detected during the first phase of growth. Then there was a burst of RNA synthesis close to the time at which glucitol utilization began. Later, the quantity of mRNA declined, although mRNA was still clearly detectable. Similar results were observed for expression of the other genes (data not shown), indicating that they are coordinately expressed. These results confirm that the gut genes in C. beijerinckii are required for glucitol metabolism and demonstrate that transcription of the gut system is repressed by glucose.
FIG. 5.
Culture growth, carbohydrate utilization, and gut gene expression in C. beijerinckii. (A) A 10-liter culture was grown in a defined medium containing glucose and glucitol. The optical density at 600 nm (OD600) (•) was monitored throughout growth. The concentrations of glucose (▴) and glucitol (▪) in the culture supernatant were determined at intervals, as described in the text. (B) Six sets of samples were taken at intervals and used to isolate RNA and prepare cell extracts as described in the text. The arrows indicate the times of sampling. (Inset) Slot blot hybridization of the RNA samples probed with a digoxigenin-labelled probe to ORF gutA2. Slots 1 through 6 in the inset correspond to sample time points 1 through 6, respectively, on the graph.
TABLE 1.
Enzyme activities in extracts of C. beijerinckii cells grown on glucitol and glucosea
| Sampleb | Glucitol 6-phosphate dehydrogenase activity (nmol · mg of protein−1 · min−1) | PTS activity (pmol · mg of protein−1 · min−1)
|
|
|---|---|---|---|
| Glucose phosphorylation | Glucitol phosphorylation | ||
| 1 | NDc | 132 | ND |
| 2 | ND | 139 | ND |
| 3 | 0.01 | 701 | ND |
| 4 | 0.03 | 770 | ND |
| 5 | 1.20 | 707 | 197 |
| 6 | 1.44 | 539 | 193 |
Extracts were prepared and assayed for each activity as described in the text.
Samples 1 through 6 correspond to sample points 1 through 6, respectively, in Fig. 5B.
ND, not detected.
DISCUSSION
By selecting for complementation of a gutD mutant of E. coli defective in the enzyme glucitol 6-phosphate dehydrogenase, we isolated a clone of C. beijerinckii which carried a 5.7-kbp insert of clostridial DNA. Sequencing of a 4.0-kbp region of the insert revealed the presence of five ORFs, four of which encoded proteins homologous to the gut gene products of E. coli and could be associated with functions involved in the uptake, phosphorylation, and oxidation of glucitol. The order of these four genes (gutA1-gutA2-gutB-gutD) was the same as the order in the gut operon of E. coli. An important difference, however, was that the gutA gene of E. coli was replaced by two genes, gutA1 and gutA2, in C. beijerinckii. Each of the two clostridial genes encoded a protein which was homologous to part of the E. coli glucitol permease, and sequence alignment showed that the two segments of the C. beijerinckii protein were contiguous. Sequence and structural comparisons of the glucitol permeases of E. coli and C. beijerinckii have shown that each consists of two membrane-bound domains (designated domains IIC-N and IIC-C to indicate the termini of the proteins) with a hydrophilic, cytoplasmic domain (domain IIB) between them. A topological model has been proposed, in which each portion of domain IIC contains four transmembrane segments and is connected to the intervening domain IIB by a flexible linker. The break in the clostridial permease occurs at the end of the domain IIC-N (18). It is believed that all bacterial PTSs arose from a single common ancestor but that extensive gene duplication and rearrangement, as well as domain shuffling, occurred throughout evolutionary time, generating the wide variety of PTS permeases known today (23). So far, the molecular architecture of the E. coli glucitol permease, which is a three-domain IICBC protein, has been unique. Furthermore, until now this permease did not exhibit significant similarity to other PTS permeases whose sequences have been determined. While clearly homologous to its counterpart in E. coli, the C. beijerinckii glucitol permease represents yet another variation because it consists of two polypeptide chains. The sequence similarity of the two permeases, in both the enzyme IICBC and IIA proteins, suggests that a glucitol-specific PTS segregated before the evolutionary divergence of the two bacteria.
While the membrane-bound IIC domains of the PTS provide a membrane channel for translocation of the substrate, the function of domains IIA and IIB appears to be to transfer the phosphoryl group to the incoming substrate. In the majority of cases, IIB domains are phosphorylated at a conserved cysteine residue (10, 23). The putative phosphorylation site in C. beijerinckii domain IIBgut is Cys-75, which corresponds to Cys-258 in the E. coli enzyme IICBCgut sequence (18). This cysteine is in a 13-amino-acid sequence that is perfectly conserved in the two proteins (Fig. 3B). The PTS IIA domains are always phosphorylated on a histidine residue (10, 23). The C. beijerinckii gutB gene product contains just one histidine (His-85), which corresponds to His-84 in the E. coli IIAgut sequence. This residue is in the region of greatest identity when the two proteins are aligned and is therefore strongly implicated as the phosphorylated amino acid in the protein. However, by comparing the enzyme IIAgut and IIAmtl domains of E. coli, which were not homologous, Saier et al. (25) identified a motif at a different position in enzyme IIAgut; Saier et al. suggested that this motif could correspond to a phosphorylation site. Identification of the actual site of enzyme IIAgut phosphorylation in both organisms will require further experimental analysis.
The function of the fifth ORF in the C. beijerinckii gut system, which is located between gutA2 and gutB, is not known. The deduced gene product exhibits similarities to transaldolase-like enzymes, but there is no evidence at present that orfX does in fact encode a transaldolase. An enzyme with transaldolase activity could, however, play an important role in cells growing on glucitol. When cells are grown on this carbon source, fructose 6-phosphate is generated as an oxidation product of glucitol 6-phosphate. While fructose 6-phosphate can be metabolized by glycolysis to generate ATP and metabolic precursors, it is also a substrate of transaldolase. Thus, if the final reaction of the pentose phosphate pathway is reversed, fructose 6-phosphate may be mobilized to produce pentose phosphates as precursors of nucleic acid synthesis. However, since orfX expression is inducible by glucitol, C. beijerinckii would have to possess a second transaldolase gene which is expressed during growth on other carbon sources.
Our results show that the C. beijerinckii gut system is both induced by glucitol and repressed by glucose, but the mechanisms by which expression is controlled have not been identified yet. Analysis of the sequence shown in Fig. 2 failed to identify any obvious promoter sequences, suggesting that transcription may be initiated some distance upstream of the sequenced genes. A number of factors may contribute to regulation of expression of the gut system. For example, it has been shown that uptake of glucitol by this organism is inhibited in the presence of glucose (13). In C. pasteurianum, similar inducer exclusion was shown to occur as a result of competition between the glucose and glucitol PTSs for a common supply of PEP, which in molecular terms equates to competition for the phospho-HPr which acts as the phosphoryl donor for the enzyme II complexes (21). A similar mechanism may be envisaged in C. beijerinckii. In addition, there is increasing evidence that the HPr protein (in particular, its phosphorylation by an ATP-dependent protein kinase) plays a pivotal role in regulation of carbohydrate metabolism in gram-positive bacteria (24). Further analysis of the clostridial PTS may therefore contribute to our understanding of the regulation of glucitol metabolism and carbohydrate metabolism in general. Industrial carbon sources are likely to contain a mixture of carbohydrates, the utilization of which often is sequential as a result of catabolite repression. The result is inefficient conversion of raw materials to the desired product. We identified a gene system in C. beijerinckii which is concerned with the metabolism of glucitol. The gene products are both necessary and sufficient to enable glucitol to serve as a carbon source for growth and are activated and repressed in response to the availability of the substrate and other carbon sources in the medium. Further elucidation of the mechanisms of control of this catabolic gene system should contribute significantly to our understanding of the physiology of the solvent-forming clostridia and in turn may help workers identify strategies by which the organisms may be manipulated to advantage in fermentation processes.
ACKNOWLEDGMENTS
We are grateful to A. J. Clark and M. Young for providing bacterial strains.
This work was supported by research grant T04089 from the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Aduse-Opoku J, Mitchell W J. Diauxic growth of Clostridium thermosaccharolyticum on glucose and xylose. FEMS Microbiol Lett. 1988;50:45–49. [Google Scholar]
- 2.Albasheri K A, Mitchell W J. Identification of two α-glucosidase activities in Clostridium acetobutylicum NCIB 8052. J Appl Bacteriol. 1995;78:149–156. [Google Scholar]
- 3.Booth I R, Mitchell W J. Sugar transport and metabolism in the clostridia. In: Reizer J, Peterkofsky A, editors. Sugar transport and metabolism in Gram-positive bacteria. Chichester, United Kingdom: Ellis-Horwood; 1987. pp. 165–185. [Google Scholar]
- 4.Chambers S P, Prior S E, Barstow D A, Minton N P. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1989;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 5.Csonka L S, Clark A J. Deletions generated by the transposon Tn10 in the srl recA region of the Escherichia coli chromosome. Genetics. 1979;93:321–343. doi: 10.1093/genetics/93.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gachelin G. A new assay of the phosphotransferase system in Escherichia coli. Biochem Biophys Res Commun. 1969;34:382–387. doi: 10.1016/0006-291x(69)90392-1. [DOI] [PubMed] [Google Scholar]
- 7.Jones D T, Keis S. Origins and relationships of industrial solvent-producing clostridial strains. FEMS Microbiol Rev. 1995;17:223–232. [Google Scholar]
- 8.Jones D T, Woods D R. Acetone-butanol fermentation revisited. Microbiol Rev. 1986;50:484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones D T, Woods D R. Solvent production. In: Minton N P, Clarke D J, editors. Clostridia. New York, N.Y: Plenum Press; 1989. pp. 105–144. [Google Scholar]
- 10.Lengeler J W, Jahreis K, Wehmeier U F. Enzymes II of the phosphoenolpyruvate-dependent phosphotransferase systems: their structure and function in carbohydrate transport. Biochim Biophys Acta. 1994;1188:1–28. doi: 10.1016/0005-2728(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 11.Minton N P, Brehm J K, Swinfield T J, Whelan S M, Mauchline M L, Bodsworth N, Oultram J D. Clostridial cloning vectors. In: Woods D R, editor. The clostridia and biotechnology. Stoneham, Mass: Butterworth-Heinemann; 1993. pp. 119–150. [Google Scholar]
- 12.Minton N P, Atkinson T, Sherwood R F. Molecular cloning of the Pseudomonas carboxypeptidase G2 gene and its expression in Escherichia coli and Pseudomonas putida. J Bacteriol. 1983;156:1222–1227. doi: 10.1128/jb.156.3.1222-1227.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell W J. Carbohydrate uptake and utilization by Clostridium beijerinckii NCIMB 8052. Anaerobe. 1996;2:379–384. [Google Scholar]
- 14.Mitchell W J. Physiology of carbohydrate to solvent conversion by clostridia. Adv Microb Physiol. 1998;39:31–130. doi: 10.1016/s0065-2911(08)60015-6. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell W J, Albasheri K A, Yazdanian M. Factors affecting utilization of carbohydrates by clostridia. FEMS Microbiol Rev. 1995;17:317–329. [Google Scholar]
- 16.Mitchell W J, Booth I R. Characterization of the Clostridium pasteurianum phosphotransferase system. J Gen Microbiol. 1984;130:2193–2200. [Google Scholar]
- 17.Mitchell W J, Shaw J E, Andrews L. Properties of the glucose phosphotransferase system of Clostridium acetobutylicum NCIB 8052. Appl Environ Microbiol. 1991;57:2534–2539. doi: 10.1128/aem.57.9.2534-2539.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reizer J, Mitchell W J, Minton N, Brehm J, Reizer A, Saier M H., Jr Proposed topology of the glucitol permeases of Escherichia coli and Clostridium acetobutylicum. Curr Microbiol. 1996;33:331–333. doi: 10.1007/s002849900123. [DOI] [PubMed] [Google Scholar]
- 19.Reizer J, Reizer A, Saier M H., Jr Novel phosphotransferase system genes revealed by bacterial genome analysis—a gene cluster encoding a unique Enzyme I and the proteins of a fructose-like permease system. Microbiology. 1995;141:961–971. doi: 10.1099/13500872-141-4-961. [DOI] [PubMed] [Google Scholar]
- 20.Rogers P, Gottschalk G. Biochemistry and regulation of acid and solvent production in clostridia. In: Woods D R, editor. The clostridia and biotechnology. Stoneham, Mass: Butterworth-Heinemann; 1993. pp. 25–50. [Google Scholar]
- 21.Roohi M S, Mitchell W J. Regulation of sorbitol metabolism by glucose in Clostridium pasteurianum: a role for inducer exclusion. J Gen Microbiol. 1987;130:2207–2215. [Google Scholar]
- 22.Saier M H., Jr A multiplicity of potential carbon catabolite repression mechanisms in prokaryotic and eukaryotic microorganisms. New Biol. 1991;3:1137–1147. [PubMed] [Google Scholar]
- 23.Saier M H, Jr, Reizer J. Proposed uniform nomenclature for the proteins and protein domains of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. J Bacteriol. 1992;174:1433–1438. doi: 10.1128/jb.174.5.1433-1438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saier M H, Jr, Chavaux S, Cook G M, Deutscher J, Reizer J, Ye J-J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 25.Saier M H, Jr, Reizer A, Pao G M, Wu L-F, Reizer J, Romano A H. Evolution of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. II. Molecular considerations. In: Mortlock R P, editor. The evolution of metabolic function. Boca Raton, Fla: CRC Press; 1992. pp. 171–204. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. pp. A1–A13. [Google Scholar]
- 27.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 107–112. [Google Scholar]
- 28.Tangney M, Smith P, Priest F G, Mitchell W J. Maltose transport in Bacillus licheniformis NCIB 6346. J Gen Microbiol. 1992;138:1821–1827. [Google Scholar]
- 29.Twigg A J, Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature (London) 1980;283:216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- 30.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson S R, Young M. Targeted integration of genes into the Clostridium acetobutylicum chromosome. Microbiology. 1994;140:89–95. [Google Scholar]
- 32.Yamada M, Saier M H., Jr Glucitol-specific enzymes of the phosphotransferase system in Escherichia coli. Nucleotide sequence of the gut operon. J Biol Chem. 1987;262:5455–5463. [PubMed] [Google Scholar]
- 33.Yamada M, Saier M H., Jr Physical and genetic characterization of the glucitol operon in Escherichia coli. J Bacteriol. 1987;169:2990–2994. doi: 10.1128/jb.169.7.2990-2994.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 35.Zamenhof S. Preparation and assay of deoxyribonucleic acid from animal tissue. Methods Enzymol. 1957;3:696–704. [Google Scholar]