Abstract
There have been relatively few small molecules developed with direct activity against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Two existing antimalarial drugs, pyronaridine and quinacrine, display whole cell activity against SARS-CoV-2 in A549 + ACE2 cells (pretreatment, IC50 = 0.23 and 0.19 μM, respectively) with moderate cytotoxicity (CC50 = 11.53 and 9.24 μM, respectively). Moreover, pyronaridine displays in vitro activity against SARS-CoV-2 PLpro (IC50 = 1.8 μM). Given their existing antiviral activity, these compounds are strong candidates for repurposing against COVID-19 and prompt us to study the structure–activity relationship of the 9-aminoacridine scaffold against SARS-CoV-2 using traditional medicinal chemistry to identify promising new analogs. Our studies identified several novel analogs possessing potent in vitro activity in U2-OS ACE2 GFP 1-10 and 1-11 (IC50 < 1.0 μM) as well as moderate cytotoxicity (CC50 > 4.0 μM). Compounds such as 7g, 9c, and 7e were more active, demonstrating selectivity indices SI > 10, and 9c displayed the strongest activity (IC50 ≤ 0.42 μM, CC50 ≥ 4.41 μM, SI > 10) among them, indicating that it has potential as a new lead molecule in this series against COVID-19.
Introduction
As of September 20, 2023, the COVID-19 pandemic has ended based on the WHO recommendation. However, there are very few small molecule treatments developed for this disease1,2 and preparation for future outbreaks of this virus or related viruses is still of great importance.3 COVID-19 is an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The pandemic caused unprecedented economic and social hardships, affecting over 770 million people and causing over 6.9 million deaths worldwide.4 Infection with SARS-CoV-2 leads to a broad range of clinical symptoms including respiratory distress, cough, immune system disruption, and loss of smell and taste.5,6 While the development of various COVID-19 vaccines has been successful at preventing infection and decreasing symptom severity,7 there is a recognized need for complementary antiviral agents, which have direct activity against circulating SARS-CoV-2 strains. Remdesivir8,9 was the only FDA-approved antiviral drug for the treatment of COVID-19, while molnupiravir and paxlovid both obtained emergency use authorizations in 2021 for treating infections with this virus in the USA,10 and the latter obtained full approval from the FDA more recently.
Current treatment strategies for COVID-19 involve either prevention of viral attachment to the host cell or disruption of viral replication.1 SARS-CoV-2 enters the host cell via a spike protein attachment to the host angiotensin converting enzyme 2 (ACE2) receptor.11 After entry, viral RNA is translated by the host ribosome to generate two polyproteins, pp1a and pp1ab. The virus encodes two cysteine proteases, papain-like protease (PLpro) and the coronavirus main protease (Mpro), which cleave pp1a and pp1ab, yielding nonstructural proteins that form complexes with the host membrane.12 These proteases are essential for viral replication, making them attractive targets for antiviral therapeutics.13 There are currently a number of Mpro inhibitors that have advanced to clinical trials; however, none have yet reached the market.14
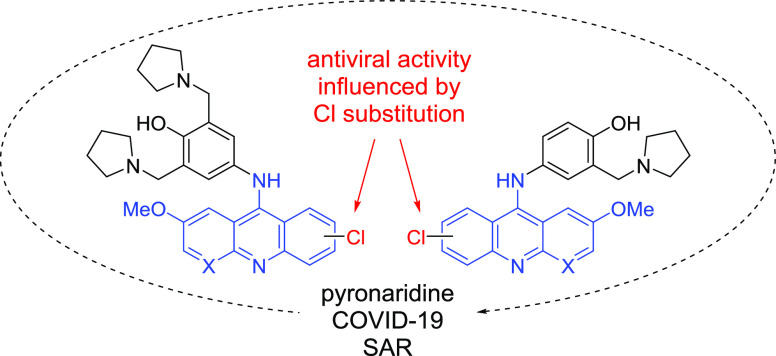
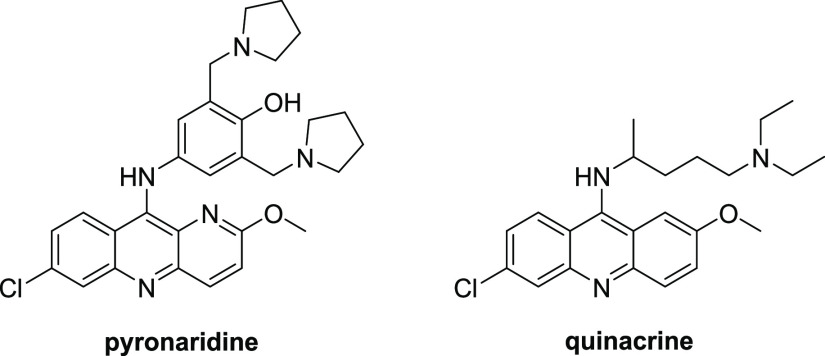
A hot topic in the identification for antiviral drug candidates is the repurposing of existing drugs for new targets.1 The above-mentioned remdesivir was originally developed for the treatment of hepatitis C and was later repositioned for COVID-19.15 Several existing drugs with a variety of biological activities have been tested against SARS-CoV-2,16 and we and others have recently found that an antimalarial agent pyronaridine (Figure 1) displays activity against a number of viruses including SARS-CoV-2.17−19 In subsequent studies, we have shown that pyronaridine reduces viral load in the lungs of SARS-CoV-2-infected mice.20 The antiviral effect of pyronaridine is most likely based on inhibition of replication because it was found to selectively inhibit SARS-CoV-2 PLpro activity in vitro.20 Another antimalarial drug popular outside of the U.S., quinacrine (Figure 1), which has similar structural motifs, also displays comparable in vitro activity against SARS-CoV-2.21,22 We have shown that quinacrine possesses activity against SARS-CoV-2 in A549 + ACE2 cells (pretreatment, IC50 = 0.19 μM, CC50 = 9.24 μM).19 These findings prompted us to study the structure–activity relationships of pyronaridine and quinacrine scaffolds against SARS-CoV-2 using a traditional medicinal chemistry approach.
Figure 1.
Structures of pyronaridine and quinacrine.
Results and Discussion
Compound Synthesis
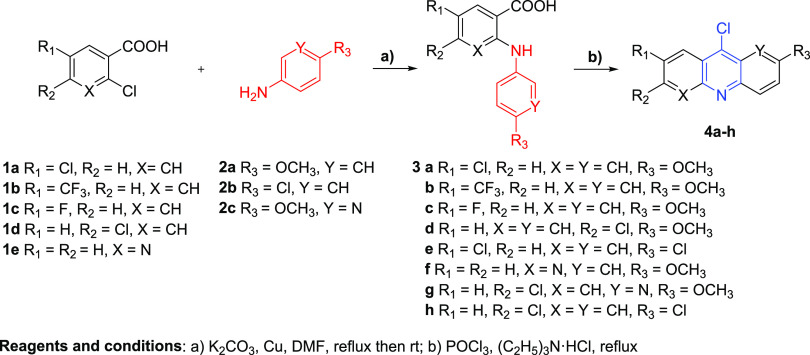
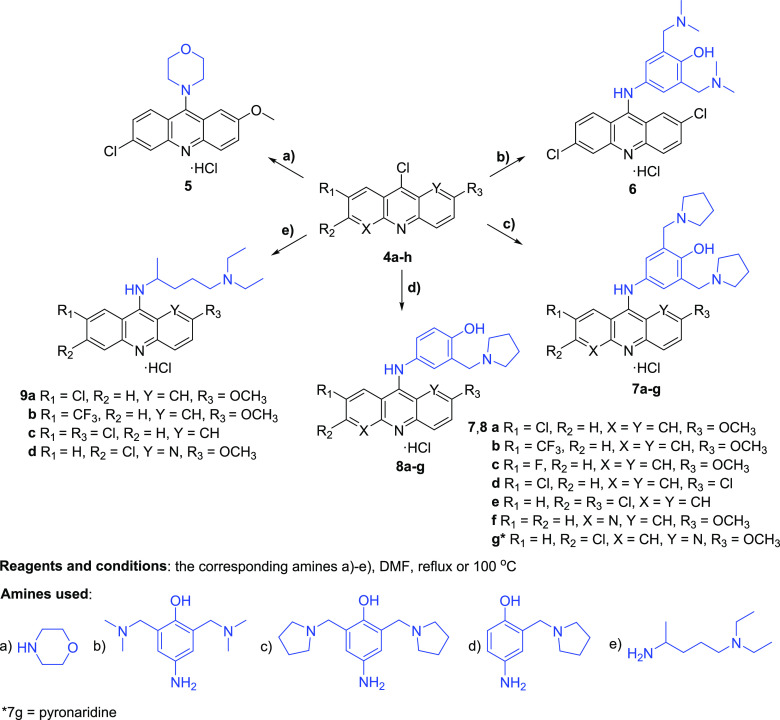
Derivatives and analogs based on the pyronaridine core were synthesized according to Schemes 1 and 2. The aryl carboxylic acids 3a–3h were obtained under Ullmann reaction conditions from commercial aryl chlorides 1a–1e and arylamines 2a–2c (Scheme 1). Subsequent cyclization of 3a–3h with phosphorus oxychloride led us to afford the desired intermediate acridines and naphthyridines 4a–4h in high yields. Further, some compounds 4a–4h were reacted with the corresponding amines listed in Scheme 2 in DMF medium at reflux or 100 °C to give the final products 7a–7g and 8a–8g.
Scheme 1. Synthesis of Acridine and Naphthyridine Derivatives 4a–4h.
Scheme 2. Synthesis of 9-Aminoacridines and Analogs 7a–7g and 8a–8g.
Structure–Activity Relationship Study of Pyronaridine and Quinacrine Analogs against SARS-CoV-2
The antiviral activity against SARS-CoV-2, cytotoxicity in U2-OS ACE2 GFP cells, and selectivity index (SI) for the synthesized compounds are summarized in Tables 1 and 2. The quinacrine derivatives in Table 1 were more active when bearing a 2-chloro substituent compared to a 3-chloro substituent (for example, quinacrine is less active than 9a and 9c). We did not see a significant difference between the substituents at R3 in this series. For the symmetric pyronaridine derivatives in Table 2 (R4 = Py), compounds bearing trifluoromethyl substituents at the 2-position (R1 as drawn) were more active than the 2-chloro compounds and had comparable antiviral activity to that of the 3-chloro derivatives (R2 as drawn). We did not observe a significant relationship regarding the 2- and 3-chloro derivatives with respect to activity. For the asymmetric pyronaridine derivatives (Table 2, R4 = H), 2-chloro substituents were generally more active than the 3-chloro derivatives, and the more electronegative fluoro- and trifluoromethyl groups did not increase the activity to a significant degree. The presence of a chloro or methoxy substituent at R3 did not have a significant effect on activity. Last, the introduction of an additional nitrogen atom into the ring system did not affect antiviral activity significantly but did result in lower cytotoxicity. In summary, there were few general trends for the SAR of the pyronaridine/quinacrine derivatives, as antiviral activity was dependent on both the identity of the side chain at the acridine 9-position and the halogenation pattern around the central ring system. The evaluation of additional derivatives in future studies may shed more light onto the specific substitution patterns, which affect potency against SARS-CoV-2.
Table 1. In Vitro Activity IC50 against SARS-CoV-2 Strain BetaCoV/France/IDF0372/2020, Cytotoxicity CC50 in U2-OS ACE2 GFP Cells, and Selectivity Index SI (Ratio of CC50 to IC50) of Quinacrine Derivatives.
| compound | Y | R1 | R2 | R3 | R4 | IC50(μM) | CC50(μM) | SI |
|---|---|---|---|---|---|---|---|---|
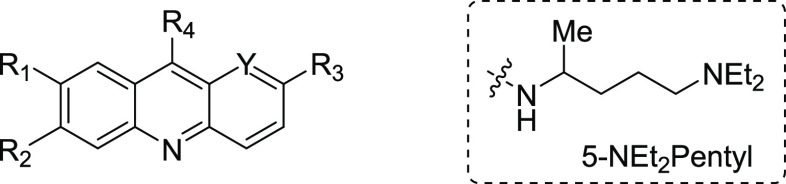
| quinacrine | CH | H | Cl | OMe | 5-NEt2pentyl | 2.57 | 2.43 | 0.95 |
| 5 | CH | H | Cl | OMe | morpholine | >50.0 | >50.0 | 1.0 |
| 9a | CH | Cl | H | OMe | 5-NEt2pentyl | 0.41 | 2.41 | 5.87 |
| 9b | CH | CF3 | H | OMe | 5-NEt2pentyl | 1.81 | 7.26 | 4.02 |
| 9c | CH | Cl | H | Cl | 5-NEt2pentyl | 0.42 | 4.41 | 10.50 |
| 9d | N | H | Cl | OMe | 5-NEt2pentyl | 1.27 | 8.25 | 6.50 |
Table 2. In Vitro Activity IC50 against SARS-CoV-22 Strain BetaCoV/France/IDF0372/2020, Cytotoxicity CC50 in U2-OS ACE2 GFP Cells, and Selectivity Index SI of Pyronaridine Derivatives.
| compound | X | Y | R1 | R2 | R3 | R4 | IC50(μM) | CC50(μM) | SI |
|---|---|---|---|---|---|---|---|---|---|
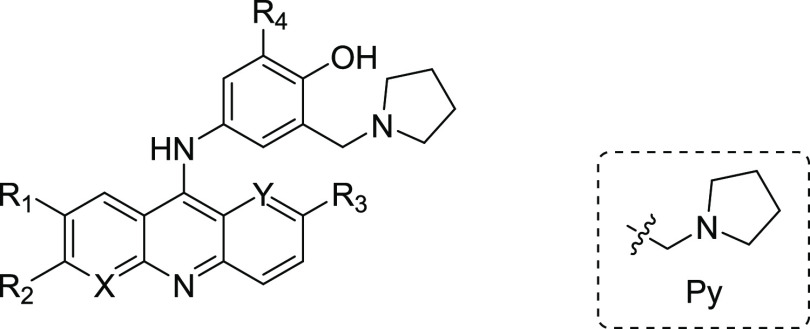
| 6a | CH | CH | H | Cl | OMe | H | 10.20 | >50.0 | >4.90 |
| 7a | CH | CH | Cl | H | OMe | Py | 1.88 | 5.19 | 2.75 |
| 7b | CH | CH | CF3 | H | OMe | Py | 0.44 | 4.36 | 9.91 |
| 7c | CH | CH | F | H | OMe | Py | 1.84 | 3.74 | 2.03 |
| 7d | CH | CH | Cl | H | Cl | Py | 1.76 | 5.46 | 3.09 |
| 7e | CH | CH | H | Cl | Cl | Py | 0.57 | 8.86 | 15.50 |
| 7f | N | CH | H | H | OMe | Py | 1.95 | 26.34 | 13.48 |
| 7gb | CH | CH | H | Cl | OMe | Py | 0.42 | 4.70 | 11.18 |
| 8a | CH | CH | Cl | H | OMe | H | 0.48 | 3.20 | 6.67 |
| 8b | CH | CH | CF3 | H | OMe | H | 1.90 | 10.08 | 5.32 |
| 8c | CH | CH | F | H | OMe | H | 1.91 | 4.03 | 2.11 |
| 8d | CH | CH | Cl | H | Cl | H | 1.75 | 4.26 | 2.44 |
| 8e | CH | CH | H | Cl | Cl | H | 1.37 | 19.30 | 14.41 |
| 8f | N | CH | H | H | OMe | H | 1.96 | 30.77 | 15.7 |
| 8g | CH | N | H | Cl | OMe | H | 1.96 | 3.62 | 1.85 |
NMe2 instead of pyrrolidine rings.
Pyronaridine.
Overall, compounds 7e, 7g, and 9c were the most active compounds, which displayed high selectivity indexes (SI > 10). Compounds 8a, 9a, and 7b were also comparable in antiviral activity but displayed higher cytotoxicity and lower selectivity indexes (SI < 10). Subsequent studies of these compounds in other common cells lines such as A549-ACE2, Huh-7, and Caco-2 will be performed in the future, as cell-type specific activity against SARS-CoV-2 has been demonstrated previously and should be addressed.19,23 However, the most active compounds are still less active than remdesivir (IC50 = 0.02–0.05 μM),8,9 suggesting the need to continue searching for compounds with increased antiviral activity. Nonetheless, these findings suggest that compounds 7e, 7g, and 9c may support further investigation of the repurposing of pyronaridine and quinacrine as antivirals against SARS-CoV-2 and illustrate how they can be used to generate new analogs with improved properties (compared to quinacrine).
While there has been considerable progress made on COVID-19 drug discovery in over 3 years since the pandemic began with preclinical and clinical molecules showing activity,1,2 there is still a need to identify promising lead compounds.3 This may be particularly important as they may be needed as the virus mutates and overcomes existing small-molecule antivirals. The 9-aminoacridines may provide a scaffold for further optimization as antivirals.17−19
Materials and Methods
Chemistry
All reagents and solvents were purchased from commercial suppliers (AlfaAesar, Acros, and Chimmed) and used without further purification. The 1H and 13C spectra were recorded on Bruker AC-300 (200 MHz, 1H) or a Bruker AC-200 (50 MHz, 13C) NMR spectrometers. Chemical shifts were measured in DMSO-d6 or CDCl3 using tetramethylsilane as an internal standard and reported as ppm values. The following abbreviations are used to indicate multiplicity: s, singlet; d, doublet; t, triplet; m, multiplet; dd, doublet of doublets; brs, broad singlet. The mass spectra were recorded on a Finnigan MAT INCOS 50 quadrupole mass spectrometer (Thermo Finnigan, San Jose, CA USA) (EI, 70 eV) with direct injection. The purity of the final compounds was analyzed by analytical high-performance liquid chromatography (HPLC) on an Elute HPLC system (Bruker Daltonik, Heidelberg, Germany) equipped with an Azura UVD 2.1S UV detector (Knauer, Berlin, Germany) with a wavelength at 254 nm and acquisition rate at 1 Hz. Chromatographic separation was carried out on an Acquity HSS T3 column (2.1 × 100 mm, 1.3 μm, 100 Å) at 30 °C and a sample injection volume of 2.0 μL. A mobile phase consisting of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) was programmed with gradient elution of 30–95% at a flow rate of 250 μL/min. Mass spectrometric detection was operated in the positive ion mode. Data were processed using Compass DataAnalysis 5.1 software (Bruker Daltonik). All final compounds were ≥95% pure. Elemental analysis (% C, H, and N) was performed on a EURO EA elemental analyzer (HEKAtech, Wegberg, Germany). Melting points were determined on an Electrothermal 9001 melting point apparatus (Electrothermal, UK) (10 °C per min) and were uncorrected. Merck KGaA silica gel 60 F254 plates were used for analytical thin-layer chromatography. Spots were detected with a UV lamp. Column chromatography was performed using silica gel, Merck 60 (70–230 mesh). Yields refer to purified products, and they were not optimized.
4-Amino-2,6-bis((dimethylamino)methyl)phenol (b), 4-amino-2,6-bis(pyrrolidin-1-ylmethyl)phenol (c), and 4-amino-2-(pyrrolidin-1-ylmethyl)phenol (d) (see Scheme 2) were prepared according to the procedures as described.24,25 The synthetic procedures are described in the Supporting Information. Compounds 5, 6, 7g, and 8g were synthesized as previously described,20 and their physicochemical properties are identical to those described in Puhl et al.20
In Vitro Testing
We have previously reported our method of generating SARS-CoV-2-infected US-OS ACE2 GFP cells.26 In short, U2-OS ACE2 GFP cells were split 1 day before infection. Compounds were resuspended in DMSO or H2O at 20 or 5 mM. These concentrations were sufficient to solubilize the molecules. The experimental compounds were incubated for 2 h at 37 °C in 100 μL of growth medium (DMEM, 10% FCS, and 1% PS). Ten microliters of SARS-CoV-2 virus (strain BetaCoV/France/IDF0372/2020) was then added to each well (final MOI, 0.1). Cells were incubated at 37 °C for 20 h, fixed with 8% PFA for 30 min at RT, and washed with PBS. To stain the nuclei and measure the viability, 100 μL of Hoechst solution was added. The plates were read with an automated confocal microscope (Opera Phoenix), which measures the number of infected cells (GFP signal) and viability (Hoechst signal). The compounds were tested from 50 μM to 0.64 nM with serial 5-fold dilutions.
Glossary
Abbreviations
- ACE2
angiotensin converting enzyme 2
- CC50
concentration of cytotoxicity 50%
- COVID-19
coronavirus disease of 2019
- DMEM
Dulbecco’s modified eagle medium
- DMF
N,N-dimethylformamide
- DMSO
dimethylsulfoxide
- Et
ethyl
- FCS
fetal calf serum
- FDA
Food and Drug Administration
- GFP
green fluorescent protein
- IC50
inhibitory concentration 50%
- Me
methyl
- MOI
multiplicity of infection
- Mpro
coronavirus main protease
- PBS
phosphate buffered saline
- PFA
paraformaldehyde
- PLpro
papain-like protease
- PS
phosphatidylserine
- rt
room temperature
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SI
selectivity index
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c05900.
Synthetic procedures, 1H and 13C NMR spectra for all new compounds, and representative HPLC traces (PDF)
Author Contributions
∥ T.J. and N.M. contributed equally to this work and should be considered cofirst authors.
Author Contributions
T.J.: data curation, writing, and review and editing; N.M.: investigation, data curation, and review and editing; F.G.-B.: investigation; A.L.: investigation and data Curation; T.B.: investigation; O.S.: investigation, methodology, and resources; T.R.L.: editing; A.C.P.: editing; V.M.: methodology, resources, supervision, and review and editing; S.E.: writing, resources, and review and editing.
S.E. kindly acknowledges NIH funding R44GM122196-02A1 from NIH NIGMS 1R43AT010585-01 and from NIH/NCCAM.
The authors declare the following competing financial interest(s): SE is CEO of Collaborations Pharmaceuticals, Inc. TJ, TRL and ACP are employees at Collaborations Pharmaceuticals, Inc. Collaborations Pharmaceuticals, Inc. has obtained FDA orphan drug designations for pyronaridine, tilorone and quinacrine for use against Ebola. CPI has also filed a provisional patent for use of these molecules against Marburg and other viruses. The other authors declare that they have no conflict of interest.
Supplementary Material
References
- Li G.; Hilgenfeld R.; Whitley R.; De Clercq E. Therapeutic strategies for COVID-19: progress and lessons learned. Nat. Rev. Drug Discovery 2023, 22, 449–475. 10.1038/s41573-023-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Delft A.; Hall M. D.; Kwong A. D.; Purcell L. A.; Saikatendu K. S.; Schmitz U.; Tallarico J. A.; Lee A. A. Accelerating antiviral drug discovery: lessons from COVID-19. Nat. Rev. Drug Discovery 2023, 22, 585. 10.1038/s41573-023-00692-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl A. C.; Lane T. R.; Ekins S. Learning from COVID-19: How drug hunters can prepare for the next pandemic. Drug Discovery Today 2023, 28 (10), 103723 10.1016/j.drudis.2023.103723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus disease (COVID-19) pandemic. 2023, https://www.who.int/europe/emergencies/situations/covid-19.
- Pan Y.; Guan H.; Zhou S.; Wang Y.; Li Q.; Zhu T.; Hu Q.; Xia L. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur. Radiol. 2020, 30, 3306. 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Naming the coronavirus disease (COVID-2019) and the virus that causes it. 2020, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it.
- Chakraborty C.; Bhattacharya M.; Dhama K. SARS-CoV-2 Vaccines, Vaccine Development Technologies, and Significant Efforts in Vaccine Development during the Pandemic: The Lessons Learned Might Help to Fight against the Next Pandemic. Vaccines 2023, 11 (3), 682. 10.3390/vaccines11030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. X. J.; Cho S.; Meyyur Aravamudan V.; Sanda H. Y.; Palraj R.; Molton J. S.; Venkatachalam I. Remdesivir in Coronavirus Disease 2019 (COVID-19) treatment: a review of evidence. Infection 2021, 49 (3), 401–410. 10.1007/s15010-020-01557-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales R.; McGovern B. L.; Rodriguez M. L.; Rai D. K.; Cardin R. D.; Anderson A. S.; Sordillo E. M.; van Bakel H.; Simon V.; García-Sastre A.; White K. M.; et al. Nirmatrelvir, Molnupiravir, and Remdesivir maintain potent in vitro activity against the SARS-CoV-2 Omicron variant. bioRxiv 2022 10.1101/2022.01.17.476685. [DOI] [Google Scholar]
- NIH Antiviral Agents, Including Antibody Products. 2023, https://www.covid19treatmentguidelines.nih.gov/therapies/antivirals-including-antibody-products/summary-recommendations/.
- Kruse R. L. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 2020, 9, 72. 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V.; Ivanov K. A.; Putics Á.; Hertzig T.; Schelle B.; Bayer S.; Weiβbrich B.; Snijder E. J.; Rabenau H.; Doerr H. W.; et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003, 84 (9), 2305–2315. 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Shen Z.; Ratia K.; Cooper L.; Kong D.; Lee H.; Kwon Y.; Li Y.; Alqarni S.; Huang F.; Dubrovskyi O.; et al. Design of SARS-CoV-2 PLpro Inhibitors for COVID-19 Antiviral Therapy Leveraging Binding Cooperativity. J. Med. Chem. 2022, 65 (4), 2940–2955. 10.1021/acs.jmedchem.1c01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberger T.; Laufer S. A.; Pillaiyar T. COVID-19 therapeutics: Small-molecule drug development targeting SARS-CoV-2 main protease. Drug Discovery Today 2023, 28 (6), 103579 10.1016/j.drudis.2023.103579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S.; Dodd L. E.; Davey R. T. Jr.; Tshiani Mbaya O.; Proschan M.; Mukadi D.; Lusakibanza Manzo M.; Nzolo D.; Tshomba Oloma A.; Ibanda A.; et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019, 381 (24), 2293–2303. 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T.; Meenakshisundaram S.; Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today 2020, 25 (4), 668–688. 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J.-Y.; Lee G. E.; Park H.; Cho J.; Kim Y.-E.; Lee J.-Y.; Ju C.; Kim W.-K.; Kim J. I.; Park M.-S.. Pyronaridine and artesunate are potential antiviral drugs against COVID-19 and influenza. bioRxiv 2020. 10.1101/2020.07.28.225102. [DOI] [Google Scholar]
- Jeon S.; Ko M.; Lee J.; Choi I.; Byun S. Y.; Park S.; Shum D.; Kim S. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents Chemother. 2020, 64, e00819–e00820. 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl A. C.; Fritch E. J.; Lane T. R.; Tse L. V.; Yount B. L.; Sacramento C. Q.; Fintelman-Rodrigues N.; Tavella T. A.; Maranhão Costa F. T.; Weston S.; et al. Repurposing the Ebola and Marburg Virus Inhibitors Tilorone, Quinacrine, and Pyronaridine: In Vitro Activity against SARS-CoV-2 and Potential Mechanisms. ACS Omega 2021, 6 (11), 7454–7468. 10.1021/acsomega.0c05996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl A. C.; Gomes G. F.; Damasceno S.; Godoy A. S.; Noske G. D.; Nakamura A. M.; Gawriljuk V. O.; Fernandes R. S.; Monakhova N.; Riabova O.; et al. Pyronaridine Protects against SARS-CoV-2 Infection in Mouse. ACS Infect. Dis 2022, 8 (6), 1147–1160. 10.1021/acsinfecdis.2c00091. [DOI] [PubMed] [Google Scholar]
- Ianevski A.; Yao R.; Fenstad M. H.; Biza S.; Zusinaite E.; Reisberg T.; Lysvand H.; Løseth K.; Landsem V. M.; Malmring J. F.; Oksenych V.; Erlandsen S. E.; Aas P. A.; Hagen L.; Pettersen C. H.; Tenson T.; Afset J. E.; Nordbø S. A.; Bjørås M.; Kainov D. E.; et al. Potential Antiviral Options against SARS-CoV-2 Infection. Viruses 2020, 12 (6), 642. 10.3390/v12060642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda B.; De la Cruz V. P.; Pando R. H.; Sotelo J. Quinacrine as a potential treatment for COVID-19 virus infection. Eur. Rev. Med. Parmacol. 2021, 25, 556–566. 10.26355/eurrev_202101_24428. [DOI] [PubMed] [Google Scholar]
- Dittmar M.; Lee J. S.; Whig K.; Segrist E.; Li M.; Kamalia B.; Castellana L.; Ayyanathan K.; Cardenas-Diaz F. L.; Morrisey E. E.; et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021, 35 (1), 108959 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlin G. B.; Ireland S. J. Potential Antimalarials. VII. Di-Mannich Bases of 4-[(7’-Trifluoromethylquinolin-4’-yl)-Aminophenol and 4-(7’-Bromo-1’,5′-naphthyridin-4’-yl)]-aminophenol via 4-Nitrophenols. Aust. J. Chem. 1988, 41 (11), 1727–1733. 10.1071/CH9881727. [DOI] [Google Scholar]
- Barlin G.; Nguyen T.; Kotecka B.; Rieckmann K. Potential Antimalarials. XVII. Di- and Mono-Mannich Bases of 2(and 4)-[2(and 8)-Trifluoromethylquinolin-4-ylamino]phenol. Aust. J. Chem. 1993, 46 (1), 21–29. 10.1071/CH9930021. [DOI] [Google Scholar]
- Buchrieser J.; Dufloo J.; Hubert M.; Monel B.; Planas D.; Rajah M. M.; Planchais C.; Porrot F.; Guivel-Benhassine F.; Van der Werf S.; et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020, 39 (23), e106267 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.