Abstract
Dextransucrase (DSR-S) from Leuconostoc mesenteroides NRRL B-512F is a glucosyltransferase that catalyzes synthesis of soluble dextran from sucrose. In the presence of efficient acceptor molecules, such as maltose, the reaction pathway is shifted toward glucooligosaccharide synthesis. Like glucosyltransferases from oral streptococci, DSR-S possesses a C-terminal glucan-binding domain composed of a series of tandem repeats. In order to determine the role of the C-terminal region of DSR-S in dextran or oligosaccharide synthesis, four DSR-S genes with deletions at the 3′ end were constructed. The results showed that the C-terminal region modulated the initial velocity of dextran synthesis but that the Km for sucrose, the optimum pH, and the activation energy were all unaffected by the deletions. The C-terminal domain modulated the rate of oligosaccharide synthesis whatever acceptor molecule was used (a good acceptor molecule such as maltose or a poor acceptor molecule such as fructose). The C-terminal domain seemed to play no role in the catalytic process in dextran and oligosaccharide synthesis. In fact, it seems that the role of the C-terminal domain of DSR-S may be to facilitate the translation of dextran and oligosaccharides from the catalytic site.
Dextransucrase (DSR-S) from Leuconostoc mesenteroides NRRL B-512F is a 1,527-amino-acid glucosyltransferase (EC 2.4.1.5) that catalyzes the synthesis from sucrose of a soluble dextran in which more than 95% of the d-glucosyl units are α(1-6) linked (21, 34). This dextran has many important industrial and medical uses (4). In the presence of efficient acceptor molecules, such as maltose, the reaction pathway is shifted toward oligosaccharide synthesis (16, 21).
Analysis of the DSR-S sequence revealed that in its N-terminal portion the region extending from amino acid 268 to amino acid 1134 is homologous to the corresponding region of glucosyltransferases (GTFs) from oral streptococci (21). In GTFs, this region is essential for maintaining glucan synthesis activity (1, 8) and includes a putative catalytic site for sucrose hydrolysis (12, 22). Essential amino acids have been identified in DSR-S (21), and structural predictions have suggested that like α-amylases, the catalytic domains of GTFs and DSR-S are members of the (β/α)8 barrel-containing protein family (19).
In GTFs the glucan-binding region is located in the carboxy-terminal portion, which contains about 300 to 400 amino acids in these enzymes (8, 35). The glucan-binding region does not participate in sucrose splitting (1, 12, 35) but strongly modulates activity (1, 8, 11, 18). It contains a series of homologous repeating units consisting of about 30 amino acids, which are also found in Streptococcus mutans glucan-binding protein (3). A number of types of repeats have been identified on the basis of sequence similarities, and these repeats have been designated A, B, C, and D repeats (8, 9, 33). All of these repeats contain the same structural element, the YG repeat, which is characterized by the presence of clusters of aromatic residues, the predominance of polar and turn-promoting residues at certain positions, and the occurrence of a glycine residue three or four residues downstream from the aromatic cluster (9). The numbers and patterns of repeats vary in different streptococcal enzymes, but no correlation between these repeats and enzyme function has been found. However, four A repeats are required for functioning of S. mutans GTF-S, which synthesizes soluble glucan composed of d-glucosyl units that are predominantly α(1-6) linked (18). In contrast, only two A repeats are necessary for S. mutans GTF-I, which synthesizes insoluble mutan, a polysaccharide composed of d-glucosyl units that are α(1-3) linked (1, 8, 11). The C terminus may also influence the structure of the glucan produced, but this has not been well studied (23, 32). Moreover, the involvement of the glucan-binding domain in the catalytic mechanism is still not clearly defined, nor is the role of the glucan-binding domain in oligosaccharide synthesis understood.
In the present paper we describe a sequence analysis of the C-terminal portion of L. mesenteroides NRRL B-512F DSR-S and the effect of sequential deletions on the biochemical properties of DSR-S. We paid particular attention to characterizing the dextran and oligosaccharide synthesis activities of mutants in order to better understand the role of the C-terminal region of DSR-S in the catalytic mechanism.
MATERIALS AND METHODS
Molecular techniques.
Escherichia coli transformations were carried out by the method of Hanahan (10). Restriction or modifying enzymes were used as described by the enzyme supplier (New England Biolabs, Inc.). DNA purification, digestion, and agarose gel electrophoresis were performed by standard procedures (20).
Construction of plasmids.
Genomic DNA from L. mesenteroides NRRL B-512F was extracted as described by Phalip et al. (25) and was used as a template to clone the full-length 4,580-bp dsr-S gene in plasmid pTrc99A in order to produce plasmid pBF7 (21) by using sequence information obtained by Wilke-Douglas et al. (34) and deposited under GenBank accession no. I09598. Briefly, a set of primers (a 5′ end primer, 5′-ATAGAAGAGAGCTCATTATAAGGAGAAAATTTATG, containing a SacI-engineered restriction site, and a 3′ end primer, 5′-TATATATCTAGAAAGCTTATGCTGACACAG, containing an XbaI-engineered restriction site) was designed to PCR amplify a 4.8-kb fragment containing the entire dsr-S gene. Plasmid pBF7 was obtained by ligating the PCR product and pTrc99A (2) that had been double digested with XbaI and SacI. The SacI cleavage site was positioned in the primer sequence so that the ATG of the vector could be used as the start codon.
In order to construct a dsr-S gene having a 255-bp deletion at the 3′ end (and coding for DSR-S1) (Fig. 1), a 4.3-kb SacI-BamHI fragment was isolated from pBF7 and cloned in pTrc99A that had been double digested with the same enzymes. To construct a dsr-S gene having a 510-bp deletion at the 3′ end (and coding for DSR-S3), a 2.7-kb SpeI-EcoRI fragment carrying the part of dsr-S lacking the 3′ end was first inserted into pBS-SK (Stratagene) to obtain plasmid pKS7. Then the 2.7-kb SpeI-KpnI fragment from pKS7 and the 1.4-kb SacI-SpeI fragment from pBF7, corresponding to the 5′ end of dsr-S, were inserted into pTrc99A.
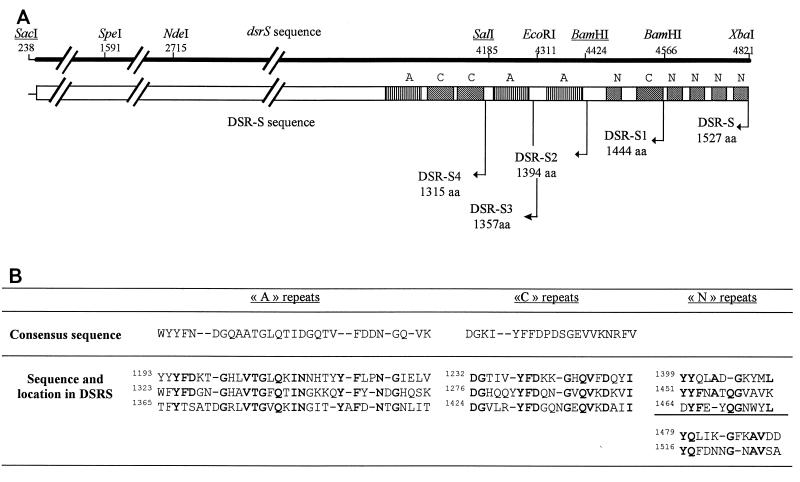
FIG. 1.
(A) Schematic representation of the structure of the C-terminal domain of DSR-S. The cleavage sites on the dsrS sequence are shown, as are the ends of truncated enzymes on the DSR-S sequence. Corresponding protein sizes are also indicated. aa, amino acids. (B) Sequences of A repeats, C repeats, and N repeats. The consensus sequences (7, 8) used to identify the A and C repeats are shown. Boldface type indicates conserved amino acid residues.
In order to construct dsr-S genes having 409- and 636-bp deletions (and coding for DSR-S2 and DSR-S4, respectively), these deleted 3′ ends of dsr-S were PCR amplified and inserted into pTrc99A with the 5′ end of dsr-S. In these two cases, the PCR consisted of annealing at 48°C, 1 min, extension at 72°C for 2 min, and denaturation at 94°C for 1 min for 4 cycles and annealing at 50°C for 1 min, extension at 72°C for 2 min, and denaturation at 94°C for 1 min for the subsequent 20 cycles. Vent polymerase (Biolabs Inc.) was used as the thermostable polymerase.
In order to clone the gene coding for DSR-S2, the 3′ end of dsr-S was PCR amplified with a 5′ end primer (5′-AGCTACAATTAGAGGATGG) located upstream from an NdeI restriction site identical to residues 2667 to 2696, which corresponded to ELQLEDG in the protein sequence, and a 3′ primer (5′-CTGATTTGGATCCTAATTGCCTGTGTT) complementary to residues 4408 to 4434, which corresponded to NTGNLITNQ in the protein sequence. An engineered BamHI restriction site was introduced into the latter primer by substituting GATCAC for GGATCC. Also, a stop codon (CTA) was inserted just after the restriction site. Following PCR, the 1.7-kb product obtained was double digested with NdeI and BamHI. It was then ligated into pTrc99A that had been double digested with SacI and BamHI along with a 2.5-kb SacI-NdeI fragment from pBF7 that corresponded to the 5′ end of dsr-S.
In order to clone the gene coding for DSR-S4, the 3′ end of dsr-S was PCR amplified with a 5′ end primer (5′-AGCTACAATTAGAGGATGG) located upstream from the NdeI restriction site and a 3′ end primer (5′-ACTATTGTCGACTTAACGTAGGATAGC) complementary to residues 4171 to 4197, which corresponded to AILRYVQNS in the protein sequence. An engineered SalI restriction site was introduced into the latter primer by substituting GTGCAA for GTCGAC. Also, a stop codon (TTA) was inserted just after the restriction site. Following amplification, the 1.5-kb fragment was digested with NdeI and SalI. It was ligated into pTrc99A that had been double digested with SacI and SalI along with a 2.5-kb SacI-NdeI fragment from pBF7 that corresponded to the 5′ end of dsr-S.
Preparation of wild-type and mutant DSR-S.
E. coli DH1 containing the dsr-S gene and E. coli DH1 containing the deleted dsr-S genes were grown as described previously (21) in 400 ml of Luria-Bertani medium supplemented with 100 mM Tris-HCl (pH 6.4) and 0.1 mg of ampicillin per ml at 30°C. Lactose was added to final concentration of 20 g/liter; this compound was used as an inducer of the trc promoter. Cells were harvested after 13 h of growth by centrifugation, resuspended in 20 mM sodium acetate buffer (pH 5.4) containing 1% (vol/vol) Triton X-100 and 1 mM phenylmethylsulfonyl fluoride, and sonicated. Debris and unbroken cells were pelleted, and the supernatant was used as the source of enzymes. Protein concentrations were determined by the method of Bradford (6) with bovine serum albumin as the standard.
Enzyme activity assays.
In order to determine dextran synthesis activity, reactions were performed at 30°C in 20 mM sodium acetate buffer (pH 5.4) containing 0.05 g of CaCl2 per liter and 100 g of sucrose per liter. Activity was assayed by the dinitrosalicylic acid method (30). One unit was defined as the amount of enzyme that catalyzed the formation of 1 μmol of fructose per min under these conditions. Activity was also determined by measuring the quantity of soluble dextran by high-performance liquid chromatography with gel permeation (type SI-100 column; Merck) by using a Hewlett-Packard series 1050 system consisting of a pump, an injector, and a model HP 1047A refractometer. The eluant was ultrapure water at a flow rate of 0.5 ml/min.
The effect of pH on activity was measured in the presence of 100 g of sucrose per liter at 30°C by using 20 mM sodium acetate buffer at pH values ranging from 4.3 to 6.6.
Oligosaccharide synthesis reactions were performed at 26°C to reduce enzyme thermal denaturation. Oligosaccharide synthesis in the presence of maltose acceptor was carried out in 20 mM sodium acetate buffer (pH 5.4) containing 0.05 g of CaCl2 per liter, 50 g of sucrose per liter, and 25 or 10 g of maltose per liter, giving ratios of sucrose concentration to maltose concentration of 2 or 5. In the presence of a fructose acceptor, sucrose was used at a concentration of 50 g/liter and fructose was used at a concentration of 50 g/liter, which gave a ratio of sucrose concentration to fructose concentration of 1. Oligosaccharides were analyzed by high-performance liquid chromatography with a type C18 column by using a Hewlett-Packard series 1050 system and ultrapure water as eluant at a flow rate of 0.5 ml/min.
Electrophoresis analysis.
Equivalent quantities of total proteins from cell extracts prepared from E. coli transformants expressing either full-length dsr-S genes or dsr-S genes having deletions or transformants simply carrying pTrc99A were denatured for 2 min at 95°C, separated by 7% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (17), and stained with Coomassie blue R-350 (Pharmacia).
Detection of reaction product in gel.
After SDS-PAGE, the gel was washed three times with 20 mM sodium acetate buffer (pH 5.4) containing 0.1% (vol/vol) Triton X-100 at 4°C to eliminate the SDS. The gel was incubated in the same buffer at 4°C, 100 g of sucrose per liter was added, and the active bands were detected by the formation of dextran as a white polymer inside the gel.
RESULTS
Construction of dsr-S genes having deletions and expression in E. coli.
Analysis of the C-terminal domain of DSR-S revealed that this region contains several types of repeats (Fig. 1). Repeats homologous to the A and C repeats identified previously in S. mutans glucan-binding protein (3) were identified at amino acids 1193 to 1396 (Fig. 1). The carboxy end of this domain was composed of a repeat homologous to the C repeat and a series of small repeats containing the characteristic elements of the YG repeats, such as a glycine residue three or four residues downstream from the aromatic cluster (9) (Fig. 1). However, these repeats are not highly conserved (Fig. 1). The fact that a C-terminal domain ends with repeating units that are not very well conserved has not been encountered previously in other glucan-binding proteins or GTFs. We have named these repeats N repeats.
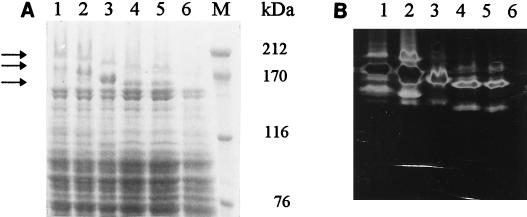
In order to determine the function of these repeating units during enzyme activity, four DSR-S deletion derivatives were constructed as described in Materials and Methods. DSR-S1 contained the first three A repeats, the first two C repeats, and only one N repeat, while DSR-S2 contained the first three A repeats and the first two C repeats. DSR-S3 contained the first two A repeats and the first two C repeats, while DSR-S4 contained the first A repeat and the first two C repeats (Fig. 1). All truncated genes were cloned into pTrc99A (2). Sonicated extracts of each deletion derivative obtained from cultures of transformed E. coli DH1 were used as sources of enzymes. SDS-PAGE staining of these extracts revealed that dsr-S and the deletions were expressed at similar levels (Fig. 2A). The typical DSR-S pattern was observed with DSR-S, and different active forms were produced, which corresponded to bands at 200, 180, and 160 kDa (21). The molecular masses of the three bands decreased with a decrease in DNA length. For DSR-S and the four deletion derivatives, these protein bands exhibited DSR-S activity that was detectable on SDS-PAGE gels (Fig. 2B).
FIG. 2.
Expression of DSR-S and truncated enzymes in E. coli DH1. (A) SDS-PAGE analysis of wild-type and truncated proteins. (B) Assay for soluble glucan synthesis following SDS-PAGE. Lanes 1, wild-type DSR-S; lanes 2, DSR-S1; lanes 3, DSR-S2; lanes 4, DSR-S3; lanes 5, DSR-S4; lanes 6, cell extract from E. coli DH1(pTrc99A); lane M, molecular weight standards.
Characterization of DSR-S and truncated DSR-S activities.
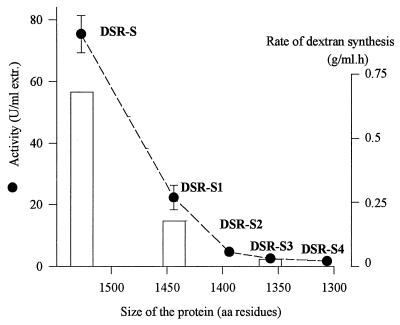
Activity assays carried out with DSR-S and the truncated enzymes indicated that removal of the C-terminal end from DSR-S resulted in a strong decrease in activity, as determined by the release of fructose (Fig. 3). The effects of deletions on dextran synthesis activity seemed to be the same; similar decreases in fructose-releasing and dextran synthesis activities were observed with DSR-S, DSR-S1, and DSR-3 (Fig. 3). The lack of the last six repeats, corresponding to a loss of only 85 amino acids, resulted in the largest decrease in activity. When DSR-S2 was compared to DSR-S4, the loss of activity was less drastic. In order to further characterize the effects of the deletions and because DSR-S2, DSR-S3, and DSR-S4 exhibited similar activities, only DSR-S1 and DSR-S3 were used in a comparison of properties with the properties of the full-length enzyme, DSR-S.
FIG. 3.
Effect of DSR-S deletions on activity in the presence of sucrose. The dashed line shows the correlation between the size of the enzyme and activity. The histogram shows the rates of dextran synthesis determined for DSR-S, DSR-S1, and DSR-S3. aa, amino acid; extr., extract.
Effects of deletions on Km and Vmax values.
In order to determine whether the deletions that altered activity also affected the binding of sucrose, the Km value for sucrose of full-length DSR-S was compared with the Km values of two truncated enzymes, DSR-S1 and DSR-S3. These values were determined from Lineweaver-Burk plots by determining initial velocities in the presence of 100 to 2.5 g of sucrose per liter. The Km values were very similar (26, 28, and 32 mM for DSR-S, DSR-S1, and DSR-S3, respectively). However the Vmax value was significantly affected by the deletions; the Vmax values for DSR-S, DSR-S1, and DSR-S3 were 75, 21.1, and 1.9 U/ml, respectively.
Effect of temperature on DSR-S activity.
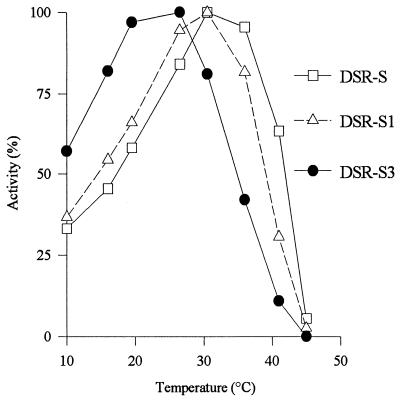
As shown in Fig. 4, DSR-S and DSR-S1 exhibited the same optimum temperature (30°C), whereas DSR-S3 had an optimum temperature of 26°C. DSR-S3, in which 170 amino acid residues in the C-terminal portion of DSR-S were deleted, was much more sensitive to temperature denaturation than DSR-S or DSR-S1. The three preparations had the same profile for activation by temperature (Fig. 4). The activation energies were determined by using Arrhenius plots and were of the same order of magnitude (39.0, 40.0, and 39.2 kJ/mol for DSR-S, DSR-S1, and DSR-S3, respectively).
FIG. 4.
Effect of temperature on DSR-S, DSR-S1, and DSR-S3 activities. The highest level of activity observed for each enzyme was defined as 100% activity for that enzyme.
Effect of pH on activity of truncated enzymes.
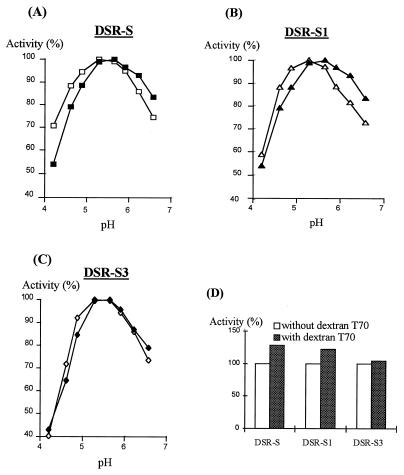
The effect of pH on activity of truncated enzymes was determined with DSR-S, DSR-S1, and DRS-S3. Exogenous soluble T70 dextran was also added at a concentration of 5 g/liter to determine if the effect on activity observed with dextransucrase from L. mesenteroides NRRL B-512F (13) was modified by deletions (Fig. 5). In the absence of exogenous dextran, the pH range for maximal activity was the same, pH 5.1 to 5.6, for the three enzymes. At low pH values, DSR-S3 was also much more sensitive to pH inhibition than DSR-S or DSR-S1 was (Fig. 5C).
FIG. 5.
Effect of pH on DSR-S, DSR-S1, and DSR-S3 activities. DSR-S (A), DSR-S1 (B), and DSR-S3 (C) activities were determined at different pH values in the presence (solid symbols) or in the absence (open symbols) of 5 g of dextran T70 per liter. The highest level of activity observed for each enzyme with or without dextran T-70 was defined as 100% activity for that enzyme. (D) Comparison of the activator effects of dextran T70 on DSR-S, DSR-S1, and DSR-S3 activities. The highest level of activity observed at the optimum pH for each enzyme was defined as 100% activity for that enzyme.
In the presence of exogenous dextran, the pH range for maximal activity was 5.3 to 5.7 for DSR-S and DSR-S1 (Fig. 5A and B). This effect was identified previously with the dextransucrase expressed from L. mesenteroides NRRL B-512F (13). In the presence of dextran, a shift in the pH range for maximal activity to higher values was not observed with DSR-S3. Also, exogenous dextran was an activator of DSR-S and DSR-S1 (Fig. 5D). With DSR-3, no activator effect was observed in the presence of dextran (Fig. 5D).
Oligosaccharide synthesis in the presence of maltose.
As previously observed with the dextran synthesis reaction, all of the deletions affected acceptor reaction kinetics (Table 1). Addition of maltose increased the initial velocity of the reaction with DSR-S. Deletions did not suppress this effect. With DSR-S1 this positive effect of maltose on activity seemed to be the same as the effect observed with DSR-S. With DSR-S3, maltose had a stronger activator effect (Table 1). With DSR-S, the yield of oligosaccharide synthesis decreased when the ratio of sucrose concentration to maltose concentration increased (Table 2). The increase in the ratio of sucrose concentration to maltose concentration also affected the yield of oligosaccharides produced by truncated enzymes. The yield was the same with all enzymes.
TABLE 1.
Effects of deletions on the initial velocities of reactions in the presence of sucrose, in the presence of sucrose plus maltose, and in the presence of sucrose plus fructose
| Sugar(s) | Ratio | Enzyme activities (U/ml)
|
||
|---|---|---|---|---|
| DSR-S | DSR-S1 | DSR-S3 | ||
| Sucrosea | 71.5 (100)b | 17.3 (100) | 4.4 (100) | |
| Sucrose + maltose | 5c | 96.9 (135) | 22.9 (132) | 10.2 (231) |
| 2d | 137.5 (192) | 32.7 (189) | 15.9 (363) | |
| Sucrose + fructose | 1e | 56.6 (79) | 14.4 (89) | 6.5 (147) |
Each reaction mixture contained 50 g of sucrose per liter (dextran synthesis reaction).
The values in parentheses are percentages; 100% was defined as the initial velocity observed in the absence of an acceptor.
Each reaction mixture contained 50 g of sucrose per liter and 10 g of maltose per liter.
Each reaction mixture contained 50 g of sucrose per liter and 25 g of maltose per liter.
Each reaction mixture contained 50 g of sucrose per liter and 50 g of fructose per liter.
TABLE 2.
Effects of deletions on oligosaccharide yields in the presence of sucrose plus maltose and on leucrose yields in the presence of sucrose plus fructose
| Sugars | Ratio | Oligosaccharide yields (%) with:
|
Leucrose yields (%) with:
|
||||
|---|---|---|---|---|---|---|---|
| DSR-S | DSR-S1 | DSR-S3 | DSR-S | DSR-S1 | DSR-S3 | ||
| Sucrose + maltose | 5a | 63 | 62 | 63 | |||
| 2b | 93 | 91 | 95 | ||||
| Sucrose + fructose | 1c | 21 | 27 | 37 | |||
Each reaction mixture contained 50 g of sucrose per liter and 10 g of maltose per liter.
Each reaction mixture contained 50 g of sucrose per liter and 25 g of maltose per liter.
Each reaction mixture contained 50 g of sucrose per liter and 50 g of fructose per liter.
All of the oligosaccharides synthesized by DSR-S are produced by DSR-S1 and DSR-S3 (Table 3). In the presence of maltose, oligosaccharides from panose to oligodextran having a degree of polymerization of 6 (OD6) were produced in all cases. However, deletions influenced the size distribution of the oligosaccharides produced. With DSR-S, the percentage of OD6 obtained when sucrose was completely consumed was found to be 16% when the ratio of sucrose concentration to maltose concentration was 5. With DSR-S3, this percentage was 30% when the ratio of sucrose concentration to maltose concentration was 5 (Table 3). The lack of the C-terminal region seemed to facilitate the production of oligosaccharides having higher degrees of polymerization.
TABLE 3.
Effects of deletions on the sizes of oligosaccharides produced in the presence of maltose and sucrose
| Enzymea | % Composition of oligosaccharides produced
|
|||
|---|---|---|---|---|
| Panose | OD4 | OD5 | OD6 | |
| DSR-S | 13.1 | 33.3 | 37.1 | 16.5 |
| DSR-S1 | 13.9 | 32.9 | 38.6 | 14.6 |
| DSR-S3 | 6.5 | 23.3 | 40.2 | 30.0 |
Each reaction mixture contained 50 g of sucrose per liter and 10 g of maltose per liter.
Effects of deletions on leucrose synthesis in the presence of fructose.
With dextranscurase produced by L. mesenteroides NRRL B-512F, fructose is a weak acceptor which slows down the overall reaction rate, and the leucrose produced [5-O-(α-d-glucopyranosyl)-d-fructopyranose] is not an acceptor (5, 14). With DSR-S, the initial velocity of the reaction in the presence of fructose was lower than the initial velocity of the reaction in the absence of fructose, (Table 1), and the leucrose yield in the presence of fructose was 21% (Table 2). With DSR-S1 and DSR-S3, the yields were greater, showing that deletions favored leucrose synthesis at the expense of dextran synthesis. Moreover, the inhibitory effect on the reaction rate was less with DSR-S1 than with DSR-S. Surprisingly, with DSR-S3, fructose, like maltose, had an activating effect (Table 1).
DISCUSSION
Like all GTFs whose sequences are known (9, 33), the C-terminal domain of DSR-S from L. mesenteroides NRRL B-512F is composed of a series of repeats homologous to A and C repeats. The terminal portion contains repeat motifs which are not highly conserved but possess the main characteristics of the YG repeats (9) that we call N repeats. This structure has not been identified in streptoccocal GTFs. In order to gain further insight into the role of C-terminal repeats in GTF activity, the effects of engineered deletions on both dextran and oligosaccharide synthesis were examined.
As with streptococcal GTFs (1, 8, 11, 18, 32), the C-terminal portion of DSR-S is crucial for maintaining a high initial rate of consumption of sucrose and a high initial rate of synthesis of dextran. There is a direct correlation between dextran synthesis and sucrose consumption, which indicates that deletions have no effect on the ratio of sucrose hydrolysis to polymer synthesis. This suggests that the C-terminal domain of DSR-S does not facilitate the transfer of glucosyl residues on the dextran chain.
The binding sites for sucrose and dextran are separate sites on DSR-S (15), and the catalytic site responsible for cleavage of sucrose is located in its N-terminal region (19, 21, 22). The fact that deletions do not have a drastic effect on the Km for sucrose suggests that they do not alter the ability of DSR-S to bind the substrate sucrose. Moreover, the activation energy of the dextran synthesis reaction is not affected by deletions, which shows that the energy level of the transition state is not modified. The optimum pH does not change. The distribution of local charges in the catalytic site of DSR-S and the distribution of these charges in the truncated enzymes are the same, which indicates that the sucrose binding site is not directly affected by deletions.
The reaction velocity is the only parameter which is strongly influenced by deletions; deletions in the C terminus of DSR-S result in decreases in the initial reaction rate. Although the truncated proteins seem to be more sensitive to thermal denaturation than DSR-S is, this difference cannot explain the decreases in the initial rate observed with the truncated enzymes. The presence of the three first repeats is sufficient to maintain a detectable dextran synthesis activity, but the last 85 amino acid residues are particularly crucial for activity. However, without additional evidence it is not possible to say whether the size of the C-terminal portion of DSR-S alone is crucial for maintaining activity or whether there is a direct correlation between the absence of a nontypical N series of repeats in the C-terminal domain and the decrease in activity.
In the case of the dextran synthesis reaction, it has been proposed that translation of the growing dextran is the limiting step in the reaction, perhaps because of steric hindrance (7). Like the initial velocity of the reaction, the glucan-binding properties of the C-terminal domain are also altered by deletions; the activator effect of dextran T70 does not occur with DSR-S3. Thus, because of its glucan-binding properties, the C-terminal domain of DSR-S could have a positive effect on the reaction velocity by making translation of the growing dextran from the catalytic site easier.
The effect of deletions on oligosaccharide synthesis has not been examined previously, but such a study could provide interesting information because mechanisms of synthesis are different; transfer of glucosyl residues occurs at the nonreducing ends of oligosaccharides, while synthesis of polymers occurs at the reducing ends (27, 28). Maltose and fructose were used as examples of good and bad acceptors, respectively. In both cases, the velocity of oligosaccharide synthesis was also dramatically affected by deletions. As in the dextran synthesis reaction, the C-terminal portion of DSR-S is crucial for maintaining a high initial rate of oligosaccharide production.
The activation of DSR-S by maltose described previously (24, 26) is even more pronounced with truncated enzymes. Paul et al. interpreted this effect as the result of a change in a limiting step of the reaction (24). In the presence of maltose, the formation of a d-glucosyl–enzyme complex before sugar is transferred to the acceptor should be the limiting step (24). This effect is observed with deleted proteins, which supports the idea that the kinetics of d-glucosyl–enzyme complex formation in the presence of maltose is not modified by deletions. The overall yields of oligosaccharides are equivalent in the presence of maltose. As previously described for dextransucrase produced by L. mesenteroides NRRL B-512F (29), only the ratio of sucrose concentration to maltose concentration had an effect on these yields. According to the proposed mechanisms for the acceptor reaction with maltose (28, 31), this supports the hypothesis that the C-terminal portion is not involved in the process which results in oligosaccharide formation.
However, the distributions of the products are not similar. A 170-amino-acid deletion in the C-terminal domain of DSR-S results in an increase in the percentage of the longest oligosaccharides produced (OD6). When the C-terminal domain is truncated, the oligosaccharides may stay longer in the microenvironment of the catalytic site, which allows the oligosaccharide chain to elongate. Thus, it seems that the role of the C-terminal domain of DSR-S in oligosaccharide synthesis is to facilitate removal of the oligosaccharides from the catalytic site. In this case, the C-terminal glucan-binding domain of DSR-S also appears to be an oligosaccharide-binding domain.
As previously described (5, 14), the presence of the poor acceptor fructose decreases the reaction rate of DSR-S. Deletions tend to suppress this inhibitory effect, and fructose is a strong acceptor with DSR-S3; both the reaction rate and the yield increase in its presence. Böker et al. (5) have proposed that the leucrose synthesis reaction is slower than the dextran synthesis reaction but inhibits dextran chain elongation. With DSR-S3, the leucrose synthesis reaction may be faster than the dextran synthesis reaction. Thus, the reaction velocity would not be limited by dextran elongation but would be limited by the step that occurs in the acceptor reaction in the presence of a strong acceptor, such as maltose.
ACKNOWLEDGMENTS
This study was supported by the European Union as part of the project “Structure-function relationships of glucosyltransferases,” by BIOTECH contract BIO2CT 943071, and by Région Midi-Pyrénées.
We thank R. R. Russell for critically reading the manuscript.
REFERENCES
- 1.Abo H, Matsumura T, Kodama T, Ohta H, Fukui K, Kato K, Kagawa H. Peptide sequences for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthetase) J Bacteriol. 1991;173:989–996. doi: 10.1128/jb.173.3.989-996.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amman E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 3.Banas J A, Russell R R B, Ferretti J J. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect Immun. 1990;58:667–673. doi: 10.1128/iai.58.3.667-673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker P E, Ganetsos G, Ajongwen N J. A novel approach to the production of clinical-grade dextran. J Chem Technol Biotechnol. 1993;57:21–26. doi: 10.1002/jctb.280570105. [DOI] [PubMed] [Google Scholar]
- 5.Böker M, Jördening H-J, Buchholz K. Kinetics of leucrose formation from sucrose by dextransucrase. Biotechnol Bioeng. 1994;43:856–864. doi: 10.1002/bit.260430904. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Ebert K H, Schenk G. Mechanism of biopolymer growth: the formation of dextran and levan. Adv Enzymol. 1968;30:179–221. doi: 10.1002/9780470122754.ch4. [DOI] [PubMed] [Google Scholar]
- 8.Ferretti J J, Gilpin M L, Russell R R B. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus Mfe28. J Bacteriol. 1987;169:4271–4278. doi: 10.1128/jb.169.9.4271-4278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giffard P M, Jacques N A. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J Dent Res. 1994;73:1133–1141. doi: 10.1177/00220345940730060201. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 11.Kato C, Kuramitsu H K. Carboxy-terminal deletion analysis of the Streptococcus mutans glucosyltransferase-I enzyme. FEMS Microbiol Lett. 1990;72:299–302. doi: 10.1016/0378-1097(90)90321-g. [DOI] [PubMed] [Google Scholar]
- 12.Kato C, Nakano Y, Lis M, Kuramitsu H K. Molecular genetic analysis of the catalytic site of Streptococcus mutans glucosyltransferases. Biochem Biophys Res Commun. 1992;189:1184–1188. doi: 10.1016/0006-291x(92)92329-v. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M, Yokoyama I, Matsuda K. Activation of dextransucrase from Leuconostoc mesenteroides by the substrate, dextran. Agric Biol Chem. 1984;48:221–223. [Google Scholar]
- 14.Kobayashi M, Yokoyama I, Matsuda K. Effectors differently modulating the dextransucrase activity of Leuconostoc mesenteroides evaluated by inhibition kinetics. Agric Biol Chem. 1985;49:3189–3195. [Google Scholar]
- 15.Kobayashi M, Yokoyama I, Matsuda K. Substrate binding sites of Leuconostoc dextransucrase evaluated by inhibition kinetics. Agric Biol Chem. 1986;50:2585–2590. [Google Scholar]
- 16.Koepsell H J, Tsuchiya H M, Hellman N N, Kasenko A, Hoffmann C A, Shape E S, Jackson R W. Enzymatic synthesis of dextran. Acceptor specificity and chain initiation. J Biol Chem. 1952;200:793–801. [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lis M, Shiroza T, Kuramitsu H K. Role of the C-terminal direct repeating units of the Streptococcus mutans glucosyltransferase-S in glucan binding. Appl Environ Microbiol. 1995;61:2040–2042. doi: 10.1128/aem.61.5.2040-2042.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacGregor A E, Jespersen H M, Svensson B. A circularly permuted α-amylase type α/β barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 1996;378:263–266. doi: 10.1016/0014-5793(95)01428-4. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 21.Monchois V, Remaud-Simeon M, Russell R R B, Monsan P, Willemot R M. Characterization of Leuconostoc mesenteroides NRRL B-512F dextransucrase (DSR-S) and identification of amino-acid residues playing a key role in enzyme activity. Appl Microbiol Biotechnol. 1997;48:465–472. doi: 10.1007/s002530051081. [DOI] [PubMed] [Google Scholar]
- 22.Mooser G, Hefta S A, Paxton R J, Shively J E, Lee T D. Isolation and sequence of an active-site peptide containing a catalytic aspartic acid from two Streptococcus sobrinus glucosyltransferases. J Biol Chem. 1991;266:8916–8922. [PubMed] [Google Scholar]
- 23.Nakano Y J, Kuramitsu H K. Mechanism of Streptococcus mutans glucosyltransferases: hybrid-enzyme analysis. J Bacteriol. 1992;174:5639–5646. doi: 10.1128/jb.174.17.5639-5646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul F, Oriol E, Auriol D, Monsan P. Acceptor reaction of a highly purified dextransucrase with maltose and oligosaccharides. Application to the synthesis of controlled-molecular-weight dextrans. Carbohydr Res. 1986;149:433–441. [Google Scholar]
- 25.Phalip V, Dartois V, Schmitt P, Divies C. Cloning of the d-lactate deshydrogenase gene from Leuconostoc mesenteroides subsp. cremoris. Biotechnol Lett. 1994;16:221–226. [Google Scholar]
- 26.Reh K-D, Jördening H J, Buchholz K. Kinetics of oligosaccharide synthesis by dextransucrase. Ann N Y Acad Sci. 1994;613:723–729. [Google Scholar]
- 27.Robyt J F, Kimble B K, Walseth T F. The mechanism of dextransucrase action. Direction of dextran biosynthesis. Arch Biochem Biophys. 1974;165:634–640. doi: 10.1016/0003-9861(74)90291-4. [DOI] [PubMed] [Google Scholar]
- 28.Robyt J F, Walseth T F. The mechanism of acceptor reaction of Leuconostoc mesenteroides NRRL B-512F dextransucrase. Carbohydr Res. 1978;61:433–445. doi: 10.1016/s0008-6215(00)84503-6. [DOI] [PubMed] [Google Scholar]
- 29.Robyt J F, Eklund S H. Relatives quantitative effects of acceptors in the reaction of Leuconostoc mesenteroides B 512-F dextransucrase action. Carbohydr Res. 1983;121:279–286. doi: 10.1016/0008-6215(83)84024-5. [DOI] [PubMed] [Google Scholar]
- 30.Sumner J B, Howell S F. A method for determination of invertase activity. J Biol Chem. 1935;108:51–54. [Google Scholar]
- 31.Tarivseven A, Robyt J F. Inhibition of dextran synthesis by acceptor reactions of dextransucrase and the demonstration of a separate acceptor binding site. Carbohydr Res. 1992;225:321–329. [Google Scholar]
- 32.Vickerman M M, Sulavik M C, Minick P E, Clewell D B. Changes in the carboxy-terminal repeat region affect extracellular activity and glucan products of Streptococcus gordonii glucosyltransferase. Infect Immun. 1996;64:5117–5128. doi: 10.1128/iai.64.12.5117-5128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Von Eichel-Streiber C, Sauerborn M, Kuramitsu H K. Evidence for a modular structure of the homologous repetitive C-terminal carbohydrate-binding sites of Clostridium difficile toxins and Streptococcus mutans glucosyltransferases. J Bacteriol. 1992;174:6707–6710. doi: 10.1128/jb.174.20.6707-6710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilke-Douglas, M., J. T. Perchorowicz, C. M. Houck, and B. R. Thomas. December 1989. Methods and compositions for altering physical characteristics of fruit and fruit products. WO 89/12386.
- 35.Wong C, Stanley A H, Paxton R J, Shively J E, Mooser G. Size and subdomain architecture of the glucan-binding domain of sucrose: 3-α-d-glucosyltransferase from Streptococcus sobrinus. Infect Immun. 1990;58:2165–2170. doi: 10.1128/iai.58.7.2165-2170.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]