Abstract
Typical hyper-IgE syndromes (HIES) are caused by autosomal-dominant-negative (DN) variants of STAT3 (Signal Transducer And Activator Of Transcription 3) or IL6ST (Interleukin 6 Cytokine Family Signal Transducer), biallelic partial loss-of-function (LOF) variants of IL6ST, or biallelic complete LOF variants of ZNF341 (Zinc Finger Protein 341). Including the two new cases described in this review, only 20 patients with autosomal-recessive (AR) ZNF341 deficiency have ever been reported. Patients with AR ZNF341 deficiency have clinical and immunological phenotypes resembling those of patients with autosomal-dominant STAT3 deficiency, but with a usually milder clinical presentation and lower NK (Natural Killer) cell counts. ZNF341-deficient cells have 50% the normal level of STAT3 in the resting state. However, as there is no clear evidence that STAT3 haploinsufficiency causes HIES, this decrease alone is probably insufficient to explain the HIES phenotype observed in the ZNF341-deficient patients. The combination of decreased basal expression level and impaired autoinduction of STAT3 observed in ZNF341-deficient lymphocytes is considered a more likely pathophysiological mechanism. We review here what is currently known about the ZNF341 gene and ZNF341 deficiency, and briefly discuss possible roles for this protein in addition to its control of STAT3 activity.
Introduction
Patients with Job’s syndrome or hyper-IgE syndrome (HIES) classically display severe bacterial infections of the skin and lungs, chronic mucocutaneous candidiasis (CMC), eczema, high-serum IgE (Immunoglobulin E) levels, poor or delayed clinical and biological inflammation, and skeletal, connective tissue and vascular abnormalities [1–4]. In 2007, more than 40 years after the first description of this syndrome, Minegishi and coworkers reported that dominant-negative (DN) monoallelic variants of the STAT3 gene, encoding signal transducer and activator of transcription 3, were responsible for most cases of autosomal-dominant (AD) HIES [5]. More than 140 heterozygous variants of STAT3 have since been reported [6]. A DN disease- causing mechanism has recently been confirmed for most (at least 95% of the 150 alleles tested) STAT3 variants [6]. The complexity of signaling via STAT3, a ubiquitously expressed transcription factor that acts downstream from a large number of cytokine or growth factor receptors, has made it difficult to understand the mechanism underlying HIES [7]. The discovery of biallelic partial loss-of-function (LOF) and monoallelic DN variants of IL6ST encoding GP130 (Glycoprotein 130), the common chain of the receptors of the IL-6 (interleukin 6) cytokine family, in patients with autosomal recessive (AR) and AD HIES, respectively, has highlighted the key role of this family of cytokines in the pathophysiology of this disease [8–10]. These variants strongly impair IL-6 and IL-11 signaling, but have a lesser effect on signaling by other IL-6 family cytokines (e.g. Leukemia Inhibitory Factor (LIF), Oncostatin-M (OSM), and IL-27). Unsurprisingly, AR IL-6R (Interleukin 6 Receptor) and AR IL-11RA (Interleukin-11 Receptor Subunit Alpha) deficiencies reproduce most of the immunological and extrahematopoietic phenotypes of HIES, respectively [11–13]. Other inborn errors of cytokines or their receptors that signal through STAT3 present with only partially overlapping or even different clinical phenotypes. Patients with AR IL-21R (Interleukin 21 Receptor) or IL-21 deficiency share some of the features of HIES, with recurrent respiratory infections, impaired humoral immune responses, and high- serum IgE levels [14–17]. However, in contrast to HIES patients, they also display severe cryptosporidiosis. In 2018, two studies reported a new inborn error of immunity underlying AR HIES: biallelic LOF variants of the zinc finger 341 (ZNF341) gene encoding a protein of previously unknown function [18,19]. These two studies demonstrated that ZNF341 is a transcription factor controlling basal and inducible STAT3 expression. This review summarizes what is currently known about the function of ZNF341. It also reports two new patients with complete ZNF341 deficiency and AR HIES.
Genetics of autosomal recessive ZNF341 deficiency
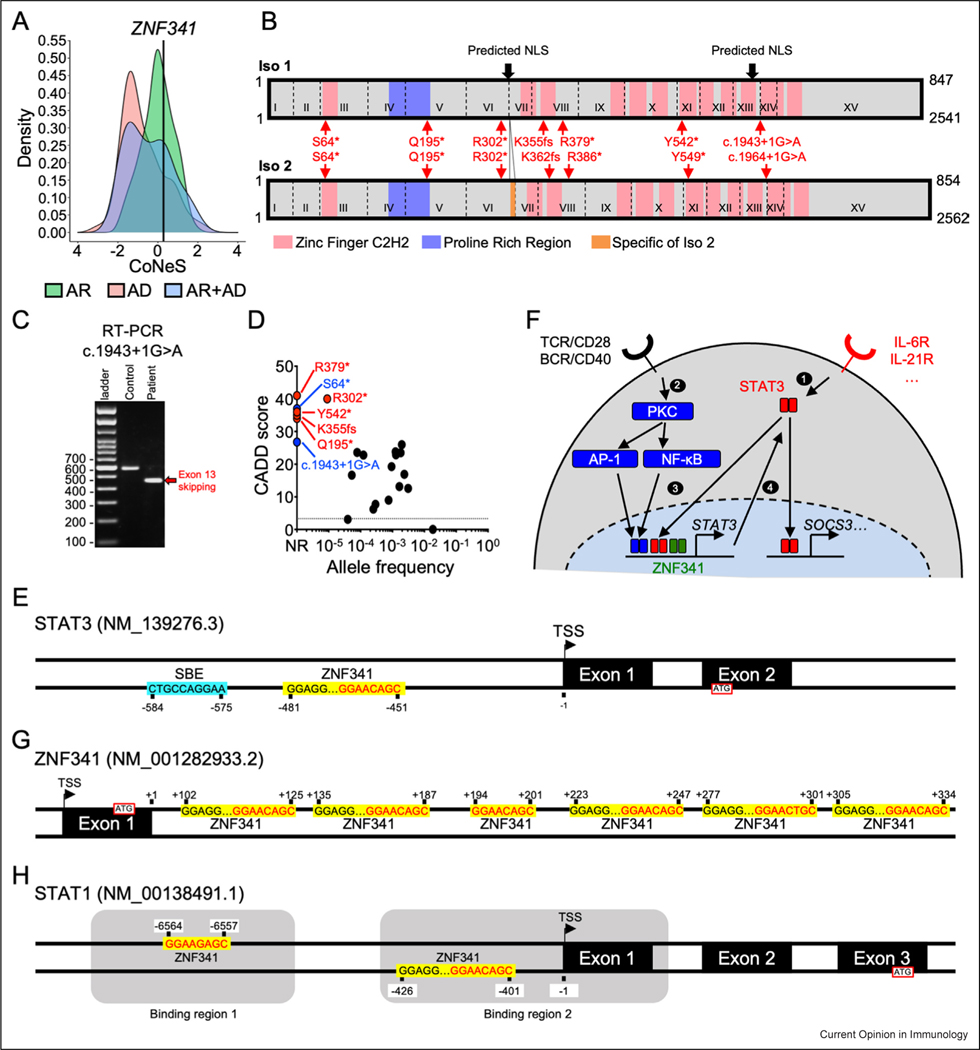
The gene encoding ZNF341 is located on chromosome 20. It contains 15 exons encoding two functional isoforms differing by only seven amino acids at the end of exon 6 [18]. No homozygous predicted loss-of-function (pLOF) variants and only 16 homozygous missense variants are reported in the gnomAD v2.1 and v3.1.2 public databases [20]. Only 18 patients (10 female and 8 male patients) from 10 kindreds with proven AR ZNF341 deficiency have been reported to date [18,19]. These patients were Moroccan, Afro-Caribbean, Iranian, Turkish, Lebanese, or Israeli Arab. All the patients with AR ZNF341 deficiency described to date were homozygous for pLOF variants, consistent with the relatively high consensus-negative selection (CoNeS) score of 0.29 (Figure 1a) [21]. All these variants induced premature stop codons (p.Gln195*, p.Arg302*, p.Lys355Serfs*28, p.Arg379*, and p.Tyr542*) and were therefore predicted to encode truncated proteins (Figure 1b). The c.904C > T/p.Arg302* variant was identified in 13 patients from six kindreds. A study of three kindreds of three different ethnicities (Iranian, African, and Turkish) showed that the inherited c.904C > T (p.Arg302*) allele belonged to different haplotypes in these patients, suggesting the existence of a mutational hotspot [18]. However, the other three families were from the same Israeli village, in which 5% of the population carried the mutated allele, suggesting a founder effect in this population [19,22]. We report here two additional patients born to consanguineous parents: P19, a 29-year-old man of African descent from the Ivory Coast living in France, and P20, a two-year-old boy from Algeria. P19 is homozygous for an essential splice-site (c.1943+1G > A) variant of ZNF341, leading to the skipping of exon 13 and predicted to result in a truncated protein, p.Gly611Glyfs*15 (Figure. 1b, c). P20 is homozygous for the c.191C > A variant of ZNF341, with a premature stop codon (p.Ser64*) (Figure 1b). Like the variants previously reported in patients with AR ZNF341 deficiency, the c.1943+1G > A/p.Gly611Glyfs*15 and the c.191C > A/p.Ser64* variants have never been reported in public databases (private variants) and have CADD scores well above the mutation significance cutoff (MSC) (Figure 1d) [23]. Of note, two additional patients with HIES and suspected ZNF341 deficiency were reported but not included in our analysis because the deleterious impact of their mutation (p.Cys352Arg) was not demonstrated [24]. In total, 20 patients with HIES and confirmed biallelic LOF variants of ZNF341 have now been identified.
Figure 1.
ZNF341 isoforms, population genetics, schematic signaling model, and DNA binding sites in STAT1, STAT3 and ZNF341 genes. (a) The CoNeS score of ZNF341 is compatible with an AR trait. (b) Schematic representation of ZNF341 isoforms 1 (top) and 2 (bottom). The exons are indicated by Roman numerals, and the exon boundaries are indicated with dashed lines. Predicted nuclear localization sequences are indicated by black arrows. The seven reported mutations underlying HIES are indicated in red for both isoforms. (c) Reverse transcription PCR (Polymerase Chain reaction) showing that the newly reported c.1943+1G > A mutation is responsible for exon-13 skipping. (d) Population genetics of ZNF341. All HIES variants are highly deleterious and only the R302* mutant has been reported in the heterozygous state in a public database. CADD: combined annotation-dependent depletion score. (e) Schematic representation of the STAT3 promoter. The SBE (STAT3 binding element) is indicated by a blue box. ZNF341-binding sites are indicated by yellow boxes. The TSS (Transcription start site) is indicated. The position of each binding site is indicated relative to the TSS. (f) Model for the ZNF341-dependent signaling pathway. BCR: B-cell receptor. (g–h) Schematic representation of the ZNF341 exon 1 and intron 1 (g), and STAT1 promoter (h), as in E. The STAT1 promoter contains two binding regions indicated by gray boxes.
Clinical features of patients with autosomal-recessive ZNF341 deficiency
The clinical phenotype of patients with AR ZNF341 deficiency is essentially a phenocopy of AD STAT3 deficiency. However, it has milder consequences, as illustrated by the median HIES NIH (National Institutes of Health) score of 28.5 for these patients, as opposed to 64 for patients with AD STAT3 deficiency (Table 1) [18,19,25–33]. Atopic dermatitis was the most common clinical presentation of patients with ZNF341 deficiency, present in all patients reported, a proportion similar to that for patients with STAT3 deficiency (> 90%). By contrast, newborn rash was rarely reported in patients with ZNF341 deficiency (15%), but was much more frequent (48%) in patients with STAT3 deficiency [25,29]. Both ZNF341-deficient and STAT3-deficient patients were highly prone to recurrent skin infections, with skin abscesses (75% vs. 73%) and CMC (60% vs. 85%). Respiratory phenotype was milder in patients with ZNF341 deficiency than in those with STAT3 deficiency. Recurrent ear, nose, and throat infections were reported in 58% of patients with ZNF341 deficiency, and 90% of those with STAT3 deficiency. Similarly, lung phenotypes were markedly less severe in patients with ZNF341 deficiency than in patients with STAT3 deficiency, with pneumonia, bronchiectasis, and pneumatocele reported in 56%, 35%, and 10%, respectively, of the ZNF341-deficient patients and 90%, 65%, and 52%, respectively of the STAT3-deficient patients. Both ZNF341-deficient and STAT3-deficient patients display skeletal and connective-tissue phenotypes, including facial dysmorphia (53% vs. 95%), high palate (45% vs. 53%), scoliosis (10% vs. 38%), joint hyperextensibility (15% vs. 50%), and the retention of deciduous teeth (25% vs. 65%). However, these extrahematopoietic manifestations were less frequent in ZNF341-deficient patients than in STAT3-deficient patients. Peripheral and brain artery abnormalities were reported in 84% of the STAT3-deficient patients, and led to vascular complications in some of these patients [34]. None of the ZNF341-deficient patients were reported to have vascular complications (e.g. aneurysm), but more detailed investigations, including full-body magnetic resonance imaging, would be required to exclude such abnormalities definitively. Overall, the clinical description of ZNF341 deficiency resembles a milder form of STAT3 deficiency.
Table 1.
Clinical/biological features of human ZNF341 and STAT3 deficiencies.
| This paper | Béziat et al [18] | Frey-Jakobs et al [19] | Total ZNF341 | STAT3 deficiency [25] | ||
|---|---|---|---|---|---|---|
|
|
||||||
| Kindred | 1 Kindred | 1 Kindred | 6 Kindred | 4 Kindred | 12 Kindred | |
| Number of patients | 1 patient | 1 patient | 7 patients | 11 patients | 20 patients | 60 patients |
| Mutant allele (isoform 1) | c.1943 + 1 G > A (essential splice site, skipping of exon 13) | p.Ser64 * | p.Gln195*(n = 1) p.Arg302*(n = 4) p.Lys355Serfs*28 (n= 1) p.Tyr542*(n = 1) |
p.Arg302 * (n = 8) p.Arg379 * (n = 3) |
||
| Age at evaluation — median (range) | 29 | 2 | 23 (14–47) | 17 (0.5–30) | 21 (2–47) | n.d. |
| Sex | m | m | 4f/3 m | 6f/5 m | 10f/10m | 30f/30m |
| HIES NIH score — median (range) | 24 | 25 | 26 (11–54) | 33 (12–62) | 28.5 (11–62) | 64 (14–86) |
| IgE levels > 1000 IU | 1/1 | 0/1 | 6/7 | 10/11 | 17/20 (85%) | 96% |
| High IgG levels (> 16 g/L) | 1/1 | 0/1 | 7/7 | 7/9 | 15/18 (83%) | 27% |
| Eosinophilia | 0/1 | 1/1 | 3/7 | 7/10 | 11/19 (58%) | 80% |
| Low NK cell (count or % of lymphocytes) | 0/1 | 0/1 | 6/7 | 4/6 | 10/15 (67%) | 8% |
| Low memory B-cell levels (% of B cells) | 0/1 | ND | 4/6 | 6/6 | 10/13 (77%) | 94,5% |
| Low Th17-cell levels | ND | ND | 5/5 | 4/6 | 9/11 (82%) | 93% |
| Allergy (food or respiratory) | 0/1 | ND | 4/7 | 0/11 | 4/19 (21 %) | 22% |
| Atopic dermatitis | 1/1 | 1/1 | 7/7 | 11/11 | 20/20 (100%) | 92% |
| Skin abscesses | 1/1 | 1/1 | 3/7 | 10/11 | 15/20 (75%) | 73% |
| Recurrent skin infection | 0/1 | 1/1 | 6/7 | 8/9 | 15/18 (83%) | 100% |
| CMC | 0/1 | 0/1 | 6/7 | 6/11 | 12/20 (60%) | 85% |
| - Oral thrush | 0/1 | 0/1 | 5/7 | 4/11 | 9/20 (45%) | 63% |
| - Onychomycosis | 0/1 | 0/1 | 3/7 | 2/11 | 5/20 (25%) | 57% |
| - Other | 0/1 | 0/1 | 3/7 | 4/11 | 7/20 (35%) | n.d. |
| Upper respiratory tract infections | 1/1 | 0/1 | 1/7 | 9/10 | 11/19 (58%) | 90% |
| Lung involvement | 1/1 | 0/1 | 4/7 | 6/7 | 11/16 (69%) | n.d. |
| - Recurrent infections | 1/1 | 0/1 | 4/7 | 4/7 | 9/16 (56%) | n.d. |
| - Pneumonia | 1/1 | 0/1 | 2/7 | 6/7 | 9/16 (56%) | 90% |
| - Bronchiectasis | 0/1 | 0/1 | 2/7 | 4/8 | 6/17 (35%) | 65% |
| - Pneumatocele | 0/1 | 0/1 | 1/7 | 1/8 | 2/20 (10%) | 52% |
| Alopecia | 0/1 | 0/1 | 1/7 | 1/11 | 2/20 (10%) | n.d. |
| Connective-tissue abnormalities | 0/1 | 0/1 | 6/7 | 9/11 | 15/20 (75%) | n.d. |
| - Facial abnormalities | 0/1 | 0/1 | 2/7 | 8/10 | 10/19 (53%) | 95% |
| - High palate | 0/1 | 0/1 | 4/7 | 5/11 | 9/20 (45%) | 53% |
| - Scoliosis | 0/1 | 1/1 | 0/7 | 1/11 | 2/20 (10%) | 38% |
| - Joint hyperextensibility | 0/1 | 0/1 | 2/7 | 1/11 | 3/20 (15%) | 50% |
| - Bone fractures with minimal trauma | 0/1 | 0/1 | 1/7 | 1/11 | 2/20 (20%) | 42% |
| - Dental abnormalities | 0/1 | 0/1 | 2/7 | 4/7 | 6/16 (38%) | n.d. |
| - Deciduous tooth retention | 0/1 | 0/1 | 0/7 | 4/7 | 4/16 (25%) | 65% |
| - Craniosynostosis | 0/1 | 0/1 | 0/7 | 0/11 | 0/20 (0%) | 3% |
| Newborn rash | 0/1 | 0/1 | 1/7 | 1/4 | 2/13 (15%) | 48% |
| Neoplasia (any type) | 0/1 | 0/1 | 0/7 | 1/11 | 1/20 (5%) | 7% |
| Growth retardation | 0/1 | 0/1 | 1/7 | 2/11 | 3/20 (15%) | n.d. |
| Mental delay | 0/1 | 0/1 | 0/7 | 7/10 | 7/19 (37%) | n.d. |
n.d.: no data.
Immunological features of patients with autosomal-recessive ZNF341 deficiency
Patients with ZNF341 deficiency have an immunological phenotype very similar to that of patients with STAT3 deficiency (Table 1). Both defects are associated with high levels of IgE (in 85% vs. 96% of patients, respectively) and eosinophilia (58% vs. 80%, respectively) [25]. However, STAT3-deficient patients may have dampened signs of inflammation with cold staphylococcal abscesses or poor or delayed clinical and biological signs of inflammation in the setting of infection. In contrast, patients with ZNF341 deficiency show appropriate clinical and biological signs of inflammation. The immunophenotyping of blood or PBMCs (Peripheral blood mononuclear cells) has shown that both deficiencies are associated with high frequencies of naive CD4+ T cells, and low frequencies of central memory CD4+ and CD8+ T cells, mucosal-associated invariant T cells, memory B cells, and innate lymphocytes (ILC) 1 and ILC2 [18,19,25,35–39]. In addition, the CD4+ T cells of these patients have a biased helper (Th (T helper)) profile, with an abnormally high frequency of Th2 cells, and abnormally low levels of T-follicular helper (Tfh) and of Th17 cells. The high proportion of Th2 cells probably underlies the high serum levels of IgE, eczema, and the other allergic manifestations seen in the patients, as suggested by the spectacular improvement of symptoms and biological parameters in a ZNF341-deficient patient treated with dupilumab, a monoclonal antibody inhibiting signaling via IL-4 and IL-13 [40]. Low proportions of Th17 cells probably underlie CMC, a clinical feature of patients with various inborn errors of IL-17 immunity [41,42]. The low proportions of Tfh and memory B cells probably at least partly account for the severe bacterial infections of the lungs observed in these patients, through the impairment of sustained humoral responses [18]. Intriguingly, despite their low frequency of memory B cells, ZNF341-deficient patients, unlike STAT3-deficient patients, have high plasma levels of IgG and a high frequency of IgG+ cells among the remaining memory B cells. Finally, unlike STAT3-deficient patients, ZNF341-deficient patients frequently display low NK cell numbers or frequencies among lymphocytes (8% vs. 67%, respectively). The remaining NK cells have a normal pattern of differentiation. Thus, ZNF341 deficiency phenocopies STAT3 deficiency in terms of the immunological phenotype of patients, except that NK cell counts are low in a majority of patients with ZNF341 deficiency but rarely in those with STAT3 deficiency.
ZNF341 controls basal levels of STAT3 expression
Before the discovery of ZNF341 defects in patients with HIES, the function of ZNF341 was unknown in both mice and humans. However, the clinical similarities between ZNF341 and STAT3 deficiencies suggested that the two genes/proteins were probably connected [18,19]. The 12 zinc finger domains and the two predicted nuclear localization sequences present in the ZNF341 protein suggested that it might act as a transcription factor. Indeed, ZNF341 was localized to the nucleus in overexpression studies, and western blots of cell extracts from primary cells and immortalized cell lines showed it to be ubiquitously expressed [18]. Chromatin immunoprecipitation sequencing (ChIP-seq) of endogenous ZNF341 from control primary T cells and Epstein–Barr virus (EBV)-transformed B (EBV-B) cells showed that ZNF341 did, indeed, bind a ZNF-like DNA-binding motif (GGAAC/GA/GGC) and a SP1-like DNA-binding motif (GGGAGG), potentially forming a high-affinity bipartite motif with the two binding sites separated by 13 or 14 nucleotides (GGGAGGn(13−14)GGAAC/GA/GGC) [18,19]. Similar DNA-binding motifs were identified in ChIP-seq data obtained from HEK293T cells transfected with a tagged wild-type (WT) ZNF341 cDNA, and ZNF341-deficient EBV-B cells stably transduced with WT ZNF341 isoform 1 or 2 [18,43]. Strikingly, the primary DNA- binding sequence most strongly targeted by ZNF341 was found in the STAT3 promoter. However, ZNF341 exerted subtle control over the expression of the STAT3 gene, which did not differ markedly between the WT and ZNF341-deficient cell lines (e.g. EBV-B cells or Herpesvirus Saimiri (HVS)-transformed T cells), due to the variability of STAT3 expression after cell immortalization. The reintroduction of WT ZNF341 in deficient EBV-B or HVS-T cells induced only a minor, but consistent, increase in STAT3 mRNA and protein levels [18]. By contrast, ZNF341 deficiency had a marked impact on basal levels of STAT3 in the patients’ primary cells. All the ZNF341-deficient cell subsets tested, including lymphocytes, monocytes, and fibroblasts, had 50% the normal level of STAT3 mRNA and protein. This resulted in 50% the normal level of STAT3 phosphorylation following cytokine stimulation, in all the primary cell subsets tested. This 50% decrease in STAT3 expression was associated with a decrease in prototypic STAT3 target gene (suppressor of cytokine signaling 3 (SOCS3)) induction upon IL-6/IL-6Rα stimulation in primary fibroblasts from patients. However, SOCS3 induction was normal in naive CD4+ T lymphocytes and monocytes after stimulation with IL-6/IL-6R and IL-10, respectively, for 2–4 h, suggesting that the 50% decrease in basal STAT3 levels had only a modest impact on downstream signaling. Thus, ZNF341 constitutively binds the STAT3 promoter and increases basal levels of STAT3, but has a modest impact on STAT3 signaling in response to cytokine stimulation only.
STAT3 autoinduction: a physiologically relevant mechanism under the control of ZNF341
At least 95% of the 150 pathogenic variants of STAT3 tested act through negative dominance [5,6], and there is no strong evidence that haploinsufficiency at the STAT3 locus causes HIES. Furthermore, ZNF341 deficiency had only a modest impact on SOCS3 induction in the primary cell types tested. The 50% decrease in STAT3 protein levels in ZNF341-deficient cells is therefore not sufficient to explain the HIES phenotype. We sought a complementary mechanism that might explain the pathogenesis of ZNF341 deficiency. Studies in mice have shown that, following stimulation with IL-6, STAT3 can bind to its own promoter, inducing the production of its own mRNA and protein in a tissue- and cell-specific manner, including in T cells [44,45]. In particular, a study of a knock-in mouse model with a homozygous mutation of the Stat3-binding element (SBE) within the Stat3 promoter showed that Stat3 autoinduction was required for the optimal induction of selected IL-6-dependent genes in specific organs, tissues, or cells [45]. Kwon et al. demonstrated that Stat3 protein induction upon IL-6 and T-cell receptor (TCR) costimulation was dependent on protein kinase C (PKC)-θ, and that stimulation with phorbol-myristate acetate (PMA), an agonist of PKC-θ, increased Stat3 protein levels [46]. This PKC-θ-mediated Stat3 transcription was dependent on the activator protein 1 (AP-1) and nuclear factor- kappa B (NF-κB) signaling pathways, through the binding of at least c-fos and p65 to the STAT3 promoter. More importantly, Th17 differentiation was found to require Stat3 autoinduction [46]. The data obtained in this study thus indicated that PKC-θ acts downstream from the TCR and Th17-priming cytokines (e.g. IL-6) to upregulate Stat3 via NF-κB and AP-1, resulting in Th17 differentiation. In humans, the STAT3-binding site within the STAT3 promoter located just upstream from the ZNF341-binding site (Figure 1e). We showed that STAT3 autoinduction in human T cells also required IL-6 and TCR costimulation [18]. STAT3 autoinduction was abolished in ZNF341-deficient naive T cells stimulated under these conditions, but not in STAT3-deficient naive T cells [18]. Consistent with the findings of Kwon et al., this defective STAT3 autoinduction was associated with low levels of Th17 cells in ZNF341-deficient patients, both ex vivo and after differentiation in vitro [18,19]. Overall, these data suggest that STAT3 autoinduction in human T cells is dependent on ZNF341 and, probably, PKC-θ signaling for Th17 differentiation (Figure 1f). Interestingly, impaired STAT3 autoinduction was also observed in ZNF341-deficient naive B cells following CD40L and IL-21 costimulation, and was associated with a severe impairment of immunoglobulin production [18]. Thus, decreased STAT3 basal levels coupled to impaired STAT3 autoinduction probably underlie the CMC and bacterial infections of the lungs observed in patients with ZNF341 deficiency. The underlying mechanism remains incompletely understood but probably involves the impairment of Th17 and B-cell differentiation, respectively.
ZNF341 functions beyond the control of STAT3 expression
ZNF341 was shown to interact significantly with UBTF, PCM1, and PAF1 in HEK293T cells transfected with a vector encoding GFP-tagged WT ZNF341, in an affinity purification and mass spectrometry experiment (AP-MS) [43]. These interactions have not been confirmed or investigated further with other techniques, but, interestingly, both UBTF and PAF1 play important roles in transcription. ChIP-seq of ZNF341 identified 1457 binding regions in primary T cells, and 5842 and 6570 binding regions in ZNF341-deficient EBV-B-cell lines transduced with vectors encoding WT ZNF341 isoforms 1 and 2, respectively [18]. In total, 228 DNA-binding regions common to primary T cells and EBV-B cells were identified [18]. Thus, in addition to controlling STAT3, ZNF341 probably controls the transcription of a large number of genes, starting with ZNF341 itself. Indeed, ChIP-seq experiments revealed a peak in ZNF341 intron 1 [18]. A detailed analysis showed that ZNF341 intron 1 contains six canonical ZNF341-binding sites spaced 4–30 nucleotides apart (Figure 1g). RNA sequencing and RT-qPCR on ZNF341-deficient B and T cells showed high levels of ZNF341 mRNA, suggesting that ZNF341 downregulates its own expression and potentially that of other genes. Two ZNF341-binding sites were identified within the STAT1 promoter, including one approximately 400 nucleotides upstream from the transcription start site (TSS), as in the STAT3 promoter (Figure 1h) [18]. ZNF341 upregulates STAT1 mRNA and protein levels, as demonstrated by the lower levels of STAT1 in ZNF341-deficient cells, and the upregulation of STAT1 expression upon WT ZNF341 overexpression in ZNF341-deficient cells [18]. AR complete and partial STAT1 deficiencies are associated with susceptibility to mycobacterial and viral infections, whereas AD STAT1 deficiency is associated with isolated susceptibility to mycobacteria [47–51]. None of the ZNF341-deficient patients described to date has been reported to have suffered unusual or severe viral diseases or to have developed adverse clinical events following vaccination with live attenuated BCG. However, at least two patients have been reported to have suffered tuberculosis [18]. Thus, despite the lack of haploinsufficiency at the STAT1 locus [52], we cannot rule out the possibility that ZNF341-deficient patients are susceptible to virulent environmental mycobacteria due to a partial impairment of STAT1 signaling. It remains unknown whether STAT1 can undergo autoinduction such as that displayed by STAT3, but STAT3 has been shown to act in synergy with nuclear EGFR to enhance STAT1 expression [53]. It would be interesting to investigate whether this STAT3-dependent induction of STAT1 expression is dependent on ZNF341, and to determine whether STAT1 can undergo autoinduction. Finally, one of the most important binding sites in all ChIP-seq datasets for ZNF341 was that within the KAT6A promoter. As for STAT3 itself, KAT6A mRNA levels were not significantly modulated by ZNF341 complementation in ZNF341-deficient EBV-transformed B cells. However, as for STAT3, it remains possible that the alterations to KAT6A levels in ZNF341-deficient cells are more visible in primary cells. Interestingly, DN variants of KAT6A are associated with Arboleda–Tham syndrome [54,55], a neurodevelopmental disorder with extrahematopoietic features also observed in AD STAT3, AR LIFR, and AR complete IL6ST deficiencies [25,56,57]. It would be interesting to determine whether KAT6A expression is controlled by STAT3/ ZNF341 in a LIF-dependent manner (or in a manner dependent on another IL-6 family cytokine). These examples highlight the elusive role of ZNF341 beyond the control of STAT3 expression, and the possible interaction between STAT3 and ZNF341 in the control of expression for various target genes remains to be tested.
Conclusions
Although generally milder, AR ZNF341 deficiency is a hardly distinguishable clinical phenocopy of AD STAT3 deficiency in the absence of genetic testing. ZNF341 and STAT3 deficiencies have several biological phenotypes in common, the exceptions being the frequently decreased NK cell counts and the appropriate clinical and biological signs of inflammation observed in patients with ZNF341 deficiency. ZNF341 is a ubiquitous nuclear protein that may repress or activate target gene expression. Its major role appears to be the control of STAT3 expression, explaining why AD STAT3 and AR ZNF341 are phenocopies. In AD STAT3 deficiency, DN STAT3 proteins interfere with the remaining activity of the WT STAT3 proteins. In AR ZNF341 deficiency, the lack of ZNF341 function reduces the basal expression of STAT3 and impairs STAT3 autoinduction ‘boost’. In both cases, the resulting impaired STAT3 signaling leads to similar B-cell and Th17 defects, underlying the bacterial and fungal infections. STAT3 autoinduction in T cells is probably dependent on cosignaling via AP-1 and NF-kB in the PKC-θ pathway. The role of ZNF341 beyond the control of STAT3 expression remains unclear. This protein has been shown to bind thousands of regions in the human genome, with cell-type specificities. ZNF341 may act as a bridge between STAT3 and other signaling pathways, by binding to promoters other than the STAT3 promoter, such as the STAT1 and KAT6A promoters. Furthermore, the interaction partners of ZNF341 remain unknown, although several excellent candidates have been proposed (e.g. UBTF, AP-1, and NF-κB). Despite the many molecular, cellular, immunological, and clinical similarities between AD STAT3 and AR ZNF341 deficiencies, ZNF341 may function beyond the control of STAT3 expression and autoinduction. For instance, ZNF341-dependent STAT1 expression may underlie susceptibility to virulent environmental mycobacteria (Mycobacterium tuberculosis) reported in some patients. Additional case reports and studies are required to fully understand the functions of this transcription factor.
Acknowledgements
We warmly thank the members of the HGID laboratory for helpful discussions. This work was supported by grants from Institut National de la Santé et de la Recherche Médicale (INSERM), the University of Paris Cité, the Integrative Biology of Emerging Infectious Diseases Laboratoire d′Excellence (ANR-10-LABX-62-IBEID), the Jeffrey Modell Foundation Translational Research Program, the French National Research Agency (ANR) (grant no. GENCMCD-ANR-11-BSV3-005-01, no. HGDIFD-ANR-14-CE15-0006-01, no. ANR-21-CE15-0034, and no. EURO-CMC-ANR-14-RARE-0005-02) and grants awarded under the ‘Investissement d′avenir’ program (grant no. ANR-10-IAHU-01), the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (grant no. U01AI109697 and no. R01AI127564), the French Society of Dermatology, ITMO Cancer of Aviesan and INCa within the framework of the 2021-2030 Cancer Control Strategy (funds administered by the Institut National de la Santé et de la Recherche Médicale), the Rockefeller University, the Howard Hughes Medical Institute, and the St. Giles Foundation.
Footnotes
Conflict of interest statement
Nothing to declare.
Data Availability
Data will be made available on request.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Davis Starkey D, Schaller J, Wedgwood Ralph J, Harvard MD: Job’s syndrome. Lancet 1966, 287:1013-1015. [DOI] [PubMed] [Google Scholar]
- 2.Buckley RH, Wray BB, Belmaker EZ: Extreme hyperimmunoglobulinemia E and undue susceptibility to infection. Pediatrics 1972, 49:59–70. [PubMed] [Google Scholar]
- 3.Zhang Q, Boisson B, Béziat V, Puel A, Casanova J-L: Human hyper-IgE syndrome: singular or plural? Mamm Genome 2018, 29:603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsilifis C, Freeman AF, Gennery AR: STAT3 Hyper-IgE syndrome—an update and unanswered questions. J Clin Immunol 2021, 41:864–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, et al. : Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 2007, 448:1058–1062. [DOI] [PubMed] [Google Scholar]
- 6.Asano T, Khourieh J, Zhang P, Rapaport F, Spaan AN, Li J, Lei W-T, Pelham SJ, Hum D, Chrabieh M, et al. : Human STAT3 variants underlie autosomal dominant hyper-IgE syndrome by negative dominance. J Exp Med 2021, 218:e20202592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu X, Li J, Fu M, Zhao X, Wang W: The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther 2021, 6:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwerd T, Twigg SRF, Aschenbrenner D, Manrique S, Miller KA, Taylor IB, Capitani M, McGowan SJ, Sweeney E, Weber A, et al. : A biallelic mutation in IL6ST encoding the GP130 co-receptor causes immunodeficiency and craniosynostosis. J Exp Med 2017, 214:2547–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahin T, Aschenbrenner D, Cagdas D, Bal SK, Conde CD, Garncarz W, Medgyesi D, Schwerd T, Karaatmaca B, Cetinkaya PG, et al. : Selective loss of function variants in IL6ST cause Hyper-IgE syndrome with distinct impairments of T-cell phenotype and function. Haematologica 2019, 104:609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Béziat V, Tavernier SJ, Chen Y-H, Ma CS, Materna M, Laurence A, Staal J, Aschenbrenner D, Roels L, Worley L, et al. : Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J Exp Med 2020, 217:e20191804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieminen P, Morgan NV, Fenwick AL, Parmanen S, Veistinen L, Mikkola ML, van der Spek PJ, Giraud A, Judd L, Arte S, et al. : Inactivation of IL11 signaling causes craniosynostosis, delayed tooth eruption, and supernumerary teeth. Am J Hum Genet 2011, 89:67-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keupp K, Li Y, Vargel I, Hoischen A, Richardson R, Neveling K, Alanay Y, Uz E, Elcioğlu N, Rachwalski M, et al. : Mutations in the interleukin receptor IL11RA cause autosomal recessive Crouzon-like craniosynostosis. Mol Genet Genom Med 2013, 1:223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer S, Bal SK, Egner W, Allen HL, Raza SI, Ma CA, Gürel M, Zhang Y, Sun G, Sabroe RA, et al. : Loss of the interleukin-6 receptor causes immunodeficiency, atopy, and abnormal inflammatory responses. J Exp Med 2019, 216:1986–1998, 10.1084/jem.20190344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotlarz D, Ziętara N, Uzel G, Weidemann T, Braun CJ, Diestelhorst J, Krawitz PM, Robinson PN, Hecht J, Puchałka J, et al. : Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J Exp Med 2013, 210:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzer E, Kansu A, Sic H, Májek P, Ikincioğullari A, Dogu FE, Prengemann NK, Santos-Valente E, Pickl WF, Bilic I, et al. : Early-onset inflammatory bowel disease and common variable immunodeficiency–like disease caused by IL-21 deficiency. J Allergy Clin Immunol 2014, 133:1651–1659 e12.. [DOI] [PubMed] [Google Scholar]
- 16.Erman B, Bilic I, Hirschmugl T, Salzer E, Çagdas D, Esenboga S, Akcoren Z, Sanal O, Tezcan I, Boztug K: Combined immunodeficiency with CD4 lymphopenia and sclerosing cholangitis caused by a novel loss-of-function mutation affecting IL21 R. Haematologica 2015, 100:e216–e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stepensky P, Keller B, Abuzaitoun O, Shaag A, Yaacov B, Unger S, Seidl M, Rizzi M, Weintraub M, Elpeleg O, et al. : Extending the clinical and immunological phenotype of human interleukin-21 receptor deficiency. Haematologica 2015, 100:e72–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Béziat V, Li J, Lin J-X, Ma CS, Li P, Bousfiha A, Pellier I, Zoghi S, Baris S, Keles S, et al. : A recessive form of hyper-IgE syndrome by disruption of ZNF341-dependent STAT3 transcription and activity. Sci Immunol 2018, 3:eaat4956. •• First description of ZNF341 deficiency (together with reference 19). Demonstrate impaired STAT3 basal expression and autoinduction in ZNF341 deficient cells.
- 19. Frey-Jakobs S, Hartberger JM, Fliegauf M, Bossen C, Wehmeyer ML, Neubauer JC, Bulashevska A, Proietti M, Fröbel P, Nöltner C, et al. : ZNF341 controls STAT3 expression and thereby immunocompetence. Sci Immunol 2018, 3:eaat4941. •• First description of ZNF341 deficiency (together with reference 18).
- 20.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. : The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rapaport F, Boisson B, Gregor A, Béziat V, Boisson-Dupuis S, Bustamante J, Jouanguy E, Puel A, Rosain J, Zhang Q, et al. : Negative selection on human genes underlying inborn errors depends on disease outcome and both the mode and mechanism of inheritance. Proc Natl Acad Sci 2021, 118:e2001248118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lachover-Roth I, Lagovsky I, Shtorch-Asor A, Confino-Cohen R, Reinstein E, Garty B-Z: Hyper IgE syndrome in an isolated population in Israel. Front Immunol 2022, 13:829–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez- Barricarte R, Scott E, Shah I, Stenson PD, Gleeson J, et al. : The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods 2016, 13:109–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.192, Hassanzadeh S, Sadeghi S, Jafari M, Najafi S, Molavi N, Sherkat R: Ciliary and immune dysfunctions and their genetic background in patients with non-cystic fibrosis bronchiectasis in Central Iran. Ir J Med Sci 2022, 192:277–283, 10.1007/s11845-022-02994-z(1971). [DOI] [PubMed] [Google Scholar]
- 25.Chandesris MO, Melki I, Natividad A, Puel A, Fieschi C, Yun L, Thumerelle C, Oksenhendler E, Boutboul D, Thomas C, et al. : Autosomal dominant STAT3 deficiency and hyper-IgE syndrome: molecular, cellular, and clinical features from a French national survey. Medicine 2012, 91:e1-e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gernez Y, Tsuang A, Smith TD, Shahjehan K, Hui Y, Maglione PJ, Cunningham-Rundles C: Hemoptysis in a patient with elevated immunoglobulin E. J Allergy Clin Immunol Pract 2016, 4:1054–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Chen J, Tian Z-Q, Zhang H, Gong R-L, Chen T-X, Hong L: Clinical manifestations and genetic analysis of 17 patients with autosomal dominant hyper-IgE syndrome in Mainland China: new reports and a literature review. J Clin Immunol 2017, 37:166–179. [DOI] [PubMed] [Google Scholar]
- 28.Tavassoli M, Abolhassani H, Yazdani R, Ghadami M, Azizi G, Abdolrahim Poor Heravi S, Moeini Shad T, Kokabee M, Movahedi M, Abdshahzadeh H, et al. : The first cohort of Iranian patients with hyper immunoglobulin E syndrome: a long-term follow-up and genetic analysis. Pediatr Allergy Immunol 2019, 30:469–478. [DOI] [PubMed] [Google Scholar]
- 29.Lorenzini T, Giacomelli M, Scomodon O, Cortesi M, Rivellini V, Dotta L, Soresina A, Dellepiane RM, Carrabba M, Cossu F, et al. : Autosomal-dominant hyper-IgE syndrome is associated with appearance of infections early in life and/or neonatal rash: Evidence from the Italian cohort of 61 patients with elevated IgE. J Allergy Clin Immunol Pract 2019, 7:2072–2075 e4.. [DOI] [PubMed] [Google Scholar]
- 30.Xiang Q, Zhang L, Liu X, Wang S, Wang T, Xiao M, Zhao X, Jiang L: Autosomal dominant hyper IgE syndrome from a single centre in Chongqing, China (2009–2018). Scand J Immunol 2020, 91:e12885. [DOI] [PubMed] [Google Scholar]
- 31.Saikia B, Rawat A, Minz RW, Suri D, Pandiarajan V, Jindal A, Sahu S, Karim A, Desai M, Taur PD, et al. : Clinical profile of hyper-IgE syndrome in India. Front Immunol 2021, 12:626593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora M, Bagi P, Strongin A, Heimall J, Zhao X, Lawrence MG, Trivedi A, Henderson C, Hsu A, Quezado M, et al. : Gastrointestinal manifestations of STAT3-deficient hyper-IgE syndrome. J Clin Immunol 2017, 37:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin L, Wang Y, Sun B, Liu L, Ying W, Wang W, Zhou Q, Hou J, Yao H, Hu L, et al. : The clinical, immunological and genetic features of 12 Chinese patients with STAT3 mutations. Allergy Asthma Clin Immunol 2020, 16:65, 10.1186/s13223-020-00462-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandesris M-O, Azarine A, Ong K-T, Taleb S, Boutouyrie P, Mousseaux E, Romain M, Bozec E, Laurent S, Boddaert N, et al. : Frequent and widespread vascular abnormalities in human signal transducer and activator of transcription 3 deficiency. Circ Cardiovasc Genet 2012, 5:25–34. [DOI] [PubMed] [Google Scholar]
- 35.Avery DT, Deenick EK, Ma CS, Suryani S, Simpson N, Chew GY, Chan TD, Palendira U, Bustamante J, Boisson-Dupuis S, et al. : B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med 2010, 207:155–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ives ML, Ma CS, Palendira U, Chan A, Bustamante J, Boisson-Dupuis S, Arkwright PD, Engelhard D, Averbuch D, Magdorf K, et al. : Signal transducer and activator of transcription 3 (STAT3) mutations underlying autosomal dominant hyper-IgE syndrome impair human CD8 T-cell memory formation and function. J Allergy Clin Immunol 2013, 132:400–411, 10.1016/j.jaci.2013.05.029e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma CS, Avery DT, Chan A, Batten M, Bustamante J, Boisson-Dupuis S, Arkwright PD, Kreins AY, Averbuch D, Engelhard D, et al. : Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood 2012, 119:3997–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, et al. : A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 2011, 35:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson RP, Ives ML, Rao G, Lau A, Payne K, Kobayashi M, Arkwright PD, Peake J, Wong M, Adelstein S, et al. : STAT3 is a critical cell-intrinsic regulator of human unconventional T cell numbers and function. J Exp Med 2015, 212:855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lévy R, Béziat V, Barbieux C, Puel A, Bourrat E, Casanova J-L, Hovnanian A: Efficacy of dupilumab for controlling severe atopic dermatitis in a patient with hyper-IgE syndrome. J Clin Immunol 2020, 40:418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lévy R, Okada S, Béziat V, Moriya K, Liu C, Chai LYA, Migaud M, Hauck F, Ali AA, Cyrus C, et al. : Genetic, immunological, and clinical features of patients with bacterial and fungal infections due to inherited IL-17RA deficiency. Proc Natl Acad Sci 2016, 113:E8277–E8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puel A: Human inborn errors of immunity underlying superficial or invasive candidiasis. Hum Genet 2020, 139:1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmitges FW, Radovani E, Najafabadi HS, Barazandeh M, Campitelli LF, Yin Y, Jolma A, Zhong G, Guo H, Kanagalingam T, et al. : Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res 2016, 26:1742–1752. • Report candidate interaction partners of ZNF341.
- 44. Ichiba M, Nakajima K, Yamanaka Y, Kiuchi N, Hirano T: Autoregulation of the Stat3 gene through cooperation with a cAMP-responsive element-binding protein. J Biol Chem 1998, 273:6132–6138. • First description of STAT3 autoinduction.
- 45. Narimatsu M, Maeda H, Itoh S, Atsumi T, Ohtani T, Nishida K, Itoh M, Kamimura D, Park S-J, Mizuno K, et al. : Tissue-specific autoregulation of thestat3 gene and its role in interleukin-6- induced survival signals in T cells. Mol Cell Biol 2001, 21:6615–6625. • Demonstrate that STAT3 autoinduction is tissue specific.
- 46. Kwon M-J, Ma J, Ding Y, Wang R, Sun Z: Protein kinase C-θ promotes Th17 differentiation via upregulation of Stat3. J Immunol 2012, 188:5887–5897. •• Demonstrate the STAT3 autoinduction is required for Th17 differentiation in mice.
- 47.Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, Holland SM, Schreiber RD, Casanova JL: Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science 2001, 293:300–303. [DOI] [PubMed] [Google Scholar]
- 48.Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, Casrouge A, Yang K, Soudais C, Fieschi C, et al. : Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet 2006, 2:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chapgier A, Kong XF, Boisson-Dupuis S, Jouanguy E, Averbuch D, Feinberg J, Zhang SY, Bustamante J, Vogt G, Lejeune J, et al. : A partial form of recessive STAT1 deficiency in humans. J Clin Invest 2009, 119:1502–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsumura M, Okada S, Sakai H, Yasunaga S, Ohtsubo M, Murata T, Obata H, Yasumi T, Kong XF, Abhyankar A, et al. : Dominant-negative STAT1 SH2 domain mutations in unrelated patients with Mendelian susceptibility to mycobacterial disease. Hum Mutat 2012, 33:1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voyer TL, Sakata S, Tsumura M, Khan T, Esteve-Sole A, Al-Saud BK, Gungor HE, Taur P, Jeanne-Julien V, Christiansen M, et al. : Genetic, immunological, and clinical features of 32 patients with autosomal recessive STAT1 deficiency. J Immunol 2021, 207:133–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al- Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, et al. : Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet 2003, 33:388–391. [DOI] [PubMed] [Google Scholar]
- 53.Han W, Carpenter RL, Cao X, Lo H-W: STAT1 gene expression is enhanced by nuclear EGFR and HER2 via cooperation with STAT3. Mol Carcinog 2013, 52:959–969, 10.1002/mc.21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arboleda VA, Lee H, Dorrani N, Zadeh N, Willis M, Macmurdo CF, Manning MA, Kwan A, Hudgins L, Barthelemy F, et al. : De Novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am J Hum Genet 2015, 96:498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tham E, Lindstrand A, Santani A, Malmgren H, Nesbitt A, Dubbs HA, Zackai EH, Parker MJ, Millan F, Rosenbaum K, et al. : Dominant mutations in KAT6A cause intellectual disability with recognizable syndromic features. Am J Hum Genet 2015, 96:507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dagoneau N, Scheffer D, Huber C, Al-Gazali LI, Rocco MD, Godard A, Martinovic J, Raas-Rothschild A, Sigaudy S, Unger S, et al. : Null leukemia inhibitory factor receptor (LIFR) mutations in Stüve-Wiedemann/Schwartz-Jampel type 2 syndrome. Am J Hum Genet 2004, 74:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y-H, Grigelioniene G, Newton PT, Gullander J, Elfving M, Hammarsjö A, Batkovskyte D, Alsaif HS, Kurdi WIY, Abdulwahab F, et al. : Absence of GP130 cytokine receptor signaling causes extended Stüve-Wiedemann syndrome. J Exp Med 2020, 217:e20191306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.