Abstract
The large intestine harbors microorganisms playing unique roles in host physiology. The beneficial or detrimental outcome of host‐microbiome coexistence depends largely on the balance between regulators and responder intestinal CD4+ T cells. We found that ulcerative colitis‐like changes in the large intestine after infection with the protist Blastocystis ST7 in a mouse model are associated with reduction of anti‐inflammatory Treg cells and simultaneous expansion of pro‐inflammatory Th17 responders. These alterations in CD4+ T cells depended on the tryptophan metabolite indole‐3‐acetaldehyde (I3AA) produced by this single‐cell eukaryote. I3AA reduced the Treg subset in vivo and iTreg development in vitro by modifying their sensing of TGFβ, concomitantly affecting recognition of self‐flora antigens by conventional CD4+ T cells. Parasite‐derived I3AA also induces over‐exuberant TCR signaling, manifested by increased CD69 expression and downregulation of co‐inhibitor PD‐1. We have thus identified a new mechanism dictating CD4+ fate decisions. The findings thus shine a new light on the ability of the protist microbiome and tryptophan metabolites, derived from them or other sources, to modulate the adaptive immune compartment, particularly in the context of gut inflammatory disorders.
Keywords: aryl hydrocarbon receptor, colitis, microbiome, regulatory T cells, tryptophan metabolites
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction
An intestinal protist Blastocystis ST7 produces indole‐3‐acetaldehyde that modulates the adaptive immune system response in the context of gut inflammation.

Introduction
The intestinal microbiome constitutes a vast collection of commensal and pathogenic organisms. Blastocystis are the only Stramenopiles, a protist group, that can infect human gut (Stensvold & Clark, 2016; Deng et al, 2021). The role of Blastocystis in the human microbiome, whether it is harmful or beneficial, is still the subject of debate, despite its widespread colonization among the population. The outcome of gastrointestinal tract colonization may be influenced by factors such as the composition of gut bacterial communities, the host's immune status, and, notably, the genetic background of various Blastocystis subtypes (STs), with subtypes ST1 and ST4 being beneficial and ST7 pathogenic (Ajjampur et al, 2016; Yason et al, 2019; Deng et al, 2023a,b). There is limited information on how these protists interact with the gut immune system during intestinal colonization.
Several elements of the Blastocystis genome were likely acquired by horizontal gene transfer, of which those related to tryptophan metabolism likely contribute to adaptation to the host environment (Denoeud et al, 2011; Eme et al, 2017). Synthesis of indole derivatives by human‐associated members of the Protista kingdom has been reported for Trypanosoma and Trichomonas (which are not members of the gut microbiome), as well as the microbiome members Blastocystis and Entamoeba (McGettrick et al, 2016; Eme et al, 2017; Moreira & López‐García, 2017).
The microbiota–aryl hydrocarbon receptor (AhR) axis is an essential element of gut immune homeostasis. Bacterial‐derived tryptophan metabolites such as indole‐3‐acetic acid, indole‐3‐aldehyde, and indole‐3‐acrylic acid are all potent bioactive compounds that affect adaptive and innate immune responses (Zelante et al, 2013; Venkatesh et al, 2014; Lamas et al, 2016; Wlodarska et al, 2017). The beneficial effect of these indole derivatives on the host immune system is thought to be due to agonistic activation of the AhR signaling pathway (Zelante et al, 2013; Wlodarska et al, 2017).
Here, we show that indolepropionic acid (IPA) and indole‐3‐acetaldehyde (I3AA) are indole derivatives produced by one of the Blastocystis subtypes, ST7. Importantly, among gut‐dwelling organisms, synthesis of IPA is known only from a few Clostridia species—C. sporogenes and C. botulinum (Dodd et al, 2017)—whereas I3AA is known only from Enterobacter cloacae (Koga et al, 1994). We found that I3AA exerts a potent effect on the CD4+ T cell compartment, skewing its reactivity toward self‐flora and contributing to a pro‐inflammatory response in the gut tissue. I3AA‐directed remodeling of CD4+ Th subsets caused a stronger than normal TCR signaling upon recognition of peptide MHC (pMHC) antigen. We found that, by affecting the CD4+ Th fate decision, I3AA destabilizes immune homeostasis, which makes this natural indole derivative a potential target in treating intestinal inflammatory disorders. We demonstrate that I3AA acts through the intrinsic AhR signaling pathway. Unlike other known tryptophan metabolites derived from the microbiome, I3AA is antagonistic to AhR. Therefore, we hypothesize that I3AA, acting as a suppressor of the AhR pathway, may play a significant role in promoting gut inflammation.
Results
TCRαβ lymphocyte compartment in the large intestinal lamina propria of Blastocystis‐infected animals
In the Blastocystis ST7 infection model, DSS treatment is followed by a recovery period, then by parasite inoculation. Blastocystis ST7 elicits pathological alterations reminiscent of ulcerative colitis (UC) within the colon (Ajjampur et al, 2016; Yason et al, 2019). To test whether this phenotype can be linked to disruption of immune homeostasis, we analyzed TCRαβ T lymphocytes in mesenteric lymph nodes (mLN) and large intestine lamina propria (LP). Lymphocytes from mLN and cecum LP showed no significant differences in distribution and total numbers of CD4+ and CD8αβ+ T cells between infected and control groups (Fig 1A and Appendix Fig S1B). In contrast, significant changes in the CD4+ compartment were observed in the colon. Colonic LP lymphocytes from groups infected with isolates ST7B or ST7H showed increased percentages of TCRαβ lymphocytes, with a substantial increase of CD4+ T cell numbers (Fig 1A). The higher proportion of CD69hi CD4+ cells revealed that the enlarged number of CD4+ T cells in the colon of mice colonized with ST7H and, to some extent, ST7B, might be a consequence of TCR stimulation and clonal T cell expansion (Fig 1B) (Testl & Lanier, 1989). The hallmark of Blastocystis infection by both isolates was a substantial decrease of CD4+ T cells expressing IL2 receptor α‐chain (CD25) (Fig 1C). Changes in CD25+ T cell numbers were not restricted to the colon but were also observed in the cecum of infected animals. CD4+ CD25+ lymphocytes are mainly Foxp3‐expressing regulatory T cells (Treg) (Fontenot et al, 2017). Analysis of Foxp3 status confirmed that the CD4+ T cell compartment from both segments of the large intestine in infected animals suffered from impaired Treg development (Fig 1D). Interestingly, LP‐derived Tregs from the control group were characterized by a “stretched” pattern of Helios expression, with approximately equal numbers of cells expressing low and high amounts of Helios (Fig 1E). In contrast, expression of Helios in Tregs from Blastocystis‐infected mice, from both cecum and colon, showed a pattern shifted toward Helioshi.
Figure 1. TCRαβ compartment in Blastocystis ST7‐infected mice.
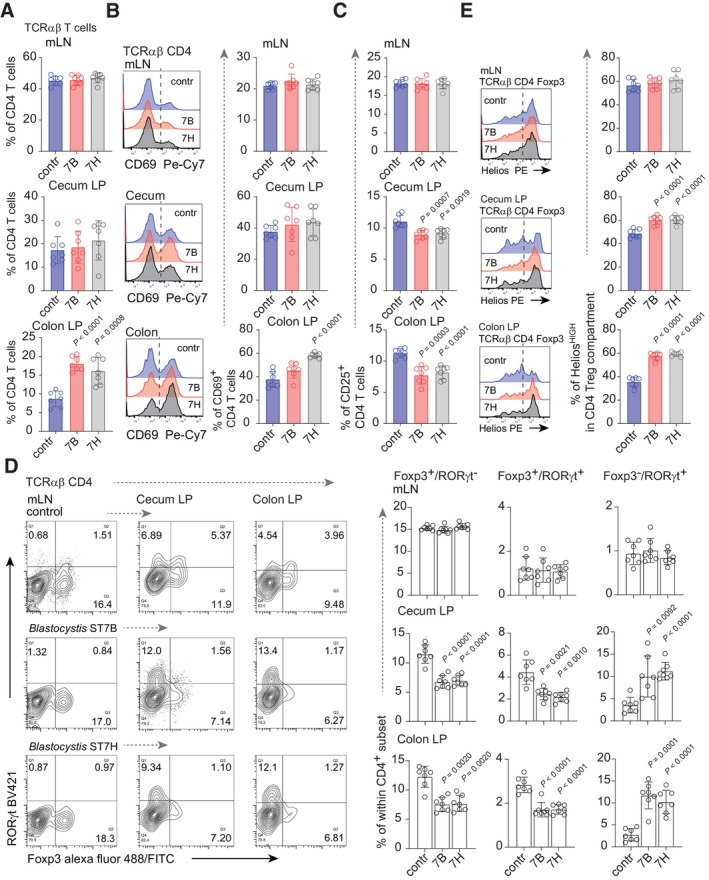
- Percentage of TCRαβ CD4+ T cells in mesenteric lymph nodes (mLN), cecum lamina propria (LP), and colon LP from control and Blastocystis ST7B‐ and ST7H‐infected groups.
- CD69 expression on CD4+ T cells from infected and non‐infected group. Left: representative histograms. The origin of T cells is indicated on the top of each histogram. Right: bar charts representing statistical analysis.
- Bar charts depicting expression of CD25 on LP and mLN CD4+ lymphocytes from control and infected cohorts.
- Left: Representative dot blots of Foxp3+ and RORγt+ distribution within CD4+ T cell compartments. Columns refer to site of CD4+ isolation while rows to the infection status. Right: bar charts representing statistical analysis. Columns refer to a particular phenotype, rows to the T cell origin.
- Helios expression in Foxp3+ Tregs from non‐infected and Blastocystis‐infected counterparts. Left: representative histograms. Right: bar charts representing percentage of HeliosHi cells within Treg subsets.
Data information: All experimental groups comprised seven animals in two separate experiments. Bar plots show mean statistic value and error bars indicate s.d., Box‐and‐whisker plot show first and third quartile with median value indicated, error bars indicate s.d. Statistical analysis was performed using a two‐tailed t‐test with Welch correction. When P < 0.05 (experimental to control groups), the values are indicated.
Source data are available online for this figure.
The LP Treg compartment comprises Foxp3+ cells with simultaneous expression of the retinoic acid‐related orphan receptor (RORγt) (Yang et al, 2016) transcription factor associated with the Th17 CD4+ subset (Ivanov et al, 2006). The development of this effector‐like Treg lineage in the gut milieu relies on interaction with the host microbiome. Interestingly, the “intersect” Th17‐Treg population, defined as Foxp3+ RORγt+, showed a substantial decline in both cecum and colon of Blastocystis‐infected cohorts, when compared to the control group (Fig 1D). In contrast to the Foxp3+ RORγt+ fraction, the CD4+ Foxp3− RORγt+ compartment, representing the majority of the Th17 effectors, was significantly expanded upon exposure to ST7B and ST7H in both segments of the large intestine (Fig 1D). To conclusively demonstrate that the observed phenotype of infected mice was associated with Blastocystis infection, we examined the large intestinal CD4+ T cell subset in uninfected animals after DSS treatment at a time point corresponding to the pre‐colonization stage. The CD4+ T cells isolated after 5 days of recovery resemble the phenotype of uninfected animals (Appendix Fig S1B). Altogether, these data suggest that Blastocystis ST7 can lead to stronger CD4+ T cell recognition of gut‐derived antigens.
Blastocystis‐derived tryptophan metabolites inhibit iTreg generation and enhance Th17 polarization in vitro
Blastocystis ST7 interacts with the host microbiome (Yason et al, 2019). Remodeling of the gut bacterial community likely changes the landscape of bacteria‐derived, biologically active metabolites in the intestine, so that Blastocystis infection might affect host immune compartments. Simultaneously, direct interaction of these protists with the host adaptive immune system could potentially occur (Denoeud et al, 2011). We therefore tested the effect of filtered supernatants from cultures of Blastocystis ST7B and ST7H on conventional CD4+ T cells (Tconv) under in vitro iTreg and Th17 polarization conditions. Treatment with Blastocystis‐derived supernatants led to a substantial decrease in iTreg number (Fig 2A upper panel). Conversely, polarization toward the Th17 phenotype was enhanced when Tconv cells were polarized in the presence of Blastocystis ST7B and ST7H metabolites (Fig 2A lower panel). Furthermore, analysis of Blastocystis cultures by PCR with bacterial‐specific 16S rDNA and Blastocystis ST7‐specific 18S rDNA primers revealed no potential microbial contamination. Hence, the biological effect on T cells in these experimental settings was due to Blastocystis‐derived metabolites (Appendix Fig S2A left). Using solid‐phase chromatography, we subdivided Blastocystis culture supernatants into three fractions (Appendix Fig S2A). Under iTreg differentiation conditions, only fractions eluted with 50% methanol (referred to hereafter as 50 m) directly affected polarization toward Foxp3+ iTreg, resulting in substantial reduction in the development of these cells (Appendix Fig S2B). The 50 m fraction was enriched for indole‐related compounds (Appendix Fig S2A). Using liquid chromatography tandem mass spectrometry (LC–MS/MS) on the 50 m fractions from Blastocystis ST7B and ST7H, we identified two indole derivatives: indolepropionic acid (IPA) and indole‐3‐acetylaldehyde (I3AA) (Fig 2B).
Figure 2. Blastocystis‐derived tryptophan metabolites negatively affect iTreg generation and enhance Th17 polarization in vitro .
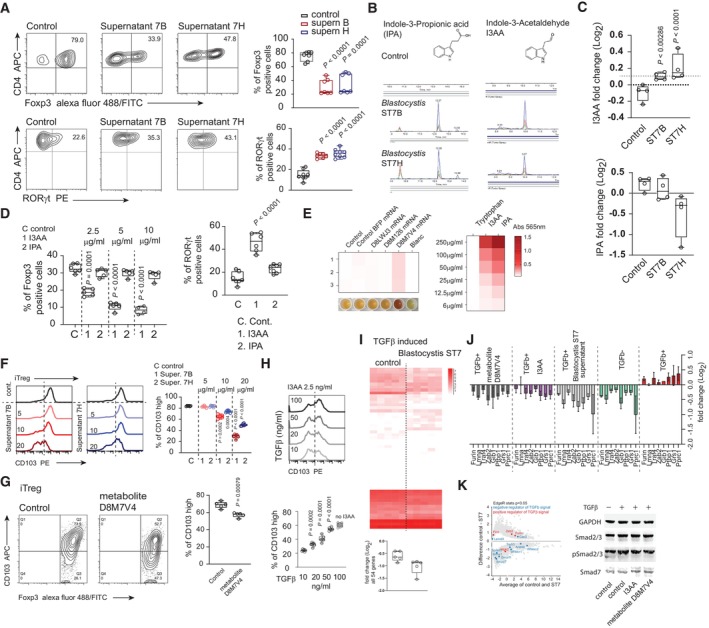
- In vitro conversion of CD4+ T cells toward iTreg (upper panel) and RORγt+ Th17 subset (lower panel) and with the presence of Blastocystis‐derived metabolites and in the control condition. Supernatants for ST7B and ST7H were derived from normalized cultures characterized by similar number of protists.
- Characterization of the Blastocystis‐derived metabolites. Using LC–MS, two indole derivatives indole‐3‐acetaldehyde (I3AA), and indole 3‐propionic acid (IPA) were identified.
- The quantitative analysis of I3AA (upper chart) and IPA (lower bar chart) metabolites in the gut content of the control and Blastocystis ST7B and ST7H‐infected animals.
- Right, Th17 in vitro conversion of naïve CD4+ T cells in the presence of I3AA and IPA. Experiments were carried out in 5 μg/ml of IPA and I3AA. Left. In vitro conversion of CD4+ T cells toward iTreg in conditions representing different concentrations of I3AA and IPA.
- Heat map representing extent of tryptophan breakdown carried out by the HEK‐293 cells transfected with different mRNAs. Heat map represents the intensity of the colored compound produced from the supernatant (left) or the compound standard (right), which is proportional to the indole derivative in the sample. Efficiency of tryptophan‐derived metabolite synthesis in culture medium was estimated colorimetrically using modified Kovac's reagent, measuring absorbance at 565 nm.
- Expression of CD103 on the surface of iTreg converted in the presence of different concentration of indole derivatives isolated from Blastocystis ST7B and ST7H culture supernatants.
- CD103 expression on the surface of iTreg converted in the presence of tryptophan metabolite produced by the D8M7V4‐transfected HEK‐293 cells.
- Expression of CD103 on the surface of iTreg converted in the presence of different concentrations of TGFβ exposed to constant concentrations of I3AA.
- The pool of 54 genes differentially expressed in the gut of Blastocystis ST7‐infected animal. TGFβ‐upregulated genes identified in the in vitro stimulation experiment were selected. Box chart below heat map represents changes in expression of the 54 TGFβ‐inducible genes in the control and Blastocystis ST7‐infected groups.
- Expression analysis (using qPCR) of the eight TGFβ‐inducible genes downregulated in the gut tissue of Blastocystis ST7‐infected animals. Analysis was carried out on cDNA synthesized from in vitro TCR stimulation of the CD4+ T cells in the presence or absence of TGFβ.
- Left, MA plot of gut tissue transcriptomes from control and Blastocystis ST7 infected animals. Expression of negative and positive TGFβ regulators are highlighted in blue and red, respectively. Right, representative Western blots of Smad2 and Smad3 phosphorylation status and total Smad7 expression in CD4+ T cells after exposure to TGFβ and I3AA or D8M7V4‐transfected HEK‐293 cell‐derived tryptophan metabolites.
Data information: Bar plots show mean statistic value and error bars indicate s.d., Box‐and‐whisker plot shows first and third quartile with median value indicated, and error bars indicate s.d. Experimental and control groups comprised n = 6 each, except n = 4 in E (two repeats of the experiment with duplicated samples per each condition). Statistical analysis performed using two‐tailed t‐test with Welch correction. P‐values are indicated when P < 0.05 (experimental to control groups).
Source data are available online for this figure.
Analysis of the large intestine content revealed that ST7B‐ and ST7H‐infected animals exhibit significantly higher concentrations of the I3AA in the gut milieu than their non‐infected counterparts (Fig 2C). The concentration of IPA was comparable between control group and ST7B‐infected animals. However, in the ST7H‐infected group, the concentration of this compound decreased. IPA is thought to be produced by some Clostridium species (Dodd et al, 2017), and according to previous reports, infection with Blastocystis ST7H negatively affects this group of beneficial bacteria (Yason et al, 2019). This indicates that the main producers of IPA (in all three groups) in the gut environment are from the bacterial component and suggests that the reduction of IPA concentration in ST7H infection is due to reshaping of the microbiome in the presence of IPA‐producing species. On the other hand, increased concentration of I3AA in both Blastocystis‐infected groups, regardless of their microbial status, strongly supports the notion that this parasite is a major source of this tryptophan metabolite. The persisting residual presence of I3AA in the intestinal contents of the control animals could be attributed to either the limited non‐enzymatic synthesis of I3AA in the gut (Smirnova et al, 2016) or its potential origin from plant‐based foods, considering that this compound serves as a substrate for auxin synthesis (McClerklin et al, 2018).
To assess whether these two metabolites could modulate the adaptive arm of the immune system during Blastocystis infection, we first analyzed their biological activity in the context of Th17 and iTreg development in vitro. Exposure of Tconv to I3AA (pure compound, commercially available) under Th17‐polarizing conditions substantially enhanced polarization toward the RORγt+ phenotype (Fig 2D and Appendix Fig S2C). Conversion to iTreg was markedly impaired in the presence of I3AA (Fig 2D and Appendix Fig S2D). The negative effect of this compound, either alone or in the complex mixture (Blastocystis culture‐derived supernatant), was associated with impaired iTreg proliferation, in a dose‐dependent manner (Appendix Fig S2E and F). Recently, I3AA has been described as a potential substrate for 6‐formylindolo[3,2‐b]carbazole (FICZ) synthesis in a non‐canonical, light‐independent process (Yang et al, 2016). However, in our experimental setting, FICZ had no effect on the efficacy of iTreg conversion (Appendix Fig S2G). This indicates that, rather than being a non‐functional intermediate for FICZ, I3AA itself exhibits biological activity in activation of intrinsic pathways.
Blastocystis‐derived aromatic amino acid aminotransferase constitutes a key enzyme involved in conversion of tryptophan to I3AA
To date, except for tryptophanase inherited by Blastocystis during lateral gene transfer from a procaryote (Eme et al, 2017), no Blastocystis genes have been linked to conversion of tryptophan into I3AA or IPA metabolites. Interestingly, according to previous reports, a Trypanosoma enzyme annotated as cytoplasmatic aspartate aminotransferase (TBsASAT) is capable of efficient deamination of aromatic amino acids (McGettrick et al, 2016). TBsASAT could thus participate in the conversion of tryptophan into indole‐3‐pyruvate, which might constitute an intermediate in the I3AA synthesis pathway.
A BLAST‐based comparative analysis of the Blastocystis genome and TBsASAT revealed three candidates whose protein sequence similarity indicated potential activity in tryptophan metabolism (Appendix Fig S3A). All three hits (UniProt accession numbers D8LWJ3, D8M126, and D8M7V4) were annotated as aspartate aminotransferases. To assess the activity of these three enzymes, we amplified D8LWJ3, D8M126, and D8M7V4 from Blastocystis mRNA, simultaneously adding T7 promoter sequence on the 5′ ends of each cDNA. In vitro generated mRNAs of the corresponding enzymes were then tested individually upon transfection of human embryonic kidney (HEK‐293) cell line. Notably, neither untransfected, BFP mRNA‐transfected, nor D8LWJ3 and D8M126 mRNA‐transfected HEK‐293 cells exhibited enzymatic activity relevant to tryptophan metabolism. However, when HEK‐293 was transfected with the D8M7V4 mRNA, noticeable tryptophan breakdown occurred, which was reflected by the positive reaction of culture supernatant with modified Kovac's reagent (Fig 2E). Precise analysis using LC–MS/MS revealed the presence of I3AA in the culture medium from the D8M7V4 mRNA‐transfected HEK‐293 cells (Appendix Fig S3C). Importantly, when CD4+ T cells were tested with HEK‐293 supernatants in Th17 and iTreg polarization conditions, only medium derived from D8M7V4 transfectants mirrored the effect on T cells exerted by I3AA. Exposure to the supernatant derived from D8M7V4‐modified HEK‐293 cells led to impaired iTreg generation and enhanced Th17 T cell progression in vitro (Fig S3D).
Interestingly, indole‐3‐pyruvic acid, a known intermediate in I3AA synthesis, was not detected in the culture medium from transfected cells. The lack of this ketoacid might indicate that after D8M7V4‐driven deamination of tryptophan, a HEK‐293‐derived enzyme continued a breakdown of indole‐3‐pyruvic acid toward I3AA. However, incubation of the HEK‐293 cells with indole‐3‐pyruvic acid alone revealed no enzymatic activity associated with metabolism of this particular compound (Appendix Fig S3E). This indicates D8M7V4 as being crucial for I3AA synthesis in Blastocystis ST7. Furthermore, in contrast to Trypanosoma‐derived orthologs, metabolism of tryptophan by D8M7V4 occurs by deamination with simultaneous decarboxylation. Finally, the phenotype of the T cells upon exposure to the D8M7V4 enzyme product (I3AA) strongly supports the hypothesis that Blastocystis‐derived tryptophan metabolites constitute a key factor in reshaping of the intestinal CD4+ T cell compartment during Blastocystis infection.
I3AA produced by Blastocystis ST7 changes sensing of the TGFβ signal by CD4+ T cells
CD103, the integrin αE, is a T cell homing molecule important for Treg localization in the intestine (Yuan et al, 2015). CD103+ Tregs have increased capacity to suppress T cell proliferation in vitro and to protect from colitis induced by CD4+ Tconv cells in lymphopenic recipients (Lehmann et al, 2002). Interestingly, exposure of T cells to Blastocystis‐derived metabolites or to I3AA under iTreg polarization conditions substantially blocked CD103 expression on iTregs, in a dose‐dependent manner (Fig 2F and Appendix Fig S2H). Similarly, a negative trend in the context of CD103 expression on the iTreg surface was observed when in vitro polarization was carried out in the presence of D8M7V4‐produced tryptophan metabolite (Fig 2G). CD103 expression on Tregs relies on TGFβ signaling (Konkel et al, 2017). To test whether I3AA might change T cell sensing of TGFβ signaling, we investigated the phenotype of iTregs generated in the presence of 2.5 μg/ml of I3AA with increasing concentrations of TGFβ (Fig 2H). The presence of TGFβ rescued the induction of CD103 expression on the iTregs in a dose‐dependent manner (Fig 2H).
Next, we analyzed transcriptomes of the gut tissue retrieved from the Blastocystis ST7‐infected animals and the control group (data are derived from Deng et al, 2023a). When we compared differentially expressed genes from this experiment to data from human T cells activated by anti‐CD3 plus anti‐CD28 in the presence of TGFβ, we found 54 genes that overlapped with the DEGs from control versus Blastocystis ST7‐infected groups (Fig 2I, left heat map). Importantly, the overall expression of these TGFβ signal signatures was decreased in the tissue from Blastocystis‐infected animals (Fig 2I, Appendix Fig S2I). To investigate whether downregulation of the TGFβ‐dependent genes in vivo could be a consequence of the I3AA produced by the Blastocystis ST7, we used qPCR to analyze expression trends of eight selected transcripts in TCR‐activated CD4+ T cells in the presence of TGFβ and Blastocystis‐derived metabolites (Fig 2J). Indeed, TGFβ leads to the upregulation of all eight signature genes. However, incubation of the T cells with Blastocystis ST7‐derived supernatant, pure I3AA, or tryptophan metabolite (I3AA) produced by D8M7V4‐transfected HEK‐293 cells, abrogated the effect of TGFβ on TCR‐stimulated T cells (Fig 2J).
One of the crucial events of intrinsic TGFβ signal transduction is phosphorylation of Smad2 and Smad3 proteins (Liu et al, 1997; Massagué, 1998), which then act in the nucleus as transcription factors. Negative regulators like Smad7 can inhibit the phosphorylation of Smad2/3. Importantly, analysis of tissue transcriptomes after Blastocystis ST7 infection revealed an increase in the expression of Smad7 as well as several other genes that serve as negative regulators of the TGFβ signaling pathway (Fig 2K left and Appendix Fig S2J). We were able to detect a different phosphorylation status of Smad2 between TGFβ+ and TGFβ− controls (Fig 2K right). However, incubation of T cells with pure I3AA or metabolite (I3AA) produced by D8M7V4‐transfected HEK‐293 cells blocked phosphorylation of this Smad2/3 and simultaneously increased expression of Smad7. All these data strongly support the hypothesis that I3AA produced by Blastocystis ST7 might impair Treg selection in the gut environment by negatively affecting the TGFβ signaling pathway (Monteleone et al, 2016; Ye et al, 2017; Troncone et al, 2018). Additionally, the previously reported positive regulation of CD103 by AhR agonist ligands in the context of T cell polarization and peripheral Treg function (Ye et al, 2017; Dean et al, 2023) supports the antagonistic properties of I3AA toward AhR.
I3AA produced by Blastocystis ST7 changes TCR signal interpretation and increases TCR‐dependent reactivity of T cells against self‐microbial flora
Recruitment of individual CD4+ T clones into specialized Th lineages is a result of intrinsic translation of the signals associated with pMHC‐TCR interactions and stimuli related to the microenvironmental context (Wojciech et al, 2020). In non‐polarizing conditions, increasing concentration of Blastocystis‐derived I3AA in a “low TCR engagement” scenario (with 1:10 anti‐CD3/CD28 beads to cell ratio) led to a linear dose‐dependent increase of CD69 expression on T cells (Fig 3A). Hence, simultaneously with Treg inhibition, exposure to I3AA may affect antigen recognition by naïve CD4+ T lymphocytes and reprogram them to be more antigen‐reactive. To address the role of I3AA in the context of microbial antigen recognition, we generated CD4+ T cell hybridomas (containing a Nur77GFP reporter for strength of T cell activation) specific to abundant members of the gut microbiome in our mouse colony (Appendix Fig S4A–C). Using an in‐house developed strategy that includes whole 16S rRNA sequencing on the MiSeq platform and species (or even strain)‐level resolution analysis of the microbiome (Appendix Fig S4A), we identified Lactobacillus johnsonii strains as being one of the most abundant residents within the large intestine. Hence, we isolated the dominant L. johnsonii strain from the gut contents and used the lysate from this strain as a source of antigens for T cell hybridoma selection and further analysis (Appendix Fig S4B and C). From the polyclonal population of CD4+ T cell hybridomas, L. johnsonii‐specific clones were single‐cell sorted (then in vitro expanded) based on expression of the Nur77GFP reporter, after challenge with APC pulsed with bacterial lysate. Analysis of 11 individual L. johnsonii‐specific hybridoma clones revealed that upon contact with cognate antigen and exposure to the Blastocystis ST7‐derived indole metabolites, these cells upregulated CD69 significantly above the level of their nominal activation status triggered by pMHC (Fig 3B). Interestingly, expression of Nur77 was not affected by the indole derivatives. To further investigate potential roles of IPA and I3AA in TCR signal interpretation, we selected three clones representing different “recognition spectra” of L. johnsonii‐associated antigens (Fig 3C). Only I3AA affected CD69 expression (Fig 3D upper panel and lower left bar plot). Importantly, the effect of this molecule on CD69 upregulation was not restricted to Lactobacillus‐specific hybridomas and was confirmed in the context of OVA recognition by OT‐II hybridomas (Appendix Fig S5A and B).
Figure 3. I3AA changes TCR signal interpretation.
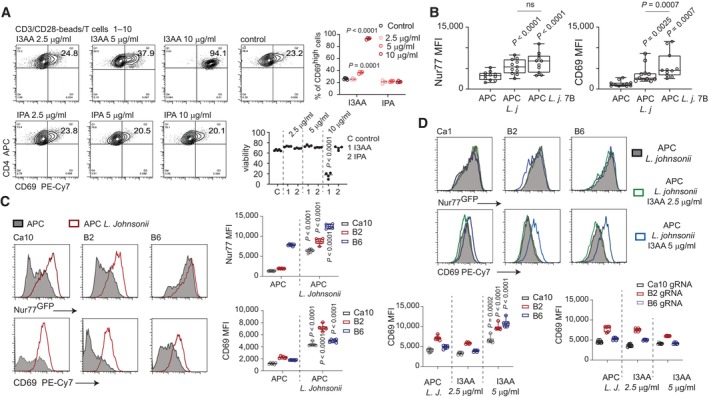
-
ACD69 expression on T cells stimulated with anti‐CD3/CD28 beads incubated with different concentrations of I3AA (upper) and IPA (lower).
-
B–DNur77GFP and CD69 expression on L. johnsonii‐specific hybridoma clones after treatment as follows: (B) Overnight incubation with APC (left), APC pulsed with bacterial lysate (middle‐APC L.j), and APC pulsed with bacterial lysate plus Blastocystis metabolites (APC L.j. 7B). Each dot represents an individual hybridoma clone. Statistical analysis was performed using two‐tailed paired t‐test. (C) Overnight incubation with APC alone or APC pulsed with bacterial lysate. S.d. calculated from four individual experiments. (D) Upper panel and left plot: Exposure to constant concentration of cognate antigen and different concentrations of I3AA. Right plot: Exposure to constant concentration of cognate antigen and different concentrations of I3AA.
Data information: Box‐and‐whisker plot show first and third quartile with median value indicated, and error bars indicate s.d. Statistical analysis was performed using a two‐tailed t‐test with Welch correction. P‐values are indicated when P < 0.05 (experimental to control groups). Experimental and control groups comprised of: n = 4 each (A), n = 11 (B), n = 6 (C, D).
Source data are available online for this figure.
I3AA is a ligand for the aryl hydrocarbon receptor (AhR). To test the potential role of AhR pathways in regulation of TCRαβ T cell reactivity orchestrated by I3AA, we developed L. johnsonii‐specific, AhR‐deficient hybridomas using CRISPR/Cas9 deletion (Appendix Fig S5C). AhR deficiency totally abrogated the effect of I3AA on CD69 expression (Fig 4D lower right and Appendix Fig S5D). These data collectively indicate that I3AA produced by Blastocystis ST7 affects both the Treg and non‐Treg CD4+ compartments. Thus, reprogramming of the flora‐specific CD4+ T cells upon intrinsic AhR‐I3AA interaction may redirect these cells toward becoming pro‐inflammatory responders and away from becoming iTregs.
Figure 4. I3AA induces hyperactivation of CD4+ T lymphocytes and prevents them from exhaustion.

- Proportion of T cells with non‐respiring mitochondria stimulated in the presence of different concentrations of indole derivatives isolated from the cultures of Blastocystis ST7B. Total indole in the given experimental condition was estimated using Sigma indole detection kit (μg/ml).
- Proportion of T cells with non‐respiring mitochondria measured in two TCR stimulation conditions, low (left) and high (right) TCR stimuli, upon incubation of T cells with different concentrations of I3AA.
- Expression of CD69 and PD‐1 by T cells experiencing different TCR stimulation conditions, incubated with different concentrations of I3AA.
- Expression of CD69 and PD‐1 by the TCR‐stimulated T cells in presence of different concentrations of tryptophan metabolite produced by D8M7V4‐transfected HEK‐293 cells.
- Expression of Blimp‐1 in cells activated with anti‐CD3/CD28 beads exposed to different doses of I3AA or IPA.
- Nuclear localization of NFATc1 (stained with PE‐conjugated antibody) visualized using IFC. Low (upper left) and high (lower left) TCR saturation were compared.
- Quantitative analysis of DNase I hypersensitivity of NFATc1‐dependent PD‐1 promoter regions. DNase I sensitivity was analyzed on nuclei from T cells treated with 5 μg of I3AA or untreated. Three experimental replicates for each condition. Non‐parametric Mann–Whitney test was utilized, * indicates statistical significance when P < 0.05.
Data information: All experimental groups comprised minimum four replicates. Box‐and‐whisker plot show first and third quartile with median value indicated, and error bars indicate s.d. Statistical analysis was performed using a two‐tailed t‐test with Welch correction. The P‐values are indicated when P < 0.05 (experimental to control groups). Experimental and control groups comprise n = 3 (F) n = 4 (B, left) n = 5 (B, right; C–E).
Source data are available online for this figure.
I3AA regulates T cell metabolic activity and prevents CD4+ T lymphocytes from TCR signaling‐induced exhaustion
Increased energy demands and glucose metabolic activity constitute a hallmark of CD4+ T cell activation (Balyan et al, 2020). Indeed, increased CD69 expression upon increasing the dose of I3AA was accompanied by increased glucose uptake by the stimulated T cells (Appendix Fig S6A). As the observed T cell phenotype indicates a switch from oxidative phosphorylation toward glycolysis (Buck et al, 2016), we looked at mitochondrial changes. By staining the cells with MitoTracker Green, to measure total mitochondrial mass, and MitoTracker Red, which labels respiring organelles (Tal et al, 2009), we estimated the proportion of cells with non‐respiring mitochondria. The Blastocystis‐derived metabolites (Fig 4B) and I3AA (Fig 4C) increased the number of T cells with dysfunctional mitochondria in a dose‐dependent manner. Furthermore, the effect was strictly dependent on the strength of the TCR stimulus (Fig 4C). Hence, to induce a similar effect on T cells activated by a lower concentration of anti‐CD3/CD28 beads, the dose of the compound had to be increased two‐fold. Increased CD69 expression and enhanced glucose uptake were accompanied by a decreased number of cells with functional mitochondria, correlating with cell viability (Fig 4A). In the most extreme condition (with 10 μg/ml of I3AA), the proportion of live cells dropped dramatically. The negative effect on cell viability of I3AA depended largely on the strength of TCR stimulus. Thus, exposure to this compound may enhance T cell immunoreactivity driven by the TCR signal, leading to activation‐induced cell death with strong TCR stimulation. In non‐polarizing conditions, gradual increase of the TCR stimuli leads to enhanced, TCR‐dependent, CD69 expression accompanied by increased expression of programmed death‐1 (PD‐1) on the T cell surface (Appendix Fig S6A). Interestingly, when T cells were stimulated together with Blastocystis‐derived metabolites, a strong increase of CD69 expression was associated with decreased expression of PD‐1 (Appendix Fig S6B). Analysis of the individual compounds revealed that, of the two characterized indole derivatives, only I3AA mirrored the biological activity of PD‐1 inhibition seen with the Blastocystis‐derived supernatants (Fig 4D). Additionally, similar effects on T cell PD‐1 expression was observed when experiments were carried out in the presence of tryptophan metabolites produced by HEK‐293 cells transduced with Blastocystis‐derived D8M7V4 (Fig 4E). T cell exhaustion constitutes an important mechanism in regulation of immunological tolerance. Elevated Blimp‐1 expression correlates with PD‐1hi status and indicates the exhausted phenotype of CD4+ T cells (Hwang et al, 2016). Importantly, when the activation of CD4+ T cells occurred in the presence of I3AA, either alone or in the context of other Blastocystis‐derived metabolites, Blimp‐1 expression was dramatically reduced (Fig 4F and Appendix Fig S6C right panel). According to previous reports, the expression of PD‐1 is in some extent regulated by NFATc1, which translocates into the nucleus and binds to the promoter regions of PD‐1 (Oestreich et al, 2008). As such, PD‐1 expression is positively regulated in a TCR‐dependent manner. Importantly, PD‐1 expression is further enhanced by positive AhR stimulation (Liu et al, 2018). Our data reveal that the nuclear translocation of NFATc1 was not disrupted during T cell stimulation after exposure to I3AA (Fig 4F). However, when we analyzed the accessibility of NFATc1‐dependent elements in the PD‐1 promoter upon I3AA treatment, using a DNase I hypersensitivity assay, we observed unchanged chromatin status in this specific promoter region (Fig 4G). These data further indicate that I3AA could act as a negative modulator of AhR, in line with previously published work regarding the AhR‐PD‐1 axis (Liu et al, 2018).
The hyper‐activated and non‐exhausted phenotype of CD4+ T lymphocytes indicates that I3AA produced by Blastocystis ST7 might modulate TCR‐dependent stimulation, simultaneously decoupling T cells from control by intrinsic inhibitory mechanisms and pathways. As such, this indole derivative restricts the plasticity of the naïve T cell subset, directing the cells toward pro‐inflammatory effectors.
I3AA mimics biological activity of a synthetic AhR inhibitor
To date, most of the identified bacterial‐derived tryptophan metabolites are categorized as beneficial in terms of host physiology. The positive impact of these indole derivatives is linked to the activation of AhR. Using the human HT29 reporter cell, we found that ST7B and ST7H‐derived isolates induced AhR nuclear translocation, but to a much lower extent than FICZ (Fig 5A left). When analyzed individually, IPA did not induce AhR, but the efficacy of AhR activation by I3AA resembled that induced by the indole derivatives isolated from Blastocystis supernatants (Fig 5A middle). Importantly, when HT29 reporter cell line was exposed to mixtures of FICZ and I3AA, an inhibitory effect of I3AA on the FICZ‐AhR interaction was detectable at 1:1 molar ratio (I3AA to FICZ), becoming undetectable at lower concentrations of I3AA (Fig 5A right). As I3AA exhibited moderate antagonistic activity to the AhR‐FICZ interaction, we tested CH‐22319, a synthetic AhR inhibitor (Kim et al, 2006; DiMeglio et al, 2014), in the context of iTreg polarization. The efficiency of in vitro Treg polarization (Fig 5B) and the decreased CD103 expression after exposure to CH‐223191, mirrored the effect of I3AA.
Figure 5. I3AA mimics biological activity of a synthetic AhR inhibitor.
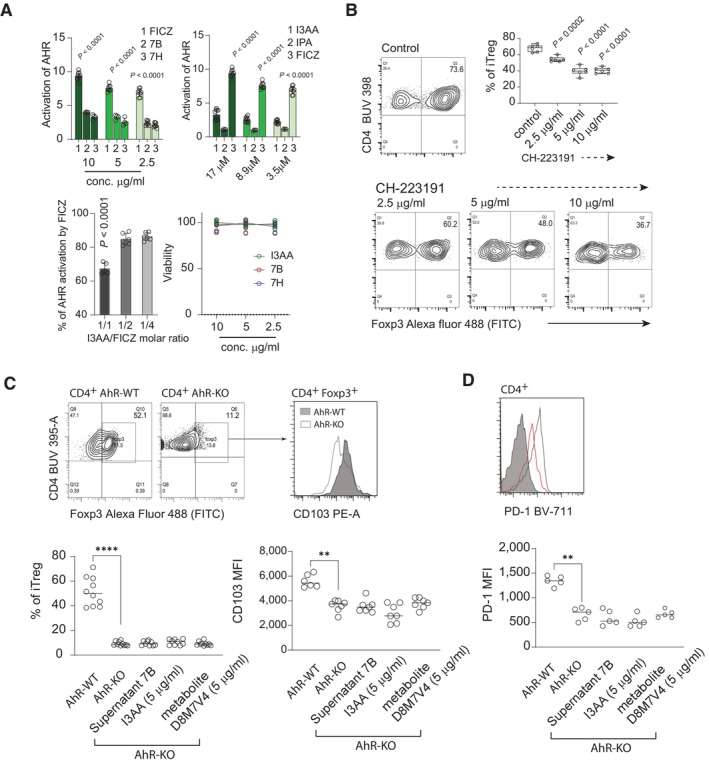
- AhR activation by FICZ and Blastocystis‐derived metabolites. Top left: AhR nuclear translocation was measured using HT29 Lucia AhR reporter cell line. Top right: AhR activation by FICZ, IPA and I3AA. Bottom left: Inhibition of the AhR‐FICZ interaction by I3AA. Bottom right: viability of HT29 cells after exposure to different concentrations of Blastocystis‐derived metabolites. All replicates (n = 8 FICZ, supernatants 7B and 7H, n = 4 IPA n = 10 I3AA, FICZ inhibition 1/1 n = 5, 1/2 n = 6, 1/4 n = 6), and statistics. P‐values are indicated from non‐parametric Mann–Whitney test.
- Efficacy of Treg polarization in the presence of the AhR antagonist CH‐223191. All replicates (n = 6) and statistics. P‐values for two‐tailed t‐test with Welch correction are indicated.
- Left: In vitro conversion of CD4+ T cells isolated from AhR‐WT and AhR‐KO mice toward iTreg. Right: CD103 expression on polarized iTreg generated from AhR‐WT and AhR‐KO CD4+ T cells. Upper plots show examples of flow cytometry data, and lower plots show all replicates (n = 10 Foxp3 staining, n = 7 CD103 staining) and statistics. P‐values are indicated with *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, from non‐parametric Mann–Whitney test.
- Expression of PD‐1 on stimulated T cells from AhR‐WT and AhR‐KO CD4+ T cells. Upper plots show examples of flow cytometry data, and lower plots show all replicates (n = 5) and statistics. P‐values are indicated with *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, from non‐parametric Mann–Whitney test.
Data information: Bar plots show mean statistic value and error bars indicate s.d., Box‐and‐whisker plot show first and third quartile with median value indicated, and error bars indicate s.d.
Source data are available online for this figure.
To further test the role AhR signaling in the context of Blastocystis‐derived I3AA, we investigated the polarization efficacy toward iTreg in vitro, using CD4+ T cells isolated from AhR‐KO mice. Similarly, to previous reports (Ye et al, 2017), AhR‐deficient T cells exhibited impaired polarization toward iTreg, with lower expression of CD103 than WT counterparts (Fig 5C and D). Importantly, neither I3AA alone, supernatant derived from Blastocystis culture, nor metabolites produced by the D8M7V4 transduced HEK‐293, changed the status of iTreg polarization of AhR‐KO CD4+ T cells. Furthermore, in non‐polarizing conditions, CD4+ T cells from AhR‐KO mice were characterized by lower PD‐1 expression than their AhR‐WT counterparts (Fig 5D). Importantly, exposure to I3AA did not change the PD‐1 status of AhR‐KO T cells (Fig 5D). All these data strongly support the notion that the CD4+ phenotype induced by I3AA is associated with antagonism of this compound toward AhR.
I3AA induces colitis by inducing Th17 and functionally skewing Tregs
From the in vitro experiments with Blastocystis‐derived metabolites, the reduced numbers of intestinal Foxp3+ RORγt+ cells (Fig 1D), and the expression of Helios in Tregs from infected mice (Fig 1E), it appeared that I3AA impaired iTreg generation. To address the role of Blastocystis‐derived indole derivatives in reshaping Treg and Tconv differentiation in vivo, but in the absence of Blastocystis, we generated a mouse colitis model. We injected mixed Tregs and CD4+ Tconv cells derived from CD45.1 and CD45.2 congenic donors, respectively, into Rag2 −/− immunocompromised recipients (Fig 6A). In these experiments, the Treg (hereafter called tTreg) population was sorted from the CD45.1 LNs based on their CD25 expression. The Tconv cells were sorted from CD45.2 counterparts according to their CD25− and Foxp3‐GFP expression status (Fig 6A).
Figure 6. I3AA produced by Blastocystis ST7 induces experimental colitis by inducing Th17 and inhibiting Treg responses.
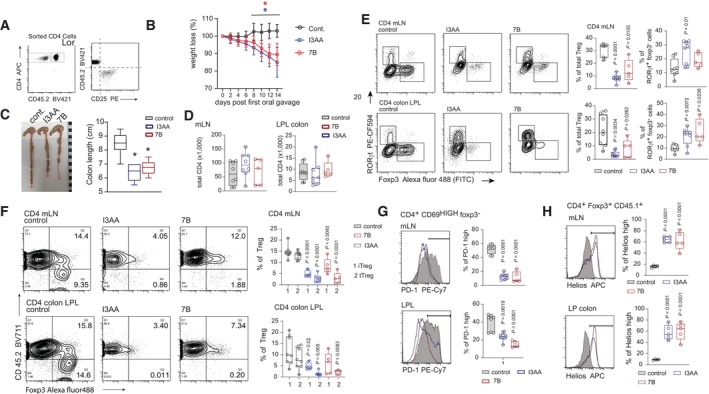
- Phenotype of T cells used for adoptive transfer into Rag2 −/− hosts. Before transfer into the host, naïve CD4+ T cells from CD45.2 and CD4+ tTreg from CD45.1 congenic strains were mixed in proportions to reconstitute the approximate natural distribution of these CD4+ compartments.
- Weight loss by the Rag2 −/− groups after treatment with I3AA alone or indole derivatives from Blastocystis cultures. Error bars indicate s.d. Non‐parametric Mann–Whitney test was utilized, * indicates statistical significance when P < 0.05.
- Colon length of control and mice treated with I3AA or Blastocystis‐derived metabolites.
- Total number of CD4+ T cells in mLN and colon LP from different groups.
- Foxp3+ and RORγt+ distribution within CD4+ T cells in mLN and colonic LP that developed in Rag2 −/− hosts from adoptively transferred cells.
- Distribution of the CD45.1+ and CD45.2+ Foxp3+ T cells developed in the Rag2 −/− hosts.
- PD‐1 expression on Foxp3−CD4+ compartment.
- Helios expression on tTreg from control and I3AA‐treated mice.
Data information: Box‐and‐whisker plot show first and third quartile with median value indicated, error bars indicate s.d. Statistical analysis was performed using two‐tailed t‐test with Welch correction (with exception of B). When P < 0.05 (experimental to control groups), values are indicated. Experimental and control groups comprised of: n = 8 for the control and I3AA‐treated groups, n = 5 for experiments with isolated metabolites from Blastocystis ST7B culture (A–G). Intracellular staining with a‐Helios antibody was performed on T cells isolated from five animals in each experimental group (H).
Source data are available online for this figure.
None of the animals from the control group developed wasting disease during the experiment (Fig 6B). This can be explained by protective tTreg from CD45.1 donors. In contrast, oral gavage of mice with I3AA and metabolites produced by Blastocystis ST7B resulted in substantial weight loss (Fig 6B). Further analysis of the control and indole‐treated mice revealed that mice exposed to I3AA (either alone or in the more complex mixture from Blastocystis culture supernatants) showed much shorter colon lengths, a hallmark of colitis driven by uncontrolled CD4+ T cell response (Fig 6C). The total number of CD4+ cells within colonic LP and mLN was similar in control and I3AA‐treated groups (Fig 6D). However, the phenotype of CD4+ subsets from groups exposed to I3AA mirrored that of Blastocystis‐infected animals (Figs 4A and 6E). Analysis of the colonic LP and mLN CD4+ T cells from mice treated with the indole derivatives revealed overrepresentation of CD4+ RORγt+ Foxp3− cells, suggesting an enhanced response to gut flora (Fig 6E). Furthermore, non‐Treg cells from colon and mLN from I3AA‐treated mice showed much lower expression of PD‐1, as seen in T cells in the in vitro assays (Fig 6G). Collectively, these results demonstrate that I3AA acted on CD4+ cells when experiments were carried out in the presence of gut microflora. This indicates that the compound is not decomposed by commensal microorganisms into non‐active metabolites upon Blastocystis infection (or oral gavage). It reaches biologically active concentrations and can affect T cell activation and polarization.
In all the experimental groups treated with I3AA, we found decreased numbers of total Treg in both mLN and colon LP (Fig 6E). The drop of Treg numbers in these animals was not simply due to impaired de novo Treg polarization. Analysis of the congenic markers indicated that tTreg expansion and iTreg progression were equally negatively affected by the indole compound (Fig 6F). Surprisingly, analysis of Helios expression in tTreg revealed that tTregs from the control group had much lower expression of Helios than their counterparts from colon and mLN of I3AA‐treated mice (Fig 6H). As previously reported, Helioshi Tregs divide much more slowly than Helioslo Tregs (Elkord et al, 2015; Thornton et al, 2019). We conclude that I3AA induces impaired activation and proliferation of Treg, contributing to the functional defect of Tregs in the large intestine during Blastocystis infection.
Discussion
The role of a diverse group of tryptophan metabolites produced by gut microbes is widely recognized in maintenance of gut homeostasis. Existing studies on the production and biological effects of indole derivatives in the gut environment suggest that bacterial‐associated tryptophan metabolism serves as the primary source for these bioactive compounds (Kaur et al, 2019; Wojciech et al, 2020). We have shown that the synthesis of some biologically active indole derivatives in the intestine is not a bacteria‐exclusive feature. We identified I3AA and IPA as Blastocystis ST7‐derived metabolites. Indole derivatives constitute an essential component of the metabolite landscape involved in the reciprocal interaction between host and microbiome, and are among those compounds that directly or indirectly affect Treg function. The beneficial effect of these indole derivatives on the host immune system is generally considered to be due to agonistic activation of the intrinsic pathway of AhR (Zelante et al, 2013; Wlodarska et al, 2017). As previously reported, gut‐derived Treg express high amounts of AhR. Additionally, positive stimulation of AhR by agonistic ligand (Ye et al, 2017) has a substantial role in promoting gut regulatory T cells. The same authors also revealed that Treg cells lacking AhR display decreased expression of CD103 a TGFβ‐dependent integrin (Konkel et al, 2017). A similar correlation between AhR, TGFβ, and CD103 has been described during CD8+ T cell polarization (Dean et al, 2023). Collectively, these findings provide evidence for a positive interaction loop between the AhR and TGFβ signaling pathways, particularly in relation to the regulation of CD103 expression. In addition, it has been reported that Smad7, a negative regulator of the TGFβ signaling pathway, exerts a negative influence on the AhR pathway. This suggests a reciprocal regulatory relationship between AhR and TGFβ signaling (Monteleone et al, 2016). Our findings indicate that, unlike other tryptophan metabolites produced by gut microbiota, I3AA exhibits distinct inhibitory effects on regulatory T cells (Treg). I3AA alters the sensitivity of Treg cells to TGFβ signals, thereby impacting their function and ability to migrate to the gut. We propose that this disruption of gut Treg development occurs through an AhR‐dependent mechanism, with I3AA acting as an antagonist of this intracellular receptor. Recognition of cognate pMHC complexes by T cells is crucial in the reprogramming and functional differentiation of individual clones (Goto et al, 2014). Besides the affinity of TCR‐pMHC interaction, downstream interpretation and physiological outcome of this signal depend on the crosstalk between partners within proximal and distal TCR signaling (Gascoigne et al, 2016). I3AA dramatically changes the TCR‐dependent activation threshold of CD4+ T cells, concomitantly securing these cells from being exhausted by downregulating PD‐1 and Blimp1. Polarization toward the pro‐inflammatory Th17 lineage requires strong TCR‐pMHC interaction (Goto et al, 2014) and at this process can be inhibited by PD‐1 expression (Zhang et al, 2020). Furthermore, PD‐1 expression positively correlates with AhR activation by agonist ligands (Liu et al, 2018). Therefore, exposure to I3AA activates intrinsic processes in an AhR‐dependent manner, which ultimately influences the interpretation of TCR signals. This leads to a shift toward the pro‐inflammatory Th17 lineage within the gut environment. This indicates that different tryptophan metabolites, regardless of their structural similarity, may exhibit diametrically different AhR–ligand relationships and could play a different role in the induction of gut inflammatory disorders.
The pro‐inflammatory nature of I3AA is striking. However, it is important to note that the synthesis of this compound by Blastocystis alone may not solely define its detrimental characteristics. As mentioned earlier, I3AA may serve as a substrate for the synthesis of indole‐3‐acetic acid (IAA) and indole‐3‐aldehyde (IAld). Importantly, the enzymes utilizing I3AA as a substrate for IAA and IAld synthesis are distributed non‐ubiquitously among different bacterial genera, and conversion of I3AA into IAA and IAld is limited to specific bacterial species (Zhang et al, 2019). Indeed, Lactobacillus reuteri is known to be a common gut‐dwelling strain that possesses the ability to produce (IAld) in the I3AA‐dependet pathway (Zelante et al, 2013). This synthesis of IAld by L. reuteri results in AhR‐dependent IL‐22 production and gut protection from inflammation, indicating IAld as a beneficial bacterial‐derived metabolite. Importantly, when two Blastocystis STs (pathogenic ST7 and commensal ST4) were compered in the context of in vivo microbiome remodeling, only pathogenic ST7 negatively affected Lactobacillus (Deng et al, 2023a). The impact of Blastocystis ST7 on the microbiome may lead to a reduction in the effectiveness of bacterial communities in clearing I3AA in the gut environment. Thus, we believe that I3AA produced by Blastocystis ST7 per se constitutes a pathogenic trait of this particular ST. At the same time, we hypothesize that the preexisting and post‐infection status of the microbiome in the context of processes targeting I3AA as a substrate for further metabolism might determine pathological outcome of this protist infection. In addition, functional annotation of the transcriptome of Blastocystis ST4 and ST7 revealed than only Blastocystis ST4 expresses the gene product of acetaldehyde dehydrogenase (Wawrzyniak et al, 2015). This specific gene product potentially plays a role in the degradation of I3AA to IAA.
In summary, we believe that our findings regarding Blastocystis ST7 tryptophan metabolism and the role of a tryptophan metabolite in pathogenesis of this subtype shed new light on indole derivatives and ecological interactions between different components of gut‐dwelling consortia during gut immune homeostasis.
Materials and Methods
Mice
C57BL/6J, B6.Cg‐Tg(TcraTcrb)425Cbn/J (OT‐II), B6.Cg‐Foxp3tm2Tch, B6.Cg‐Rag2 tm1.1Cgn/J (Rag2 −/−), B6.SJL‐Ptprc a Pepc b /BoyJ (CD45.1), and AhR−/− on C57BL/6J genetic background (AhR‐KO) mice were bred in restricted flora (RF) facilities at Comparative Medicine, NUS. Mice were treated under NUS Institutional Animal Care and Use Committee‐approved guidelines in accordance with approved protocols.
Antibodies and probes
T cells and hybridomas were stained with the following primary antibodies: TCRβ (clone H57‐597, BioLegend and BD), CD4 (clone GK15 eBioscence and BD), CD4 (clones RM4‐5 eBioscence and BD), CD8α (clone 53–6.7, eBioscence and BD), CD25 (clone 7D4, BD), CD69 (clone H1,2F3, BioLegend and eBioscence), CD45.1 (clone A20, eBioscence), CD45.2 (clone 104, BioLegend and eBioscence), CD103 (clone 2E7, Biolegend), PD‐1 (clone J43, ebioscence), RORγt (clone AFKJS‐9, eBioscence), Foxp3 (clone MF14, BioLegend), Helios (clone 22F6, eBioscence), AhR (clone 4MEJJ, eBioscence), Blimp1 (clone 5E7 BD), CD62L (clone MEL‐14, eBioscence), CD122 (clone 5H4, eBioscence), CD44 (clone IM7, eBioscence), NFATc1 (clone 7A6, Biolegend), and Hoechst 33342 (Invitrogen). For plate coating and in vitro T cell stimulation, anti‐mouse CD28 functional grade purified (eBioscience) and anti‐mouse CD3ε purified (eBioscience) antibodies were used.
Tryptophan derivatives and AhR ligands
For the in vitro and in vivo experiments, addressing tryptophan metabolite synthesis pathways and their biological role in the context of T cell polarization: synthetic tryptophan, indole‐3‐acetaldehyde (I3AA), indole‐3‐pyruvic acid, indole‐3‐propionic acid (I4AA), 6‐formylindolo[3,2‐b]carbazole, and CH‐223191 (all obtained from Sigma‐Aldrich) were used.
Blastocystis cultures and acute infection protocol
Human Blastocystis ST7 isolates B and H were axenized from patients at the Singapore General Hospital (Ho et al, 1993). Blastocystis isolates were cultured in 8 ml pre‐reduced Iscove's modified Dulbecco's medium (IMDM) (Gibco) supplemented with heat‐inactivated 10% horse serum (Gibco) in anaerobic jars (Oxoid) with Anaerogen gas packs (Oxoid) at 37°C and subcultured every 3–4 days. To analyze Blastocystis cultures in the context of microbial contamination, we collected two milliliters of the cultures and centrifuged at 2,655 g, 10 min. Culture pellets were used for total DNA isolation using PureLinkTM Microbiome DNA Purification Kit (Invitrogen). Isolated DNA was further utilized as a PCR template with Blastocystis‐specific primers targeting 18S rDNA and primers specific to the bacterial 16S rDNA gene. 18S primer sequences were as follows: forward GCTAGGGGATCGAAGAGGAT, reverse TGGTAAGTTTCCCCGTGTTG with a product length 229 bp. Sequences of bacterial 16S rDNA primers are described in detail in Materials and Methods section regarding whole 16S rDNA sequencing. For each pair of primers, negative contamination controls were introduced with deionized water instead of template DNA. As a positive amplification control for bacterial 16S primers, we used total DNA isolated from the large intestine content of SPF‐reared mice uninfected with Blastocystis.
Blastocystis infection was performed as described in detail previously (Yason et al, 2019). Briefly, all infection experiments were carried out on 6–week‐old C57BL/6 mice. The experimental scheme included induction of colitis by 2% DSS in the drinking water for 4 days, followed by a recovery period of 5 days. Then, 50 μl PBS suspension containing 105 Blastocystis cysts purified direct from in vitro cultures was injected intracecally. DSS‐treated animals representing surgical control were injected intracecally with 50 μl PBS. The mice were euthanized on day 3 post‐infection, and the mLN, colon, and cecum were extracted for T cell isolation.
Isolation and identification of Blastocystis‐derived tryptophan metabolites
Supernatants from Blastocystis cultures were mixed with absolute ethanol to a final concentration of 50% alcohol and kept at −20°C, at least overnight before further steps. Precipitated debris was then pelleted upon centrifugation at 13,000 g for 30 min. Supernatants were collected and filtered using Amicon® Ultra 15 ml Centrifugal Filter Units 10 kDa according to the manufacturer's protocol. To remove alcohol, flowthrough fractions were concentrated using vacuum concentrators. Pellets were resuspended in deionized water. For further processing, the sample was adjusted to pH3 by formic acid (Thermo Scientific). Tryptophan metabolite enrichment was conducted using solid‐phase chromatography (HyperSep™ C18 Cartridges, Thermo Scientific). Briefly, before indole derivative extraction, C18 columns were pre‐conditioned with 5 ml of methanol, 5 ml of 80% methanol, 5 ml of 50% methanol, 5 ml of 20% methanol, and 5 ml of water at pH3 (adjusted by formic acid). Three fractions were eluted from the C18 column. Fraction 10 m eluted with 10% methanol, 50 m eluted with 50% methanol, and 80 m eluted with 80% methanol. The presence and concentration of indole derivatives in the individual eluted fractions was estimated according to the manufacturer's protocol.
The LC–MS–MS analysis was performed using an Agilent 1290 Infinity LC system (Agilent Technologies, Santa Clara, CA, USA) interfaced with an AB Sciex QTRAP 3500 tandem mass spectrometer (MS/MS) (AB SCIEX, Framingham, MA) equipped with a Turbo Ion Spray source (Applied Biosystems, Foster City, CA). Data acquisition and processing were performed using AB Sciex OS 1.5 (Applied Biosystems, Foster City, CA, USA). Mobile phase A was composed of 0.1% formic acid in water; mobile phase B was 0.1% formic acid in acetonitrile. Chromatographic separation was achieved using an Acquity™ Premier HSS T3 1.8 μM, VanGuard™ Fit, 2.1 mm × 150 mm column at a flow rate of 0.3 ml/min under the following condition: isocratic 100% B (0.00–1.00 min), linear gradient 100% to 90% B (1.00–5.00 min), linear gradient 90 to 70% B (5.00–10.00 min), linear gradient 70% to 5% B (10.00–19.00 min), linear gradient 5–100% B (19.00–20.00 min), and isocratic 100% B (20.00–25.00 min). Using an autosampler thermostatted at 4°C, 1 μl of each sample was injected onto the column maintained at 45°C. The needle was flushed with 50% water with 50% acetonitrile post‐injection to minimize carry‐over effect. Tandem mass spectrometry was operated in Multiple Reaction Monitoring (MRM) approach in positive mode (see Table 1). Electrospray ionization parameters were optimized for 0.3 ml/min flow rate and are summarized as follows: electrospray voltage of −4,500 V, temperature of 500°C, curtain gas of 30, CAD gas of 9, and gas 1 and 2 of 50 and 50 psi, respectively.
Table 1.
MRM Transition.
| Analyte | Q1 Mass (Da) | Q3 Mass (Da) | Time (min) | Declustering potential (V) | Collision energy (V) | Collision exit potential (V) | Entrance potential (V) |
|---|---|---|---|---|---|---|---|
| Indole‐3‐Acetaldehyde | 160.031 | 117.00 | 200 | 131.00 | 33.00 | 8.00 | 10.00 |
| 160.031 | 88.90 | 200 | 131.00 | 57.00 | 8.00 | 10.00 | |
| 160.031 | 90.10 | 200 | 131.00 | 43.00 | 8.00 | 10.00 | |
| Indole‐3‐Propionic | 190.038 | 130.00 | 200 | 86.00 | 21.00 | 12.00 | 10.00 |
| 190.038 | 103.00 | 200 | 86.00 | 49.00 | 8.00 | 10.00 | |
| 190.038 | 77.10 | 200 | 86.00 | 63.00 | 8.00 | 10.00 |
Cloning and de novo expression of Blastocystis orthologs associated with I3AA synthesis
Comparative analysis of the Trypanosoma cruzi strain CL Brener aspartate aminotransferase (accession number XP_807788.1) with Blastocystis ST7B translated CDs data base (accession number GCA_000151665.1) was done locally using BLAST software. The lcl|FN668638.1, lcl|FN668644.1, and lcl|FN668683.1 (UniProt numbers D8LWJ3, D8M126, D8M7V4, respectively) represented the closest sequence similarity to the Trypanosoma cruzi‐derived aspartate aminotransferase. To amplify all three cDNAs, DNase‐treated RNA, isolated from Blastocystis ST7B, was used for RT reaction using SuperScript IV cDNA synthesis kit (Invitrogen). Target genes were amplified with following pairs of primers:
| D8LWJ3 | f‐TAATACGACTCACTATAGGGATGAATAGAGAATCTTGGTACA |
| r‐TTACTGCTTTCCATTGAGCTTCCACTCGACCACCTC. | |
| D8M126 | f‐TAATACGACTCACTATAGGGATGTTCGCTAAGGTTGTTAAGTTCGCC |
| r‐TTAGTTCTTGGTAACCTCATGGAAAGCCTTAGC | |
| D8M7V4 | f‐TAATACGACTCACTATAGGGATGCTGTCAAAGTTTACTCCCGTTGCT |
| r‐CTACTTCGTCACCTCGTGCATCGCCTTGGCCAGATAC |
where f and r indicate forward and reverse primers, respectively, and TAATACGACTCACTATAGGG sequence of forward primers constitutes T7 promoter. Amplicons were used for in vitro mRNA synthesis using HiScribe™ T7 ARCA mRNA Kit (with tailing) (NEB). For transfection of HEK‐293 cells seeded in 6‐well plates and cultured in IMDM medium supplemented with tryptophan (1 mM), 2.5 μg of RNAClean XP (Beckman Coulter) cleaned mRNA was used (TransIT®‐mRNA Transfection Kit, Mirus). For tryptophan breakdown analysis and functional assays, medium from transfected cells was collected 72 h after transfection. As a positive control for HEK‐293 cells transfection, we used BFP mRNA synthesized on a DNA template amplified from pLV‐hCAS9‐mTag vector using PCR pLV‐hCAS9‐mTag with primers: f‐TAATACGACTCACTATAGGGATGAGCGAGCTGATTAAGGAGAACA and r‐CTATTAATTAAGCTTGTGCCCCAGTTTGCTAGGGAGGTCGCAG.
I3AA‐induced colitis mouse model
Six‐ to eight‐week‐old Rag2 −/− males were intravenously injected with a mixture of naïve CD25− Foxp3GFP– CD4+ T cells isolated from the LN of B6.Cg‐Foxp3tm2Tch donors and CD25+ CD4+ T cells (representing bulk Treg cells) isolated from the LN of CD45.1 donors. Each Rag2 −/− mouse received 106 CD4+ T cells. The T cells were mixed in 10:1 proportion of naïve to Treg T cells, respectively. Two weeks later, mice were bled and screened for T cell reconstitution. For I3AA treatment, 3 weeks after T cell injection, 500 μg of I3AA dissolved in sterile drinking water was orally administered (gavage) for 14 consecutive days. Control groups were administrated with similar volume of sterile drinking water. During the period of I3AA administration, treated and control mice were weighed daily. After 14 days from the start of I3AA treatment, animals from the experimental and control groups were sacrificed. The mLN and colon tissues were collected for further investigation, including measurements of colon length and phenotypic analysis of the adaptive immune compartments using flow cytometry. To minimize bias and ensure objective data analysis, a single blinding approach was implemented.
Isolation of CD4+ T cells from LN and large intestine LP
T cells from lymph nodes were isolated by meshing entire LN through 70 μm cell strainer. Cells were then washed twice in RPMI and resuspended in RPMI supplemented with 5% fetal bovine serum (FBS). The single‐cell suspension was stained with appropriate panel of antibodies and used directly for analysis by flow cytometry or sorted and used in downstream experiments (adoptive transfer, in vitro stimulation). LP cells from colon and cecum were isolated separately. The intestines were washed with ice‐cold PBS to remove luminal contents and opened longitudinally. The intestines were cut in pieces and incubated in RPMI (Sigma‐Aldrich) containing 1 mM EDTA (Sigma‐Aldrich) and 1 mM DTT (Sigma‐Aldrich) with gentle agitation at room temperature for 15 min, then transferred to RPMI supplemented with 5% FBS, 2 mM EDTA, 1 mM DTT, and 20 mM HEPES and incubated for 40 min at 37°C. Tissues were then washed twice with RPMI without additives. Intestinal tissues were digested using Liberase™ TL (Roche) in RPMI for 40 min at 37°C. Enzymatic reaction was terminated by adding FBS to obtain 5% final concentration of serum. Digested tissues were then meshed using 70 μm cell strainer and passed through glass wool filters (Sigma‐Aldrich). Isolated LP cell fractions were centrifuged and washed once with washing buffer (PBS with 1% FBS), stained with the appropriate antibodies, and analyzed using flow cytometry. To minimize bias and ensure objective data analysis, a single blinding approach was implemented.
In vitro T cell stimulation and differentiation
For in vitro polarization, non‐Treg (CD25− Foxp3GFP– CD4+ T) cells were sorted from B6.Cg‐Foxp3tm2Tch donors. Sorted cells were stimulated using anti‐CD3 and anti‐CD28 beads (Dynabeads™ Mouse T‐Activator CD3/CD28 for T‐Cell Expansion and Activation, GIBCO), with 1/1 beads to cell proportion according to the manufacturer's protocol. Experiments were carried out in U‐bottom 96‐well plates with 50,000 cells/well in a total volume of 200 μl. The in vitro Treg polarization was carried out in complete Iscove's modified Dulbecco's medium (IMDM) containing 10% FBS (GIBCO), 100 U/ml penicillin–streptomycin (GIBCO), 1 mM sodium pyruvate (GIBCO), and 2 mM glutamine (GIBCO) supplemented with 40 ng/ml of recombinant human TGFβ (Peprotech) and 5 ng/ml recombinant mouse IL‐2 (Peprotech). Polarization efficacy was estimated 4 days later using flow cytometry. Cells were first stained using LIVE/DEAD™ Fixable Near‐IR Dead Cell Stain Kit (Molecular Probes) accordingly to the manufacturer's protocol. After one wash with PBS supplemented with 1% FBS (washing buffer) cells were surface stained with antiCD4 and again washed with washing buffer. Using Foxp3 Transcription Factor Staining Buffer Set (eBioscience™), cells were first fixed and permeabilized. Intracellular staining with anti‐Foxp3 was carried out overnight in 1× permeabilization buffer. Before flow cytometric acquisition, cells were washed two times with washing buffer.
The Th17 polarization was carried out in the same culture (cell number, U‐bottom 96‐well plate) and stimulation conditions. Complete IMDM was supplemented with 50 ng/ml recombinant mouse IL‐6, 1 ng/ml recombinant human TGFβ1, and 10 μg/ml each of anti‐mouse IL‐2, anti‐mouse IL‐4, and anti‐mouse IFNγ. The conversion of the T cells was estimated after 5 days. Surface and intracellular staining protocol were as described for Treg polarization. Permeabilized cells were incubated overnight with anti‐RORγt antibody resuspended in 1× permeabilization buffer. After washing, cells were analyzed using flow cytometry. In the non‐polarizing condition, sorted cells were stimulated with anti‐CD3/CD28 beads in ratios 1/1, 1/5, or 1/10 beads to cells in complete IMDM without cytokines. After 2 days, cells were stained using LIVE/DEAD™ Fixable Near‐IR Dead Cell Stain Kit, washed once in washing buffer and either surface stained for CD4, CD69, and PD‐1 or stained with MitoTracker Green FM (Invitrogen) and MitoTracker Red TM (Invitrogen) probes according to the manufacturer's protocol. Glucose uptake was measured using fluorescent D‐glucose analog 2‐[N‐(7‐nitrobenz‐2‐oxa‐1,3‐diazol‐4‐yl) amino]‐2‐deoxy‐D‐glucose (2‐NBDG) (Invitrogen). Cells cultured in non‐polarizing conditions were stained with LIVE/DEAD probe, washed twice in RPMI without glucose (GIBCO), and incubated in the same RPMI medium supplemented with 2 μM 2‐NBDG for 30 min (5% CO2 37°C in dark). Uptake of 2‐NBDG by live T cells was estimated using flow cytometry. In experiments with total Blastocystis‐derived metabolites, the culture medium was supplemented with 10 μl/well of filtered supernatants from Blastocystis cultures or supernatant fractions containing indole derivatives eluted from C18 column. Experiments were carried out at 20, 10, or 5 μg/ml final concentration of total indole, respectively.
Generation of CD4+ T cell hybridomas
To generate immortalized T cell clones, we used as the fusion partner a variant of BW5147 thymoma containing Nur77GFP reporter (a kind gift from Prof. Leszek Ignatowicz. Cell line was mycoplasma tested before fusion). Cas9 was introduced to this cell line using a retroviral system with expression vector pMIGII with cloned cDNA for the Cas9 nuclease. After transduction with pMIG.Cas9 vector, cells were single‐cell sorted. The progeny of single clones expressing GFP and Cas9 were selected and used for further fusion experiments. CD4+ T cell hybridomas were generated as previously described (Wojciech et al, 2014, 2018). In brief, sorted mLN CD4+ T cells were stimulated in vitro with antiCD3/CD28 antibodies and expanded for 3 days. Expanded T cells were fused with the Nur77GFP and Cas9 expressing variant of BW5147 and selected according to the previously described protocol. Specificity of selected hybridomas was investigated after incubation with bone marrow‐derived dendritic cells pulsed with bacterial lysate (Wojciech et al, 2018) and clones specific to Lactobacillus johnsonii were selected and used in further experiments. As a specificity readout, we analyzed Nur77GFP reporter expression and upregulation of CD69 expression (measured using surface staining with anti‐CD69 antibody) by flow cytometry.
Generation of AhR‐deficient Lactobacillus johnsonii‐specific CD4+ T cell hybridomas
AhR‐specific gRNA sequences were selected using CHOPCHOP online tool. The AhR‐targeting gRNAs were synthesized in‐house using PCR and HiScribe™ Quick T7 High Yield RNA Synthesis Kit (NEB). To generate the DNA template for the T7 transcription, two rounds of amplification were carried out. To generate functional gRNA with sequence specific to the AhR in a first round of amplification on the template of in‐house generated pLVTHM vector, an irrelevant gRNA sequence (21 bp) was swapped with AhR‐specific sequence. The primers were as follows:
| Forward primers | |
| AhRgRNA1 | CGAAATCCTGACCTACGTGCAGGGTTTTAGAGCTAGAAATAG |
| AhRgRNA2 | ATTGGTAGGGGATCCATTATGGGGTTTTAGAGCTAGAAATAG |
| AhRgRNA3 | GTATAATAGACTGCTGCCTGTGGGTTTTAGAGCTAGAAATAG |
| Common reverse primer | |
| AAAAGCACCGACTCGGTGCCAC | |
During the second amplification, T7 promoter sequence was incorporated into the 5′ end of the amplicons. The primers were as follows:
| T7AhRgRNA1 | AAGCTAATACGACTCACTATAGGCGAAATCCTGACCTACGTGCAGG |
| T7AhRgRNA2 | AAGCTAATACGACTCACTATAGGATTGGTAGGGGATCCATTATGGG |
| T7AhRgRNA3 | AAGCTAATACGACTCACTATAGGGTATAATAGACTGCTGCCTGTGG |
A common reverse primer was used. Amplicons were purified using AMPure XP beads (Beckman Coulter Life Sciences) according to the manufacturer's protocol and 2 μl of the DNA was used for in vitro RNA synthesis using HiScribe™ Quick T7 High Yield RNA Synthesis Kit (NEB). Synthesis of the RNA was carried out overnight at 37°C. RNA was digested with TURBO™ DNase (Invitrogen) and cleaned using RNAClean XP beads (Beckman Coulter). T cell hybridomas were transfected with the mixture of generated gRNAs using TransIT‐TKO® Transfection Reagent (Mirus). Efficacy of AhR gene editing was assessed by intracellular staining of the transfected and non‐transfected hybridomas with AhR monoclonal antibody (eBioscence).
AhR signaling reporter assay
Analysis of indole derivatives in the context of AhR activation was carried out using HT29‐Lucia™ AhR cells (Invivogen) according to the manufacturer's protocol. HT29 cells were plated at 105 cells/well in a 96‐well plate 1 day before exposure to the tryptophan metabolites. Cells were cultured in 200 μl/well of complete IMDM medium. Next day, the culture medium was changed, and appropriate amounts of indole derivatives were added to the wells. Each exposure condition was triplicated on the single plate, and the experiments were repeated 2–3 times. Lucia luciferase activity was measured in the luminescence plate reader using QUANTI‐Luc™ (Invivogen) assay according to the protocol guidelines.
Isolation of large intestine‐dwelling Lactobacillus strains
The large intestine content was collected and resuspended in 5 ml of sterile PBS buffer, then filtered through sterile glass wool. The flowthrough was spun down at 3,000 g for 10 min at room temperature. The pellet was washed twice with 5 ml PBS under the same centrifugation conditions. Finally, intestinal isolates were resuspended in 5 ml of sterile PBS, serially diluted up to 1/10,000 in sterile PBS and 100 μl of each dilution was used for plating on De Man, Rogosa, and Sharpe agar plates (MRS) (Sigma‐Aldrich) supplemented with Tween‐80 (Sigma‐Aldrich). The MRS plates were incubated 48 h at 32°C. Single colonies were passaged four times on the MRS plates in the same culture conditions. Finally, individual colonies were transferred into 4 ml of liquid MRS medium supplemented with Tween‐80 and cultured for 24 h at 32°C. For banking purposes, 2 ml of overnight culture was mixed with equal volume of 50% glycerol (Sigma‐Aldrich) stock solution, aliquoted in Eppendorf tubes, and transferred into −80°C. One milliliter of overnight culture was used for genomic DNA isolation (PureLink™ Microbiome DNA Purification Kit, Invitrogen). Isolated DNA was then used as a template for amplification of the whole 16S rDNA. The amplicons were cloned into the TOPO vector (Invitrogen) and sequenced. The DNA sequences were compared to the global database using a basic local alignment search tool (BLAST) software.
RNA extraction, RNA sequencing, and bioinformatics analysis
Briefly, RNA from tissue was isolated using RNAzol RT (Sigma‐Aldrich) and from in vitro stimulated T cells using RNeasy Micro Kit (Qiagen) according to the manufacturer's protocols. Synthesis of the libraries was performed using MagMAX™ DNA Multi‐Sample Ultra Kit and sequencing of the obtained genetic material was performed on an Illumina Novaseq6000 instrument for sequencing using a 2 × 150 paired‐end configuration according to the manufacturer's instructions. The raw reads were aligned to the reference genome using Hisat2 v2.1.0 (Kim et al, 2015). SeqMonk v1.46.0 data analyzer was used to generate a raw reads expression matrix. Further bioinformatic analysis was performed using RStudio software (R Core Team, 2020). Differential gene expression analysis was done using EdgeR package (Robinson et al, 2009) with the significance cut‐off FDR < 0.05. Availability of raw sequencing data is described in Deng et al (2023a).
DNase I hypersensitivity assay
DNase I hypersensitivity assay was conducted on the Pdcd1 promoter region, which includes NFATc1 responsive elements, applying modified protocols based on previous publications (Lu & Richardson, 2004; Oestreich et al, 2008). Briefly, harvested cells were washed twice with ice‐cold phosphate‐buffered saline (PBS). Then, nuclei extraction was carried out in hypotonic lysis buffer (10 mM Tris–HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA‐630). Nuclei from each experiment (control and T cells exposed to I3AA) were split into equal aliquots and treated with increasing concentrations of DNase I enzyme (0 as control, 2, 4, and 6 units of DNase I). After digestion, DNA was extracted using phenol/chloroform. DNA concentration was quantified using Qubit 1× dsDNA HS Assay Kit (Invitrogen). Quantitative analysis of DNase I digestion was performed using qPCR. Each qPCR utilized 5 ng of previously purified DNA as a template. Amplification of the Pdcd1 promoter region containing NFATc1 responsive elements was carried out using the KAPA SYBR FAST qPCR Master Mix with the following primers: PD‐1_CR‐CF (GAGGTCCTTTCACTCTCCACG) and dnase_PD‐1_CR‐C (CGACTTGTGTGCATGCATAGTACC). The relative change was normalized to the non‐DNase I‐digested control. These assays were performed on three individually prepared sets of DNA.
RNA isolation, RT reaction, and qPCR
The total RNA was isolated from overnight in vitro T cell culture using RNeasy Micro Kit (Qiagen) according to the manufacturer's protocols. Reverse transcription was carried out using SuperScript™ IV First‐Strand Synthesis System (Invitrogen). qPCR was performed using SsoFast™ qPCR Supermixes with EvaGreen (Bio‐Rad) on StepOnePlus Real‐Time PCR Systems (Applied biosystem). The primers were used as follows:
Pprc1: f‐CAG GAG AAG AAG CCC TTA GAC C, r‐CTT TCG CCA AGA GTG AGA CAG
Grs1: f‐CCC TCC GTT TCC CAT CTC TAT TT, r‐CTG CCT GAG TAC GAG TTG AGC
Ptbp1: f‐CAC ACC CCA AAG CCT CTT TAT, r‐ATC TGC ACA AGT GCG TTC TCC
Gib1: f‐TGG TGC TCC GCA GGT ATC T, r‐TGA AGT CTG GGC CTT TTG TTC
Zab2: f‐ATT GCA CAT CAG ACT TTG AGG AA, r‐ATA ATG GCC GTG TCG CTT CG
Traf4: f‐CCC GGC TTC GAC TAC AAG TTC, r‐TCA GGG CAT TTG AAG ACT CCT
Lmna: f‐ACC CCG CTG AGT ACA ACC T, r‐TTC GAG TGA CTG TGA CAC TGG
Furin: f‐TCG GTG ACT ATT ACC ACT TCT GG, r‐CTC CTG ATA CAC GTC CCT CTT
Whole 16S rDNA sequencing
To analyze the distribution of the bacterial species in the large intestines, colon contents were collected and resuspended in 5 ml of sterile PBS. As in the protocol described above, samples were filtered and washed 2× in PBS. Pellets were used directly for genomic DNA isolation using Microbiome DNA Purification Kit and whole 16S rDNA
V1: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGRGTTTGATYMTGGCTCAG.
V9: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGYTACCTTGTTACGACTT
primers. The obtained amplicons were purified using AMPure XP beads. DNA concentration was quantified using Qubit DNA quantification assays (Invitrogen). The tagmentation, library barcoding, and amplification were carried out using Nextera XT DNA Library Preparation Kit (Illumina), and libraries were sequenced on MiSeq instrument using MiSeq Reagent Kit v3 (600‐cycle) (Illumina). The raw reads were de novo assembled using MATAM (Pericard et al, 2018) and aligned to the SSU database (SILVA 138.1 release).
SDS–PAGE and western blot
T cell protein contents were extracted in lysis buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.5% Nonidet P‐40, 0.5% sodium deoxycholate, 5 mM EDTA, 2 mM DTT, 1 mM sodium orthovanadate, 10 mM β‐glycerophosphate, 1 mM sodium fluoride, 1 μM microcystin, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 10 μg/ml aprotinin] at 4°C. Cell lysates were clarified by microcentrifugation at 28,672 g at 4°C for 30 min. The supernatants were recovered, and total protein concentrations were measured using Bio‐Rad protein assay reagent. The equivalent amounts of total cellular proteins were heated at 95°C for 3 min and then separated by SDS–PAGE, followed by electrotransfer to polyvinylidene difluoride membranes. The membranes were subsequently blocked for 1 h with Intercept® (TBS) Blocking Buffer (LI‐COR) and then incubated overnight at 4°C with appropriate antibodies (total rabbit Smad3/2, rabbit pSmad2/3, and GAPDH (Cell Signaling)). Primary antibody binding was detected using Alexa Fluor 647‐conjugated anti‐rabbit immunoglobulin G secondary antibody (Invitrogen) visualized with Odyssey instrument (LI‐COR).
Imaging flow cytometry (IFC)
Analysis of the NFATc1 nuclear localization was conducted using Imaging Flow Cytometry (IFC) on a Cytek® Amnis® Imaging Flow Cytometer. Data acquisition was done according to a previously described protocol (Gautam et al, 2018). The nuclear localization of NFATc1 was determined by calculation of the similarity in distribution of staining between Hoechst 33342 (nucleus) and NFATc1 PE.
Author contributions
Nicholas RJ Gascoigne: Conceptualization; supervision; funding acquisition; writing – original draft; writing – review and editing. Lukasz Wojciech: Supervision; investigation; visualization; methodology; writing – original draft; writing – review and editing. Chin Wen Png: Investigation; visualization; methodology. Eileen Y Koh: Investigation; methodology. Dorinda Yan Qin Kioh: Investigation; visualization; methodology. Lei Deng: Investigation; methodology. Ziteng Wang: Investigation; visualization; methodology. Liang‐zhe Wu: Resources. Maryam Hamidinia: Visualization; methodology. Desmond WH Tung: Investigation. Wei Zhang: Resources. Sven Pettersson: Resources. Eric Chun Yong Chan: Methodology; writing – review and editing. Yongliang Zhang: Writing – review and editing. Kevin SW Tan: Conceptualization; supervision; funding acquisition; writing – original draft; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank Dr. P. Hutchinson and Mr. G. Teo (NUS Immunology Program Flow Cytometry Laboratory) for helping with cell sorting, Chua Yen Leong for help with mouse colony maintenance, Ng Geok Choo for helping with Blastocystis culture, and Steven Santino Leonardi for helping with Blastocystis culture and Blastocystis‐specific primer design. We also would like to thank Prof Leszek Ignatowicz (Georgia State University, GA, USA) for providing the BW cell line variant expressing Nur77GFP reporter. This work was founded by Singapore Ministry of Education NUHS seed grants NUHSRO/2019/049/T1/SEED‐MAR/02 and T1‐NUHS Joint Grant Call FY17‐1st call‐03. Lukasz Wojciech is the recipient of an NUSMed Postdoctoral Fellowship. Lei Deng was the recipient of a scholarship from the China Scholarship Council (CSC).
The EMBO Journal (2023) 42: e112963
Contributor Information
Kevin SW Tan, Email: mictank@nus.edu.sg.
Nicholas RJ Gascoigne, Email: micnrjg@nus.edu.sg.
Data availability
The data and reagents are available from the corresponding authors on request.
References
- Ajjampur SSR, Png CW, Chia WN, Zhang Y, Tan KSW (2016) Ex vivo and in vivo mice models to study Blastocystis spp. adhesion, colonization and pathology: closer to proving Koch's postulates. PloS One 11: e0160458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balyan R, Gautam N, Gascoigne NRJ (2020) The ups and downs of metabolism during the lifespan of a T cell. Int J Mol Sci 21: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MDD, O'Sullivan D, Klein Geltink RII, Curtis JDD, Chang CH, Sanin DEE, Qiu J, Kretz O, Braas D, van der Windt GJJW et al (2016) Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166: 63–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JW, Helm EY, Fu Z, Xiong L, Sun N, Oliff KN, Muehlbauer M, Avram D, Zhou L (2023) The aryl hydrocarbon receptor cell intrinsically promotes resident memory CD8+ T cell differentiation and function. Cell Rep 42: 111963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wojciech L, Gascoigne NRJ, Peng G, Tan KSW (2021) New insights into the interactions between Blastocystis, the gut microbiota, and host immunity. PLoS Pathog 17: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wojciech L, Png CW, Kioh DYQ, Gu Y, Aung TT, Malleret B, Chan ECY, Peng G, Zhang Y et al (2023a) Colonization with two different Blastocystis subtypes in DSS‐induced colitis mice is associated with strikingly different microbiome and pathological features. Theranostics 13: 1165–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wojciech L, Png CW, Kioh YQD, Ng GC, Chan ECY, Zhang Y, Gascoigne NRJ, Tan KSW (2023b) Colonization with ubiquitous protist Blastocystis ST1 ameliorates DSS‐induced colitis and promotes beneficial microbiota and immune outcomes. NPJ Biofilms Microbiomes 9: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Roussel M, Noel B, Wawrzyniak I, Da Silva C, Diogon M, Viscogliosi E, Brochier‐Armanet C, Couloux A, Poulain J et al (2011) Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol 12: R29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMeglio P, Duarte JH, Ahlfors H, Owens NDL, Li Y, Villanova F, Tosi I, Hirota K, Nestle FO, Mrowietz U et al (2014) Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity 40: 989–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA et al (2017) A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551: 648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkord E, Al Samid MA, Chaudhary B (2015) Helios, and not FoxP3, is the marker of activated Tregs expressing GARP/LAP. Oncotarget 6: 20026–20036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L, Gentekaki E, Curtis B, Archibald JM, Roger AJ (2017) Lateral gene transfer in the adaptation of the anaerobic parasite Blastocystis to the gut. Curr Biol 27: 807–820 [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY (2017) Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. J Immunol 198: 986–992 [PubMed] [Google Scholar]
- Gascoigne NRJ, Rybakin V, Acuto O, Brzostek J (2016) TCR signal strength and T cell development. Annu Rev Cell Dev Biol 32: 327–348 [DOI] [PubMed] [Google Scholar]
- Gautam N, Sankaran S, Yason JA, Tan KSW, Gascoigne NRJ (2018) A high content imaging flow cytometry approach to study mitochondria in T cells: MitoTracker Green FM dye concentration optimization. Methods 134–135: 11–19 [DOI] [PubMed] [Google Scholar]
- Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II (2014) Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40: 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Singh M, Suresh G, Ng G, Yap E (1993) Axenic culture of Blastocystis hominis in Iscove's modified Dulbecco's medium. Parasitol Res 7: 614–616 [DOI] [PubMed] [Google Scholar]
- Hwang SJ, Cobb DA, Bhadra R, Youngblood B, Khan IA (2016) Blimp‐1‐mediated CD4 T cell exhaustion causes CD8 T cell dysfunction during chronic toxoplasmosis. J Exp Med 213: 1799–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR (2006) The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL‐17+ T helper cells. Cell 126: 1121–1133 [DOI] [PubMed] [Google Scholar]
- Kaur H, Bose C, Mande SS (2019) Tryptophan metabolism by gut microbiome and gut‐brain‐axis: an in silico analysis. Front Neurosci 13: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Henry EC, Kim D, Kim Y, Shin KJ, Han MS, Lee TG, Kang J, Gasiewicz TA, Ryu SH et al (2006) Novel compound 2‐methyl‐2h‐pyrazole‐3‐carboxylic acid aryl hydrocarbon receptor. Mol Pharmacol 69: 1871–1878 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements Daehwan HHS Public Access. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga J, Syono K, Ichikawa T, Adachi T (1994) Involvement of L‐tryptophan aminotransferase in indole‐3‐acetic acid biosynthesis in Enterobacter cloacae. Biochim Biophys Acta 1209: 241–247 [DOI] [PubMed] [Google Scholar]
- Konkel JE, Zhang D, Zanvit P, Chia C, Zangarle‐Murray T, Jin W, Wang S, Chen WJ (2017) Transforming growth factor‐β signaling in regulatory T cells controls T helper‐17 cells and tissue‐specific immune responses. Immunity 46: 660–674 [DOI] [PubMed] [Google Scholar]
- Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM et al (2016) CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 22: 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann J, Huehn J, De La Rosa M, Maszyna F, Kretschmer U, Krenn V, Brunner M, Scheffold A, Hamann A (2002) Expression of the integrin αEβ7 identifies unique subsets of CD25+ as well as CD25− regulatory T cells. Proc Natl Acad Sci U S A 99: 13031–13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF (1997) Transforming growth factor β‐induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci U S A 94: 10669–10674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liang X, Dong W, Fang Y, Lv J, Zhang T, Fiskesund R, Xie J, Liu J, Yin X et al (2018) Tumor‐repopulating cells induce PD‐1 expression in CD8+ T cells by transferring kynurenine and AhR activation. Cancer Cell 33: 480–494 [DOI] [PubMed] [Google Scholar]
- Lu Q, Richardson B (2004) Methods for analyzing the role of DNA methylation and chromatin structure in regulating T lymphocyte gene expression. Biol Proced Online 6: 189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J (1998) TGF‐β signal transduction. Annu Rev Biochem 67: 753–791 [DOI] [PubMed] [Google Scholar]
- McClerklin SA, Lee SG, Harper CP, Nwumeh R, Jez JM, Kunkel BN (2018) Indole‐3‐acetaldehyde dehydrogenase‐dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog 14: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettrick AF, Corcoran SE, Barry PJG, McFarland J, Crès C, Curtis AM, Franklin E, Corr SC, Mok KH, Cummins EP et al (2016) Trypanosoma brucei metabolite indolepyruvate decreases HIF‐1α and glycolysis in macrophages as a mechanism of innate immune evasion. Proc Natl Acad Sci U S A 113: E7778–E7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone I, Marafini I, Zorzi F, Di Fusco D, Dinallo V, Rizzo A, Sileri P, Sica G, Monteleone G (2016) Smad7 knockdown restores aryl hydrocarbon receptor‐mediated protective signals in the gut. J Crohns Colitis 10: 670–677 [DOI] [PubMed] [Google Scholar]
- Moreira D, López‐García P (2017) Protist evolution: stealing genes to gut it out. Curr Biol 27: R223–R225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Yoon H, Ahmed R, Boss JM (2008) NFATc1 regulates PD‐1 expression upon T cell activation. J Immunol 181: 4832–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericard P, Dufresne Y, Couderc L, Blanquart S, Touzet H (2018) MATAM: reconstruction of phylogenetic marker genes from short sequencing reads in metagenomes. Bioinformatics 34: 585–591 [DOI] [PubMed] [Google Scholar]
- R Core Team (2020) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.r-project.org/ [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2009) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova A, Wincent E, Vikström Bergander L, Alsberg T, Bergman J, Rannug A, Rannug U (2016) Evidence for new light‐independent pathways for generation of the endogenous aryl hydrocarbon receptor agonist FICZ. Chem Res Toxicol 29: 75–86 [DOI] [PubMed] [Google Scholar]
- Stensvold CR, Clark CG (2016) Current status of Blastocystis: a personal view. Parasitol Int 65: 763–771 [DOI] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A (2009) Absence of autophagy results in reactive oxygen species‐dependent amplification of RLR signaling. Proc Natl Acad Sci U S A 106: 2774–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testl R, Lanier LL (1989) T cell activation via Leu‐23 (CD69). J Immunol 23: 1123–1128 [PubMed] [Google Scholar]
- Thornton AM, Lu J, Korty PE, Kim YC, Martens C, Sun PD, Shevach EM (2019) Helios+ and Helios− Treg subpopulations are phenotypically and functionally distinct and express dissimilar TCR repertoires. Eur J Immunol 49: 398–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncone E, Marafini I, Stolfi C, Monteleone G (2018) Transforming growth factor‐β1/Smad7 in intestinal immunity, inflammation, and cancer. Front Immunol 9: 4–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS et al (2014) Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll‐like receptor 4. Immunity 41: 296–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyniak I, Courtine D, Osman M, Hubans‐Pierlot C, Cian A, Nourrisson C, Chabe M, Poirier P, Bart A, Polonais V et al (2015) Draft genome sequence of the intestinal parasite Blastocystis subtype 4‐isolate WR1. Genomics Data 4: 22–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Luo C, Kolde R, d'Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA et al (2017) Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 22: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciech L, Ignatowicz A, Seweryn M, Rempala G, Singh Pabla S, McIndoe RA, Kisielow P, Ignatowicz L (2014) The same self‐peptide selects conventional and regulatory CD4+ T cells with identical antigen receptors. Nat Commun 5: 5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciech L, Szurek E, Kuczma M, Cebula A, Elhefnawy WR, Pietrzak M, Rempala G, Ignatowicz L (2018) Non‐canonicaly recruited TCRαβCD8αα IELs recognize microbial antigens. Sci Rep 8: 10848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciech L, Tan KSW, Gascoigne NRJ (2020) Taming the sentinels: microbiome‐derived metabolites and polarization of T cells. Int J Mol Sci 21: 7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, Föhse L, Prinz I, Pezoldt J, Suerbaum S et al (2016) Foxp3+ T cells expressing RORγt represent a stable regulatory T‐cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol 9: 444–457 [DOI] [PubMed] [Google Scholar]
- Yason JA, Liang YR, Png CW, Zhang Y, Tan KSW (2019) Interactions between a pathogenic Blastocystis subtype and gut microbiota: in vitro and in vivo studies. Microbiome 7: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Qiu J, Bostick JW, Ueda A, Schjerven H, Li S, Jobin C, Chen Z m E, Zhou L (2017) The aryl hydrocarbon receptor preferentially marks and promotes gut regulatory T cells. Cell Rep 21: 2277–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Dee MJ, Altman NH, Malek TR (2015) IL‐2Rβ–dependent signaling and CD103 functionally cooperate to maintain tolerance in the gut mucosa. J Immunol 194: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, DeLuca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi‐Benedetti C, Fallarino F et al (2013) Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin‐22. Immunity 39: 372–385 [DOI] [PubMed] [Google Scholar]
- Zhang P, Jin T, Sahu SK, Xu J, Shi Q, Liu H, Wang Y (2019) The distribution of tryptophan‐dependent indole‐3‐acetic acid synthesis pathways in bacteria unraveled by large‐scale genomic analysis. Molecules 24: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Rong HM, Li T, Zhai K, Tong ZH (2020) PD‐1 deficiency promotes macrophage activation and T‐helper cell type 1/T‐helper cell type 17 response in pneumocystis pneumonia. Am J Respir Cell Mol Biol 62: 767–782 [DOI] [PubMed] [Google Scholar]


