Abstract
The denitrifying strain T1 is able to grow with toluene serving as its sole carbon source. Two mutants which have defects in this toluene utilization pathway have been characterized. A clone has been isolated, and subclones which contain tutD and tutE, two genes in the T1 toluene metabolic pathway, have been generated. The tutD gene codes for an 864-amino-acid protein with a calculated molecular mass of 97,600 Da. The tutE gene codes for a 375-amino-acid protein with a calculated molecular mass of 41,300 Da. Two additional small open reading frames have been identified, but their role is not known. The TutE protein has homology to pyruvate formate-lyase activating enzymes. The TutD protein has homology to pyruvate formate-lyase enzymes, including a conserved cysteine residue at the active site and a conserved glycine residue that is activated to a free radical in this enzyme. Site-directed mutagenesis of these two conserved amino acids shows that they are also essential for the function of TutD.
The aromatic hydrocarbon toluene is a hazardous substance that poses health risks to humans, particularly to the central nervous system. Individuals with long-term exposures may exhibit memory loss, attention and concentration deficiencies, and poor abstracting abilities (17, 21). Toluene has also been shown to serve as the sole carbon source for a number of denitrifying (12, 14, 19, 33, 34), sulfate-reducing (3, 30), and iron-reducing (24) microorganisms. Recently, the mechanism of toluene metabolism under anaerobic conditions has attracted much interest. The mechanism is expected to be different from the aerobic pathways (which all require oxygen for metabolism) and may shed light on the anaerobic metabolism of other aromatic compounds.
A number of novel mechanisms have been proposed for the anaerobic degradation of toluene. All of the suggested pathways have benzoyl-coenzyme A (CoA) serving as a key intermediate, although the initial steps that generate this compound vary in the different proposals (2, 5, 9, 13, 20, 24). Initial reactions proposed include hydroxylation of the aromatic ring to form p-cresol (20, 24, 35), oxidation of the methyl group to form benzyl alcohol (20), attack of the methyl group by fumarate to form benzylsuccinate (2, 5), and attack of the methyl group by acetyl-CoA to form phenylpropionyl-CoA (hydrocinnamoyl-CoA) (9, 13). These proposals have been based primarily on the observation of intermediates or side reaction products appearing in the culture media. Biochemical evidence in strain T and Thauera aromatica K172 (both denitrifying strains) have strongly suggested that a fumarate attack to form benzylsuccinate occurs (2, 5).
Based on the observation that strain T1 accumulates the presumed side reaction products initially identified as benzylsuccinic acid and benzylfumaric acid, Evans et al. (13) have proposed a toluene metabolic pathway for the denitrifying strain T1 (tentatively identified as a Thauera sp.). This pathway proposes that the side reaction products benzylsuccinic acid and benzylfumaric acid are produced by a succinyl-CoA attack on the methyl group of toluene, while productive metabolism occurs via an acetyl-CoA attack on the methyl group to form phenylpropionyl-CoA. In an effort to definitively elucidate the toluene utilization pathway of strain T1, we have undertaken a combined genetic and molecular approach as an alternative to the more biochemical and physiological approaches reported thus far. Previously, we isolated mutants of strain T1 that were unable to grow with toluene serving as the sole carbon source (11). Characterization of one of these mutants led to the identification and cloning of two putative regulatory genes involved in the toluene metabolism of T1 (11).
In the current study, we report the characterization of two additional toluene utilization mutants that are blocked early in the metabolic pathway and the cloning, nucleotide sequences, and predicted protein sequences of a total of four genes. The predicted tutE gene product has homology to pyruvate formate-lyase-activating enzymes (which require iron for function) and ferredoxins, suggesting that iron binding is needed for function of TutE. The predicted tutD gene product has homology to pyruvate formate-lyases (acetyl-CoA–formate C-acetyltransferase [EC 2.3.1.54]) and contains conserved amino acid residues that have been shown to be necessary for function in Escherichia coli pyruvate formate-lyase (29, 31, 36). Site-directed mutagenesis of TutD shows that these residues are also essential for function of this enzyme in the toluene utilization pathway.
MATERIALS AND METHODS
Strains and plasmids.
Isolation and characterization of strain T1, a gram-negative peritrichously flagellated denitrifying organism, have been described previously (14). The E. coli strains HB101 (8), XL-1 Kan Blue (Stratagene, LaJolla, Calif.), and XL-1 Blue (Stratagene), which were used to propagate and transfer DNA, were transformed by the calcium chloride technique (25, 26) or were purchased from the company as competent cells. Strain HB101(pRK2013) (Kanr) (16) contains a helper plasmid that permitted mobilization of cosmids and plasmids into the T1 strain background.
Plasmids used in this study include pRK415 (23) for construction of subclones and matings and the pBluescript vector (Stratagene) for sequencing, subcloning, and preparation of DNA fragments.
Media.
Strain T1 and all strains derived from T1 were grown on either brain heart Infusion (BHI; Difco Laboratories, Detroit, Mich.) medium or a mineral salts medium (15) (vitamins and yeast extract omitted). Unless otherwise specified, toluene (0.3 to 0.5 mM) or pyruvic acid (5 mM) was used as the carbon source to supplement the minimal medium. Nitrate was supplied at a concentration of 10 to 20 mM unless otherwise specified. The plates always contained 2% Agar Noble (Difco Laboratories). Liquid medium was prepared and placed in serum bottles, which were then tightly stoppered with Teflon-coated butyl rubber and aluminum crimp seals. Anaerobic conditions were generated by evacuation and subsequent filling of the bottles with argon. This process was performed a total of four times. E. coli was grown in Luria-Bertani agar or broth (LB) (4) or on BHI agar plates.
The antibiotics kanamycin (used at 50 μg/ml) and tetracycline (used at 25 μg/ml) were supplied to solid or liquid medium when needed. A 12.5-mg/ml stock of tetracycline was made in ethanol. Upon addition to liquid minimal medium, the tetracycline served to select for the cosmid, while the ethanol (final concentration of approximately 17 mM) served as the carbon source for the transconjugant strains.
DNA preparation.
In general, DNA plasmid minipreps were performed by the boiling method of Holmes and Quigley (22) or the QIAprep spin miniprep kit (Qiagen, Santa Clarita, Calif.). When larger-scale preparations were needed, Qiagen maxipreps were carried out according to the manufacturer’s instructions.
Restriction mapping and subcloning.
DNA manipulations were carried out as described by Maniatis et al. (26). All enzymes were obtained from New England Biolabs (Beverly, Mass.).
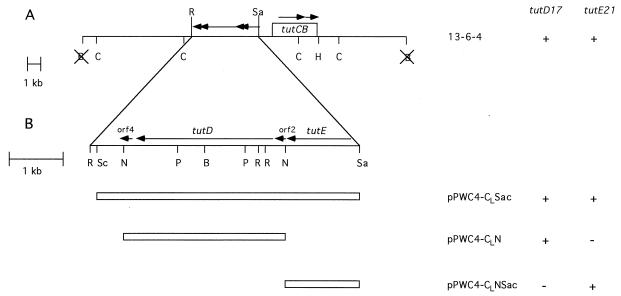
Restriction mapping was carried out with fragments inserted into the pBluescript vector to facilitate identification of restriction sites and to help place the sites on a restriction map. Digests were run on various percentages of agarose gels with size standards to estimate the sizes of the fragments and to locate restriction sites. Fragments were also subcloned into pRK415 to transfer via triparental mating into the mutant strains. Plasmid pPWC4-CLSac was constructed by inserting the 4.9-kb SacI/SacII fragment of cosmid clone 13-6-4 (11) into the SacI site of pRK415 (see Fig. 1). Plasmids pPWC4-CLNSac and pPWC4-CLN were constructed by inserting the 1.3-kb NcoI/SacII fragment and the 3.0-kb NcoI fragment (respectively) of pPWC4-CLSac into the Kpn/SacI site of pRK415 (see Fig. 1). Plasmid pPWC3-uCLΔSacII was constructed by inserting the approximately 9-kb ClaI fragment of cosmid 13-6-4 into the ClaI site of the pBluescript vector (Stratagene) and then deleting the DNA between the SacII sites. The resulting plasmid contains all of the DNA from the ClaI site to the SacII site as shown in Fig. 1.
FIG. 1.
Restriction map of the cosmid clone 13-6-4 (A) and a more detailed map of the region that contains the tutD and tutE genes (B). Four open reading frames contained in the SacII/EcoRI region are indicated by arrows. Subclones tested for complementation are included. The abilities of cosmid clone 13-6-4 and the subclones to complement the tutD17 and tutE21 mutations are also indicated: +, full complementation; −, no complementation. Abbreviations: B, BamHI; C, ClaI; H, HindIII; N, NcoI; P, PstI; R, EcoRI; Sa, SacII; Sc, SacI. Not all sites are shown in panel A. Sites blocked by methylation are omitted. X, the site is lost in the construct.
Triparental mating.
Triparental matings were carried out essentially as described previously (10). Mutants of strain T1 were grown for 3 days in minimal medium containing nitrate and pyruvic acid. HB101 (or XL-1 Kan Blue) carrying the donor cosmid or plasmid was grown in LB-tetracycline overnight. HB101(pRK2013) was grown in LB-kanamycin overnight. One milliliter of each culture was centrifuged, and the pellet was resuspended in 1 ml of 100 mM phosphate buffer (pH 7). Ten microliters of each culture was spotted (one on top of the other) onto a BHI-nitrate plate. After a 3-day incubation at 30°C in an anoxic environment, the resulting growth was scraped off the plate; resuspended in phosphate buffer; spotted onto a minimal agar plate containing pyruvic acid, nitrate, ethanol, and tetracycline to select for transconjugants; and incubated for an additional 3 days at 30°C in an anoxic environment. Cells from the resultant growth were streaked onto the same medium and again incubated in the absence of oxygen. After 3 more days, single transconjugant colonies were isolated from these plates and tested for complementation.
Testing for complementation.
Cosmid clones and subclones constructed in pLAFR3 or plasmid subclones constructed in pRK415 were mated into the mutant backgrounds via the triparental mating technique. The resultant transconjugant strain was tested to determine if the subclone complements the mutation. First, the transconjugants were streaked onto minimal medium containing nitrate in which toluene was supplied in the vapor phase (14). After 5 to 7 days of anaerobic incubation (30°C), the subclones were scored for the ability to restore growth on toluene to the mutants. The transconjugants were also grown in sealed 50-ml serum bottles of liquid minimal medium containing nitrate (10 mM), pyruvic acid (1 mM), and toluene (0.4 mM) with an argon headspace. After 3 to 4 days of incubation (30°C), samples were withdrawn and assayed for toluene utilization and for production of benzylsuccinic acid and a monounsaturated derivative (see below). If the subclone restored the ability of the mutant to grow on toluene, restored the ability to utilize toluene (in the presence of pyruvate) in liquid culture, and restored the ability to produce wild-type levels of benzylsuccinic acid and a monounsaturated derivative, the subclone was considered to complement the mutation.
Analysis of toluene utilization and production of benzylsuccinic acid and a monounsaturated derivative.
Samples of the culture were withdrawn anaerobically with a sterile syringe flushed with argon. The samples were centrifuged (10 min in a Microfuge), and the supernatant was filtered through a 0.45-μm-pore-size filter (Millipore, Bedford, Mass.) into a sample vial. For analysis of benzylsuccinic acid and a monounsaturated derivative, samples were analyzed by high-pressure liquid chromatography (HPLC) with a System Gold HPLC (Beckman, Fullerton, Calif.) equipped with an autosampler and a C18 column (250 by 4.6 mm; particle size, 5 μm; Beckman) with UV detection at 260 nm. The mobile phase was methanol-water-acetic acid (30:68:2) (18) (vol/vol) at a flow rate of 1 ml/min. Peaks were identified by comparison to the external standards benzylmaleic acid and benzylsuccinic acid (13). The cultures consistently produced less benzylsuccinic acid than the monounsaturated derivative, as judged by HPLC peak height. For toluene analysis, samples were analyzed in the same manner with the exception that UV detection was at 254 nm and the mobile phase was acetonitrile-water (55:45 [vol/vol]) at a flow rate of 1 ml/min. Toluene was used as the external standard for peak identification.
Site-directed mutagenesis.
The QuickChange site-directed mutagenesis kit (Stratagene) was used to make mutations in the tutD gene. To change the glycine at position 828 to an alanine, primers G828AF (GTGCGCGTTTCCGCCTACAGCGCTC) and G828AR (GAGCGCTGTAGGCGGAAACGCGCAC) were synthesized and used as directed. Plasmid pPWC3-CLΔSacII (see restriction mapping and subcloning, above) served as the target for mutagenesis. The resulting plasmids were sequenced to identify those containing the desired mutation. The 4.9-kb SacI/SacII fragments of three plasmids with the correct change were subcloned into plasmid pRK415 and used to test for complementation of the tutD17 mutation. To change the cysteine at position 492 to an alanine, primers C492AF (CAACGTGCTGGCCATGTCGCCCGGCATCC) and C492AR (GGATGCCGGGCGACATGCCCAGCACGTTG) were synthesized and used in the manner described above.
DNA sequence analysis.
DNA was sequenced (both strands) by the dideoxy method of Sanger et al. (32) with α-35S-dATP serving as the label. Sequenase enzyme (modified T7 polymerase) and reagents were obtained in a Sequenase kit from U.S. Biochemicals (Cleveland, Ohio). The Bluescript vector and some primers used for sequence analysis were obtained from Stratagene. An Erase-a-Base System (Promega, Madison, Wis.) was used to generate deletions of the cloned DNA inserted in the Bluescript vector for sequence analysis. Synthetic oligonucleotide primers were also purchased so that sequence data could be obtained to fill in gaps not covered by the deletions.
Computer analysis.
Searches for protein sequence similarity were carried out against the nonredundant GenBank protein database using the BLAST 2.0.2 program (1). The Motif program (28) was used to identify patterns in the protein sequences that could have a functional role. Multiple sequence alignments were performed with the Lasergene software package from DNASTAR (Madison, Wis.).
Nucleotide sequence accession number.
The nucleotide sequence reported here has been submitted to the GenBank data base and assigned accession no. AF036765.
RESULTS
Identification and cloning of the tutD and tutE genes.
The isolation of a number of nitrosoguanidine-generated mutants of strain T1 which are deficient in their ability to metabolize toluene has previously been reported (11). One class of mutants, the tutB class, was unable to grow with toluene serving as the sole carbon source but was able to grow when provided with benzoate. These mutants were also unable to metabolize (at wild-type levels) toluene when provided with pyruvate and were unable to produce (at wild-type levels) benzylsuccinic acid and a monounsaturated derivative (13) from toluene in liquid media. Hence, it was determined that this class of mutants was blocked early in the toluene utilization pathway. A cosmid with a genomic insert of approximately 20 kb (cosmid 13-6-4) was isolated for its ability to complement the tutB16 mutation (11). This original cosmid clone, along with a number of subclones generated in the characterization of the tutB gene, were tested for their abilities to complement the mutations referred to as tutB17 and tutB21, which have phenotypes similar to the tutB16 mutation (11). The original cosmid complemented both the tutB17 and tutB21 mutations, while a number of subclones that complemented tutB16 did not (data not shown). Hence, these mutations were placed in new complementation groups and designated tutD17 and tutE21 (see below).
In an effort to determine where on the cosmid the fragments that complement the tutD17 and tutE21 mutations were located, a series of subclones were constructed. Subclones were made in plasmid pRK415, a broad-host-range tetracycline-resistant vector that can be conjugatively transferred into the T1 background (see Materials and Methods). Figure 1A shows a restriction map of cosmid 13-6-4 and a schematic representation of three of the subclones. Each subclone was tested for its ability to complement the tutD17 and tutE21 mutations. Complementation was assayed in three ways: (i) the ability to grow with toluene serving as the sole carbon source on solid media, (ii) the ability to metabolize toluene in the presence of pyruvic acid in liquid media, and (iii) the ability to produce benzylsuccinic acid and a monounsaturated derivative (13) from toluene in liquid media. Restoration of the wild-type phenotype in all three assays was required in order for the subclones to be considered as complementing the mutation.
As shown in Fig. 1B, the tutD17 mutation and the tutE21 mutation were complemented by mutually exclusive subclones. The 3.0-kb NcoI fragment of 13-6-4 (pPWC4-CLN) was able to complement the tutD17 mutation but not the tutE21 mutation. Conversely, the adjacent 1.3-kb NcoI/SacII fragment (pPWC4-CLNSac) was able to complement the tutE21 mutation but not the tutD17 mutation. We conclude from these results that the 3.0-kb NcoI fragment is sufficient to replace the missing activity in the tutD17 mutant strain and that the 1.3-kb NcoI/SacII fragment is sufficient to replace the missing activity in the tutE21 mutant strain. Thus, the two mutations indeed belong to different complementation groups.
Sequence analysis of the SacII/EcoRI region.
The complete nucleotide sequence of the 4,905-bp SacII/EcoRI fragment of cosmid 13-6-4 (containing the tutD and tutE genes) was determined for both strands and has been deposited in the GenBank database (accession no. AF036765). Analysis of this sequence revealed the presence of four open reading frames on the same strand of DNA. The first open reading frame, present between the SacII and NcoI sites (subclone pPWC4-CLNSac) and corresponding to the tutE gene, is a sequence of 375 amino acids. The TutE protein has a calculated molecular mass of 41,300 Da and a predicted pI of 6.8.
Two open reading frames were identified on the 3.0-kb NcoI fragment immediately downstream of the tutE gene (subclone pPWC4-CLN). The first of these two open reading frames (designated open reading frame 2) consists of a 60-amino-acid sequence which would code for a protein with a calculated molecular mass of 6,900 Da and a predicted pI of 5.2. The translational start begins at the NcoI restriction site, and hence no upstream transcriptional regulatory sites or ribosome binding sites for this open reading frame are included on this fragment. Therefore, it is highly unlikely that this open reading frame is responsible for the complementation of the tutD17 mutation observed with this subclone. This observation, along with evidence from the site-directed mutagenesis experiments (see below), indicates that ORF2 is not the tutD gene. The significance of this open reading frame is not known.
The second open reading frame in this fragment is 864 amino acids in length, with a calculated molecular mass of 97,600 Da. The predicted pI of this protein is 6.0. Results from the site-directed mutagenesis clearly show that this open reading frame corresponds to the tutD gene (see below).
The fourth open reading frame (designated open reading frame 4) identified in the SacII/EcoRI fragment consists of a sequence of 81 amino acids with a calculated molecular mass of 9,300 Da and a predicted pI of 7.8. The pPWC4-CLN subclone removes approximately 50% of the C-terminal end of this protein. This result, in conjunction with the evidence presented regarding the third open reading frame, indicates that this 81-amino-acid protein is not the tutD gene product. The significance of this open reading frame is not known.
Protein sequence similarity.
The protein sequence of the tutD gene product was used to search the GenBank nonredundant database for proteins with homology to TutD. The BLAST program identified a number of similar proteins, all of which are identified as either pyruvate formate-lyases (formate acetyltransferases) or pyruvate formate-lyase homologs. Interestingly, the sequences showing the highest degree of similarity with TutD are the E. coli proteins f810 (27% identical to TutD as calculated by the BLAST program) (7) and PflD (26% identical to TutD) (6), both pyruvate formate-lyase homologs. The sequence similarities between TutD and these two proteins plus PflB (22% identical to TutD), a pyruvate formate-lyase from E. coli (31, 37), are shown in Fig. 2. As can be seen in the figure, the most conserved region is in the carboxyl end of these proteins. There is a highly conserved region around the glycine residue at position 828 of TutD (marked with an asterisk). In the E. coli pyruvate formate-lyase, this glycine has been shown to form a free radical which is essential for enzymatic function (36). Additionally, in a less-conserved region, there is a cysteine residue at position 492 of TutD (marked with a dagger) that has been shown to transiently form a covalent bond with the acetyl group that is being transferred, an action which is also essential to enzyme function (29, 31). The results of this protein sequence similarity analysis suggest a mechanism for TutD where glycine-828 forms a free radical which is necessary for the transient formation of a covalent bond between cysteine-492 and the compound (possibly acetate or fumarate) that is being transferred to the methyl group of toluene (or a toluene metabolite). This mechanism may involve a transient cysteine radical at an undetermined location, as proposed in the E. coli pyruvate formate-lyase system (37).
FIG. 2.
Comparison of the predicted amino acid sequence of the TutD protein with the predicted sequences of the f810 protein of E. coli (7), the PflD protein of E. coli (6), and the PflB protein of E. coli (31, 37). The conserved glycine residue that is proposed to form a free radical is indicated by an asterisk (position 828 of TutD). The conserved cysteine residue that is proposed to form a transient covalent bond is indicated by a dagger (position 492 of TutD). Amino acids identical to the tutD translation are shaded, and conserved amino acids are boxed. Dashes indicate gaps introduced by the computer program to maximize the alignment score.
A similar search was performed with the protein sequence of the tutE gene product. The proteins with the highest degree of homology were identified as pyruvate formate-lyase-activating enzymes or pyruvate formate-lyase-activating enzyme homologs. The sequence similarities between TutE and f308 (34% identical to TutE as calculated by the BLAST program) (7), PflC (32% identical to TutE) (6), and PflA (28% identical to TutE) (31) (all from E. coli) are shown in Fig. 3. In addition, the BLAST search identified a number of ferredoxins with weaker homology scores (data not shown). Subsequent subjection of the TutE protein sequence to a Motif analysis (28) identified a radical activating region from amino acids 60 to 81 (Fig. 3). This region which contains potential Fe binding sites (as identified by the Motif analysis) is conserved in the pyruvate formate-lyase activating enzymes. Additionally, the analysis revealed a 4Fe-4S binding domain typically found in ferredoxins (amino acids 98 to 109 [Fig. 3]). This region is not very well conserved in the E. coli pyruvate formate-lyase activating enzyme and homologs. PflA is missing this region, and both f308 and PflC have alterations to the spacing or sequence. The results of this protein sequence similarity analysis are consistent with the predicted role of TutE serving as the activator for TutD and suggest that the activation may involve iron and/or iron-sulfur binding.
FIG. 3.
Comparison of the predicted amino acid sequence of the TutE protein with the predicted sequences of the f308 protein of E. coli (7), the PflC protein of E. coli (6), and the PflA protein of E. coli (31). The region defined as a radical-activating region is indicated by a line (positions 60 to 81 of TutE). The region defined as a 4Fe-4S binding domain typical of ferredoxins is indicated by an open box (positions 98 to 109 of TutE). This motif was identified only in TutE. Amino acids identical to the tutE translation are shaded, and conserved amino acids are boxed. Dashes indicate gaps introduced by the computer program to maximize the alignment score.
Site-directed mutagenesis of the TutD protein.
In an effort to determine if the conserved glycine and cysteine residues of TutD play an essential role in the enzymatic function of the protein, as has been shown for PflB (29, 31, 36), both amino acids were individually changed to an alanine as described in Materials and Methods. Three independent isolates of the resulting plasmids (pPWC4-CLSac-G828A and pPWC4-CLSac-C492A) were mated into the strain carrying the tutD17 mutation, and the resulting transconjugants were then tested for their abilities to complement the mutation. The plasmid carrying the unaltered clone (pPWC4-CLSac) fully complements the tutD17 mutation (utilizes 100% of the toluene provided in the presence of pyruvate and produces wild-type levels of benzylsuccinic acid and a monounsaturated derivative). Neither of the altered plasmids pPWC4-CLSac-G828A and pPWC4-CLSac-C492A was able to fully complement the tutD17 mutation (Table 1). Both of these strains utilized about the same amount of toluene as that utilized by the mutant carrying plasmid pRK415, the vector alone. Likewise, they produced significantly less benzylsuccinic acid and a monounsaturated derivative than the tutD17 mutant strain carrying the unaltered plasmid pPWC4-CLSac. The mutant carrying plasmid pPWC4-CLSac-C492A produced about the same amount of these compounds as the mutant carrying plasmid pRK415, while the strain carrying plasmid pPWC4-CLSac-G828A showed higher levels of these compounds than the vector alone but levels much lower than those observed with the unaltered plasmid. Since the E. coli pyruvate formate-lyase is known to be a homodimer which requires the formation of only one glycine free radical (37), the small amount of activity observed in the mutant carrying plasmid pPWC4-CLSac-G828A may be due to mixed dimers in which the free radical forms on the defective chomosomally encoded TutD protein. The results in Table 1 clearly demonstrate that glycine 828 and cysteine 492 are essential for function of the TutD protein. Thus, the role of a glycine free radical and a covalent substrate-cysteine bond are proposed to be key mechanistic features of the TutD protein in its role in toluene metabolism by strain T1.
TABLE 1.
Complementation of the tutD17 mutation by the site-directed mutations constructed in the tutD gene.
| Plasmid | % Toluene utilized | % Monounsaturated, benzylsuccinic acid-derived, compound producedc |
|---|---|---|
| pPWC4-CLSaca | 100 | 100 |
| pRK415b | 23.5 ± 6.4 | 1.3 ± 0.1 |
| pPWC4-CLSac-G828A | 34.2 ± 9.7 | 13.0 ± 3.8 |
| pPWC4-CLSac-C492A | 17.7 ± 5.4 | 1.8 ± 0.1 |
The plasmid carrying the unaltered clone, serving as a positive control.
The vector alone, serving as a negative control.
Normalized to 100% for pPWC4-CLSac, the positive control.
DISCUSSION
We have isolated two mutants defective in an early step of toluene degradation and cloned the region of DNA that complements both mutations. Analysis of the complementing region indicates that these mutations lie in two different complementation groups and hence two different genes. Sequence analysis of this region has revealed four open reading frames, two of which correspond to the two mutated genes (see results). The open reading frame corresponding to the gene named tutD has sequence similarity to pyruvate formate-lyase and pyruvate formate-lyase homologs. Correspondingly, the tutE gene product has sequence similarity to pyruvate formate-lyase-activating enzymes and their homologs as well as to ferredoxins.
Although suggestive, the sequence similarities do not by themselves provide sufficient evidence for the mechanism for the TutD and TutE proteins. However, for the E. coli pyruvate formate-lyase, several key amino acids which are essential for function have been identified. The results from the site-directed mutagenesis studies of the TutD protein indicate that the conserved glycine residue at position 828 is essential for protein function, suggesting that TutD may form a free radical necessary for enzymatic function. Similarly, the cysteine residue at position 492 of TutD is also essential for protein function, as shown in the site-directed mutagenesis studies. These results suggest a mechanism for TutD whereby glycine-828 forms a stable free radical which is necessary for the transient covalent attachment to cysteine-492 of the compound that is being transferred to the methyl group of toluene (or a toluene metabolite). This mechanism may involve a transient cysteine radical at an undetermined location, as proposed in the E. coli pyruvate formate-lyase system (37).
Based on sequence similarity, we also propose that the toluene utilization protein TutE functions in a manner similar to that of the pyruvate formate-lyase activator proteins such that the TutE protein is necessary for the formation of the glycine free radical at position 828 of TutD. This mechanism may require the putative radical activation domain found in TutE. However, since no single amino acid has yet been determined to be essential for the pyruvate formate-lyase activator proteins, the site-directed mutagenesis experiments of tutE are less straightforward. It is interesting to note that the TutE protein also has a ferredoxin-like domain which is not found in any of the pyruvate formate-lyase activator/pyruvate formate-lyase activator homologs (as identified by the Motif search). The glycine free radical activation domain is believed to bind iron which would be necessary to form the free radical. Ferredoxins also bind iron via a 4Fe-4S binding domain. The presence of this 4Fe-4S binding domain in TutE may indicate that both regions are necessary to form the free radical in TutD.
Although the toluene utilization enzymes of strain T1 may have mechanistic similarities to the pyruvate formate-lyases, it must be kept in mind that the compounds being metabolized are structurally different. Clearly the tutD gene product is not required for anaerobic pyruvate metabolism, since strains carrying the tutD17 mutation are able to grow as well as the wild type under anaerobic conditions with pyruvate as the sole carbon source. This suggests that the TutD protein is unlikely to be a pyruvate formate-lyase per se but has a similar catalytic mechanism. Indeed, an examination of Fig. 2 shows that there is little sequence similarity between TutD and the pyruvate formate-lyases over the amino-terminal half of the protein. This would suggest that although there appear to be mechanistic similarities between the proteins, there are significant differences, perhaps because of the need to bind different substrates. Additionally, the TutD protein must act early in toluene metabolism, since benzoic acid metabolism is not affected in the tutD17 mutant strain (11).
The isolation of mutants demonstrates that the tutD and tutE genes are necessary for the toluene utilization of strain T1, but other enzymes are undoubtedly involved in this pathway as well. For example, it is not known what role, if any, the two open reading frames flanking the tutD gene play in this metabolism. The protein products could possibly play a direct enzymatic role or could be part of a protein complex involved in toluene metabolism. Alternatively, they could have a regulatory role or no metabolic role at all. By obtaining mutations in these genes, it could be determined if these putative proteins have a function in toluene metabolism. Additionally, another mutant unable to grow on toluene was isolated in the previously reported screen that generated tutD17 and tutE21 (11). This gene product appears to be necessary for toluene utilization, since the mutant strain is also unable to grow with toluene serving as the sole carbon source. However, this mutation is not complemented by the cosmid clone 13-6-4. Cloning and characterization of this gene will lead to further insights into the toluene utilization pathway of strain T1. Continued molecular characterization of the tut genes and use of these genes for biochemical studies should more clearly define the mechanistic steps involved in the toluene utilization pathway of strain T1.
ACKNOWLEDGMENTS
This work was supported in part by NSF grant MCB9507132 and DARPA grant N00014-92-J-1888. We acknowledge support from the Center for Agricultural Molecular Biology, Rutgers University, the New Jersey Commission on Science and Technology (L.Y.Y.), and Ohio University (P.W.C.).
We thank David Scheraga for help in generating the site-directed mutations, Don Kobayashi for supplying plasmids, and Gerben Zylstra for supplying strains. We also thank Gerben Zylstra, Mike Logan, Olivia Harriott, Anne Frazer, and Karen Coschigano for helpful discussion and comments on the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Lipman W M D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beller H R, Spormann A M. Anaerobic activation of toluene and o-xylene by addition to fumarate in denitrifying strain T. J Bacteriol. 1997;179:670–676. doi: 10.1128/jb.179.3.670-676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beller H R, Spormann A M, Sharma P K, Cole J R, Reinhard M. Isolation and characterization of a novel toluene-degrading sulfate-reducing bacterium. Appl Environ Microbiol. 1996;62:1188–1196. doi: 10.1128/aem.62.4.1188-1196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1952;167:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biegert T, Fuchs G, Heider J. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur J Biochem. 1996;238:661–668. doi: 10.1111/j.1432-1033.1996.0661w.x. [DOI] [PubMed] [Google Scholar]
- 6.Blattner F R, Burland V, Plunkett G, Sofia H J, Daniels D L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993;21:5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science (Washington, DC) 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 9.Chee-Sanford J C, Frost J W, Fries M R, Zhou J, Tiedje J M. Evidence for acetyl coenzyme A and cinnamoyl coenzyme A in the anaerobic toluene mineralization pathway in Azoarcus tolulyticus Tol-4. Appl Environ Microbiol. 1996;62:964–973. doi: 10.1128/aem.62.3.964-973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coschigano P W, Häggblom M M, Young L Y. Metabolism of both 4-chlorobenzoate and toluene under denitrifying conditions by a constructed bacterium. Appl Environ Microbiol. 1994;60:989–995. doi: 10.1128/aem.60.3.989-995.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coschigano P W, Young L Y. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl Environ Microbiol. 1997;63:652–660. doi: 10.1128/aem.63.2.652-660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolfing J, Zeyer J, Binder-Eicher P, Schwarzenbach R P. Isolation and characterization of a bacterium that mineralizes toluene in the absence of molecular oxygen. Arch Microbiol. 1990;154:336–341. doi: 10.1007/BF00276528. [DOI] [PubMed] [Google Scholar]
- 13.Evans P J, Ling W, Goldschmidt B, Ritter E R, Young L Y. Metabolites formed during anaerobic transformation of toluene and o-xylene and their proposed relationship to the initial steps of toluene mineralization. Appl Environ Microbiol. 1992;58:496–501. doi: 10.1128/aem.58.2.496-501.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans P J, Mang D T, Kim K S, Young L Y. Anaerobic degradation of toluene by a denitrifying bacterium. Appl Environ Microbiol. 1991;57:1139–1145. doi: 10.1128/aem.57.4.1139-1145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans P J, Mang D T, Young L Y. Degradation of toluene and m-xylene and transformation of o-xylene by denitrifying enrichment cultures. Appl Environ Microbiol. 1991;57:450–454. doi: 10.1128/aem.57.2.450-454.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foo S C, Phoon W O, Lee J. Neurobehavioral symptoms among workers occupationally exposed to toluene. Asia Pac J Public Health. 1988;2:192–197. doi: 10.1177/101053958800200310. [DOI] [PubMed] [Google Scholar]
- 18.Frazer A C, Ling W, Young L Y. Substrate induction and metabolite accumulation during anaerobic toluene utilization by the denitrifying strain T1. Appl Environ Microbiol. 1993;59:3157–3160. doi: 10.1128/aem.59.9.3157-3160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fries M R, Zhou J, Chee-Sanford J, Tiedje J M. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grbic-Galic D, Vogel T M. Transformation of toluene and benzene by mixed methanogenic cultures. Appl Environ Microbiol. 1987;53:254–260. doi: 10.1128/aem.53.2.254-260.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hänninen H, Eskelinen L, Husman K, Nurminen M. Behavioral effects of long-term exposure to a mixture of organic solvents. Scand J Work Environ Health. 1976;4:240–255. doi: 10.5271/sjweh.2805. [DOI] [PubMed] [Google Scholar]
- 22.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 23.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 24.Lovley D R, Lonergan D J. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism GS-15. Appl Environ Microbiol. 1990;56:1858–1864. doi: 10.1128/aem.56.6.1858-1864.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel M, Higa A. Calcium dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 27.Migaud M E, Chee-Sanford J C, Tiedje J M, Frost J W. Benzylfumaric, benzylmaleic, and Z- and E-phenylitaconic acids: synthesis, characterization, and correlation with a metabolite generated by Azoarcus tolulyticus Tol-4 during anaerobic toluene degradation. Appl Environ Microbiol. 1996;62:974–978. doi: 10.1128/aem.62.3.974-978.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogiwara A, Uchiyama I, Takagi T, Kanehisa M. Construction and analysis of a profile library characterizing groups of structurally known proteins. Protein Sci. 1996;5:1991–1999. doi: 10.1002/pro.5560051005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plaga W, Frank R, Knappe J. Catalytic-site mapping of pyruvate formate lyase. Eur J Biochem. 1988;178:445–450. doi: 10.1111/j.1432-1033.1988.tb14468.x. [DOI] [PubMed] [Google Scholar]
- 30.Rabus R, Nordhaus R, Ludwig W, Widdel F. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol. 1993;59:1444–1451. doi: 10.1128/aem.59.5.1444-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rödel W, Plaga W, Frank R, Knappe J. Primary structure of Escherichia coli pyruvate formate-lyase and pyruvate formate-lyase activating enzyme deduced from the DNA nucleotide sequences. Eur J Biochem. 1988;177:153–158. doi: 10.1111/j.1432-1033.1988.tb14356.x. [DOI] [PubMed] [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schocher R J, Seyfried B, Vazquez F, Zeyer J. Anaerobic degradation of toluene by pure cultures of denitrifying bacteria. Arch Microbiol. 1991;157:7–12. doi: 10.1007/BF00245327. [DOI] [PubMed] [Google Scholar]
- 34.Su J-J, Kafkewitz D. Utilization of toluene and xylenes by a nitrate-reducing strain of Pseudomonas maltophilia under low oxygen and anoxic conditions. FEMS Microbiol Ecol. 1994;15:249–258. [Google Scholar]
- 35.Vogel T M, Grbic-Galic D. Incorporation of oxygen from water into toluene and benzene during anaerobic fermentative transformation. Appl Environ Microbiol. 1986;52:200–202. doi: 10.1128/aem.52.1.200-202.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volker-Wagner A F, Frey M, Neugebauer F A, Schäfer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner A F, Frey M, Neugebauer F A, Schäfer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]