Abstract
OBJECTIVE
To investigate gastrointestinal infection episodes (GIEs) in relation to the appearance of islet autoantibodies in The Environmental Determinants of Diabetes in the Young (TEDDY) cohort.
RESEARCH DESIGN AND METHODS
GIEs on risk of autoantibodies against either insulin (IAA) or GAD (GADA) as the first-appearing autoantibody were assessed in a 10-year follow-up of 7,867 children. Stool virome was characterized in a nested case-control study.
RESULTS
GIE reports (odds ratio [OR] 2.17 [95% CI 1.39–3.39]) as well as Norwalk viruses found in stool (OR 5.69 [1.36–23.7]) at <1 year of age were associated with an increased IAA risk at 2–4 years of age. GIEs reported at age 1 to <2 years correlated with a lower risk of IAA up to 10 years of age (OR 0.48 [0.35–0.68]). GIE reports at any other age were associated with an increase in IAA risk (OR 2.04 for IAA when GIE was observed 12–23 months prior [1.41–2.96]). Impacts on GADA risk were limited to GIEs <6 months prior to autoantibody development in children <4 years of age (OR 2.16 [1.54–3.02]).
CONCLUSIONS
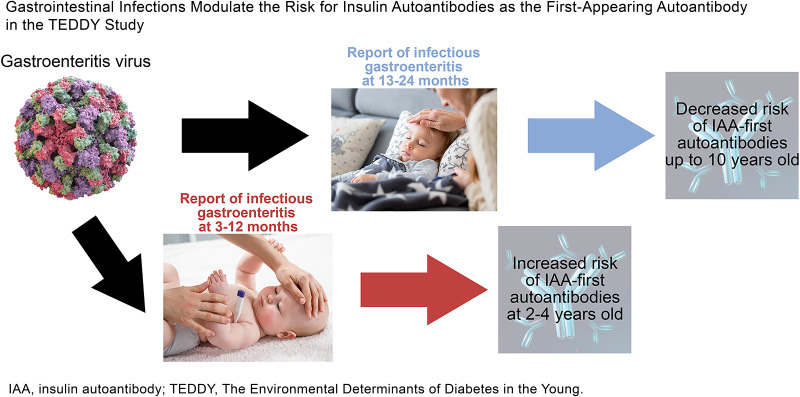
Bidirectional associations were observed. GIEs were associated with increased IAA risk when reported before 1 year of age or 12–23 months prior to IAA. Norwalk virus was identified as one possible candidate factor. GIEs reported during the 2nd year of life were associated with a decreased IAA risk.
Graphical Abstract
Introduction
The Environmental Determinants of Diabetes in the Young (TEDDY) study is a large, multinational, prospective birth cohort aimed at identifying and characterizing environmental factors at the onset of islet autoimmunity (IA) and its progression to clinical type 1 diabetes (T1D) in subjects at increased genetic risk for T1D (1–3). Respiratory infections, the most common infection type reported in TEDDY subjects (4), have been associated with the onset of IA, as well as with male subjects’ progression to clinical T1D (5,6). Other studies have also found respiratory infections to be associated with IA (7–9). Furthermore, gestational respiratory infections modified the association between CTLA-4 gene polymorphism and the risk of insulin autoantibody (IAA) seroconversion in offspring (10). In line with this, certain viruses causing respiratory symptoms, particularly enteroviruses, have been linked to the risk of T1D (11–13). In addition, viruses that replicate both in the respiratory and gastrointestinal (GI) tract, such as enterovirus, or in the GI tract, such as rotavirus, have been linked to IA and T1D (14–16). The mechanisms of these associations are not known. It is possible that enteral viruses can spread from the intestinal mucosa and gut-associated immune system to the closely located pancreas via common lymphatic networks and infect islet cells. Transmission to the pancreas may also occur via blood, since enteral viruses have been detected in the blood during acute infection. The frequent detection of enterovirus protein and RNA in the pancreas of patients with T1D supports the possible role of virus in infecting pancreatic islet cells (17–19).
The current study sought to identify associations between prospective reports of GI infections, the second most common type of infections in TEDDY subjects (4), and onset of IA defined by the first-appearing islet autoantibody during the first 10 years of life. The prospective study design’s frequent monitoring of autoantibodies, comprehensive collection of questionnaire data on infections, and use of modern sequencing technologies to detect viruses in longitudinal stool sample series (13) allowed us to identify time-dependent associations at different ages and correlate these associations with genetic and other host factors in relation to which islet autoantibody was first detected.
Research Design and Methods
Participants
Details of the study design can be found in previous publications (1–3). Six clinical research centers: three in the U.S. (Colorado, Georgia/Florida, and Washington State) and three in Europe (Finland, Germany, and Sweden) participated in a population-based HLA screening of newborns between 2004 and 2010. After screening, 8,676 children with HLA haplotypes conferring an increased risk of T1D were enrolled between 3 and 4.5 months of age (20). The children are followed until 15 years of age, and the study is ongoing. Study clinic visits include a blood draw every 3 months until 4 years of age, and every 6 months thereafter. Stool samples are collected monthly between 3 and 48 months of age and quarterly thereafter until 10 years of age (21). Written informed consent was obtained from the primary caregiver separately for genetic screening and participation in the follow-up. TEDDY has been approved by local institutional ethics boards and is monitored by an external evaluation committee. The current study with data frozen as of August 2020 included 7,867 children who were prospectively followed for the development of IA until their 10th birthday. Participants missing four or more consecutive visits were considered withdrawn after the date of their last visit. Virome was analyzed in a subset of the cohort: a nested case-control study included 383 case children who presented with IA by 31 May 2012 (median age 21 months, interquartile range [IQR] 13–33 months), along with one matching control child for each case child selected from risk sets that did not develop IA by at least 6 months of age after the date of the case event (13).
Islet Autoantibodies
Islet autoantibodies to insulin (IAA), GAD (GADA), and IA2 (IA-2A) were analyzed from blood samples (1,2). Persistent IA was defined by the presence of an islet autoantibody (GADA, IA-2A, or IAA) at each of the two TEDDY reference laboratories on two or more consecutive visits. The U.S. reference laboratory was the Barbara Davis Center for Childhood Diabetes at the University of Colorado Denver, and the European reference laboratory was the University of Bristol in the U.K. All autoantibody-positive samples, as well as 5% of negative samples, were reanalyzed by the other reference laboratory. Both laboratories showed high sensitivity, specificity, and concordance (22). Onset of IA was the age at which a confirmed and persistent autoantibody was first detected (23).
Reported GI Infections
Illnesses were recorded by the parents at home in a diary. At each clinic visit, illnesses reported since the previous visit were translated into ICD-10 diagnosis codes by study nurses (5). Infectious disease data processing and the infection episode approach have been previously described (4). In this study, GI infection episodes (GIEs) were identified as a record of an ICD-10 code for an infective gastroenteritis.
Stool Virome
The virome of stool samples collected monthly from 3 months of age until IA development was characterized using Illumina mass sequencing, which can identify widely different kinds of human viruses as previously described (13). Virome analyses were originally performed as part of the nested case-control study with 383 IA case children and risk set control children who were additionally matched by study site, sex, and family history of T1D. Stool samples from 751 children (379 case-control pairs) were eligible and available for the current study. Altogether, 7,202 stool samples of children up to 24 months of age collected monthly prior to IA onset were analyzed for association of common viruses (present in >2.5% of samples) with GIE reports within 2 weeks of stool sample collection.
Other Factors
TEDDY has previously identified other predictors of IA, including gestational respiratory infections (10), upper respiratory infections before 3 months of age (6), respiratory infectious episodes (5), and the use of probiotics before 28 days of age (24). Therefore, HLA and other genetic risk factors as described elsewhere were considered as potential confounders or effect modifiers, as some of these factors were related to age of seroconversion, particularly with type of first-appearing islet autoantibody (IAA or GADA) (5,9). Single nucleotide polymorphism (SNP) genotyping was performed by the Center for Public Health Genomics at the University of Virginia using the Illumina immunochip, a custom array for genotyping selective SNPs from regions of the human genome associated with autoimmune diseases (6,25).
Statistical Methods
GIEs and IA were examined across discrete time intervals at scheduled visits when both a blood draw sample and a diary of reported infections since the last visit were obtained. Time-varying correlations of GIEs with first-appearing islet autoantibodies were assessed using discrete cause-specific hazards models. The models were specified by multinomial logistic regression models with four categories (IAA-first, GADA-first, other IA, and a category for survival beyond the discrete event time) (26,27). The effect measures are reported as odds ratios (ORs) with 95% CIs for IAA or GADA as the first-appearing islet autoantibody. The effect of GIEs was modeled prospectively by several time-varying predictors on two different timescales: child’s age (in years) at infectious exposure and year lag from infection to IA. The age of GIE was modeled as a step function, and GIEs prior to IA were modeled as a lag function. Prior infections were defined as the time prior to the last scheduled visit before the child developed IA. Secondary analysis further examined associations by age of seroconversion, by season of GIE report and autoantibody development, and for clustering over shorter time periods. P values were Bonferroni adjusted to correct for familywise error rate. All discrete time survival models were adjusted for sex, HLA-DR-DQ genotype, family history of T1D, country, and age of child, with age modeled as a quadratic polynomial. The time-varying GIE predictors were examined separately in relation to either IAA or GADA as the first-appearing autoantibody, as well as later with single islet autoantibody overall (IAA-first, GADA-first, IA-2A-first) and multiple islet autoantibodies at seroconversion, which represented two or more islet autoantibodies appearing for the first time since the last blood draw.
A generalized linear mixed model with logit link was fitted to examine how common viruses (>2.5%) in stool were related to a child’s specific odds of having a GIE reported within 2 weeks of stool sample collection. The model included all viruses and adjusted for site, sex, family history of T1D, and age of the child when the sample was taken. The aim of this analysis was to identify which viruses were independently related to the likelihood of reporting a GIE. Based on results in the full cohort, it was intended for specific viruses associated with reported GIEs to be examined at important times of interest in relation to IA development using the nested case-control design. All viruses were individually examined in relation to IA by conditional logistic regression models, with models adjusted for matching factors (site, sex, and family history of T1D) and HLA-DR-DQ genotype. Results from conditional logistic regression models and generalized mixed models using the nested case-control study were described as ORs with 95% CIs. To examine potential confounding, the final models were adjusted for factors that have previously shown an association with GIE or the appearance of autoantibodies in TEDDY. Unless otherwise stated, P < 0.05 was considered significant. All statistical analyses were performed using the software SAS 9.4 (SAS Institute Inc., Cary, NC), R, and GraphPad Prism 5.03 (GraphPad Software Inc., San Diego, CA) for figures.
Data and Resource Availability
The data sets generated and analyzed during the current study will be made available in the National Institute of Diabetes and Digestive and Kidney Diseases Central Repository at https://repository.niddk.nih.gov/studies/teddy.
Results
Incidence of First-Appearing Islet Autoantibodies and GIEs
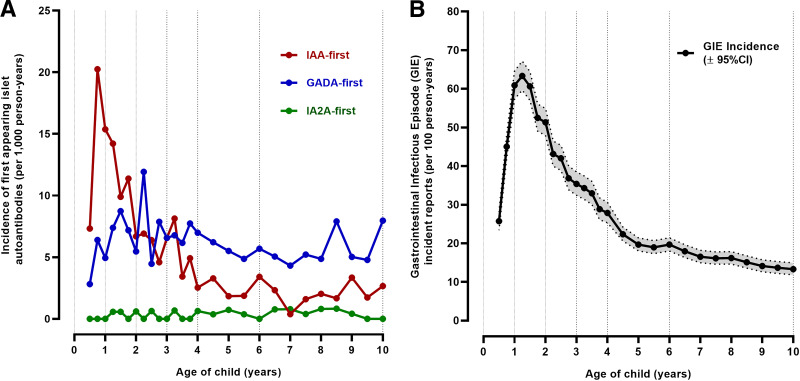
Islet autoantibodies developed in 763 (9.7%) children. IAA was the first-appearing autoantibody in 283 (37.1%) children; 333 (43.6%) had GADA-first, 20 (2.6%) had IA-2A-first, 92 (12.1%) had both IAA and GADA with no determination of which was first, and 35 (4.6%) had other autoantibody combinations. The low number of children with IA-2A-first did not allow meaningful statistical analyses in this subgroup. The peak incidence of IAA-first was between 6 and 9 months of age (20 per 1,000 person-years), with a rapid decline in incidence between 9 and 24 months of age and a median age of seroconversion at 2.0 years (IQR 1–3.9 years) (Fig. 1A). Incidence of GADA-first rose to 12 per 1,000 person-years at age 27 months and then stabilized to 5–7 per 1,000 person-years from 2 to 9 years of age (median seroconversion age 4.5 years, IQR 2.3–7.4 years). While children were still observed at risk for IA, incidence of GIEs peaked at 15 months of age (63.3 per 100 person-years) (Fig. 1B).
Figure 1.
Age-specific incidence of IAA, GADA, and IA-2A as the first-appearing autoantibody (A) and GIEs (B).
Age-Specific GIE Reports on IAA and GADA Risk
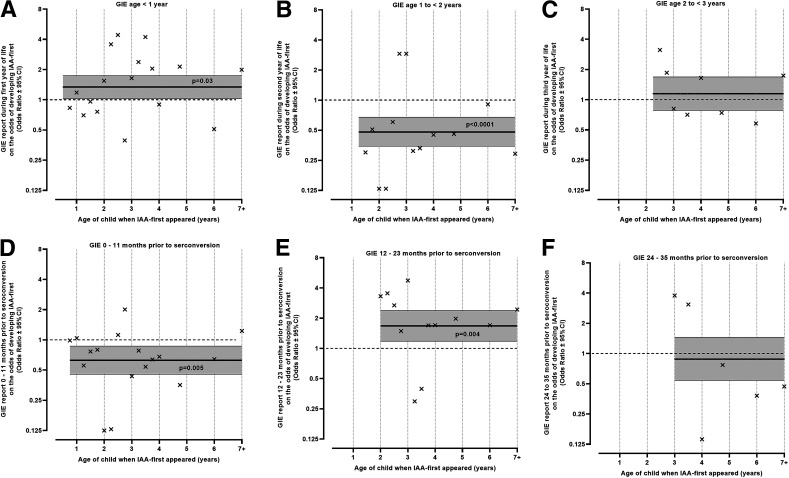
A GIE reported at 3–12 months of age was associated with a higher odds of IAA (P = 0.03), especially at 25–60 months of age (OR 2.17 [95% CI 1.39–3.39], P = 0.0006) (Fig. 2A). A GIE reported at 13–24 months of age was associated with a lower risk of IAA-first (OR 0.48 [0.35–0.68], P < 0.0001) (Fig. 2B). This inverse association was strongest when IAA-first appeared between 13 and 24 months of age (OR 0.30 [0.14–0.63], P = 0.002) but was still observed when IAA appeared after 24 months of age (OR 0.57 [0.39–0.84], P = 0.0004). No seasonal correlation between the age-specific GIE reports and IAA risk was observed. No significant associations were observed between age-specific reports of GIEs and GADA (Supplementary Fig. 1A–C).
Figure 2.
IAA risk irrespective of age (solid horizontal line, OR [95% CI]) and by age (×, OR) when GIE was reported at <1 year of age (A), 1 to <2 years of age (B), and 2 to <3 years of age (C). Time lag correlation at 0–11 months (D) 12–23 months (E), and 24–35 months (F) between GIEs and subsequent risk of IAA-first development irrespective of age (solid horizontal line, OR [95% CI]) and at the age when children developed IAA (×, OR). Period of autoantibody development is defined as time after a scheduled blood draw visit up until the next blood draw visit when autoantibodies can be first detected.
Lag Correlations Between GIE Reports and IAA and GADA Risk
GIEs were associated with an immediate lower risk of IAA-first (OR 0.63 if a GIE was reported 0–11 months prior [95% CI 0.45–0.87], P = 0.005), followed by an increased risk of IAA-first (OR 1.67 if GIE was reported 12–23 months prior [1.18–2.38], P = 0.004) (Fig. 2D and E). When these lag correlations were examined by age of IAA appearance (× signs in Fig. 2D–F), strong inverse associations were observed at ∼2, 3, and 4 years of age in (Fig. 2D, E, and F, respectively). These inverse associations were linked with the strong age-specific inverse effect of GIEs at 13–24 months of age on subsequent IAA risk and explained the immediate (0–11 months prior) inverse lag effect (Fig. 2D) and reduced the 12–23 months prior lag effect (Fig. 2E). No significant lag correlations between GIEs and GADA were observed (Supplementary Fig. 1D–F), although a lag of 0–11 months was close to significant (P = 0.05). When this lag was examined further, a significant lag influence was found for GIEs 6 months prior to GADA development in children <48 months of age (OR 2.16 [1.54–3.02], P < 0.0001) (Supplementary Fig. 2). The lag correlations between GIE reports and IAA or GADA risk were not seasonal.
Stool Viruses and GIEs
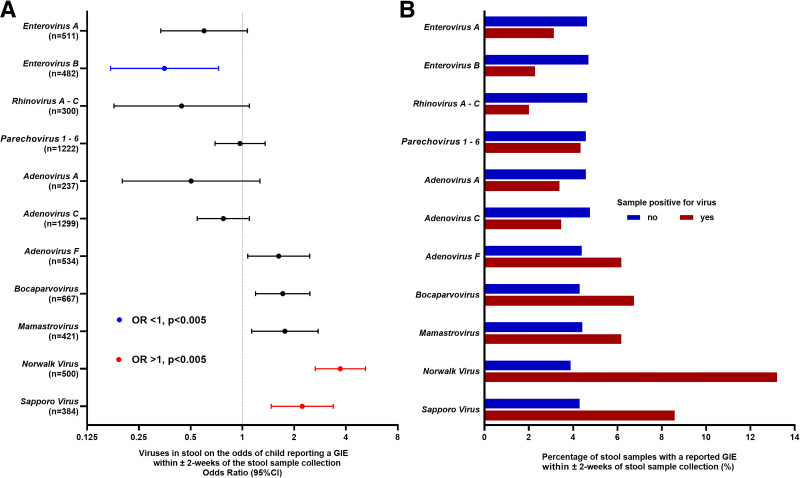
Viruses from stool samples collected monthly until 24 months of age while at risk for IA were examined in relation to the likelihood of a GIE being reported within ±2 weeks of a stool sample collection (Fig. 3). The number of stool samples included 4,450 between 3 and 12 months and 2,752 between 13–24 months of age. Figure 3 shows common viruses found in stool (prevalence >2.5% of samples), including viruses causing frequent and/or intense GI symptoms (lower part of y-axis: adenovirus F, bocaparvovirus, mamastrovirus, Norwalk virus, sapporo virus) and viruses causing infrequent and/or mild GI symptoms (upper part of y-axis: enterovirus A and B, rhinovirus A–C, parechovirus 1–6, adenovirus A and F). Norwalk virus (OR 3.71 [95% CI 2.65–5.19], P < 0.0001) and sapporo virus (OR 2.23 [1.47–3.37], P = 0.0002) were associated with a higher odds of a GIE report. Adenovirus F, bocaparvovirus, and mamastrovirus correlated with an elevated odds of a GIE report. In contrast, enterovirus B (OR 0.35 [– 0.17 to 0.73], P = 0.005) was associated with a lower odds of a GIE report. There were no significant interactions between viruses and age of stool sample.
Figure 3.
Common viruses in stools (n = 7,202) collected monthly from 751 children 3–24 months of age and association with the odds of GIE report within ±2 weeks from stool sample collection date (A). Percentage of stool samples (B) (separate bars for virus-positive and virus-negative samples) with a reported GIE within ±2 weeks from stool sample collection date.
Stool Viruses and First-Appearing Islet Autoantibodies
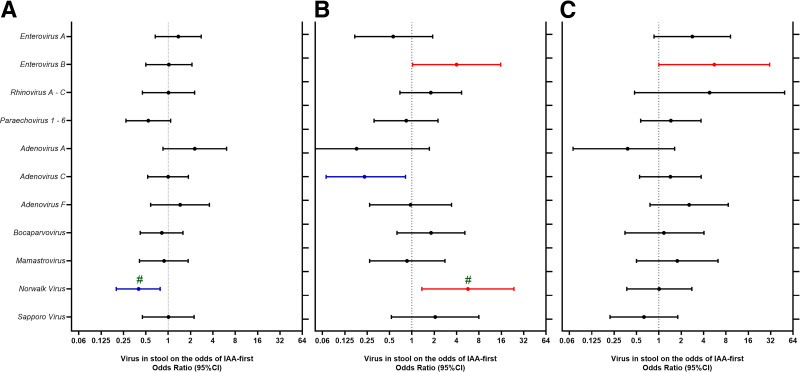
Next, viruses in stools were examined in relation to the subsequent risk of IAA-first (Fig. 4A–C). Norwalk virus during the 1st year of life (≤12 months) correlated with a significant age-dependent association with IAA-first (interaction P < 0.001). Norwalk virus detected during the 1st year of life was associated with a reduced risk of IAA appearing until 24 months of age (OR 0.40 [95% CI 0.20–0.78) and an increased risk of IAA-first appearing between 25 and 60 months of age (OR 5.69 [1.36–23.7]). Enterovirus B up to 24 months of age was strongly associated with risk of IAA-first between 25 and 60 months of age (OR 9.34 [1.88–46.5], P = 0.006), regardless of whether the virus was detected during the 1st (OR 4.00 [1.02–15.7]) or 2nd year of life (OR 5.57 [1.00–31.0]). Norwalk virus between 13 and 24 months of age was not associated with IA.
Figure 4.
Common viruses (>2.5% of samples) in stools of children between 3 and 12 months of age (A and B) and 13–24 months of age (C) in relation to risk of IAA-first appearance between the 6 and 24 months of age (A) (n = 127 IAA-first case and match control pairs) and between 25 and 60 months of age (B and C) (n = 52 IAA-first case and match control pairs). Viruses in relation to IAA-first were examined separately, adjusting for HLA-DR genotype and matching factors (sex, site, and family history of T1D). Associations with P < 0.05 are shown in color. #Significant interaction between virus and age of seroconversion.
FUT2 Variant
SNP rs601338G>A in the FUT2 gene with the nonsecretor status (genotype A/A) confers resistance against Norwalk virus. This was validated in the current study. Children with the A/A genotype had fewer reported GIEs before 2 years of age (Supplementary Fig. 3A), and their stools were less likely to be positive for Norwalk virus (Supplementary Fig. 3B). Correlations with first-appearing islet autoantibodies were reexamined for dependence on the FUT2-A/A genotype (Supplementary Table 1). An inverse association between GIE reports between 13 and 24 months of age and risk of IA overall depended on the FUT2-A/A genotype. A GIE report during the 2nd year of life was associated with a lower risk of IA only among children carrying the nonsecretor FUT2-A/A genotype (OR 0.33 [95% CI 0.19–0.57], P = 0.0001), regardless of the first islet autoantibody to appear (IAA-first: OR 0.26 [0.09–0.73]; GADA-first: OR 0.47 [0.22–1.03]; IAA and GADA at first appearance of IA: OR 0.19 [0.04–0.80]).
Stratification by Country and Adjusting for Other Factors
The associations between the major time-dependent GIE variables and IAA-first were found relatively consistent in direction and magnitude across countries (Supplementary Fig. 4). Associations between Norwalk virus in the 1st year of life and IAA risk at 2–4 years of age was stronger in Europe (OR 9.02 [95% CI 1.31–62.3]) than in the U.S. (OR 1.38 [0.11–17.4]), matching what was observed with GIEs during the 1st year and IAA-first (Supplementary Fig. 4A).
The major observed GIE associations also remained after adjusting for previously reported factors associated with GIE or IA in the TEDDY cohort (Supplementary Tables 2 and 3). A GIE reported between 13 and 24 months of age was associated with a decreased risk of IAA from 2 years of age onward (OR 0.49 [95% CI 0.33–0.71], P = 0.0002). A GIE before 1 year of age correlated with an increased risk of IAA from 2 years of age onward (OR 1.70 [1.17–2.46], P = 0.005).
Conclusions
GI infections showed a clear association with the appearance of IA. The impact depended on the type of the first-appearing islet autoantibody (IAA-first or GADA-first) and the age of infection. GIEs before 1 year of age were associated with increased IAA risk with a relatively long delay, and Norwalk virus seemed like a possible factor for this phenomenon. The observed time lag between infection and increased IAA risk suggests slowly operating mechanisms. It can also be a sign of other nonviral factors operating. The observed associations remained after adjusting for potential confounders, but this does not exclude the possibility of such additional factors. Viral infection may induce a sustained shift in the infant’s immune response pattern and make the child susceptible for onset of autoimmunity by additional factors. Furthermore, acute GI infection may sometimes be followed by persistent low-level virus replication. For example, signs of a slow, persistent enterovirus infection (coxsackievirus B) have been found in β-cells, and this may lead to chronic inflammation promoting onset of β-cell autoimmunity (28).
GIE during the 2nd year of life showed a clear correlation with a reduced risk of IAA as the first-appearing autoantibody. This inverse association was significant up to 10 years of age, but it was strongest up to 12 months after the infection. The reason why infections at this particular age could be important risk modifiers is not known. The fact that GI infections peak at this age can be one factor since frequent exposure increases statistical power to detect such an association. In addition, children at this age are susceptible to the virus since breastfeeding or maternally acquired virus antibodies are no longer protecting them. This leads to efficient virus replication in gut mucosa, which, in turn, may stimulate immunoregulatory pathways in the gut immune system. For example, both rotavirus and Norwalk virus infections have been shown to induce strong interleukin 10 responses in man (29,30), and rotavirus infection leads to strong regulatory T-cell activation in gnotobiotic pigs (31). Murine Norwalk virus infection protects NOD mice from the development of diabetes and is associated with an expansion of regulatory T cells and reduced proinflammatory T cells (32). Such effects could downregulate autoimmune reactions and IAA.
Detection of viruses that replicate in the gut and typically cause GI symptoms were associated with GIE reports (Fig. 3). However, while GIEs between 13 and 24 months of age were associated with a decreased IAA risk, Norwalk virus found in stool at the same age showed no association with IAA risk. Additionally, while the inverse association between GIEs and IAA risk was most pronounced among children with FUT2 genetic resistance against Norwalk virus, the association between GIEs at the same age and GADA risk was similarly impacted by this FUT2 genetic resistant group. These findings suggest that viruses other than Norwalk virus may account for the inverse association between GIEs and IAA risk in such a way as to have an impact on risk of IA in general. Other viruses that were detected in stool and typically cause GI infections were not associated with this phenomenon. One should note, however, that the low detection rate of rotavirus (n = 42) made it impossible to study its possible contribution. Further virus serology studies are in progress to evaluate the possible role of rotavirus and other enteral viruses.
Associations between GIEs and onset of IA with GADA-first were limited. The primary analyses showed no significant associations (Supplementary Fig. 1). Only after a secondary analysis of shorter lags did we find that GADA risk was increased by GIEs reported 0–6 months prior to GADA seroconversion in children <4 years of age. Also, respiratory infections have been reported to increase IA risk (both GADA and IAA) when reported shortly prior (0–9 months) to islet autoantibody seroconversion (5). Infections a few months prior to onset of IA may act as triggering or precipitating factors for IA, but they can also be due to reverse causation; that is, immune dysregulation or low-level autoimmunity may increase susceptibility to infections. We tried to minimize the possibility of reverse causation by not considering infections all the way up to the age of first detection of autoantibodies but only up to the previous scheduled blood draw. The blood draw interval was 3 months in children <4 year of age and 6 months in older children. Accordingly, infections were considered only up to 3 or 6 months before first detection of autoantibodies.
Virus interference (type 1 interferons induced by ongoing viral infection protect against other viruses) may explain some of the findings in the current study (33). The decreased frequency of GIE reports at the time when enterovirus B was detected in stools may reflect virus interference where ongoing replication of enteroviruses could protect against other viruses. Furthermore, the inverse association between GIEs at 13–24 months of age and risk for IAA-first may also be due to virus interference where GIEs may provide protection against onset of IA simply by blocking concomitant infections that could promote autoimmunity, such as enterovirus B infections (13).
This study has major advantages. The prospective study design, large cohort size, multinational subjects, and daily recording of GI infections in a diary have allowed for powerful statistical analyses and general applicability of results. An additional strength was the use of stool virome data to validate and characterize the GIE reports. Stool was collected in monthly intervals, which allowed diligent observation of viral exposures over time and helped with combining stool virome results with diary data. Furthermore, serum samples were drawn every 3 months, which allowed recognition of subjects with either IAA or GADA onset of autoimmune process and analysis of GIE association separately for these two pathogenic pathways.
A limitation of the study is that despite the extensive data and sample collection, the study still did not capture all GI infections because part of these infections may be symptomless and, thus, are not recorded by the parents. Also, there may be viruses that replicate in the gut only briefly and are not captured on the day of stool sampling (e.g., rotavirus detection rate was low). The role of rotaviruses as well as mild, symptomless infections could be evaluated by measuring virus antibody levels in serum. Finally, we cannot rule out the possibility of a bystander effect of some other environmental factor linking with GIEs.
To our knowledge, this study is the first to show that overt GI infections may modulate the risk of IA when the autoimmune process starts with IAA as the first-appearing autoantibody. It is possible that viral influence on the risk of developing IA is strongest in IAA-first cases, as also enterovirus B exposure is linked with an increased risk of particularly IAA-first (34). We observed that GI infections were associated with either increased or decreased IAA risk depending on the age and timing of infections. Norwalk virus was identified as one potential factor that increases the IAA risk, whereas other viruses seem to account for decreased IAA risk. These results open new opportunities and directions to identify risk-modifying viruses and the mechanisms mediating their effect.
This article contains supplementary material online at https://doi.org/10.2337/figshare.23823507.
Article Information
Acknowledgments. The authors thank Sarah Austin-Gonzalez with the Health Informatics Institute at the University of South Florida for assistance with editing and preparing the graphical abstract. The authors especially acknowledge the TEDDY families for continued participation in this wonderful study.
Funding. The TEDDY study is funded by National Institute of Diabetes and Digestive and Kidney Diseases grants U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, U01 DK124166, and U01 DK128847 and contract HHSN267200700014C and by the National Institute of Allergy and Infectious Diseases, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute of Environmental Health Sciences, Centers for Disease Control and Prevention, and JDRF. This work is also supported in part by the National Institutes of Health/National Center for Advancing Translation Science clinical and translational science awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR002535).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.L., K.F.L., and H.H. made the research plan, analyzed and evaluated the data, and wrote the manuscript. M.R., Å.L., K.V., B.A., W.H., J.K., R.A.M., J.T., A.-G.Z., J.F.P., and R.L. reviewed and edited the manuscript. M.L. and K.F.L. are the guarantors of this work, and as such, had full access to all the data in the study and take responsibility for the integrity of data and the accuracy of data analysis.
Funding Statement
The TEDDY study is funded by National Institute of Diabetes and Digestive and Kidney Diseases grants U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, U01 DK124166, and U01 DK128847 and contract HHSN267200700014C and by the National Institute of Allergy and Infectious Diseases, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute of Environmental Health Sciences, Centers for Disease Control and Prevention, and JDRF. This work is also supported in part by the National Institutes of Health/National Center for Advancing Translation Science clinical and translational science awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR002535).
Footnotes
Members of the TEDDY Study Group are listed in the supplementary material online.
Contributor Information
TEDDY Study Group:
Marian Rewers, Kimberly Bautista, Judith Baxter, Daniel Felipe-Morales, Brigitte I. Frohnert, Marisa Stahl, Isabel Flores Garcia, Patricia Gesualdo, Sierra Hays, Michelle Hoffman, Rachel Karban, Edwin Liu, Leila Loaiza, Jill Norris, Holly O’Donnell, Loana Thorndahl, Andrea Steck, Kathleen Waugh, Jorma Toppari, Olli G. Simell, Annika Adamsson, Suvi Ahonen, Mari Åkerlund, Sirpa Anttila, Leena Hakola, Anne Hekkala, Tiia Honkanen, Heikki Hyöty, Jorma Ilonen, Sanna Jokipuu, Taru Karjalainen, Leena Karlsson, Jukka Kero, Jaakko J. Koskenniemi, Miia Kähönen, Mikael Knip, Minna-Liisa Koivikko, Katja Kokkonen, Merja Koskinen, Mirva Koreasalo, Kalle Kurppa, Salla Kuusela, Jarita Kytölä, Jutta Laiho, Tiina Latva-aho, Siiri Leisku, Laura Leppänen, Katri Lindfors, Maria Lönnrot, Elina Mäntymäki, Markus Mattila, Maija Miettinen, Teija Mykkänen, Tiina Niininen, Sari Niisistö, Noora Nurminen, Sami Oikarinen, Hanna-Leena Oinas, Paula Ollikainen, Zhian Othmani, Sirpa Pohjola, Solja Raja-Hanhela, Jenna Rautanen, Anne Riikonen, Minna Romo, Juulia Rönkä, Nelli Rönkä, Satu Simell, Päivi Tossavainen, Mari Vähä-Mäkilä, Eeva Varjonen, Riitta Veijola, Irene Viinikangas, Silja Vilmi, Suvi M. Virtanen, Richard McIndoe, Desmond Schatz, Diane Hopkins, Michael Haller, Risa Bernard, Melissa Gardiner, Ashok Sharma, Laura Jacobsen, Jennifer Hosford, Kennedy Petty, Leah Myers, Chelsea Salmon, Anette G. Ziegler, Ezio Bonifacio, Cigdem Gezginci, Willi Grätz, Anja Heublein, Eva Hohoff, Sandra Hummel, Annette Knopff, Melanie Köger, Sibylle Koletzko, Claudia Ramminger, Roswith Roth, Jennifer Schmidt, Marlon Scholz, Joanna Stock, Katharina Warncke, Lorena Wendel, Christiane Winkler, Åke Lernmark, Daniel Agardh, Carin Andrén Aronsson, Rasmus Bennet, Corrado Cilio, Susanne Dahlberg, Ulla Fält, Malin Goldman Tsubarah, Emelie Ericson-Hallström, Lina Fransson, Emina Halilovic, Gunilla Holmén, Susanne Hyberg, Berglind Jonsdottir, Naghmeh Karimi, Helena Elding Larsson, Marielle Lindström, Markus Lundgren, Marlena Maziarz, Jessica Melin, Caroline Nilsson, Kobra Rahmati, Anita Ramelius, Falastin Salami, Anette Sjöberg, Evelyn Tekum Amboh, Carina Törn, Ulrika Ulvenhag, Terese Wiktorsson, Åsa Wimar, William A. Hagopian, Michael Killian, Claire Cowen Crouch, Jennifer Skidmore, Trevor Bender, Megan Llewellyn, Cody McCall, Arlene Meyer, Jocelyn Meyer, Denise Mulenga, Nole Powell, Jared Radtke, Shreya Roy, Preston Tucker, Dorothy Becker, Margaret Franciscus, MaryEllen Dalmagro-Elias Smith, Ashi Daftary, Mary Beth Klein, Chrystal Yates, Jeffrey P. Krischer, Rajesh Adusumali, Sarah Austin-Gonzalez, Maryouri Avendano, Sandra Baethke, Brant Burkhardt, Martha Butterworth, Nicholas Cadigan, Joanna Clasen, Kevin Counts, Laura Gandolfo, Jennifer Garmeson, Veena Gowda, Christina Karges, Shu Liu, Xian Liu, Kristian Lynch, Jamie Malloy, Lazarus Mramba, Cristina McCarthy, Jose Moreno, Hemang M. Parikh, Cassandra Remedios, Chris Shaffer, Susan Smith, Noah Sulman, Roy Tamura, Dena Tewey, Henri Thuma, Michael Toth, Kendra Vehik, Ponni Vijayakandipan, Melissa Wroble, Jimin Yang, Kenneth Young, Liping Yu, Dongmei Miao, Kathleen Gillespie, Kyla Chandler, Ilana Kelland, Yassin Ben Khoud, Matthew Randell, Stephen S. Rich, Wei-Min Chen, Suna Onengut-Gumuscu, Emily Farber, Rebecca Roche Pickin, Jonathan Davis, Jordan Davis, Dan Gallo, Jessica Bonnie, Paul Campolieto, Joseph Petrosino, Nadim J. Ajami, Richard E. Lloyd, Matthew C. Ross, Jacqueline L. O’Brien, Diane S. Hutchinson, Daniel P. Smith, Matthew C. Wong, Xianjun Tian, Tulin Ayvaz, Auriole Tamegnon, Nguyen Truong, Hannah Moreno, Lauren Riley, Eduardo Moreno, Tonya Bauch, Lenk Kusic, Ginger Metcalf, Donna Muzny, Harsha V. Ardhan Doddapaneni, Richard Gibbs, Chris Deigan, Beena Akolkar, Thomas Briese, Todd Brusko, Teresa Buckner, Suzanne Bennett Johnson, Eoin McKinney, Tomi Pastinen, Steffen Ullitz Thorsen, and Erick Triplett
References
- 1. TEDDY Study Group . The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes 2007;8:286–298 [DOI] [PubMed] [Google Scholar]
- 2. TEDDY Study Group . The Environmental Determinants of Diabetes in the Young (TEDDY) study. Ann N Y Acad Sci 2008;1150:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rewers M, Hyöty H, Lernmark Å, et al.; TEDDY Study Group . The Environmental Determinants of Diabetes in the Young (TEDDY) study: 2018 update. Curr Diab Rep 2018;18:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lönnrot M, Lynch K, Larsson HE, et al.; TEDDY Study Group . A method for reporting and classifying acute infectious diseases in a prospective study of young children: TEDDY. BMC Pediatr 2015;15:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lönnrot M, Lynch KF, Elding Larsson H, et al.; TEDDY Study Group . Respiratory infections are temporally associated with initiation of type 1 diabetes autoimmunity: the TEDDY study. Diabetologia 2017;60:1931–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krischer JP, Lynch KF, Lernmark Å, et al.; TEDDY Study Group . Genetic and environmental interactions modify the risk of diabetes-related autoimmunity by 6 years of age: the TEDDY study. Diabetes Care 2017;40:1194–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rasmussen T, Witsø E, Tapia G, Stene LC, Rønningen KS. Self-reported lower respiratory tract infections and development of islet autoimmunity in children with the type 1 diabetes high-risk HLA genotype: the MIDIA study. Diabetes Metab Res Rev 2011;27:834–837 [DOI] [PubMed] [Google Scholar]
- 8. Beyerlein A, Wehweck F, Ziegler AG, Pflueger M. Respiratory infections in early life and the development of islet autoimmunity in children at increased type 1 diabetes risk: evidence from the BABYDIET study. JAMA Pediatr 2013;167:800–807 [DOI] [PubMed] [Google Scholar]
- 9. Beyerlein A, Donnachie E, Jergens S, Ziegler AG. Infections in early life and development of type 1 diabetes. JAMA 2016;315:1899–1901 [DOI] [PubMed] [Google Scholar]
- 10. Lynch KF, Lee HS, Törn C, et al.; TEDDY Study Group . Gestational respiratory infections interacting with offspring HLA and CTLA-4 modifies incident β-cell autoantibodies. J Autoimmun 2018;86:93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ 2011;342:d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hyöty H. Viruses in type 1 diabetes. Pediatr Diabetes 2016;17(Suppl. 22):56–64 [DOI] [PubMed] [Google Scholar]
- 13. Vehik K, Lynch KF, Wong MC, et al.; TEDDY Study Group . Prospective virome analyses in young children at increased genetic risk for type 1 diabetes. Nat Med 2019;25:1865–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Honeyman MC, Coulson BS, Stone NL, et al. Association between rotavirus infection and pancreatic islet autoimmunity in children at risk of developing type 1 diabetes. Diabetes 2000;49:1319–1324 [DOI] [PubMed] [Google Scholar]
- 15. Perrett KP, Jachno K, Nolan TM, Harrison LC. Association of rotavirus vaccination with the incidence of type 1 diabetes in children. JAMA Pediatr 2019;173:280–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rogers MAM, Basu T, Kim C. Lower incidence rate of type 1 diabetes after receipt of the rotavirus vaccine in the United States, 2001-2017. Sci Rep 2019;9:7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oikarinen S, Krogvold L, Edwin B, et al. Characterisation of enterovirus RNA detected in the pancreas and other specimens of live patients with newly diagnosed type 1 diabetes in the DiViD study. Diabetologia 2021;64:2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG. The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia 2009;52:1143–1151 [DOI] [PubMed] [Google Scholar]
- 19. Krogvold L, Leete P, Mynarek IM, et al. Detection of antiviral tissue responses and increased cell stress in the pancreatic islets of newly diagnosed type 1 diabetes patients: results from the DiViD study. Front Endocrinol (Lausanne) 2022;13:881997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagopian WA, Erlich H, Lernmark A, et al.; TEDDY Study Group . The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes 2011;12:733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018;562:583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bonifacio E, Yu L, Williams AK, et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for National Institute of Diabetes and Digestive and Kidney Diseases consortia. J Clin Endocrinol Metab 2010;95:3360–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krischer JP, Lynch KF, Schatz DA, et al.; TEDDY Study Group . The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia 2015;58:980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uusitalo U, Liu X, Yang J, et al.; TEDDY Study Group . Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr 2016;170:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma A, Liu X, Hadley D, et al.; TEDDY Study Group . Identification of non-HLA genes associated with development of islet autoimmunity and type 1 diabetes in the prospective TEDDY cohort. J Autoimmun 2018;89:90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tutz G, Pritscher L. Nonparametric estimation of discrete hazard functions. Lifetime Data Anal 1996;2:291–308 [DOI] [PubMed] [Google Scholar]
- 27. Tutz G, Schmid M. Modeling Discrete Time-to-Event Data. New York, Springer, 2016 [Google Scholar]
- 28. Nekoua MP, Alidjinou EK, Hober D. Persistent coxsackievirus B infection and pathogenesis of type 1 diabetes mellitus. Nat Rev Endocrinol 2022;18:503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Newman KL, Moe CL, Kirby AE, Flanders WD, Parkos CA, Leon JS. Human norovirus infection and the acute serum cytokine response. Clin Exp Immunol 2015;182:195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen SM, Ku MS, Lee MY, Tsai JD, Sheu JN. Diagnostic performance of serum interleukin-6 and interleukin-10 levels and clinical predictors in children with rotavirus and norovirus gastroenteritis. Cytokine 2012;59:299–304 [DOI] [PubMed] [Google Scholar]
- 31. Wen K, Li G, Yang X, et al. CD4+ CD25- FoxP3+ regulatory cells are the predominant responding regulatory T cells after human rotavirus infection or vaccination in gnotobiotic pigs. Immunology 2012;137:160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pearson JA, Tai N, Ekanayake-Alper DK, et al. Norovirus changes susceptibility to type 1 diabetes by altering intestinal microbiota and immune cell functions. Front Immunol 2019;10:2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu A, Mihaylova VT, Landry ML, Foxman EF. Interference between rhinovirus and influenza A virus: a clinical data analysis and experimental infection study. Lancet Microbe 2020;1:e254–e262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sioofy-Khojine AB, Lehtonen J, Nurminen N, et al. Coxsackievirus B1 infections are associated with the initiation of insulin-driven autoimmunity that progresses to type 1 diabetes. Diabetologia 2018;61:1193–1202 [DOI] [PubMed] [Google Scholar]