Abstract
Drug resistance underpins poor outcomes in many malignancies including refractory and relapsed acute myeloid leukemia (R/R AML). Glucuronidation is a common mechanism of drug inactivation impacting many AML therapies, e.g., cytarabine, decitabine, azacytidine and venetoclax. In AML cells, the capacity for glucuronidation arises from increased production of the UDP-glucuronosyltransferase 1A (UGT1A) enzymes. UGT1A elevation was first observed in AML patients who relapsed after response to ribavirin, a drug used to target the eukaryotic translation initiation factor eIF4E, and subsequently in patients who relapsed on cytarabine. UGT1A elevation resulted from increased expression of the sonic-hedgehog transcription factor GLI1. Vismodegib inhibited GLI1, decreased UGT1A levels, reduced glucuronidation of ribavirin and cytara-bine, and re-sensitized cells to these drugs. Here, we examined if UGT1A protein levels, and thus glucuronidation activity, were targetable in humans and if this corresponded to clinical response. We conducted a phase II trial using vismodegib with ribavirin, with or without decitabine, in largely heavily pre-treated patients with high-eIF4E AML. Pre-therapy molecular assessment of patients’ blasts indicated highly elevated UGT1A levels relative to healthy volunteers. Among patients with partial response, blast response or prolonged stable disease, vismodegib reduced UGT1A levels, which corresponded to effective targeting of eIF4E by ribavirin. In all, our studies are the first to demonstrate that UGT1A protein, and thus glucuronidation, are targetable in humans. These studies pave the way for the development of therapies that impair glucuronidation, one of the most common drug deactivation modalities. Clinicaltrials.gov: NCT02073838.
Introduction
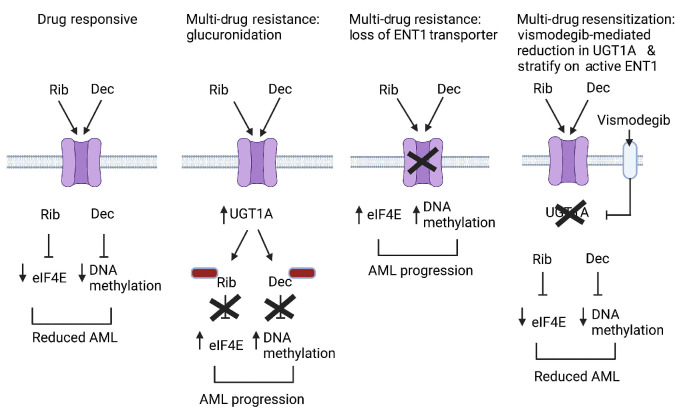
Drug resistance remains a major challenge in the treatment of many malignancies and is responsible for reduced overall survival in settings such as refractory and relapsed (R/R) acute myeloid leukemia (AML). Clinical response following initial chemotherapy for most AML patients is about one year.1-4 After first relapse, outcomes are worse, with overall survival (OS) lasting approximately 4-5 months and at second relapse approximately two months with chemotherapy.5,6 The outlook is similarly dismal at first relapse with venetoclax plus hypomethylating agents (HMA), with a median OS of about 2.4 months.4 By understanding the molecular bases of clinical resistance, it is possible to design new therapeutic strategies to overcome them. Drivers of clinical resistance include impaired drug entry into the cell, inactivation of drugs through chemical modification, genetic re-wiring, and/or enhanced efflux of drugs from cells.7 These events can elicit multi-drug resistance even to therapies for which there were no prior exposures, which thereby impacts outcomes of subsequent regimens. For example, the commonly used AML drugs cytarabine, azacytidine and decitabine all employ the equilibrative nucleoside transporter 1 (ENT1) to enter AML cells, and reduced ENT1 levels elicit resistance to all these drugs8-11 (Figure 1). One of the most common forms of drug resistance in pharmacology is the covalent addition of glucuronic acid to drugs, which results in inactivation, thereby driving drug resistance. Glucuronidation occurs both in the liver and extrahepatically,12-15 and AML cells can develop the capacity to glucuronidate multiple drugs simultaneously via elevated levels of UDP-glucuronosyltransferase 1A (UGT1A) enzymes.8,16 These events occur independently of hepatic glucuronidation. Glucuronidation impacts approximately 50% of prescribed drugs including cytarabine, venetoclax, decitabine and azacytidine8,12,16-18 (Figure 1). These observations highlight the importance of developing inhibitors that overcome drug resistance to produce durable responses in R/R AML patients.
This ability of AML blasts to glucuronidate drugs was first revealed during clinical studies targeting the eukaryotic translation initiation factor eIF4E with ribavirin.8,12,16,17 eIF4E is elevated in a subset of de novo and R/R AML patients, and its elevation alone is correlated with worse outcomes.19 High-eIF4E AML specimens are typically characterized by highly elevated and often nuclear-enriched eIF4E.20-22 Here, eIF4E drives the production of factors that support malignancy through its impacts on several nuclear RNA metabolism steps, including capping, splicing, and RNA export, as well as on translation in the cytoplasm through association with their m7G RNA cap.19,20,23-29 In this way, eIF4E serves as an exemplar for targeting RNA metabolism in malignancies. Ribavirin acts as a m7G RNA cap competitor, and thereby represses the biochemical and oncogenic activities of eIF4E.8,30-42 In a phase II ribavirin monotherapy and subsequent ribavirin plus low-dose cytarbine (LDAC) phase I/II trial, treatments reduced high-eIF4E AML cell numbers and re-localized eIF4E from the nucleus to the cytoplasm, corresponding to repressed oncogenic capacity and objective clinical responses, including complete remissions.21,22,43 Ribavirin also targeted eIF4E in other cancers, including those of the prostate and head and neck.44,45
Despite robust responses, all patients relapsed on these trials. We noted at relapse the emergence of populations of high-UGT1A cells.8,16 This correlated with increased glucuronidation and subsequent deactivation of ribavirin and cytarabine in cells.8,16 Some specimens additionally had reduced ENT1 levels, and thus impaired uptake of ribavirin and cytarabine, indicative of the development of multiple forms of resistance simultaneously.8,21,22 High-UGT1A cell populations also emerged at relapse in patients treated with standard of care (cytarabine with an anthracycline), indicating that glucuronidation-driven resistance is not restricted to ribavirin-based therapies.8 This glucuronidation led to drug resistance to several AML drugs, including cytarabine, venetoclax, azacytidine and decitabine.8,16 Increased UGT1A protein levels arose due to elevation of the hedgehog transcription factor GLI1, but can likely occur by other means as well.8,16 GLI1-inducible glucuronidation was repressed by vismodegib which indirectly targets GLI1 via the Smoothened extracellular receptor (SMO).8,16 Vismodegib reduced UGT1A levels, decreased glucuronidation, and resensitized cell lines to these drugs.8,16
Figure 1.
Schematic of forms of drug resistance relevant to this trial. Glucuronidation of ribavirin (Rib) and decitabine (Dec) is indicated by red boxes; the ENT1 transporter is depicted as the purple channel. While not shown, drug resistance can also occur via the simultaneous loss of ENT1 and elevated UGT1A (and thus increased glucuronidation). Model created using Biorender.
We sought to examine the capacity of vismodegib to lower UGT1A levels, and thereby reduce drug glucuronidation, in largely heavily pre-treated high-eIF4E AML patients in a phase II clinical trial. Given that vismodegib binds an extracellular receptor, it escapes cellular glucuronidation, providing an additional rationale for its use here. We also included decitabine in our combination, as ribavirin combined with HMA provided better inhibition of high-eIF4E AML patient specimen growth ex vivo than either agent alone.32 Finally, both ribavirin and decitabine employ the ENT1 drug transporter,8,11-13,46 and thus we ensured patients entered the trial with active ENT1. This also enabled us to study flux in ENT1 levels as a predictor of relapse in these patients in parallel with monitoring UGT1A levels. In all, we sought to develop strategies to target heavily pre-treated AML patients focusing on UGT1A. Our studies demonstrated for the first time that glucuronidation can be targeted in humans and that this is associated with clinical benefit.
Methods
Study design
This was a multi-center, open-label, randomized phase II study of ribavirin and vismodegib with or without decitabine in AML. The primary objective was to determine the efficacy of ribavirin, vismodegib with or without decitabine using overall response rate defined as the rate of complete remission (CR), complete remission with incomplete blood count recovery (CRi), partial remission (PR), morphologic leukemia-free state (MLFS) or blast response (BR). Correlative studies were included to assess relevant molecular targets.
Patients randomized to the VRD arm (vismodegib/ribavirin/decitabine) were administered decitabine at 20 mg/m2 intravenously daily on days -7 to -3 for cycle 1 and days 1 to 5 on subsequent cycles. Ribavirin at 1400 mg orally was taken twice daily and vismodegib was taken orally at 150 mg once a day starting on day 1. Treatment cycles were 28 days long. Patients randomized to the VR arm (vismodegib/ribavirin) received ribavirin 1400 mg twice a day and vismodegib 150 mg once a day. Ribavirin and decitabine were donated by Pharmascience Inc., Montreal, Quebec, Canada; vismodegib was donated by Hoffmann-La Roche Ltd., Mississauga, Ontario, Canada.
A run-in phase of three patients in each arm was performed to ensure both the safety of these combinations and adequate ribavirin plasma levels. Based on the respective toxicity profiles, no safety signal was observed and adequate plasma levels of ribavirin were found. The VR arm of the trial was terminated early due to futility as per the Simon two-stage design (see Online Supplementary Methods). Enrolment on the VRD arm closed prior to reaching 21 participants due to overall challenges in recruiting evaluable participants during the COVID pandemic and the understanding that the treatment landscape for AML was evolving.
Patient selection
Patients at least 18 years of age with AML were eligible to participate in this study. All patients must have failed primary therapy (defined as two induction chemotherapies), must have relapsed, or must not have been suitable candidates for intensive induction chemotherapy. All patients reviewed and signed an appropriate informed consent which had been approved by the institutional review boards (CIUSSS West-Central Montreal Research Review Office, approval 14-046, and University Health Network Research Ethics Board, approval 15-9586C) and Health Canada in accordance with the Declaration of Helsinki. Eligibility criteria are detailed in the Online Supplementary Methods.
Acute myeloid leukemia primary specimens
Blasts were isolated from peripheral blood or bone marrow by flow cytometry using forward and side scatter and CD45, as described previously.22 Cells were sorted on a BD FACSAria flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). For comparison, normal CD34+ cells were obtained from Lonza (Hayward, CA, USA) or ATCC (Manassas, VA, USA).
Immunofluorescence
Immunostaining of cells with eIF4E (BD Biosciences, Mississauga, ON, Canada) and UGT1A antibodies (Antibodies-online, further purified in-house) is described in detail in the Online Supplementary Methods, along with a description of the purification of the commercial UGT1A antibody to yield a single band on western blot.
Western bloting, quantitative polymerase chain reaction and RNA interference
Detailed methodology for immunoblotting, RNAi and reverse transcription-quantitative polymerase chain reaction (RT-qPCR), including antibodies, primers and RNAi sequences used, is presented in the Online Supplementary Methods.
Results
Patients' characteristics and clinical results
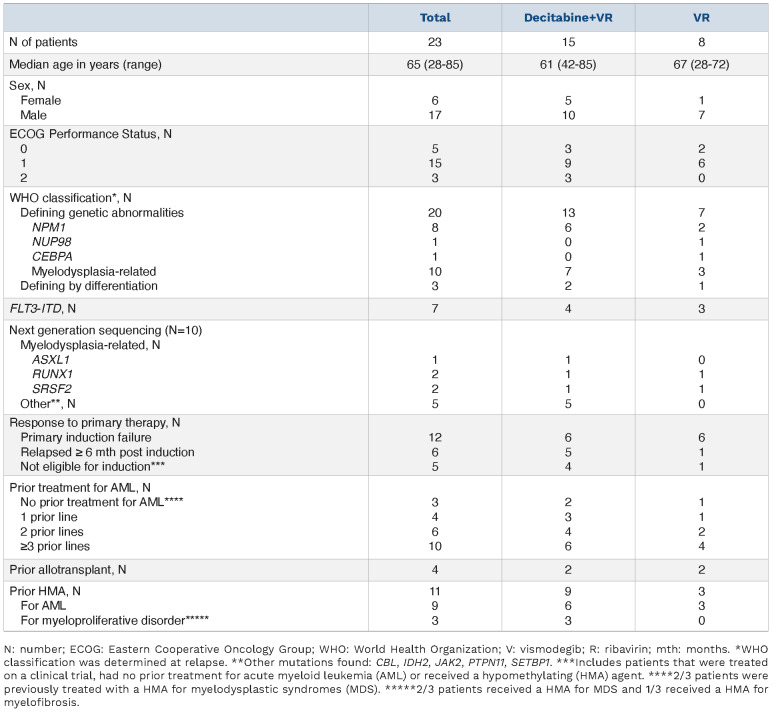
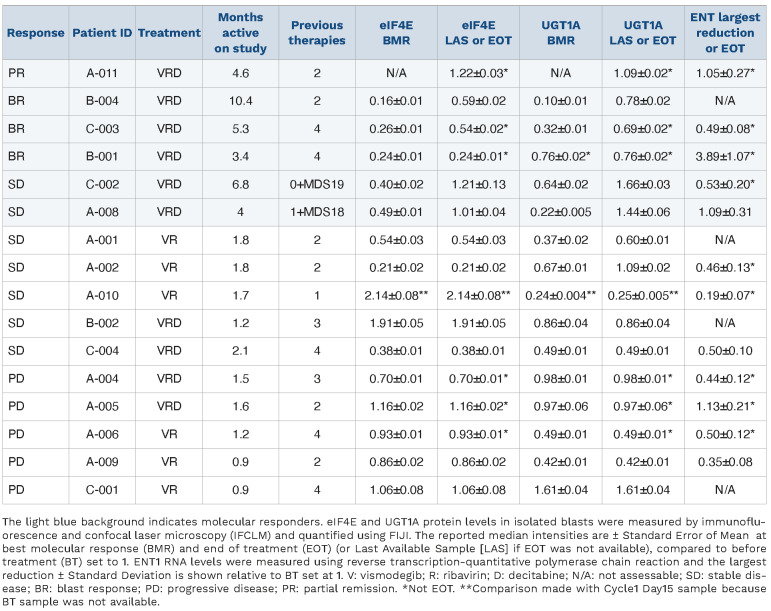
Between May 2015 and February 2021, 23 patients were enrolled onto the study. To ascertain eligibility, we performed molecular screening on 47 patients, examining AML blasts isolated as described.22 For inclusion, patients’ blasts had to have ≥3-fold elevated eIF4E levels versus CD34+ cells from healthy volunteers as described,21,22 and functional ENT1 as assessed by 3H-ribavirin uptake equivalent to, or higher than, CD34+ cells from healthy volunteers. Fourteen patients failed molecular screening: seven due to impaired ribavirin uptake, two without elevated eIF4E, and five due to insufficient material to screen (Online Supplementary Figure S1). Baseline patients' characteristics for the enrolled patients are detailed in Table 1. Median age was 65 years (range 28-85), patients were heavily pre-treated with 43% of patients having received >3 lines of therapy and 52% of the patients having failed primary induction. Testing for FLT3-ITD and NPM1 mutations was performed in all patients and were found in 30% and 35% of the patients, respectively. Post-hoc next generation sequencing (NGS) performed on ten patient baseline samples, did not identify TP53 mutations (see Table 1 for list of mutations). The median duration of treatment was 1.6 months (range 0.4-10.4). The most common treatment-emergent adverse events regardless of causality were febrile neutropenia (65%; grade ≥3: 65%), nausea (61%; grade ≥3: 9%), diarrhea (52%; grade ≥3: 4%), vomiting (48%; grade ≥3: 0%), and fatigue (43%; grade ≥3: 13%) (Online Supplementary Table S1). Seventeen patients were evaluable for clinical response, 16 of whom were heavily pre-treated for AML and/or myelodysplastic syndromes with a median of three prior therapies for the VRD arm and two for the VR arm; 8/17 patients relapsed from prior HMA, 1/17 prior venetoclax plus HMA, and 15/17 prior cytarabine.
Table 1.
Patients' baseline characteristics.
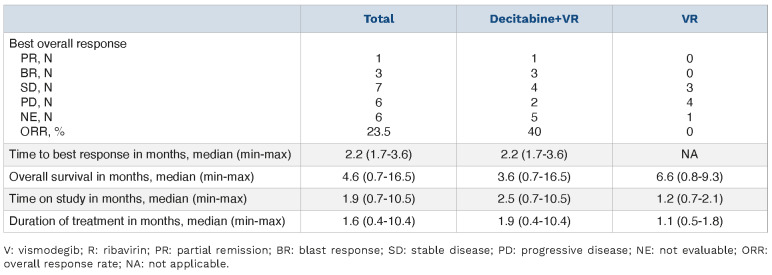
Overall, 4/10 patients in the VRD arm achieved objective responses: one PR and three BR (treatment range 5-10 cycles); two durable SD (treatment range 4-6 cycles); two SD and two PD (Table 2). Median time to response was 2.2 months (range 1.7-3.6). In comparison to an earlier study with decitabine alone, median time to response was 157 days (5.2 months) and only untreated or patients receiving second-line therapy responded.47 Thus, decitabine alone is not likely to have driven the observed responses in this trial given the short time to response, but rather the HMA-ribavirin combination was associated with response, as observed in ex vivo studies.32 Among responding patients, 4/6 had relapsed while on HMA, suggesting re-sensitization to decitabine by vismodegib and/or added benefit by targeting eIF4E with ribavirin. Responses in the VR arm were 3/7 SD and 4/7 PD, and this arm was closed. Pharmacokinetic studies indicated that vismodegib and ribavirin were not affected by decitabine exposure (Online Supplementary Figure S2).
Molecular assessment reveals correlation between UGT1A and eIF4E targeting and clinical response
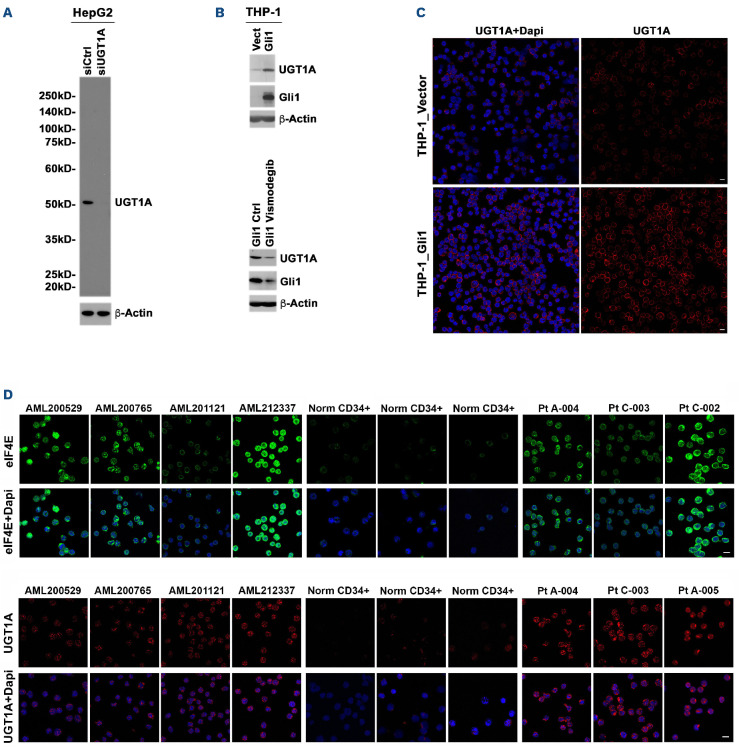
To study the impact of glucuronidation in both treated and treatment naïve patients, we examined UGT1A protein levels relative to healthy volunteers (Figure 2). For this purpose, we employed immunofluorescence and confocal laser microscopy (IFCLM). The commercially available panUGT1A antibodies available were found to have extraneous bands and thus were further affinity purified prior to use. The resulting antibody revealed a single band on the western blot, and RNAi reduction using a pan-UGT1A RNAi versus luciferase RNAi controls demonstrated reduced levels in the endogenous UGT1A band in liver HepG2 cells, thereby indicating the antibody is specific (Figure 2A). Actin indicates equal protein loading and is not impacted by the RNAi, as expected. Consistent with previous studies, we observed that UGT1A levels were elevated in high-eIF4E AML THP-1 cells over-expressing GLI1 versus vector controls using IFCLM and western blot, and that addition of vismodegib reduced both GLI1 and UGT1A, further validating this purified antibody8,16 (Figure 2B, C). For patient specimens, we observed that our heavily pretreated cohort was characterized by elevated UGT1A prior to exposure to study drugs relative to healthy volunteers (Figure 2D). For comparison, eIF4E staining is shown for some of the same specimens and demonstrates there is no correlation between eIF4E and UGT1A levels; it also confirms elevated eIF4E levels (Figure 2D). Interestingly, 3/4 treatment naïve AML patients (not participants in this trial) had elevated UGT1A at baseline, potentially predicting future treatment failure. Our previous studies demonstrated that the other major UGT family, UGT2B, were not elevated in the AML patients examined and thus were not monitored here.8 In all, patients in our trial were characterized by elevated UGT1A levels as well as elevated eIF4E levels relative to healthy volunteers.
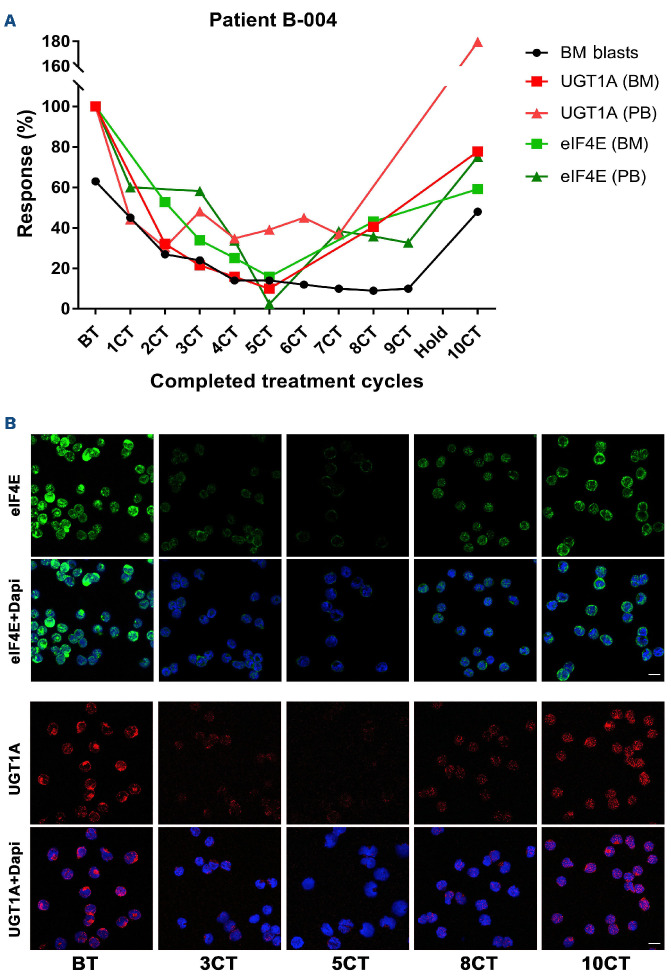
We next measured median intensity of UGT1A protein levels to ascertain whether levels decreased in patients upon treatment with vismodegib and if that corresponded to ribavirin targeting of eIF4E (Table 3). We note that only 16/17 evaluable patients had material available for analysis. Staining was quantified on a per cell basis using FIJI measuring more than 35 cells per condition. Median intensities ± Standard Error of Mean (SEM) at best molecular re sponse (BMR) and end of treatment (EOT) (or last available sample [LAS] if EOT was not available) were relative to before treatment (BT) which was set to 1. For patients on trial for one cycle, the same value is provided as both BMR and EOT/LAS relative to BT. Importantly, neither vismodegib nor HMA impact eIF4E activity or levels.8,32 We observed a median 3.1-fold reduction in UGT1A and median 3.8-fold reduction in eIF4E levels relative to BT in patients who achieved PR, BR or durable SD (patients B-001, A-008, C-002, B-004, A-011, C-003) (Table 3, Figures 3 and 4). As an example, patient B-004 bone marrow blasts had a 6.25fold and 10-fold reduction in eIF4E and UGT1A levels, respectively, at BMR relative to BT and this correlated with reduction of blast count to <10% (Table 3, Figure 3). During response, patients generally had re-localization of eIF4E to the cytoplasm, as we had observed in our previous ribavirin trials, whereby eIF4E entry into the nucleus is prevented by ribavirin interference with eIF4E interaction with its importin.21,22,43 At relapse, eIF4E and UGT1A levels were elevated, nearing BT levels, which corresponded with increased blasts, and increased eIF4E levels and its nuclear re-entry were evident, as observed in our previous trials (Table 3, Figures 3 and 4). In parallel, we measured ENT1 and adenosine kinase (ADK) since its loss is also associated with ribavirin resistance.8,48 ENT1 and ADK RNA levels were measured using RT-qPCR and the largest reduction is shown relative to BT set at 1 (Table 3, Online Supplementary Table 2). We observed that 2/6 of these patients (C-002 and C-003) had reduced ENT1 levels which likely contributes to drug resistance in parallel to elevation of UGT1A relative to BT. We note that ribavirin did not impact on UGT1A protein expression8,49 and, moreover, patients in both the VR and VRD arms achieved reduced UGT1A levels, indicating that this reduction was not driven by decitabine (Table 3).
Table 2.
Efficacy in treated patients.
Figure 2.
Characterization of UGT1A levels in cells and primary acute myeloid leukemia specimens. (A) UGT1A antibody purification is described in the Online Supplementary Methods. The resulting antibody revealed a single band when monitoring endogenous UGT1A in liver HepG2 cells on the western blot with reduction for pan-UGT1A RNAi versus luciferase RNAi controls. Actin blot is provided for loading. Pan-UGT1A antibody purified in (A) recognized elevated UGT1A in THP-1 cells over-expressing GLI1 as assessed both by western blot (B) and confocal microscopy (C). In addition, UGT1A levels are reduced by vismodegib in these cells (B). (D) UGT1A are elevated in AML patients, including some treatment naïve patients, relative to healthy volunteers. UGT1A levels were observed to be elevated in all the AML patient specimens examined including 3/4 treatment naïve AML patients relative to healthy volunteers. Scale bar: 10 µm.
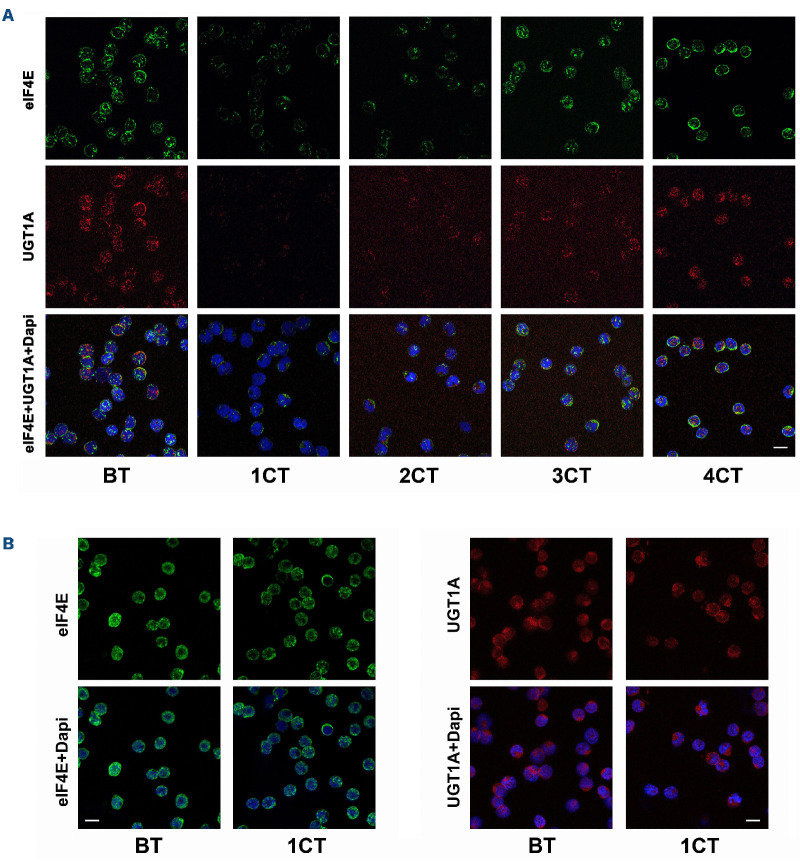
For patients with short SD, most achieved targeting of either UGT1A or eIF4E, or only very transient targeting of both (e.g., patients A-001 and A-010) (Table 3). For most PD patients, there was no targeting of UGT1A protein levels, eIF4E protein levels or eIF4E localization; furthermore, ENT1 or ADK were generally reduced relative to BT (Table 3, Figure 4B, Online Supplementary Table S2). For one SD and one PD patient, there was reduced UGT1A but no eIF4E targeting whereby resistance likely resulted from reduction in ENT1 relative to BT (Table 3). In all, simultaneous targeting of UGT1A and eIF4E correlated with objective clinical response or durable SD, while loss of eIF4E targeting corresponded to resistance and/or relapse via increased UGT1A protein levels and/or decreased ENT1 levels.
Table 3.
Changes in protein levels of eIF4E and UGT1A or RNA levels of ENT upon treatment.
Discussion
While glucuronidation has been a well-established impediment to the development of therapeutics in humans for more than 70 years,15 there have been no modalities reported to overcome this deactivation mechanism in patients. This is despite the observation that an estimated >50% of FDA-approved drugs,18 including many of the drugs used in the treatment of AML, are deactivated in this manner.18 In more recent years, it has become clear that glucuronidation is not limited to the liver, but also occurs in other tissues, indicating that drugs that bypass liver metabolism will not a priori escape this modification.8,13-15,50 Indeed, recent studies indicate that cancer cells, including AML blasts, can develop the capacity to glucuronidate drugs through the elevation of UGT1A proteins to evade the impacts of chemotherapies.8,13-15,50 We had previously identified vismodegib to target UGT1A levels in AML cells resistant to ribavirin and cytarabine, whereby vismodegib correlated with reduced drug glucuronidation as observed by mass spectrometry and restored drug sensitivity.8,16 Herein, we report on the first clinically feasible avenue to reduce glucuronidation and re-sensitize cells to drugs in humans and demonstrate, for the first time, that UGT1A protein levels could be targeted in patients. Indeed, vismodegib resulted in an up to 10-fold reduction in UGT1A protein levels in patients which persisted for months. Given the central role that glucuronidation plays in endogenous metabolite metabolism, we note that this reduction in UGT1A levels did not cause severe toxicity, a longstanding concern for the development of clinically useful glucuronidation inhibitors. In patients who had reduced UGT1A levels, we observed ribavirin targeting of eIF4E as demonstrated by nuclear to cytoplasmic re-localization of eIF4E and reduction in eIF4E protein levels up to 6-fold in AML cells. While targeting DNA hypomethylation by decitabine could not be directly measured due to insufficient material, it could be inferred from our previous studies into decitabine glucuronidation in cells.16 In all, simultaneous reduction in UGT1A protein levels and eIF4E targeting correlated with objective clinical benefits including a partial remission, blast responses, and durable SD.
Figure 3.
Molecular response of eIF4E and UGT1A for patient B-004 who achieved a blast response. (A) Samples for completed treatment cycles (CT) 1 to 10 (1CT-10CT) were collected at the end of each cycle; the patient had a 28-day treatment interruption after 9CT. Percentage of bone marrow blasts is shown (in black). Changes in protein expression of eIF4E (green) and UGT1A (red) in AML blasts isolated by FACS and stained as described in the Online Supplementary Appendix. Graph generated using GraphPad Prism 7. (B) Representative confocal micrographs of blasts used for quantification in (A). eIF4E (green), UGT1A (red) and DAPI (blue) in sorted bone marrow blasts are shown. Note also that a higher fraction of eIF4E is in the nucleus before treatment (BT) and at end of treatment (EOT) than during response, as observed previously in patients treated with ribavirin.21,22 Dapi provided as a nuclear marker. Scale bars: 10 µm.
Figure 4.
UGT1A and eIF4E levels in responding and non-responding patients A-008 and A-004. Representative micrographs used for quantifications in Table 3. eIF4E (green), UGT1A (red) and DAPI (blue) in sorted bone marrow blasts are shown. AML blasts isolated by FACS and stained as described in the Online Supplementary Appendix. Patient A-008 (A) achieved a 4-month stable disease (SD) and patient A-004 (B) a progressive disease (PD). BT: before treatment; CT: completed treatment cycle. Dapi provided as a nuclear marker. Scale bars: 10 µm.
At clinical relapse, we observed that both UGT1A and eIF4E protein levels became elevated and eIF4E re-localized to the nucleus, suggesting that acquired resistance to vismodegib provided conditions whereby ribavirin glucuronidation led to reduced ribavirin-targeting of eIF4E and, by inference, deactivation of decitabine (Table 3, Figure 3).8,16 Indeed, re-emergence of UGT1A levels is correlated with multi-drug resistance.8,12-16,50,51 There are several precedents for vismodegib resistance arising in cancer patients which could be at play here. For example, vismodegib failure occurs in basal cell carcinoma due to SMO mutations which arise in as little as two months of treatment.52 Thus, SMO mutations could be related to the loss of response here; this will be tested in future studies. In addition, FLT3-ITD drives GLI1 production independently of SMO thereby circumventing vismodegib action.53 In our study, 3/5 PD patients harbored FLT3-ITD mutations which could explain why UGT1A protein levels were not impacted by vismodegib in these patients; this is consistent with the FLT3-ITD mutations harbored by PD patients A-005 and C-001 who had no change in UGT1A, while additional reasons underlie patient A-009 who had FLT3-ITD mutations but still had reduced levels of UGT1A. Previous studies indicated that vismodegib monotherapy had modest effects in AML,54 suggesting that responses observed here are co-operative, i.e., impaired drug glucuronidation with vismodegib restored sensitivity to ribavirin and decitabine. The appearance of SMO mutations and/or alternative means to elevate UGT1A protein levels suggest it would be beneficial to develop therapeutics that directly target selected UGT1A proteins. In this way, the rapid adaptation of upstream signaling pathways like SMO-GLI1 can be evaded. Early-stage inhibitors which directly bind to UGT1A were identified, and these reduced glucuronidation and restored sensitivity to ribavirin and cytarabine in cell lines.49 This presents a promising future direction for next generation therapeutics to target glucuronidation for longer in patients.
To date, ribavirin has been the only means to target eIF4E in cancer patients that has led to objective clinical responses.21,22,44,45 In contrast, the antisense oligonucleotide strategies to lower eIF4E levels did not effectively reduce eIF4E protein levels in patients and did not produce objective clinical responses.55 The phosphorylation status of eIF4E is often linked to its oncogenicity and this can be targeted with MNK1/2 inhibitors in AML cells.56 Notably MNK1/2 inhibitors also target phosphorylation of many other proteins, including hnRNPA1 and PSF,57 and thus could be an interesting combination with ribavirin to produce more robust impacts on eIF4E and, through MNK, more multifaceted effects. Moreover, whether or not MNK1/2 inhibitors are targets of inducible glucuronidation in patients has yet to be studied, and thus like many other drugs, combining these with effective glucuronidation inhibitors could be clinically relevant. eIF4E has been targeted with the 4GI-1 inhibitor which allosterically reduces the interaction of eIF4E with eIF4G to reduce translation and this could also be considered to enhance eIF4E inhibition.58 Finally, the nuclear fraction of eIF4E has recently been found to play a role in splicing.19 Thus the combination of ribavirin with splicing inhibitors, which would be predicted to severely impair splicing in AML cells, could improve efficacy. Clinical trials will be required to assess the utility of these combinations.
These and previous studies position UGT1A and glucuronidation as important factors to consider for the development of AML therapeutics. In support of this, many of the mainstays of AML treatment are deactivated through glucuronidation in cell-line models, including venetoclax and azacytidine, and in patients, for instance, cytarabine and ribavirin.8,16 Here, we demonstrated that both treatment naïve and heavily pre-treated AML patients had substantial elevation of UGT1A protein levels relative to healthy volunteers (Figure 2), suggesting glucuronidation is a widespread barrier to effective AML treatment. Quantifying the prevalence of UGT1A protein dysregulation in AML patients on a global level is warranted, particularly given that UGT1A mRNA and protein levels do not always correlate,8,16 and, thus, RNA levels will not always be an accurate surrogate for UGT1A protein levels.
Aside from glucuronidation, we also monitored the loss of the ENT1 transporter as another contributor to multi-drug resistance and relapse in these patients. While all patients who entered the trial passed functional screens for active ENT1, we observed that several patients manifested substantial reductions in ENT1 RNA levels during treatment. ENT1 is required for cellular entry of ribavirin and decitabine as well as many other AML drugs, including cytarabine and azacytidine. Reduced ENT1 corresponded to a loss of eIF4E targeting by ribavirin and clinical resistance even in the face of reduced UGT1A protein levels. There appears to be substantial selection on this transporter which likely influences responses to subsequent salvage regimens. In all, clinical responders in the trial required targeting of both UGT1A and eIF4E, and functional ENT1.
In this third trial using ribavirin in high-eIF4E AML, we continue to observe robust targeting of eIF4E in patients corresponding to objective responses supporting the further clinical development of ribavirin, and, in the future, next generation eIF4E inhibitors. In prior ribavirin monotherapy and ribavirin with LDAC clinical trials, we observed higher overall response rates than those observed here with 6/15 and 5/14, respectively, including complete responses in both trials.21,22 In comparison to the current trial, these patients had had a median of one prior therapy; patients in our trial were much more heavily pre-treated, with a median of three prior therapies in the VRD group, likely contributing to the observed differences in responses.
In conclusion, this study demonstrated that UGT1A protein levels can be decreased in patients, and this is associated with objective clinical benefit. We also showed that an up to 10-fold reduction in UGT1A levels can be achieved in patients without substantial toxicity over the course of months (Table 3). These observations have implications far beyond AML, ribavirin and decitabine. Given the large number of FDA-approved drugs that can be deactivated via glucuronidation,17,51 these findings pave the way for the development of pharmaceuticals targeting glucuronidation in patients, one of the most widespread and longstanding problems in therapeutic development.
Supplementary Material
Acknowledgments
We are grateful for the gifts of ribavirin and decitabine from Pharmascience and vismodegib from Roche.
Funding Statement
Funding: This work was funded by the Leukemia and Lymphoma Society USA TRP (R6478) and the Jewish General Hospital Foundation. KLBB holds a Canada Research Chair in Molecular Biology of the Cell Nucleus. SA is a Chercheur Clinicien de Merite of the FRSQ.
References
- 1.de Lima M, Roboz GJ, Platzbecker U, Craddock C, Ossenkoppele G. AML and the art of remission maintenance. Blood Rev. 2021;49:100829. [DOI] [PubMed] [Google Scholar]
- 2.DiNardo CD, Rausch CR, Benton C, et al. Clinical experience with the BCL2-inhibitor venetoclax in combination therapy for relapsed and refractory acute myeloid leukemia and related myeloid malignancies. Am J Hematol. 2018;93(3):401-407. [DOI] [PubMed] [Google Scholar]
- 3.DiNardo CD, Jonas BA, Pullarkat V, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med. 2020;383(7):617-629. [DOI] [PubMed] [Google Scholar]
- 4.Maiti A, Rausch CR, Cortes JE, et al. Outcomes of relapsed or refractory acute myeloid leukemia after frontline hypomethylating agent and venetoclax regimens. Haematologica. 2021;106(3):894-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangan JK, Luger SM. Salvage therapy for relapsed or refractory acute myeloid leukemia. Ther Adv Hematol. 2011;2(2):73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva WFD, Rosa LID, Seguro FS, et al. Salvage treatment for refractory or relapsed acute myeloid leukemia: a 10-year singlecenter experience. Clinics (Sao Paulo). 2020;75:e1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zahreddine H, Borden KL. Mechanisms and insights into drug resistance in cancer. Front Pharmacol. 2013;4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511(7507):90-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda K, Hosokawa M, Iwakawa S. Cellular uptake of decitabine by equilibrative nucleoside transporters in HCT116 cells. Biol Pharm Bull. 2015;38(8):1113-1119. [DOI] [PubMed] [Google Scholar]
- 10.Hummel-Eisenbeiss J, Hascher A, Hals PA, et al. The role of human equilibrative nucleoside transporter 1 on the cellular transport of the DNA methyltransferase inhibitors 5-azacytidine and CP-4200 in human leukemia cells. Mol Pharmacol. 2013;84(3):438-450. [DOI] [PubMed] [Google Scholar]
- 11.Wohnsland A, Hofmann WP, Sarrazin C. Viral determinants of resistance to treatment in patients with hepatitis C. Clin Microbiol Rev. 2007;20(1):23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borden KL. When will resistance be futile? Cancer Res. 2014;74(24):7175-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landmann H, Proia DA, He S, et al. UDP glucuronosyltransferase 1A expression levels determine the response of colorectal cancer cells to the heat shock protein 90 inhibitor ganetespib. Cell Death Dis. 2014;5(9):e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber M, Bellemare J, Hoermann G, et al. Overexpression of uridine diphosphoglucuronosyltransferase 2B17 in high-risk chronic lymphocytic leukemia. Blood. 2013;121(7):1175-1183. [DOI] [PubMed] [Google Scholar]
- 15.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581-616. [DOI] [PubMed] [Google Scholar]
- 16.Zahreddine HA, Culjkovic-Kraljacic B, Gasiorek J, Duchaine J, Borden KLB. GLI1-inducible glucuronidation targets a broad spectrum of drugs. ACS Chem Biol. 2019;14(3):348-355. [DOI] [PubMed] [Google Scholar]
- 17.Rowland A, Miners JO, Mackenzie PI. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol. 2013;45(6):1121-1132. [DOI] [PubMed] [Google Scholar]
- 18.Allain EP, Rouleau M, Levesque E, Guillemette C. Emerging roles for UDP-glucuronosyltransferases in drug resistance and cancer progression. Br J Cancer. 2020;122(9):1277-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghram M, Morris G, Culjkovic-Kraljacic B, et al. The eukaryotic translation initiation factor eIF4E reprogrammes the splicing machinery and drives alternative splicing. EMBO J. 2023;42(7):e110496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Topisirovic I, Guzman ML, McConnell MJ, et al. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol Cell Biol. 2003;23(24):8992-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assouline S, Culjkovic-Kraljacic B, Bergeron J, et al. A phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia with elevated eIF4E. Haematologica. 2015;100(1):e7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Assouline S, Culjkovic B, Cocolakis E, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114(2):257-260. [DOI] [PubMed] [Google Scholar]
- 23.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3'UTR. J Cell Biol. 2005;169(2):245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175(3):415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topisirovic I, Siddiqui N, Lapointe VL, et al. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009;28(8):1087-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Culjkovic-Kraljacic B, Fernando TM, Marullo R, et al. Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood. 2016;127(7):858-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borden KL. The eukaryotic translation initiation factor eIF4E wears a "cap" for many occasions. Translation (Austin). 2016;4(2):e1220899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis MR, Delaleu M, Borden KLB. Nuclear eIF4E stimulates 3’-end cleavage of target RNAs. Cell Rep. 2019;27(5):1397-1408.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Culjkovic-Kraljacic B, Skrabanek L, Revuelta MV, et al. The eukaryotic translation initiation factor eIF4E elevates steady-state m(7)G capping of coding and noncoding transcripts. Proc Natl Acad Sci U S A. 2020;117(43):26773-26783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101(52):18105-18110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kentsis A, Volpon L, Topisirovic I, et al. Further evidence that ribavirin interacts with eIF4E. RNA. 2005;11(12):1762-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraljacic BC, Arguello M, Amri A, Cormack G, Borden K. Inhibition of eIF4E with ribavirin cooperates with common chemotherapies in primary acute myeloid leukemia specimens. Leukemia. 2011;25(7):1197-1200. [DOI] [PubMed] [Google Scholar]
- 33.Volpon L, Osborne MJ, Zahreddine H, Romeo AA, Borden KL. Conformational changes induced in the eukaryotic translation initiation factor eIF4E by a clinically relevant inhibitor, ribavirin triphosphate. Biochem Biophys Res Commun. 2013;434(3):614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Culjkovic-Kraljacic B, Fernando TM, Marullo R, et al. Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood. 2016;127(7):858-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersson F, Yau C, Dobocan MC, et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin Cancer Res. 2011;17(9):2874-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersson F, del Rincon SV, Emond A, et al. Genetic and pharmacolgic inhibition of eIF4E reduces breast cancer cell migration, invasion and metastasis. Cancer Res. 2015;75(6):1102-1112. [DOI] [PubMed] [Google Scholar]
- 37.Zahreddine H, Kraljacic-Culjkovic B, Edmond A, et al. The eukaryotic translation initiation factor eIF4E harnesses hyaluronan production to drive its malignant activity. Elife. 2017;6:e29830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bollmann F, Fechir K, Nowag S, et al. Human inducible nitric oxide synthase (iNOS) expression depends on chromosome region maintenance 1 (CRM1)- and eukaryotic translation initiation factor 4E (elF4E)-mediated nucleocytoplasmic mRNA transport. Nitric Oxide. 2013;30:49-59. [DOI] [PubMed] [Google Scholar]
- 39.Shi F, Len Y, Gong Y, et al. Ribavirin inhibits the activity of mTOR/eIF4E, ERK/Mnk1/eIF4E signaling pathway and synergizes with tyrosine kinase inhibitor imatinib to impair Bcr-Abl mediated proliferation and apoptosis in Ph+ leukemia. PLoS One. 2015;10(8):e0136746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zismanov V, Attar-Schneider O, Lishner M, Heffez Aizenfeld R, Tartakover Matalon S, Drucker L. Multiple myeloma proteostasis can be targeted via translation initiation factor eIF4E. Int J Oncol. 2015;46(2):860-870. [DOI] [PubMed] [Google Scholar]
- 41.Volpin F, Casaos J, Sesen J, et al. Use of an anti-viral drug, ribavirin, as an anti-glioblastoma therapeutic. Oncogene. 2017;36(21):3037-3047. [DOI] [PubMed] [Google Scholar]
- 42.Wang G, Li Z, Li Z, et al. Targeting eIF4E inhibits growth, survival and angiogenesis in retinoblastoma and enhances efficacy of chemotherapy. Biomed Pharmacother. 2017;96:750-756. [DOI] [PubMed] [Google Scholar]
- 43.Volpon L, Culjkovic-Kraljacic B, Osborne MJ, et al. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc Natl Acad Sci U S A. 2016;113(19):5263-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunn LA, Fury MG, Sherman EJ, et al. Phase I study of induction chemotherapy with afatinib, ribavirin, and weekly carboplatin and paclitaxel for stage IVA/IVB human papillomavirus-associated oropharyngeal squamous cell cancer. Head Neck. 2017;40(2):233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kosaka T, Maeda T, Shinojima T, Nagata H, Mizuno R, Mototsugu O. A clinical study to evaluate the efficacy and safety of docetaxal with ribavirin in patients with progressive castration resistant prostate cancer who have previously received docetaxol alone. J Clin Oncol. 2017;35(15 Suppl):e14010. [Google Scholar]
- 46.Zimmerman EI, Huang M, Leisewitz AV, Wang Y, Yang J, Graves LM. Identification of a novel point mutation in ENT1 that confers resistance to Ara-C in human T cell leukemia CCRF-CEM cells. FEBS Lett. 2009;583(2):425-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder T, Rautenberg C, Krüger WH, et al. Decitabine as salvage therapy for relapse of AML and MDS after allogeneic stem cell transplantation - a retrospective multicenter analysis on behalf of the German Cooperative Transplant Study Group. Blood. 2016;128(22):3446. [Google Scholar]
- 48.Mori K, Hiraoka O, Ikeda M, et al. Adenosine kinase is a key determinant for the anti-HCV activity of ribavirin. Hepatology. 2013;58(4):1236-1244. [DOI] [PubMed] [Google Scholar]
- 49.Osborne MJ, Coutinho de Oliveira L, Volpon L, Zahreddine HA, Borden KLB. Overcoming drug resistance through the development of selective inhibitors of UDP-glucuronosyltransferase enzymes. J Mol Biol. 2019;431(2):258-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dellinger RW, Matundan HH, Ahmed AS, Duong PH, Meyskens FL Jr. Anti-cancer drugs elicit re-expression of UDP-glucuronosyltransferases in melanoma cells. PLoS One. 2012;7(10):e47696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guillemette C, Levesque E, Rouleau M. Pharmacogenomics of human uridine diphospho-glucuronosyltransferases and clinical implications. Clin Pharmacol Ther. 2014;96(3):324-339. [DOI] [PubMed] [Google Scholar]
- 52.Pricl S, Cortelazzi B, Dal Col V, et al. Smoothened (SMO) receptor mutations dictate resistance to vismodegib in basal cell carcinoma. Mol Oncol. 2015;9(2):389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wellbrock J, Latuske E, Kohler J, et al. Expression of hedgehog pathway mediator GLI represents a negative prognostic marker in human acute myeloid leukemia and its inhibition exerts antileukemic effects. Clin Cancer Res. 2015;21(10):2388-2398. [DOI] [PubMed] [Google Scholar]
- 54.Bixby D, Noppeney R, Lin TL, et al. Safety and efficacy of vismodegib in relapsed/refractory acute myeloid leukaemia: results of a phase Ib trial. Br J Haematol. 2019;185(3):595-598. [DOI] [PubMed] [Google Scholar]
- 55.Hong DS, Kurzrock R, Oh Y, et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin Cancer Res. 2011;17(20):6582-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suarez M, Blyth GT, Mina AA, et al. Inhibitory effects of tomivosertib in acute myeloid leukemia. Oncotarget. 2021;12(10):955-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joshi S, Platanias LC. Mnk kinase pathway: cellular functions and biological outcomes. World J Biol Chem. 2014;5(3):321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papadopoulos E, Jenni S, Kabha E, et al. Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. Proc Natl Acad Sci U S A. 2014;111(31):E3187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.