Abstract
Purpose
Protein arginine methyltransferase 7 (PRMT7) is a member of a family of enzymes that catalyzes the methylation of arginine residues on several protein substrates. Biallelic pathogenic PRMT7 variants have previously been associated with a syndromic neurodevelopmental disorder characterized by short stature, brachydactyly, intellectual developmental disability, and seizures. To our knowledge, no comprehensive study describes the detailed clinical characteristics of this syndrome. Thus, we aim to delineate the phenotypic spectrum of PRMT7-related disorder.
Methods
We assembled a cohort of 51 affected individuals from 39 different families, gathering clinical information from 36 newly described affected individuals and reviewing data of 15 individuals from the literature.
Results
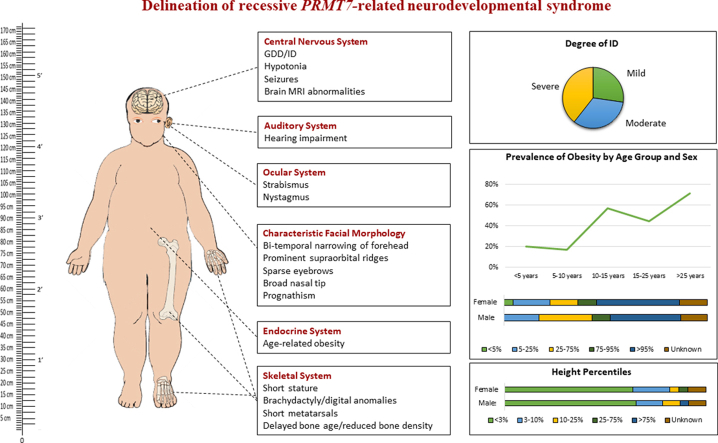
The main clinical characteristics of the PRMT7-related syndrome are short stature, mild to severe developmental delay/intellectual disability, hypotonia, brachydactyly, and distinct facial morphology, including bifrontal narrowing, prominent supraorbital ridges, sparse eyebrows, short nose with full/broad nasal tip, thin upper lip, full and everted lower lip, and a prominent or squared-off jaw. Additional variable findings include seizures, obesity, nonspecific magnetic resonance imaging abnormalities, eye abnormalities (i.e., strabismus or nystagmus), and hearing loss.
Conclusion
This study further delineates and expands the molecular, phenotypic spectrum and natural history of PRMT7-related syndrome characterized by a neurodevelopmental disorder with skeletal, growth, and endocrine abnormalities.
Keywords: Chromatinopathy, Mendelian disorders of the epigenetic machinery, PRMT7, Syndromic neurodevelopmental disorder, Syndromic obesity
Graphical abstract
Introduction
Protein arginine methyltransferase 7 (PRMT7) is a member of a family of enzymes that catalyze the methylation of arginine residues on several protein substrates. Arginine methylation plays a crucial role in various biological pathways, influencing chromatin, RNA biology, and phase separation.1,2 The PRMT family regulates physiological functions, and its dysfunction is linked to pathologies as diverse as cancer, inflammation, and neurodegeneration. PRMT7 is a unique, evolutionarily conserved PRMT family member that catalyzes the monomethylation of arginine.3 Biallelic pathogenic variants in PRMT7 (OMIM ∗ 610087) have been associated with a syndromic neurodevelopmental disorder characterized by short stature, brachydactyly, intellectual developmental disability, and seizures, which is currently known as SBIDDS syndrome4, 5, 6, 7, 8, 9 (OMIM 617157). Since the identification of PRMT7 as a disease-associated gene in 2015, 15 cases from 9 families have been published in 6 separate reports. Until now, to our knowledge, there have been no comprehensive studies describing the characteristics and phenotypic spectrum of the PRMT7-related disorder.
Here, we report a large cohort of 51 affected individuals from 39 different families, assembled by gathering clinical information from 36 newly described patients and reviewing data of 15 affected individuals from the literature.
Materials and Methods
The affected individuals were identified through data sharing with collaborators and screening the databases of several diagnostic and research genetic laboratories worldwide, as well as using GeneMatcher.10 Next-generation sequencing was performed on genomic DNA extracted from blood in different diagnostic or research laboratories worldwide. The candidate variants were confirmed, and segregation analysis was performed by Sanger sequencing. Detailed clinical data and family history were collected for new and published cases in the form of completing a clinical proforma by the recruiting clinicians. Detailed assessment of facial morphology was performed on clinical photographs by a clinical geneticist expert in dysmorphology (M.Su.). Where permission to share clinical photographs was not given, information on the dysmorphological features was provided by clinical assessment performed by the family’s clinicians.
Results
Clinical characterization
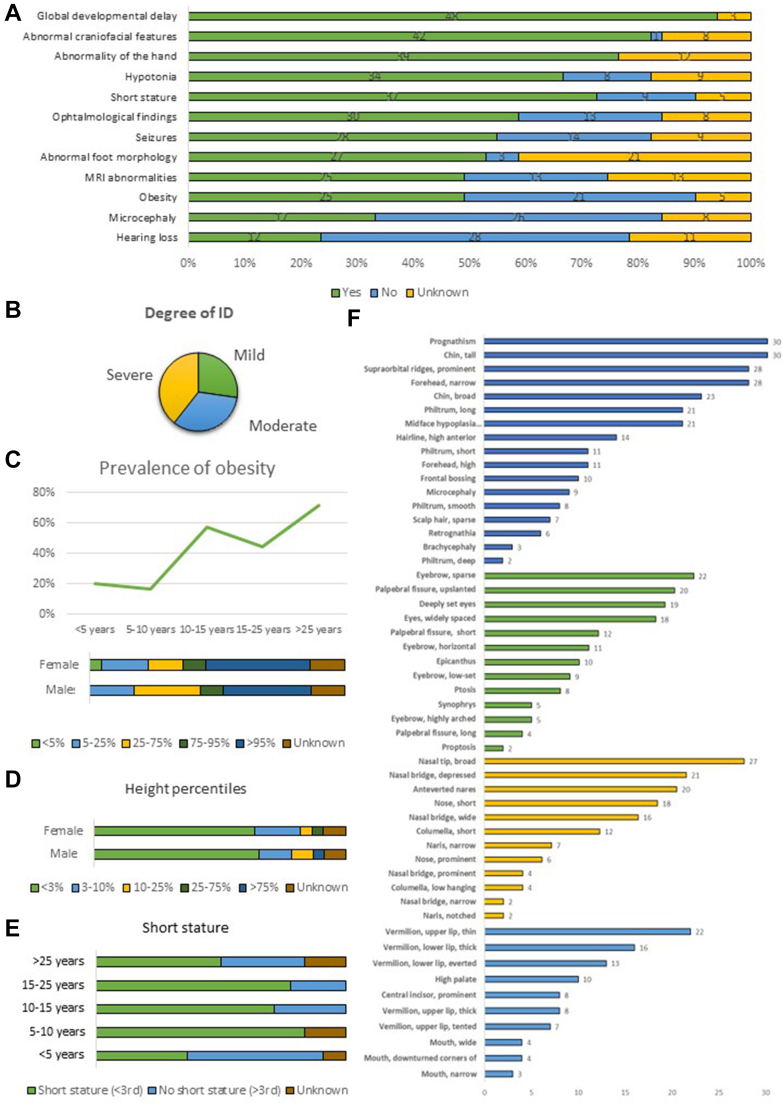
The cohort consisted of 23 males and 28 females whose age at last evaluation ranged between 34 weeks of gestation to 55 years (median 9.5 years, interquartile range 5.4-17.8 years). Consanguinity was reported in 16 families (16 of 36, 44%). Figure 1 displays the facial appearance of individuals for whom photographs were available. Figure 2 gives an overview of the core clinical findings, summarized in Supplemental Table 1.
Figure 1.
Phenotypic presentation of individuals with PRMT7-related syndrome. From I to XVI: craniofacial features of affected individuals, front view. From XVIIa to XXIVd: evolution of facial features among different ages. From XXV to XL: craniofacial features of affected individuals, sagittal view. From XLI to LIII: digital abnormalities. From LIV to LIX: stature of affected individuals. From LX to LXVIII: foot abnormalities.
Figure 2.
Overview of the main clinical features of PRMT7-related disorder. A. Occurrence of the main phenotypic features in the cohort. Green: Phenotype present. Blue: Phenotype not present. Yellow: Information not available. B. Degree of intellectual disability. C. Prevalence of obesity by age group and sex. D. Distribution of height percentile throughout the cohort. E. Prevalence of short stature among different ages. F. Deep characterization of the main dysmorphological features of the craniofacial (blue), periorbital (green), nasal (yellow), and perioral (light blue) areas.
Most individuals were born at term. Pregnancy was uneventful with unremarkable perinatal period for 20 individuals (20 of 43, 45%), whereas 23 individuals (22 of 40, 55%) presented with intrauterine growth restriction (14 of 31, 45%) and/or other variable prenatal manifestations (20 of 41, 48%). Nineteen newborns were described as small for gestational age (19 of 41, 46%). Weight at birth was within the normal range for 20 children, less than the 5th percentile for 19 children, and greater than the 95th percentile for 1 child. Decreased head circumference at birth was described in 6 children (6 of 23, 26%). At last evaluation, microcephaly was reported in 17 individuals (17 of 43, 40%). When specified, failure to thrive was documented in 18 individuals (18 of 35, 51%), and most individuals presented short stature (37 of 46, 80%). If available, findings from x-ray imaging included decreased bone density (4 of 10, 40%) and/or delayed bone age (5 of 10, 50%). Twenty-five affected individuals were either overweight (n = 5) or obese (n = 20) at last evaluation (25 of 46, 54%). Interestingly, obesity was more commonly reported in individuals older than 10 years, suggesting a delayed onset.
Notably, all individuals (48 of 48, 100%) showed global developmental delay (GDD) in early infancy and intellectual disability (ID). Where specified (n = 33), the degree of ID was mild in 9 (27%), moderate in 11 (33%), and severe in 13 (40%). Hypotonia was reported in most individuals (37 of 42, 88%, where specified n = 10 generalized, n = 5 axial), often manifesting in the neonatal period. Seizures developed in 28 individuals (28 of 42, 67%) with a median age of seizure onset of 3 years (range: 7 months-45 years, interquartile range: 1-4.8). Seizure onset varied from generalized (n = 18) to focal (n = 4). Generalized-onset seizures include tonic-clonic (n = 9), absence (n = 6), atonic (n = 5), and myoclonic (n = 2) seizures. Febrile seizures were reported in 2 individuals. Only 4 patients (4 of 25, 16%) presented with intractable seizures, whereas most showed good response to treatments (ie, topiramate, sodium valproate) or were seizure-free without therapy. Some individuals presented with strabismus (19 of 42, 45%) and hearing impairment (12 of 40, 30%). Of the 37 brain magnetic resonance imaging scans for which a clinical report was available, 13 were described as unremarkable. Magnetic resonance imaging findings included nonspecific changes, such as enlargement of the ventricles or subarachnoid spaces, white matter abnormalities, and mild parenchymal atrophy.
Dysmorphology assessment
Photographs were available from 28 new affected individuals from 23 families. These included childhood and adult photographs from 3 patients, including 2 siblings from 1 family (patients 6, 7, and 8). Facial features of 12 previously reported patients were also reviewed.4, 5, 6, 7,9 These included 3 adults, 1 adolescent, and 8 children. Photographs of the hand(s) and feet were only available for 19 new affected individuals from 15 families and 10 previously reported patients from 7 families.4, 5, 6, 7,9 There was no consent to share photographs of individuals 13, 15, 19, 20, 29, 30, and 36, and information on their dysmorphological features was retrieved from the clinical assessment performed by their clinicians.
Based on the available photographs, the most recurrent facial dysmorphic features were frontal bossing/prominent supraorbital ridges (70%), bitemporal (bifrontal) narrowing of forehead (67.5%), prognathism (72.5%), sparse eyebrows (55%), broad nasal tip (62.5%), and tall (prominent) chin (75%). Less frequently seen features included mid-face hypoplasia (47.5%), deep-set and widely spaced eyes (42.5%), upslanted palpebral fissures (45%), short nose (40%), depressed nasal bridge (47.5%), short columella (30%), long philtrum (45%), thin upper lip vermilion (47.5%), thick lower lip vermilion (40%), everted lower lip vermilion (32.5%), and a broad chin (52.5%). Another commonly reported feature was short neck. Many affected individuals also had changes affecting their fingers and toes. These included short fingers (brachydactyly) (70%), broad thumbs (37.5%), and short toes or short metatarsals (45.9%). The characteristic facial dysmorphism is not seen in very young children but can be seen in older children and adults. Detailed dysmorphological features of each individual can be found in Supplemental Table 2.
PRMT7 variants
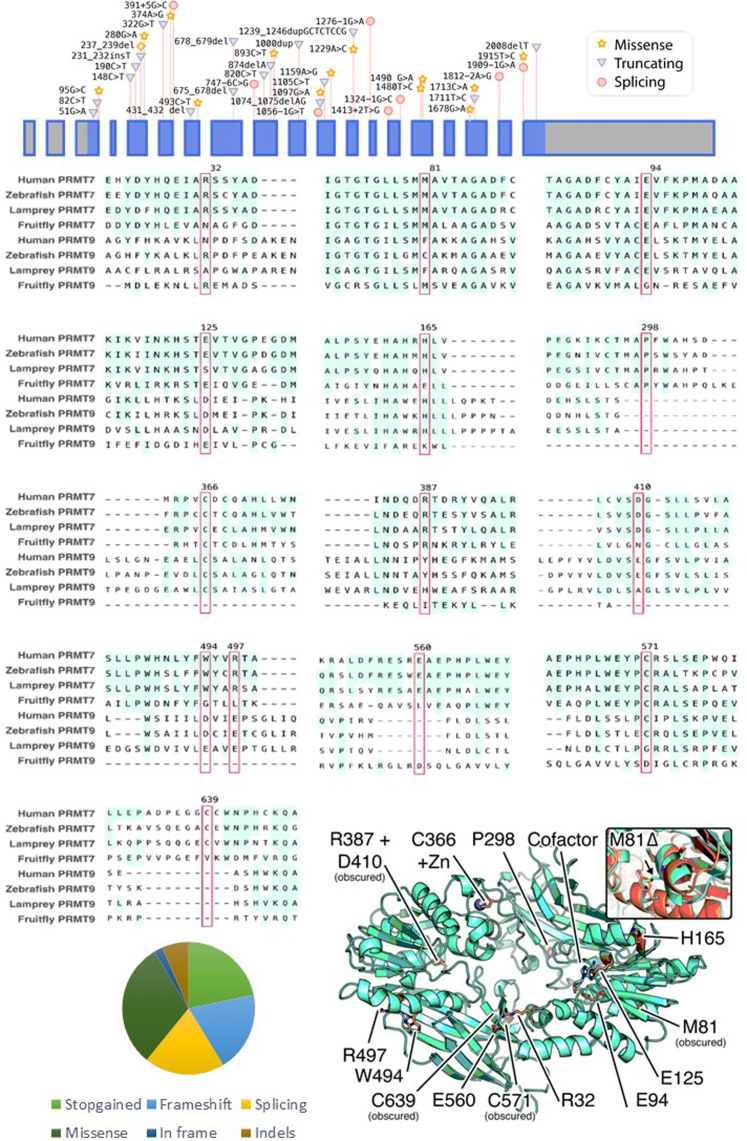
A total of 51 individuals with either compound heterozygous (n = 25, biallelic inheritance from unaffected parents) or homozygous (n = 26, biallelic inheritance from unaffected parents) PRMT7 variants were included in this cohort. Forty-six variants were detected, of which 34 were not previously reported in the literature. This includes 19 truncating variants (n = 9 frameshift, n = 10 nonsense), 1 in-frame deletion, 9 splicing, 14 missense variants, and 3 large indels. All identified variants are absent or found at very low allele frequencies in several variant databases (range 0.0–0.00002), are predicted to be damaging across a suite of in silico tools, and affect highly evolutionarily conserved residues. According to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology classification,11,12 28 variants were classified as pathogenic, 3 variants as likely pathogenic, and 12 variants were of uncertain significance. Variants of uncertain significance were then reclassified as likely pathogenic/pathogenic based on striking similarity to other cases and results of co-segregation studies. The characteristics of the variants are summarized in Supplemental Table 3 and shown in Supplemental Figure 1 and interactively at https://michelanglo.sgc.ox.ac.uk/r/prmt7
Discussion
We report on the molecular and phenotypic spectrum of 51 individuals with biallelic PRMT7 variants, representing the most comprehensive study to date. The phenotypic spectrum of PMRT7-related disorder includes GDD/ID, short stature, distinct craniofacial and digital defects, epilepsy, and obesity.
PRMTs are a family of enzymes that catalyze the methylation of arginine residues on several protein substrates. Arginine methylation has been largely studied for its key role in post-translational modification, influencing several biological processes, such as messenger RNA splicing, DNA repair, transcription regulation, and signal transduction.1,2 Known substrates methylated by PRMT7 include core histones (H2A, H2B, H3, and H4), Wnt signaling molecules, and transcription factors, involving PRMT7 in a wide range of biological processes, such as gene expression and epigenetic regulation, cell differentiation, muscle and neuron development, and adipogenesis.3,13,14 A significant number of genes involved in chromatin regulation have been associated with neurodevelopmental disorders, collectively known as chromatinopathies.15 PRMT7-related disorder shares some overlapping phenotypes with chromatin-related neurodevelopmental disorders, particularly short stature and growth delay and presence of major craniofacial dysmorphisms and skeletal and digital abnormalities (eg, Wiedemann-Steiner syndrome and Chung-Jansen syndrome). Given its known importance in histone methylation and genome regulation, PRMT7-related syndrome could potentially be considered as one of the Mendelian disorders of the epigenetic machinery.16
As further confirmation of its crucial function, PRMT7 has hitherto been the sole member of this family to be associated with a monogenic disorder. Prmt7-deficient mice generated by Jeong et al17 in 2016 exhibited defects in bone and skeletal muscle mass, delayed or impaired neuronal development, and increased adipogenesis, resembling many aspects of the human phenotype.
PRMT7 plays an important role in neuronal development and differentiation, caused by interaction with MLL418 and Wnt signaling molecules, and in HCN (hyperpolarization-activated, cyclic-nucleotide-gated) channel functioning and neuronal excitability, through regulation of SHANK3 and NALCN, respectively.19 Consistent with such a role, all the affected individuals in our cohort presented with GDD/ID, and 70% of them developed seizures.
Although PRMT7’s role in bone remodeling and development is still uncertain, adult mutated mice showed limb bone anomalies and reduced length. Likewise, 80% of the affected individuals of our cohort were diagnosed with short stature and, if available, findings from x-ray imaging included decreased bone density or delayed bone age and metacarpal and metatarsal shortening.
There is increasing evidence of the pivotal role of PRMT7 in adipogenesis and muscle development. PRMT7 negatively regulates adipocyte differentiation through modulation of C/EBP-β and therefore PPAR-γ2.20 Prmt7 knockout mice showed decreased energy expenditure and developed age-related obesity with excessive body fat accumulation at middle age. Similarly, half of our cohort exhibited obesity, with a higher prevalence after puberty. Half of the newborns were small for gestational age, consistent with the finding that the Prmt7-deficient mice displayed reduced body size and weight at birth. The same mice showed a switch from oxidative to glycolytic fibers in muscle before obesity development, suggesting that the consequent imbalance in energy homeostasis might be the cause of the obesity in mice and implying PRMT7 as an important co-factor in adult muscle development. Similarly, some of our patients reported reduced endurance exercise capacities. We hereby confirm the importance of PRMT7 function in human adipogenesis, suggesting that it might be a potential target for intervention and treatment of obesity. We suggest that PRMT7-related disorder should be considered in the differential diagnosis of monogenic syndromic obesity. Syndromic forms of obesity that may share overlapping features with PRMT7-related disorder include Borjeson-Forssman-Lehmann syndrome, CHOPS syndrome, Chung-Jansen syndrome, Cohen syndrome, and TRAPPC9-related disorder.21
After performing systematic review of the facial features of our cohort, we observed that PRMT7-related disorder shows a recognizable facial gestalt that is distinguishable from the other conditions mentioned above. This disorder should be suspected, in the context of a likely recessive inheritance, in patients with DD/ID who have digital abnormalities, bifrontal narrowing, prominent supraorbital ridges, sparse eyebrows, short nose with full/broad nasal tip, thin upper lip, full and everted lower lip, and a prominent or squared-off jaw. Some other important points in the differential diagnosis have been discussed and included in Supplemental Table 4.
There is also a considerable overlap with the phenotype associated with pseudohypoparathyroidism type 1A and 1C (Albright hereditary osteodystrophy), acrodysostosis, and chromosome 2q37 deletion syndrome (including brachydactyly-mental retardation syndrome because of loss-of-function variants in the HDAC4). This includes not only DD/ID but also short stature, similar facial dysmorphic features, short fingers and toes with broad thumbs, metacarpal and/or metatarsal shortening, and the tendency to excessive weight gain. However, unlike PRMT7-related disorder, these are all autosomal dominant disorders caused by heterozygous variants in GNAS (PHP 1A and 1C), PRKAR1A and PDE4D (acrodysostosis), and/or 2q37 deletion resulting in loss of the HDAC4. Furthermore, patients with these disorders can have normal neurodevelopment and can show evidence of multiple hormone resistance.22
In conclusion, we delineate and expand the phenotypic spectrum and natural history of the disease associated with pathogenic biallelic PRMT7 variants, providing a comprehensive review of the associated clinical phenotype. In addition, we characterize the distinct craniofacial morphology of this syndrome. This study provides a valuable resource for clinicians for the accurate diagnosis, assessment, counseling, and better management of affected individuals with this rare syndrome. Further studies will be needed to deeply understand PRMT7 regulatory functions and possibly identify strategies for therapy.
Data Availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.
Conflict of Interest
Megan Li is an employee of Invitae. Erin Torti, Amber Begtrup, and Rhonda E Schnur are employees of GeneDx, Inc. All other authors declare no conflicts of interest.
Acknowledgments
The authors thank all patients and families for participation in this study. Part of this research was possible thanks to the Deciphering Developmental Disorders study. The Deciphering Developmental Disorders study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003). This study makes use of DECIPHER (http://www.deciphergenomics.org), which is funded by Wellcome. See www.ddduk.org/access.html for full acknowledgment. This study was also supported by the Wellcome Trust (WT093205MA and WT104033AIA to H.H. and 203141/Z/16/Z to M.P.F. and J.C.T.), Medical Research Council (H.H.), European Community’s Seventh Framework Programme (FP7/2007-2013, under grant agreement No. 2012-305121 to H.H.), the National Institute for Health Research (NIHR), University College London Hospitals, Biomedical Research Centre, and Fidelity Foundation. The Yale Center for Mendelian Genomics (UM1HG006504) is funded by the National Human Genome Research Institute. D.B. is supported by NIHR Research Professorship (RP-2016-07-011). F.L. and A.G. received funding from European Union and Région Normandie in the context of Recherche Innovation Normandie 2018. Europe gets involved in Normandie with the European Regional Development Fund. The authors thank the families and KFMC Research Centre for the partial support (Intramural Research Fund; Demography of Recessive Diseases in KSA; Grant No. 019-052). This work was also supported by King Salman Center for Disability research through Research Group RG-2022-010.
Author Information
Conceptualization: E.C., M.Su., R.M.; Data Curation: E.C., R.M.; Formal Analysis: E.C., H.H., R.M.; Methodology: E.C., M.Sc., M.Su., R.M.; Funding Acquisition: H.H., D.B., F.L., A.G., J.C.T.; Investigation - Computational Methods: M.P.F., J.C.T.; Recruitment, Clinical, and Diagnostic Evaluations: M.Sa., S.A., E.K.B., M.H., K.M.W., D.B., M.Y.V.M., J.S., K.P., K.S., M.J., D.M.N., J.J., R.O.L., D.W., N.Z., L.R., A.G., F.L., M.D.-C., G.H., Y.D., M.D., S.M.S., A.B., N.O., E.A.F., S.B., A.T.V.-v.S., Y.H., A.P.A.S., M.E.N., M.G., A.G., Z.F., M.M., S.E., E.G.K., A.R., M.R., A.L., G.A.-S., M.L., M.B., A.J.M., M.C.D., H.T., D.N.S., A.K., I.V., S.D.M., C.P., M.Sa., A.B., P.S., V.S., R.E.S., E.T., T.B.H., C.P., F.S.A.; Writing-original draft: E.C., M.Su., R.M.; Writing-review and editing: all authors.
Ethics Declaration
Individuals (and/or their legal guardians) recruited in a research setting gave informed consent for their research participation. Those individual research studies received approval from the Review Boards and Bioethics Committees at University College London Hospital (project 06/N076) and the other institutions involved in this study. Permission for inclusion of their anonymized medical data in this cohort, including photographs, was obtained using standard forms at each local site by the responsible referring physicians.
Footnotes
Additional Information
The online version of this article (https://doi.org/10.1016/j.gim.2022.09.016) contains supplementary material, which is available to authorized users.
Additional Information
Supplementary Figure S1.
References
- 1.Guccione E., Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol. 2019;20(10):642–657. doi: 10.1038/s41580-019-0155-x. Published correction appears in Nat Rev Mol Cell Biol. 2019;20(9):567. [DOI] [PubMed] [Google Scholar]
- 2.Lorton B.M., Shechter D. Cellular consequences of arginine methylation. Cell Mol Life Sci. 2019;76(15):2933–2956. doi: 10.1007/s00018-019-03140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halabelian L., Barsyte-Lovejoy D. Structure and function of protein arginine methyltransferase PRMT7. Life (Basel) 2021;11(8):768. doi: 10.3390/life11080768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akawi N., McRae J., Ansari M., et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat Genet. 2015;47(11):1363–1369. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agolini E., Dentici M.L., Bellacchio E., et al. Expanding the clinical and molecular spectrum of PRMT7 mutations: 3 additional patients and review. Clin Genet. 2018;93(3):675–681. doi: 10.1111/cge.13137. [DOI] [PubMed] [Google Scholar]
- 6.Valenzuela I., Segura-Puimedon M., Rodríguez-Santiago B., et al. Further delineation of the phenotype caused by loss of function mutations in PRMT7. Eur J Med Genet. 2019;62(3):182–185. doi: 10.1016/j.ejmg.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Kernohan K.D., McBride A., Xi Y., et al. Loss of the arginine methyltranserase PRMT7 causes syndromic intellectual disability with microcephaly and brachydactyly. Clin Genet. 2017;91(5):708–716. doi: 10.1111/cge.12884. [DOI] [PubMed] [Google Scholar]
- 8.Birnbaum R., Yosha-Orpaz N., Yanoov-Sharav M., et al. Prenatal and postnatal presentation of PRMT7 related syndrome: expanding the phenotypic manifestations. Am J Med Genet A. 2019;179(1):78–84. doi: 10.1002/ajmg.a.6. [DOI] [PubMed] [Google Scholar]
- 9.Poquérusse J., Whitford W., Taylor J., et al. Novel PRMT7 mutation in a rare case of dysmorphism and intellectual disability. J Hum Genet. 2022;67(1):19–26. doi: 10.1038/s10038-021-00955-5. [DOI] [PubMed] [Google Scholar]
- 10.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biesecker L.G., Harrison S.M. ClinGen Sequence Variant Interpretation Working Group. The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet Med. 2018;20(12):1687–1688. doi: 10.1038/gim.2018.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B., Zhang M., Liu Z., Mu Y., Li K. PRMT7: A pivotal arginine methyltransferase in stem cells and development. Stem Cells Int. 2021;2021 doi: 10.1155/2021/6241600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma T., Li L., Chen R., et al. Protein arginine methyltransferase 7 modulates neuronal excitability by interacting with NaV1.9. Pain. 2022;163(4):753–764. doi: 10.1097/j.pain.0000000000002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valencia A.M., Pașca S.P. Chromatin dynamics in human brain development and disease. Trends Cell Biol. 2022;32(2):98–101. doi: 10.1016/j.tcb.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: postnatal malleability and therapeutic prospects. Hum Mol Genet. 2019;28(R2):R254–R264. doi: 10.1093/hmg/ddz174. Published correction appears in Hum Mol Genet. 2020;29(5):876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong H.J., Lee H.J., Vuong T.A., et al. Prmt7 deficiency causes reduced skeletal muscle oxidative metabolism and age-related obesity. Diabetes. 2016;65(7):1868–1882. doi: 10.2337/db15-1500. [DOI] [PubMed] [Google Scholar]
- 18.Dhar S.S., Lee S.H., Kan P.Y., et al. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012;26(24):2749–2762. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S.Y., Vuong T.A., So H.K., et al. PRMT7 deficiency causes dysregulation of the HCN channels in the CA1 pyramidal cells and impairment of social behaviors. Exp Mol Med. 2020;52(4):604–614. doi: 10.1038/s12276-020-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leem Y.E., Bae J.H., Jeong H.J., Kang J.S. PRMT7 deficiency enhances adipogenesis through modulation of C/EBP-β. Biochem Biophys Res Commun. 2019;517(3):484–490. doi: 10.1016/j.bbrc.2019.07.096. [DOI] [PubMed] [Google Scholar]
- 21.Kehinde T.A., Bhatia A., Olarewaju B., Shoaib M.Z., Mousa J., Osundiji M.A. Syndromic obesity with neurodevelopmental delay: opportunities for targeted interventions. Eur J Med Genet. 2022;65(3) doi: 10.1016/j.ejmg.2022.104443. [DOI] [PubMed] [Google Scholar]
- 22.Mantovani G., Spada A., Elli F.M. Pseudohypoparathyroidism and Gsα-cAMP-linked disorders: current view and open issues. Nat Rev Endocrinol. 2016;12(6):347–356. doi: 10.1038/nrendo.2016.52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.