Abstract
Improved nitrogen-fixing inoculum strains for leguminous crops must be able to effectively compete with indigenous strains for nodulation, enhance legume productivity compared to the productivity obtained with indigenous strains, and maintain stable expression of any added genes in the absence of selection pressure. We constructed a transposable element containing the tfx region for expression of increased nodulation competitiveness and the par locus for plasmid stability. The transposon was inserted into tetA of pHU52, a broad-host-range plasmid conferring the H2 uptake phenotype. The resulting plasmid, pHUTFXPAR, conferred the plasmid stability, trifolitoxin production, and H2 uptake phenotypes in the broad-host-range organism Sinorhizobium sp. strain ANU280. The broad applications of a transposon conferring plasmid stability are discussed.
An important goal in nitrogen fixation research is genetic improvement of inoculum strains of root nodule bacteria sold commercially for the formation of nitrogen-fixing root nodules on leguminous crops. Three obstacles have prevented the achievement of this goal (25). First, genes that enhance nitrogen fixation must be identified. Second, mechanisms which enhance the ability of an inoculum strain to compete for infection sites with indigenous root nodule bacteria must be discovered. And third, sufficient knowledge of these genes must be available in order to make constructs that allow stable expression of the genes in the absence of selection pressure. The progress made in these areas was recently reviewed by Maier and Triplett (25).
Hydrogen is an obligate product of the nitrogenase reaction (37). One phenotype expressed by microsymbionts that has been shown to increase legume yield is the H2 uptake phenotype (commonly referred to as the Hup phenotype). With the Hup phenotype, the root nodule bacteria recover the energy lost in the production of the H2 evolved during the nitrogenase reaction (12, 46). The increases in soybean yields are as great as 17% (11). However, most indigenous strains of root nodule bacteria that infect leguminous crops do not possess uptake hydrogenase activity. In Bradyrhizobium japonicum, less than 25% of the isolates collected from the soybean-growing region of the northeastern United States were Hup+ (17, 44). Very few strains of the alfalfa microsymbiont, Sinorhizobium meliloti, are Hup+, and even the strains that are Hup+ are not efficient at recovering the energy obtained from hydrogen oxidation (22, 23). In addition, very few strains of Rhizobium leguminosarum bv. viceae, the pea microsymbiont, are Hup+, and most of these strains are unable to efficiently couple H2 oxidation to ATP formation (27, 28, 34).
Transfer of the yield-enhancing Hup phenotype to Rhizobium, Sinorhizobium, and Bradyrhizobium inoculum strains requires the isolation of the genes involved in this process. Significant progress has been made in elucidating the genetics of the uptake hydrogenases of B. japonicum and R. leguminosarum (for a review, see reference 25). Lambert et al. (19) isolated a cosmid clone, pHU52, from a B. japonicum 122DES gene bank, and this cosmid clone conferred uptake hydrogenase activity and chemolithotrophic growth to Hup− strains of B. japonicum, S. meliloti, R. leguminosarum bv. viceae, and R. leguminosarum bv. trifolii. However, as pHU52 is not stable in the absence of selection pressure, legume plants inoculated with pHU52-containing root nodule bacteria expressed low levels of hydrogen uptake activity (20, 21).
As a result, pHU52 has no commercial value for enhancing legume productivity despite the fact that it contains all of the genes necessary for full Hup phenotype expression in Bradyrhizobium, Rhizobium, and Sinorhizobium strains. Weinstein et al. have described a set of genes from RK2 that is capable of conferring complete plasmid stability in the absence of selection pressure in both free-living and nodule bacteroids of S. meliloti (47). We describe here the construction and use of a transposon that includes the plasmid stability locus from RK2. This transposon was used to stabilize pHU52 both in free-living cells and in nodule bacteroids in the absence of selection pressure. Similarly, we constructed a broad-host-range plasmid that includes genes that enhance nodulation competitiveness, as well as hydrogen uptake and plasmid stability.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are shown in Table 1. Escherichia coli strains were maintained on Luria-Bertani medium (35) at 37°C, while Rhizobium and Sinorhizobium strains were cultured on the synthetic medium of Bergersen et al. (1) at 28°C. Antibiotics were used at the following concentrations: 100 μg/ml for ampicillin, 50 μg/ml for kanamycin and spectinomycin, 20 μg/ml for nalidixic acid and chloramphenicol, and 2.5 μg/ml for tetracycline. Streptomycin was used at a concentration of 25 μg/ml for E. coli strains and at a concentration of 50 μg/ml for Rhizobium and Sinorhizobium strains.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| C2110nal | Nalr derivative of C2110, polA | 10 |
| DH5 | Nalr, recA | Bethesda Research Laboratories |
| HB101 | Smr, recA | 4 |
| R. leguminosarum bv. viceae 128C1 | TFXs strain used in trifolitoxin assays | LiphaTech, Inc. |
| Sinorhizobium sp. strain ANU280 | Smr derivative of Sinorhizobium sp. strain NGR234 | 7 |
| Plasmids | ||
| pTR102 | par, Tcr | 47 |
| pTFX24 | pBluescript II KS+::7.1-kb MluI fragment of pTFX1 | 5 |
| pHoKmGus | Tn3HoHo derivative, Tn3 (Kmr AprtnpA, promoterless uidA) | 2 |
| pSShe | pACYC184::tnpA, Cmr | 38 |
| pRK2013 | ColE1 mobilization helper (Tra+ Mob+), Kmr | 13 |
| pRK2073 | ColE1 mobilization helper (Tra+ Mob+), Spr Smr | 10 |
| pLAFR1 | RK2-derived broad-host-range cosmid vector, Tcr | 14 |
| pHU52 | pLAFR1 with 30-kb insert containing the hup locus, Tcr | 19 |
| pTn3PAR | 3.2-kb par locus cloned into the ClaI site of pHoKmGus | This study |
| pTn3TFXPAR | 10.4-kb fragment containing tfx operon and par locus cloned into the ClaI site of pHoKmGus | This study |
| pLAFR1::Tn3 | pLAFR1 carrying Tn3Gus within tetA, Tcs Kmr | This study |
| pLAFR1::PAR | pLAFR1 carrying Tn3PAR within tetA, Tcs Kmr | This study |
| pTFXPAR | pLAFR1 carrying Tn3TFXPAR within tetA, Tcs Kmr | This study |
| pHUTn3 | pHU52 carrying Tn3Gus within tetA, Tcs Kmr | This study |
| pHUPAR | pHU52 carrying Tn3PAR within tetA, Tcs Kmr | This study |
| pHUTFXPAR | pHU52 carrying Tn3TFXPAR within tetA, Tcs Kmr | This study |
Construction of transposons.
The transposable par locus of pTn3PAR was constructed by excising par from pTR102 on a 3.2-kb KpnI-BamHI fragment and cloning this fragment into the ClaI site of pHoKmGus by using blunt-end ligation. pTn3TFXPAR carries a transposable element which includes both tfx and par. This transposon was constructed by cloning the 3.2-kb par locus into the XhoI site of pTFX24 by using blunt-end ligation. A 10.4-kb ApaI-SacI fragment containing both the par locus and the tfx operon was then inserted into the ClaI site of pHoKmGus.
Transposition into tetA of pHU52 and pLAFR1.
To accomplish transposition into the tetA gene of the cosmids, pHoKmGus, pTn3PAR, and pTn3TFXPAR were each transformed into HB101(pSshe, pHU52) and HB101(pSshe, pLAFR1). To enhance the transposition frequency of Tn3Gus, Tn3PAR, and Tn3TFXPAR, the transformation reaction mixtures were incubated at 28°C (18, 43). Transformants were selected on plates containing only kanamycin. Kanamycin-resistant transformants from each transformation were pooled and used as the donors in triparental matings in which E. coli C2110 was used as the recipient and HB101(pRK2073) was used as the helper strain. Transconjugants were selected for Nalr and Kmr and then screened for Tcs. Since pHoKmGus and its derivatives are not able to replicate in a polA strain, such as C2110, this procedure allows isolation of pLAFR1 and pHU52 with an insertion in the tet region.
The transpositions of pTn3Par and pTn3TFXPAR into tetA of pHU52 and pLAFR1 were verified by performing PCR that included primers for tetA found on the cosmids and uidA found on the transposon. Each reaction mixture contained primer uidA-R (5′-TTGGGGTTTCTACAGGACG-3′) and either primer tetA-F (5′-GTGAAACCCAACATACCC-3′) or primer tetA-R (5′-CGGCTCGTTGCCCTGCG-3′). Amplification products that were 0.3 to 1.2-kb long indicated that successful transposition into tetA had occurred. This procedure also allowed us to determine the orientation of each transposon within tetA. The identities of the hup-containing plasmids were verified by performing PCR for the hupS region of each cosmid with primers hupS-F (5′-ATGGGCGCGGCGACGGAAAC-3′) and hupS-R (5′-TCAGCTGTTGTGGTCGGCGT-3′).
Plant culture, hydrogen evolution, and nitrogenase assays.
Seeds were germinated in Leonard jars and were inoculated 3 days after planting. Plants were cultured in a growth chamber as described by Datta et al. (8). Hydrogen evolution by whole root systems of Vigna unguiculata (L.) Walp. cv. Blackeye was determined by gas chromatography 24 days after inoculation with Sinorhizobium sp. strain ANU280 or a derivative of this strain carrying pHU52, pHUTn3, pHUPAR, pHUTFXPAR, pLAFR1, pLAFR1::Tn3, pLAFR1::PAR, or pLAFR1::TFXPAR. Gas chromatography was performed as described by Hanus et al. (15) with a Shimadzu gas chromatograph, and N2 was used as the carrier gas. Nitrogenase assays were performed by the acetylene reduction method as described by Rasche and Arp (30) by using the same root systems used for the H2 evolution determination. The relative efficiency of root nodule nitrogenase activity was determined as described by Van Kessel and Burris (45) by measuring C2H2 reduction and H2 evolution during acetylene reduction, as well as in the absence of C2H2.
Plasmid stability and trifolitoxin production assays.
Sinorhizobium sp. strain ANU280 was the background strain used for the plasmids used in the plasmid stability and trifolitoxin production assays. Plasmid stability was determined in free-living cells as described by Weinstein et al. (47), except that cells were grown in hydrogen uptake medium broth (24). Trifolitoxin production was determined by the plate assay described by Breil et al. (5) by using R. leguminosarum bv. viceae 128C1 as the sensitive strain, except that the assay was done on hydrogen uptake medium. Plasmid maintenance in planta was assessed by collecting nodules from the root systems used for hydrogen evolution assays and determining the percentages of cells that contained the plasmid found in the inoculum strain.
RESULTS
Cloning and transposition strategies.
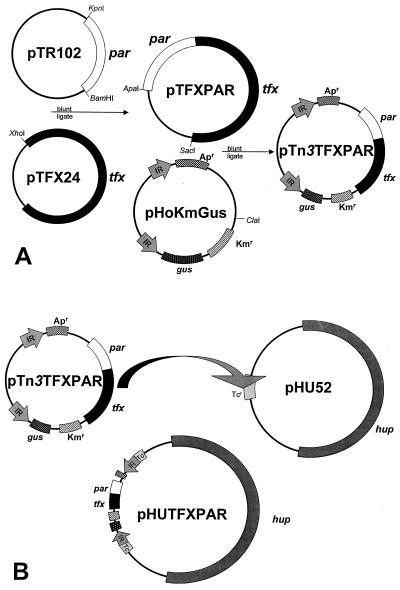
The cloning and transposition strategies used in this study are shown in Fig. 1. The strategy which we used to stabilize hydrogenase expression involved inserting the 3.2-kb par locus into pHU52. This was accomplished by inserting the par locus into the inverted repeats of a Tn3 transposon and then transposing the par locus into the tetracycline resistance gene of pHU52, tetA. The possibility that the par locus would transpose out of pHU52 after insertion was greatly reduced by providing a transposase in trans. In addition, as the Tn3 transposon which we used for this study contains kanamycin and ampicillin resistance genes, selection for pHU52 remained after tetA was interrupted. The resulting plasmid is referred to below as pHUPAR.
FIG. 1.
Strategy used to construct pHUTFXPAR. The par locus from pTR102 was cloned into a unique XhoI site of pTFX24. An ApaI-SacI fragment containing tfx and par was then cloned into a unique ClaI site of pHoKmGus to create pTn3TFXPAR (A). The Tn3 derivative containing tfx and par was transposed into tetA of pHU52 to generate pHUTFXPAR (B). pHUPAR was constructed in a similar fashion by using a transposon containing par but not tfx.
The transposable partitioning system also allowed us to insert other genes of interest into the inverted repeats of the Tn3 transposon and then transpose them into pHU52. Of particular interest was the cassette of the tfxABCDEFG genes from R. leguminosarum bv. trifolii T24 that confers trifolitoxin production and resistance (5). Trifolitoxin is a peptide antibiotic that inhibits the growth of a specific group of the α subclass of the Proteobacteria (42). Rhizobium strains with the ability to produce trifolitoxin have been shown to exhibit increased nodulation competitiveness (33, 39–41). Thus, by adding both tfxABCDEFG and the partitioning locus in the inverted repeats of Tn3GUS, we were able to add the trifolitoxin production phenotype to pHU52 and to stabilize pHU52 by creating pHUTFXPAR. As in the construction of pHUPAR, we screened for transposition events within tetA.
Insertion of the transposons into tetA had a number of purposes. First, it allowed us to easily screen for insertion into a specific gene within pHU52. Second, it ensured that the genes of interest had not transposed into a gene essential for plasmid replication or hydrogen uptake. And third, it removed tetracycline resistance from this plasmid, eliminating a potential regulatory hurdle to the commercialization of this technology.
Plasmid stability in vitro and in planta.
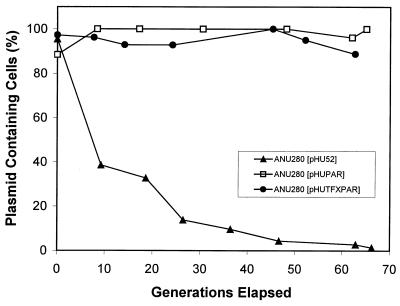
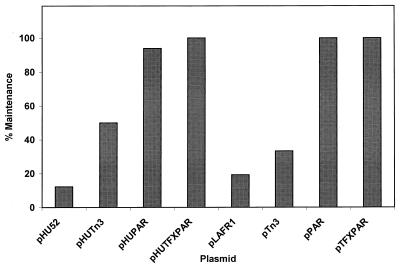
The stability of each plasmid was assessed in vitro by serial passage of each strain for more than 60 generations without selection for the plasmid marker. The addition of the par locus from RK2 by transposition into pHU52 completely stabilized this plasmid in ANU280. The resulting plasmid, pHUPAR, was stable in the absence of selection pressure for more than 60 generations (Fig. 2). The addition of the tfx region had no effect on this stabilization since both pHUPAR and pHUTFXPAR were very stable in the absence of selection pressure (Fig. 2). In contrast, pHU52 was absent from 90 and 99% of the ANU280 cells after 30 and 66 generations, respectively (Fig. 2). The stability of the plasmids in nodule bacteroids was characterized by determining the percentage of the bacteria in a nodule that contained the plasmid present in the inoculum. The presence of the transposeable par locus conferred plasmid stability throughout the infection process (Fig. 3).
FIG. 2.
Proportions of free-living Sinorhizobium sp. strain ANU280 cultured in the absence of selection pressure that expressed the appropriate antibiotic resistance characteristics for the plasmid present in the inoculum strain.
FIG. 3.
Proportions of Sinorhizobium sp. strain ANU280 recovered from nodules that expressed the appropriate antibiotic resistance characteristics for the plasmid present in the inoculum strain.
Trifolitoxin production.
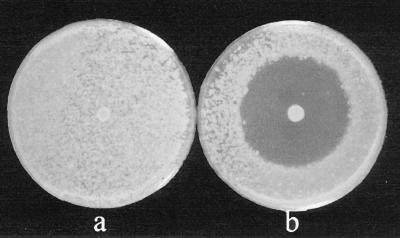
Trifolitoxin was produced by any Rhizobium or Sinorhizobium strain carrying either pHUTFXPAR or pLAFR1::TFXPAR. Trifolitoxin production by ANU280 (pHUTFXPAR) is illustrated in Fig. 4. Strains lacking the tfx region failed to inhibit the trifolitoxin-sensitive strain R. leguminosarum bv. viceae 128C1.
FIG. 4.
Lack of trifolitoxin production by ANU280(pHUPAR) (a) and trifolitoxin production by ANU280(pHUTFXPAR) (b). In both cases the inhibited strain was R. leguminosarum bv. viceae 128C1. Identical results were obtained with these plasmids with several other Sinorhizobium and Rhizobium strains.
Hydrogen evolution and acetylene reduction by root nodules.
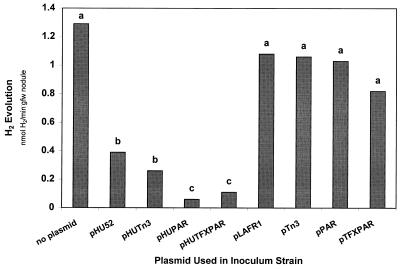
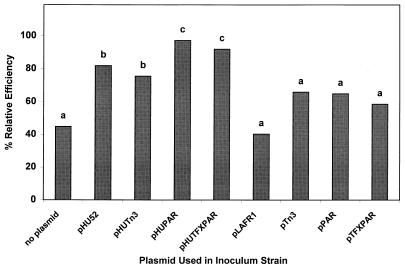
Hydrogen is an obligate product of the nitrogenase reaction. Bacteroid hydrogen oxidation has been shown to improve the efficiency of nitrogen fixation (46). This phenotype is often referred to as the Hup phenotype. Cowpea nodules infected with any strain lacking the uptake hydrogenase region from pHU52 evolved significant amounts of H2 (Fig. 5). As expected, pHU52-infected nodules also evolved significant amounts of H2 as a result of plasmid loss during the infection and nodule development processes. However, nodules infected with either ANU280(pHUPAR) or ANU280(pHUTFXPAR) evolved very low amounts of H2 (Fig. 5) since each of these strains harbors a plasmid that contains both the uptake hydrogenase genes from B. japonicum 122DES and the plasmid stability locus from RK2. As the stabilization of pHU52 reduced H2 evolution in root nodules to nearly undetectable levels, the relative efficiency of electron allocation in bacteroids of ANU280 containing either pHUPAR or pHUTFXPAR as defined by Van Kessel and Burris (45) was significantly enhanced compared with the relative efficiency of electron allocation in ANU280 or its derivatives lacking either the uptake hydrogenase genes or the stability locus (Fig. 6).
FIG. 5.
Hydrogen evolution from cowpea root systems. The values for bars with the same letter were statistically similar at the 5% confidence level. Each value is the mean of the values from three replicates. gfw, gram (fresh weight).
FIG. 6.
Relative efficiency of nitrogenase electron allocation. The values for bars with the same letter were statistically similar at the 5% confidence level. Each value is the mean of the values from three replicates. Relative efficiency (RE) was determined by the formula of Van Kessel and Burris (45): RE = 1 − [(H2 evolution in air)/(C2H2 reduction + H2 evolution in 10% C2H2)].
DISCUSSION
Rationale for the cloning and transposition strategies used.
We used a transposable partitioning locus to stabilize a large cosmid clone in gram-negative bacteria. Our strategy was based on the discovery of a plasmid partitioning locus in the broad-host-range plasmid RK2 (47). A 3.2-kb par locus from RK2, which contains five genes in two divergently transcribed operons, forces complete plasmid partitioning to occur during cell division. Each operon codes for an independent plasmid stability mechanism. The parCBA operon includes a resolvase mechanism; the parDE operon encodes a toxin (ParE)-antitoxin (ParD) mechanism (16). The par locus confers plasmid stability regardless of the replicon containing the 3.2-kb region (9, 31, 32). This region has also been shown to confer complete plasmid stability in S. meliloti during root nodule development (47). The ability of this par region to confer plasmid stability in other species of root nodule bacteria has been confirmed by workers in our laboratory (33, 42).
The hup-containing cosmid pHU52 is unstable in the absence of selection and does not increase legume productivity in agricultural situations. Addition of the par locus to this cosmid allows stable expression of the Hup phenotype to occur.
Benefits of the approach presented here.
Stabilization of a cosmid clone harboring commercially useful genes is important in many situations. For our purposes, we were faced with the dilemma of finding a method to transfer and stably express a large set of genes that confer a yield-enhancing phenotype present in certain root nodule bacteria for leguminous crops. As the complete nucleotide sequence of the uptake hydrogenase region is not known, it is very difficult to identify unique restriction sites for the cloning of this large (30-kb) set of genes into a vector useful for marker exchange in the chromosome. In addition, cloning these genes into a chromosome or megaplasmid would require identification of symbiotically silent sites in the genome of every species in which the uptake hydrogenase phenotype is desired.
Identification of symbiotically silent sites is difficult. During the 1980s, geneticists at Biotechnica International identified such sites in S. meliloti by cloning antibiotic resistance genes into regions thought to be symbiotically silent. Alfalfa greenhouse yield trials were performed to determine the effects of the insertions on the symbiosis (3). In these greenhouse experiments, no effect was observed when cloning into either the inositol catabolism locus or a site between the nif and fix regions called P3 was performed. However, multiple-year and multiple-site alfalfa yield trials performed in the field revealed significantly altered yields after insertion into these sites (36). Thus, at least for alfalfa, the effects of gene deletions or insertions in a microsymbiont on the crop yield of a rhizobium-legume symbiotic system can often be observed only if the plants are grown to maturity or under proper agricultural conditions (11, 36).
In addition to the practical problems of identifying symbiotically silent insertion sites, such sites have to be identified for every microsymbiont species of interest. A symbiotically silent site in S. meliloti may not be present or may not be symbiotically silent in R. leguminosarum. Thus, it may be necessary to develop new insertion strategies for each species.
To avoid these problems, we decided to construct a stably maintained, broad-host-range plasmid which could be used to improve the symbiotic properties of root nodule bacteria. pHUTFXPAR can be transferred easily to any Kms or Aps strain of root nodule bacteria by conjugation. In addition, it is stably maintained in the absence of selection pressure and provides two phenotypes that provide improved efficacy to an inoculum strain. First, as a result of the presence of the trifolitoxin production and resistance genes, this multicopy plasmid confers high levels of trifolitoxin production. Trifolitoxin production has been shown to increase nodulation competitiveness in soil (33). The presence of the tfx genes on a multicopy plasmid, such as pHUTFXPAR, results in much higher levels of trifolitoxin production than the level of production that would occur if single copies of the genes were present on the chromosome (39, 40). The addition of nodulation competitiveness on a multicopy plasmid is better than chromosomal integration of multiple copies of competitiveness genes in tandem, as described recently (26). Concatamers containing repeated regions of nodulation competitiveness genes can easily recombine with one another and be lost over time. This is especially true of many Rhizobium genomes that are known to be unstable as a result of recombination events (6, 29).
Second, the presence of the uptake hydrogenase genes on pHUTFXPAR is expected to enhance legume yield compared to the legume yield obtained after infection with rhizobia that lack this phenotype. The presence of these genes on pHUTFXPAR and pHUPAR was confirmed by the significant reductions in H2 evolution from ANU280 (pHUTFXPAR)- and ANU280(pHUPAR)-infected nodules; H2 evolution was reduced by 91.5 and 95.3%, respectively. Thus, the addition of pHUTFXPAR to a wide variety of root nodule bacteria can be used to enhance the productivity of many legumes worldwide. All of this was done without the need for complex cloning strategies that include symbiotically silent sites as described previously (3).
Limitations to the current approach and their solutions.
The limitations of the approach described here are few, and all of them can be overcome. For example, pTn3PAR is limited in that it confers kanamycin resistance. Thus, our approach is limited to target plasmids that lack kanamycin resistance. This limitation is easily overcome by excising the kanamycin resistance gene in pHoKmGus and replacing it with either a gene that confers resistance to a different antibiotic or a reporter gene, such as the gene for the green fluorescent protein. Also, pTn3PAR does include a promoterless reporter gene, uidA, that is expressed if it is transposed in the proper orientation into an expressed gene.
Another limitation of the technology described here is also one of its advantages. This limitation is the fact that the genes provided to root nodule bacteria by pHUTFXPAR are present on a broad-host-range plasmid rather than inserted into the chromosome. The advantages of this are described above. The primary disadvantage is that plasmid pHUTFXPAR can be mobilized from one strain to another in the environment. In our specific example, this may not necessarily be a disadvantage as the improved symbiotic properties of the plasmid may be transferred to indigenous rhizobia, albeit at a low frequency, and improve their characteristics. However, a way to restrict the movement of a plasmid such as pHUTFXPAR can be envisioned with one modification. One of the two mechanisms of plasmid stability conferred by the par locus of RK2 is a toxin-antitoxin system expressed by parE and parD. Spatial separation of these two genes such that parD is on the chromosome and parE is on the broad-host-range plasmid should substantially reduce the transfer of the plasmid to other bacteria, as the plasmid would express only production of the toxin.
Thus, any shortcomings of the strategy described here can be overcome with additional refinements of the system. These refinements should also enhance the number of useful applications of this system.
Other applications for the transposable stability locus.
The approach described here for stabilization of pHU52 has broad applications for any plasmid used in biotechnology for which providing antibiotic selection pressure is very expensive or, as is the case in our example, impossible. The transposable stability locus constructed here, pTn3PAR, can provide stability to any plasmid maintained in any bacterium which can serve as a host for RK2. As RK2 has the broadest host range of any known plasmid, the applicability of this approach is extensive. Once stability is provided to a plasmid, the transposon remains stable since the transposase is provided in trans on a nonreplicating plasmid (2, 38).
As we did here with pHUTFXPAR, additional valuable genes can be added to a plasmid along with the plasmid stability region by simply cloning into the transposon another set of genes along with the par locus from RK2. The entire cassette can then be transposed into the plasmid of interest.
ACKNOWLEDGMENTS
We thank the USDA NRICGO, LiphaTech, Inc. (Milwaukee, Wis.), and the University of Wisconsin-Madison College of Agricultural and Life Sciences for providing support for this work.
REFERENCES
- 1.Bergersen F J, Gibson A H, Licis I. Growth and N2-fixation of soybeans inoculated with strains of Bradyrhizobium japonicum differing in energetic efficiency and Phb utilization. Soil Biol Biochem. 1995;27:611–616. [Google Scholar]
- 2.Bonas U, Stall R E, Staskawicz B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol Gen Genet. 1989;218:127–136. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- 3.Bosworth A H, Williams M K, Albrecht K A, Hankinson T R, Kwiatkowski R, Beynon J, Ronson C W, Cannon F, Wacek T J, Triplett E W. Alfalfa yield response to inoculation with recombinant strains of Rhizobium meliloti carrying an extra copy of dct and/or modified nifA expression. Appl Environ Microbiol. 1994;60:3815–3832. doi: 10.1128/aem.60.10.3815-3832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 5.Breil B T, Ludden P W, Triplett E W. DNA sequence and mutational analysis of genes involved in the production and resistance of the antibiotic peptide trifolitoxin. J Bacteriol. 1993;175:3693–3702. doi: 10.1128/jb.175.12.3693-3702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brom S, Garcia los Santos A, de Lourdes-Girard M, Davila G, Palacios R, Romero D. High-frequency rearrangements in Rhizobium leguminosarum bv. phaseoli plasmids. J Bacteriol. 1991;173:1344–1346. doi: 10.1128/jb.173.3.1344-1346.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Batley M, Redmond J, Rolfe B G. Alteration of the effective nodulation properties of a fast-growing broad host range Rhizobium due to changes in exopolysaccharide synthesis. J Plant Physiol. 1985;120:331–350. [Google Scholar]
- 8.Datta D B, Wong P P, Triplett E W. Immunocytochemical localization of glutamine synthetase in organs of Phaseolus vulgaris L. Plant Physiol. 1991;96:507–512. doi: 10.1104/pp.96.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis T L, Helinski D R, Roberts R C. Transcription and autoregulation of the stabilizing functions of the broad-host-range plasmid RK2 in Escherichia coli, Agrobacterium tumefaciens, and Pseudomonas aeruginosa. Mol Microbiol. 1992;6:1969–1979. doi: 10.1111/j.1365-2958.1992.tb01371.x. [DOI] [PubMed] [Google Scholar]
- 10.Ditta G. Tn5 mapping of Rhizobium nitrogen fixation genes. Methods Enzymol. 1986;118:519–528. [Google Scholar]
- 11.Evans H J, Hanus F J, Haugland R A, Cantrell M A, Xu L S, Russell F J, Lambert G R, Harker A R. World Soybean Research Conference. III. Boulder, Colo: Westview Press; 1985. Hydrogen recycling in nodules affects nitrogen fixation and growth of soybeans; pp. 935–942. [Google Scholar]
- 12.Evans H J, Harker A R, Papen H, Russell F J, Hanus F J, Zuber M. Physiology, biochemistry, and genetics of the uptake hydrogenase in rhizobia. Annu Rev Microbiol. 1987;41:335–361. doi: 10.1146/annurev.mi.41.100187.002003. [DOI] [PubMed] [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 15.Hanus F J, Carter K R, Evans H J. Techniques for measurement of hydrogen evolution by nodules. Methods Enzymol. 1980;69:731–739. [Google Scholar]
- 16.Johnson E P, Ström A R, Helinski D R. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J Bacteriol. 1996;178:1420–1429. doi: 10.1128/jb.178.5.1420-1429.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keyser H H, Weber D F, Uratsu S L. Rhizobium japonicum serogroup and hydrogenase phenotype distribution in 12 states. Appl Environ Microbiol. 1984;47:613–615. doi: 10.1128/aem.47.4.613-615.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kretschmer P J, Cohen S N. Effects of temperature on translocation frequency of the Tn3 element. J Bacteriol. 1979;139:515–519. doi: 10.1128/jb.139.2.515-519.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert G R, Cantrell M A, Hanus F J, Russell S A, Haugland R A, Haddad K R, Evans H J. Intra- and interspecies transfer and expression of Rhizobium japonicum hydrogen uptake genes and autotrophic growth capability. Proc Natl Acad Sci USA. 1985;82:3232–3236. doi: 10.1073/pnas.82.10.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert G R, Harker A R, Cantrell M A, Hanus F J, Russell S A, Haugland R A, Evans H J. Symbiotic expression of cosmid-borne Bradyrhizobium japonicum hydrogenase genes. Appl Environ Microbiol. 1987;53:422–428. doi: 10.1128/aem.53.2.422-428.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert G R, Harker A R, Zuber M, Dalton D A, Hanus F J, Russell S A, Evans H J. Characterization, significance and transfer of hydrogen uptake genes from Rhizobium japonicum. In: Evans H J, Bottomley P J, Newton W E, editors. Nitrogen fixation research progress. Dordrecht, The Netherlands: Nijhoff; 1985. pp. 209–215. [Google Scholar]
- 22.Lentzsch P, Miksch G. Detection of uptake hydrogenase in Rhizobium leguminosarum and Rhizobium meliloti. Zentralbl Mikrobiol. 1988;143:269–274. [Google Scholar]
- 23.Lentzsch P, Miksch G. Effect of different Hup phenotypes of Rhizobium meliloti on symbiotic properties of alfalfa. Zentralbl Mikrobiol. 1989;144:67–71. [Google Scholar]
- 24.Maier R J, Campbell N E R, Hanus F J, Simpson F B, Russell S A, Evans H J. Expression of hydrogenase activity in free-living Rhizobium japonicum. Proc Natl Acad Sci USA. 1978;75:3258–3262. doi: 10.1073/pnas.75.7.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier R J, Triplett E W. Toward more productive, efficient, and competitive nitrogen-fixing symbiotic bacteria. Crit Rev Plant Sci. 1996;15:191–234. [Google Scholar]
- 26.Mavingui P, Flores M, Romero D, Martinez-Romero E, Palacios R. Generation of Rhizobium strains with improved symbiotic properties by random DNA amplification. Nat Biotechnol. 1997;15:564–569. doi: 10.1038/nbt0697-564. [DOI] [PubMed] [Google Scholar]
- 27.Nelson L M. Hydrogen recycling by Rhizobium leguminosarum isolates and growth and nitrogen contents of pea plants (Pisum sativum L.) Appl Environ Microbiol. 1983;45:856–861. doi: 10.1128/aem.45.3.856-861.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson L M, Salminen S O. Uptake hydrogenase activity and ATP formation in Rhizobium leguminosarum bacteroids. J Bacteriol. 1982;151:989–995. doi: 10.1128/jb.151.2.989-995.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios R, Castillo M, Flores M, Hernandez G, Mavingui P, Romero D. Dynamics of the Rhizobium genome. Curr Plant Sci Biotechnol Agric. 1995;27:353–357. [Google Scholar]
- 30.Rasche M E, Arp D J. Hydrogen inhibition of nitrogen reduction by nitrogenase in isolated soybean nodule bacteroids. Plant Physiol. 1989;91:663–668. doi: 10.1104/pp.91.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts R C, Helinski D R. Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. J Bacteriol. 1992;174:8119–8132. doi: 10.1128/jb.174.24.8119-8132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts R C, Ström A, Helinski D R. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J Mol Biol. 1994;237:35–52. doi: 10.1006/jmbi.1994.1207. [DOI] [PubMed] [Google Scholar]
- 33.Robleto E A, Scupham A J, Triplett E W. Trifolitoxin production in Rhizobium etli strain CE3 increases competitiveness for rhizosphere growth and root nodulation of Phaseolus vulgaris in soil. Mol Plant Microbe Interact. 1997;10:228–233. [Google Scholar]
- 34.Ruiz-Argueso T, Maier R J, Evans H J. Hydrogen production and uptake by pea nodules as affected by strains of Rhizobium leguminosarum. Arch Microbiol. 1978;116:113–118. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Scupham A J, Bosworth A H, Ellis W R, Wacek T J, Albrecht K A, Triplett E W. Inoculation with recombinant strains of Sinorhizobium meliloti increases alfalfa yield compared to the wild-type strain. Appl Environ Microbiol. 1996;62:4260–4262. doi: 10.1128/aem.62.11.4260-4262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson F B, Burris R H. A nitrogen pressure of 50 atmospheres does not prevent evolution of hydrogen by nitrogenase. Science. 1984;224:1095–1097. doi: 10.1126/science.6585956. [DOI] [PubMed] [Google Scholar]
- 38.Stachel S E, Gynheung A, Floes C, Nester E W. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Triplett E W. Construction of a symbiotically effective strain of Rhizobium leguminosarum bv. trifolii with increased nodulation competitiveness. Appl Environ Microbiol. 1990;56:98–103. doi: 10.1128/aem.56.1.98-103.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Triplett E W. Isolation of genes involved in nodulation competitiveness from Rhizobium leguminosarum bv. trifolii T24. Proc Natl Acad Sci USA. 1988;85:3810–3814. doi: 10.1073/pnas.85.11.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Triplett E W, Barta T M. Trifolitoxin production and nodulation are necessary for the expression of superior nodulation competitiveness by Rhizobium leguminosarum bv. trifolii T24. Plant Physiol. 1987;85:335–342. doi: 10.1104/pp.85.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Triplett E W, Breil B T, Splitter G A. Expression of tfx and sensitivity to the rhizobial peptide antibiotic trifolitoxin in a taxonomically distinct group of α-proteobacteria including the animal pathogen Brucella abortus. Appl Environ Microbiol. 1994;60:4163–4166. doi: 10.1128/aem.60.11.4163-4166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner A K, de la Cruz F, Grinsted J. Temperature sensitivity of transposition class II transposons. J Gen Microbiol. 1990;136:65–67. doi: 10.1099/00221287-136-1-65. [DOI] [PubMed] [Google Scholar]
- 44.Uratsu S L, Keyser H H, Weber D F, Lim S T. Hydrogen uptake (HUP) activity of Rhizobium japonicum from major U.S. soybean production areas. Crop Sci. 1982;22:600–602. [Google Scholar]
- 45.Van Kessel C, Burris R H. Effect of H2 evolution on 15N2 fixation, C2H2 reduction and relative efficiency of leguminous symbionts. Plant Physiol. 1983;59:329–334. [Google Scholar]
- 46.Van Soom C, Rumjanek N, Vanderleyden J, Neves M C P. Hydrogenase in Bradyrhizobium japonicum: genetics, regulation and effect on plant growth. World J Microbiol Biotechnol. 1993;9:615–624. doi: 10.1007/BF00369567. [DOI] [PubMed] [Google Scholar]
- 47.Weinstein M, Roberts R C, Helinski D R. A region of the broad-host-range plasmid RK2 causes stable in planta inheritance of plasmids in Rhizobium meliloti cells isolated from alfalfa in root nodules. J Bacteriol. 1992;174:7486–7489. doi: 10.1128/jb.174.22.7486-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]