Abstract
The presence or absence of two DNA modification systems, XorI and XorII, in 195 strains of Xanthomonas oryzae pv. oryzae collected from different major rice-growing countries of Asia was assessed. All four possible phenotypes (XorI+ XorII+, XorI+ XorII−, XorI− XorII+ and XorI− XorII−) were detected in the population at a ratio of approximately 1:2:2:2. The XorI+ XorII+ and XorI− XorII+ phenotypes were observed predominantly in strains from southeast Asia (Philippines, Malaysia, and Indonesia), whereas strains with the phenotypes XorI− XorII− and XorI+ XorII− were distributed in south Asia (India and Nepal) and northeast Asia (China, Korea, and Japan), respectively. Based on the prevalence and geographic distribution of the XorI and XorII systems, we suggest that the XorI modification system originated in northeast Asia and was later introduced to southeast Asia, while the XorII system originated in southeast Asia and moved to northeast Asia and south Asia. Genomic DNA from all tested strains of X. oryzae pv. oryzae that were resistant to digestion by endonuclease XorII or its isoschizomer PvuI also hybridized with a 7.0-kb clone that contained the XorII modification system, whereas strains that were digested by XorII or PvuI lacked DNA that hybridized with the clone. Size polymorphisms were observed in fragments that hybridized with the 7.0-kb clone. However, a single hybridization pattern generally was found in XorII+ strains within a country, indicating clonal maintenance of the XorII methyltransferase gene locus. The locus was monomorphic for X. oryzae pv. oryzae strains from the Philippines and all strains from Indonesia and Korea.
Xanthomonas oryzae pv. oryzae causes bacterial blight, the most important bacterial disease of rice in Asia (15, 16). Compared to the long history of rice cultivation, the deployment of genes for resistance to X. oryzae pv. oryzae in commercial rice cultivars is relatively recent. The introduction of these genes for resistance into rice is correlated with a change in the pathogenic diversity of X. oryzae pv. oryzae populations, that is, new races of the pathogen emerge and overcome deployed resistance (15, 17). These observations have stimulated much curiosity concerning the contribution of host genotype and other factors to the genetic diversity of the pathogen.
Multilocus molecular markers have been used in conjunction with virulence typing to evaluate the diversity and structure of X. oryzae pv. oryzae populations within and between countries in Asia (1, 3, 6, 11, 12, 21, 27). In general, regionally defined pathogen populations in Asia were found to be distinct (1). This finding could be due either to slow pathogen migration or dispersal or to spatial partitioning of host genotypes (different cultivar preferences between regions). Although populations within a region generally were similar, in some cases genetically similar strains were detected in different regions, suggesting the migration of strains between countries, possibly as a consequence of germ plasm exchange (1, 6). Within one country, the Philippines, significant differentiation in populations was observed between different agroecosystems; that is, populations of X. oryzae pv. oryzae in the cool, mountainous highlands, where one crop of traditional varieties per year is grown, were different from those in the tropical lowlands, where two or three crops of semidwarf, early-maturing rice varieties per year are grown (3). Since several ecological factors in addition to host genotype likely influenced the genetic diversity of the pathogen population collected in that study (3), the magnitude of the contribution of any one of those factors to pathogen diversity is not clear.
To more critically evaluate the genetic structure and movement of X. oryzae pv. oryzae populations throughout Asia and to understand some of the factors that influence the population structure of this pathogen, we have been investigating two X. oryzae pv. oryzae DNA restriction modification (R-M) systems (XorI and XorII) previously shown to be present in the pathogen (5, 28). R-M systems are particularly interesting for such studies, because they are thought to protect the bacterial genome from invasion by introduced bacteriophage or plasmid DNA (9) and thus may inhibit genomic variability due to DNA exchange on uptake. We cloned and sequenced two genes that are part of the XorII R-M system (xorIIM and xorII-vsr) (5). Not all X. oryzae pv. oryzae strains contained the XorI and XorII modification systems (5, 24), indicating that the systems might prove useful for developing an understanding of the origins of X. oryzae pv. oryzae genetic lineages and their distribution patterns. In this study, we determined the distribution of the two modification systems in a group of X. oryzae pv. oryzae strains collected from throughout Asia. Based on this information, we formulated a hypothesis on the geographic origin and historical migration of the DNA modification systems. In addition, we discuss the potential influence of these modification systems on the genetic stability of X. oryzae pv. oryzae populations.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The 195 strains of X. oryzae pv. oryzae used in this study were obtained from Korea (18 strains collected from five provinces between 1987 and 1989), the Philippines (64 strains collected from a wide range of ecosystems on three different islands, Luzon, Mindanao, and Visayas [see reference 11] between 1972 and 1990), India (10 strains from five states, collected in 1987, 1990, and 1991), Malaysia (7 strains from three provinces, collected between 1982 and 1989), Indonesia (14 strains from two islands, collected in 1976, 1990, and 1992), and Nepal (33 strains from 15 districts, collected between 1987 and 1989) and were provided by K. S. Jin (National Institute of Agricultural Sciences and Technology, Rural Development Administration, Suweon, Korea), T. W. Mew (International Rice Research Institute, Los Baños, the Philippines), S. Gnanamanickam (University of Madras, Madras, India), K. S. Lum (Malaysian Agriculture Research and Development Institute, Serdang, Selangor, Malaysia), R. H. Hartini (Bogor Research Institute of Food Crops, Bogor, Indonesia), and T. Adhikari (Institute of Agriculture and Animal Science, Kathmandu, Nepal), respectively. DNAs of strains from China (43 strains from 15 provinces, collected from 1981 to 1986) and Japan (6 strains from five prefectures, collected in 1968, 1971, and 1985) were provided by Q. Zhang (Institute of Crop Breeding and Cultivation, Chinese Academy of Agricultural Science, Beijing, China) and M. Watabe (Sekisui Chemical Co., Ltd., Osaka, Japan), respectively. A summary of data relevant to this communication is included in Table 1; data for each strain, including the site of collection within a country, the year of collection, the race (pathotype), and the restriction fragment length polymorphism (RFLP) type with multilocus probes, are available by request from J. E. Leach.
TABLE 1.
Distribution of XorI and XorII DNA modification systems of X. oryzae pv. oryzae in Asiaa
| Region of Asia and country of origin | No. of strains | No. of strains with the following DNA modification system patternb:
|
Genetic diversityc | |||
|---|---|---|---|---|---|---|
| XorI+XorII+ | XorI+XorII− | XorI−XorII+ | XorI−XorII− | |||
| South | ||||||
| India | 10 | 0 | 0 | 0 | 10 | 0.98 |
| Nepal | 33 | 0 | 0 | 6 | 27 | 0.92 |
| Southeast | ||||||
| Philippines | 64 | 18 | 0 | 33 | 13 | 0.97 |
| Malaysia | 7 | 2 | 1 | 4 | 0 | 0.72 |
| Indonesia | 14 | 4 | 0 | 10 | 0 | 0.59 |
| Northeast | ||||||
| China | 43 | 2 | 38 | 1 | 2 | 0.93 |
| Korea | 18 | 4 | 14 | 0 | 0 | 0.89 |
| Genetic diversity in each DNA modification system groupd | 0.95 | 0.74 | 0.97 | 0.95 | ||
The distribution of the four different R-M systems was nonrandom within the three geographic regions of Asia (south, southeast, and northeast), based on an analysis by Fisher’s exact test (P = 0.00001).
The DNA modification system phenotype was determined by sensitivity or resistance to PstI (isoschizomer of XorI) and XorII and/or its isoschizomer PvuI as well as hybridization with xorIIM and xorII-vsr genes in pE7.0. Resistance to digestion indicates that the DNA modification system is present (+), and sensitivity to digestion indicates that the system is absent (−).
Genetic diversity was calculated with the formula [n/(n − 1)][1 − ΣXi2], where Xi is the proportion of the ith RFLP type within a group and n is the number of strains tested in each group. Data are from RFLP analysis performed with two probes, pJEL101 and pBSavrXa10 (1). Diversity was calculated from the combined data for each country. Data for Japan are not reported because RFLP data were not available for these strains.
Genetic diversity determined with RFLP data as described in footnote c but partitioned for DNA modification system group.
X. oryzae pv. oryzae strains were cultured in peptone-sucrose broth (26) or nutrient broth (Difco Laboratories, Detroit, Mich.) at 28°C with shaking at 200 rpm. Bacterial genomic DNA was isolated by a lysozyme-sodium dodecyl sulfate lysis procedure (22) modified as described previously (10).
Strains of X. oryzae pv. oryzicola (159.14m from China and BLS335 from the Philippines) and strains from various pathovars of X. campestris (pathovars vesicatoria [65-2], alfalfae [KX-1], malvacearum [28], holcicola [123], pisi [NCPPB762], pruni [ATCC 19316], cucurbitae [NZ2299], fragariae [ICPBXF122], and phleipratensis [PDDCC5744]) were from L. Claflin, Kansas State University.
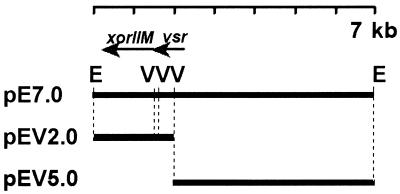
Plasmid pE7.0 contains a 7.0-kb fragment from X. oryzae pv. oryzae which includes the gene encoding XorII methyltransferase (xorIIM) and a vsr-like gene (very-short-patch-repair endonuclease gene; xorII-vsr) in the vector pBluescript KS+ (Fig. 1) (5). Plasmid pEV2.0 contains xorIIM and the 3′ end of xorII-vsr in a 2.0-kb insert, whereas plasmid pEV5.0 contains the 5′ portion of xorII-vsr and 5′-flanking regions in a 5.0-kb fragment. These plasmids were used as probes for hybridization of X. oryzae pv. oryzae genomic DNA that had been digested with EcoRV, EcoRI, and BamHI (see below). Plasmid DNA from Escherichia coli was prepared by the alkaline lysis method as described by Morelle (18).
FIG. 1.
Restriction digestion maps of clones that contain the XorII methyltransferase gene from X. oryzae pv. oryzae JW89011. The direction of transcription of xorIIM, which codes for XorII methyltransferase, and vsr, which has sequence identity with a very-short-patch-repair endonuclease gene, is indicated by the arrowheads. V, EcoRV; E, EcoRI.
DNA analysis.
The presence of XorI and XorII modification systems in X. oryzae pv. oryzae was determined by the resistance of genomic DNA to digestion with PstI (isoschizomer of XorI) and XorII and/or its isoschizomer PvuI, respectively. Digestion conditions were as described by the enzyme manufacturer (Promega), except that a severalfold excess of each enzyme was added and the mixtures were incubated for 3 h.
RFLP analysis.
To analyze the genome organization around the xorIIM and xorII-vsr loci, genomic DNA was digested to completion with EcoRI, BamHI, or EcoRV, fractionated by gel electrophoresis, and transferred to nylon membranes (Magna NT; MSI, Westboro, Mass.) as described previously (4). The blot was probed with pE7.0 (Fig. 1). For diversity analysis, RFLP analysis of genomic DNA was performed with two multilocus markers as described previously (1). The probes were plasmids containing a mobile, repetitive DNA element, IS1112 (in plasmid pJEL101; 10), and a member of a multicopy avirulence gene family, avrXa10 (in plasmid pBSavrXa10; 7). Genomic DNA of each bacterial strain was digested to completion with BamHI (for hybridization with pBSavrXa10) or EcoRI (for hybridization with pJEL101). A 1-kb ladder (Bethesda Research Laboratories) was included in gels as a size standard. The plasmids were labeled with [32P]dCTP by use of a nick translation kit (Bethesda Research Laboratories); vector (pBluescript II and pUC18) DNAs did not hybridize with X. oryzae pv. oryzae genomic DNA (data not shown). High-stringency hybridization, washing conditions, and autoradiography were as described previously (10).
Data analysis.
Genetic diversity determined by RFLP analysis with multilocus markers for X. oryzae pv. oryzae subpopulations with the four possible DNA modification system phenotypes was estimated with Nei and Tajima’s haplotypic diversity index (19, 20). The genetic diversity within each R-M group was estimated with the formula [n/(n − 1)][1 − ΣXi2], where Xi is the proportion of the ith RFLP type within each DNA modification system group and n is the number of strains tested in each group. The distributions of the DNA modification system phenotypes in the three regions (south, southeast, and northeast) of Asia were compared with Fisher’s exact test as previously described (3); analysis was performed with the program StatXact (version 1.00; Cytel Software Corp., Cambridge, Mass.).
RESULTS
DNA modification systems in X. oryzae pv. oryzae and their distribution in Asia.
Genomic DNAs from a collection of 195 strains of X. oryzae pv. oryzae were evaluated for the presence or absence of the XorI and XorII modification systems (summarized in Table 1). The presence of the XorI modification system was assumed if genomic DNA was resistant to digestion with PstI, an isoschizomer of XorI. The XorII modification system was identified by resistance of DNA to digestion with XorII or PvuI (isoschizomer of XorII) and by hybridization of the genomic DNA to clone pE7.0, which contains the XorII R-M system (5). DNA from all strains that were resistant to digestion with XorII and PvuI hybridized with pE7.0, whereas DNA from strains that were digested with the two enzymes did not hybridize with pE7.0.
As determined by Fisher’s exact test, the distribution of the XorI and XorII modification systems in Asia is not random and is differentiated by geographic region (Table 1). Of the four possible phenotypes (XorI+ XorII+, XorI+ XorII−, XorI− XorII+, and XorI− XorII−), only the XorI− XorII− phenotype was found in the 10 strains from the northern, central, and eastern parts of India (Table 1). Most Nepalese strains (27 of 33), also collected from widely separated geographic regions within the country, exhibited the XorI− XorII− phenotype. The XorI+ XorII+ and XorI− XorII+ phenotypes were predominantly observed in strains from southeast Asia (Philippines, Malaysia, and Indonesia). Both phenotypes were present at different sampling sites in the Philippines, indicating that the two phenotypes were widely distributed within the country. The XorI− XorII− phenotype was found in the Philippines in only one group of strains, race 6, which were detected only at a single site. The strains from northeast Asia (north China, Korea, and Japan) exhibited both the XorI+ XorII+ and the XorI+ XorII− phenotypes, although the most prevalent phenotype in north China (38 of 43 strains) and Korea (14 of 18 strains) was XorI+ XorII−. The DNA modification systems (XorI− XorII+ and XorI− XorII−) of the three strains from south China were different from those of the majority of strains collected in north and northeast China and were more similar to those of strains from southeast (XorI− XorII+) and south (XorI− XorII−) Asia (Table 1).
Genetic diversity of X. oryzae pv. oryzae with different DNA modification systems.
The genomic diversity in X. oryzae pv. oryzae strains with each of the four DNA modification system phenotypes was assessed with RFLP data obtained from an analysis with two multicopy DNA markers, avrXa10 and IS1112 (1). Analysis with these markers resulted in a combined total of 64 potential band positions (1). The RFLP data were partitioned for country of origin and for each of the four DNA modification system phenotypes, and the genetic diversity was calculated. Diversity was high when the data were partitioned by country of origin, ranging from 0.59 for Malaysia to 0.98 for India. Diversity was high in all DNA modification system groups, ranging from 0.74 for XorI+ XorII− to 0.97 for XorI− XorII+ (Table 1).
Genomic organization of X. oryzae pv. oryzae around the XorII modification locus.
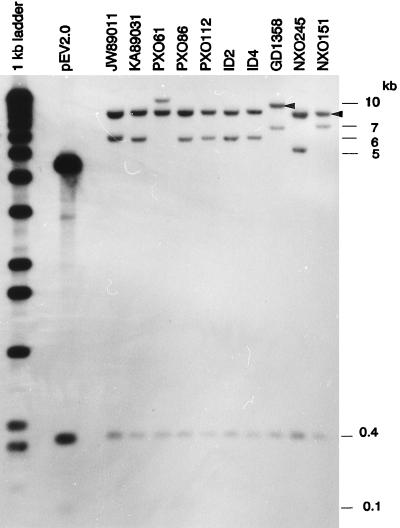
The genomic organization around the XorII modification locus was determined by RFLP analysis with the probes pEV2.0 and pEV5.0, which are subclones of a 7.0-kb region (pE7.0) that contains the locus. A total of 33 X. oryzae pv. oryzae strains originating from different countries and whose DNA was resistant to digestion with XorII were selected at random. EcoRV fragments of DNA from the strains were separated by electrophoresis, blotted to membranes, and hybridized with pEV2.0 or pEV5.0 (Fig. 1). Two EcoRV fragments that hybridized with plasmid pEV2.0 (0.1 and 0.4 kb) were found in DNA from all tested strains (Fig. 2 and 3). In addition, pEV2.0 hybridized with a fragment of approximately 5, 6, 7, or 10 kb, depending on the strain. Plasmid pEV5.0 hybridized with fragments of approximately 8.5 or 9 kb (Fig. 2). The 0.1-kb fragment contains sequences for conserved domains II and III of XorII methyltransferase (Fig. 1) (also see references 5 and 23). The 0.4-kb fragment contains sequences for the conserved domain I of XorII methyltransferase as well as the C terminus of the putative Vsr peptide. Other fragments that hybridized with pEV2.0 included the remainder of the xorIIM gene and downstream (3′) flanking regions (Fig. 1 and 3). Fragments that hybridized with pEV5.0 contained the 5′ portion of the xorII-vsr gene and sequences upstream (5′) of the gene (Fig. 1 to 3).
FIG. 2.
Composite blot showing RFLPs in DNA from X. oryzae pv. oryzae strains from different countries. PXO, strains from the Philippines; ID, strains from Indonesia; NXO, strains from Nepal. Strain GD1358 is from China, and strains JW89011 and KA89031 are from Korea. Fragments of genomic DNA generated by digestion with EcoRV were separated by electrophoresis, transferred to nylon membranes, and hybridized with either pEV2.0 or pEV5.0. Note that this blot shows hybridization to both probes. Bands labeled with sizes in kilobases hybridized with pEV2.0; note that bands at approximately 0.1 kb are not visible at this exposure. Bands at approximately 8.5 and 9.0 kb (marked with arrowheads) hybridized with pEV5.0.
FIG. 3.
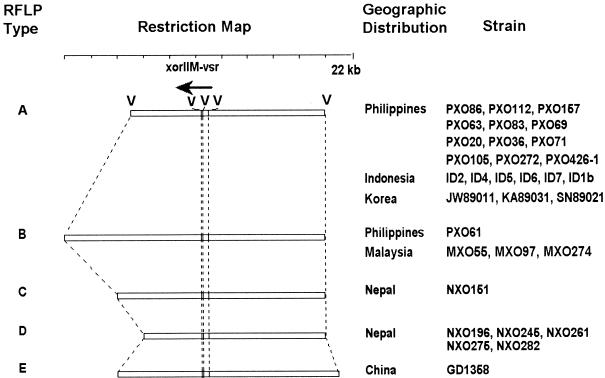
Map of randomly selected X. oryzae pv. oryzae strains with the XorII modification system from different countries. The map is based on an analysis with pEV5.0 and pEV2.0 as probes. Fragment size differences of less than 0.5 kb in regions flanking the XorII methyltransferase gene were seen in some strains grouped in RFLP type D. xorIIM, XorII methyltransferase gene; vsr, putative very-short-patch-repair endonuclease gene; V, EcoRV.
Figure 3 summarizes the organization of the XorII modification locus in 33 X. oryzae pv. oryzae strains collected from different countries. The regions upstream of the xorII-vsr gene were highly conserved (a fragment of approximately 8.5 kb), except in one strain from China (GD1358; RFLP type E), while the regions downstream of xorIIM were more polymorphic, with fragments of approximately 5, 6, 7, or 10 kb (Fig. 3). RFLP pattern A was found in DNA from Korean strain JW89011 (from which the XorII methyltransferase gene was isolated) and from many strains originating in the Philippines, Indonesia, and Korea (Fig. 3). Strains exhibiting both the XorI− XorII+ and the XorI+ XorII+ phenotypes were represented in pattern A. RFLP pattern B was characteristic of one Philippine strain (PXO61; XorI+ XorII+) and all Malaysian strains (XorI+ XorII+ and XorI− XorII+). RFLP types C and D were found only in the six Nepalese strains with the XorI− XorII+ phenotype (Fig. 3).
Absence of xorII-related DNA in other xanthomonads.
The presence or absence of DNA sequences related to the XorII R-M system in other Xanthomonas species was determined by high-stringency DNA hybridization analysis with pE7.0 as a probe. DNA from strains of X. oryzae pv. oryzicola from the Philippines and China did not hybridize with pE7.0. DNA from strains of various X. campestris pathovars (vesicatoria, alfalfae, malvacearum, holcicola, pisi, pruni, cucurbitae, fragariae, and phleipratensis), which are pathogenic in a wide variety of hosts, also did not hybridize with pE7.0.
DISCUSSION
Based on the nonrandom geographic distribution of XorI and XorII modification systems in X. oryzae pv. oryzae strains, the regions of origin of the modification systems were inferred. The similarity in genome organizations around the xorIIM and xorII-vsr genes indicated that the XorII R-M loci that are widely distributed in Asia are from a common ancestor. The XorII system is present in most strains from southeast Asia (79% contain DNA that is not digested with XorII or PvuI and that hybridizes with the fragment containing the genes for the XorII modification system), whereas this system is absent from most strains isolated in other parts of Asia (88% contain DNA that is digested with XorII and PvuI and that does not hybridize with the XorII modification system genes). Therefore, we suggest that the XorII system originated in southeast Asia (the Philippines, Indonesia, and Malaysia) and was distributed from there to south Nepal and northeast Asia (China, Korea, and Japan) (Fig. 4). Although more widely distributed, the XorI system likely originated in northeast Asia (DNA from 95% of the total strains from China and Korea is not digested by XorI or PstI) and moved from there to southeast Asia. Strains containing these systems may have moved between parts of Asia by weather systems, such as typhoons, or on seed during the movement of rice germ plasm (Fig. 4). The lack of either modification system in most strains from south Asia (Nepal and India) is of interest. We speculate that fewer strains with the two systems are found in south Asia because (i) this area is isolated from weather affecting northeast and southeast Asia, such as typhoons originating in the Pacific Ocean, and/or (ii) rice germ plasm was not until recently moved from other parts of Asia into south Asia.
FIG. 4.
Proposed model for geographic migration of XorI and XorII modification systems of X. oryzae pv. oryzae in major rice-growing countries in Asia. The positions of circles containing I or II indicate the proposed geographic origins of the XorI and XorII modification systems, respectively. The broken and solid arrows indicate possible geographic migration patterns for XorI and XorII, respectively.
Two of the three lineages for modification systems that were found in the Philippines (XorI+ XorII+ and XorI− XorII+) were distributed throughout the country. This finding suggests that each R-M group of X. oryzae pv. oryzae was maintained as a clone. Within each clonal group, there was considerable variation based on the high genetic diversity (Table 1) and pathotypic differentiation (data not shown).
Adhikari et al. (1) and George et al. (6) also found evidence of geographic migration of X. oryzae pv. oryzae by using multicopy genetic markers. They suggested that the distribution might be through germ plasm exchange. This assumption is reasonable, especially in the case of the Philippine race 6 strains (XorI− XorII−) (Table 1), which were distributed in a very limited area of the Philippines and only for a short period of time (1979 to 1982) (17). All other Philippine strains contain at least XorII. The limited distribution within the Philippines of this unusual phenotype and the similarity of the genomic DNAs of these Philippine XorI− XorII− strains to those of strains from south Asia, as compared by RFLP analysis with multilocus markers (1), further suggest that these strains were introduced into the Philippines from south Asia.
Prior to the introduction of rice cultivars with the Xa4 bacterial blight resistance gene, race 1 was the predominant pathotype of X. oryzae pv. oryzae in the lowland areas of the Philippines (17). After the release of Xa4, the pathogen population structure changed from one where race 1 was the prevalent type to one dominated by races that are virulent to rice with Xa4 (races 2 and 3). Previous RFLP analyses suggested that the virulent races which were prevalent after the deployment of Xa4 were from a different genetic lineage than race 1 (1, 10, 21). Our work provides compelling additional evidence that race 1 was not the genetic ancestor of the new races detected in the early 1970s to mid 1980s, since race 1 strains collected from that time period had the XorI+ XorII+ phenotype, while strains from all other races (besides race 6) had the XorI− XorII+ phenotype. Thus, the lineage containing the first population virulent to rice with Xa4, predominantly race 2 (XorI− XorII+), likely displaced the XorI+ XorII+ race 1 population.
Recently, Ardales et al. (3) and Vera Cruz et al. (27) demonstrated that over the last 10 to 15 years, the X. oryzae pv. oryzae population structure has shifted again, such that a race 3 population is now the predominant population in Luzon, the Philippines. Based on multilocus analysis using RFLPs and repetitive-sequence-based PCR (rep-PCR), Vera Cruz et al. (27) suggested that the race 3 population arose from at least two different genetic lineages, one corresponding to the lineage containing race 2 and the other corresponding to the early race 1 lineage. The predominant race 3 population found from the late 1980s to the early 1990s was derived from the genetic lineage that contained race 1. However, the R-M content of this prevalent race 3 group is largely XorI− XorII+, suggesting that the loss of XorI activity coincided with the increase in this population in the Philippines and an increase in host range (now virulent to rice with Xa4). Intriguingly, in the last 10 years, the X. oryzae pv. oryzae population in Korea has shifted from XorI− XorII+ to XorI+ XorII+, and this shift is correlated with a decrease in pathogenic host range (4a). Although pathotypic variation is strongly affected by the host (17), there is no a priori reason to expect that DNA modification systems are anything but neutral to host plant selection pressures. However, the observations from the Philippine and Korean population analyses suggest that the two phenotypes (race and R-M system content) may somehow be related. Another intriguing explanation is that where host resistance selection pressure is strong (as for the Xa4 gene in the Philippines), the effects of R-M systems on the structure of the population may be masked, whereas when host resistance selection pressure is weak (as in Korea), the effects of R-M systems on the pathogen population structure may be greater.
The XorII methyltransferase has 10 conserved polypeptide domains that are commonly found in the 5-methylcytosine methyltransferase family among a wide variety of prokaryotes (5, 23). Likewise, the amino acid sequence of the produce of xorII-vsr shows similarity to the deduced amino acid sequences of other vsr homologs that are associated with m5cytosine methyltransferase systems (5, 8, 25). Based on these facts, it is possible that the XorII R-M loci originated from a common ancestor and were transmitted to X. oryzae pv. oryzae through horizontal gene transfer (2). However, the XorII modification genes from X. oryzae pv. oryzae did not hybridize with DNAs from two different X. oryzae pv. oryzae bacteriophage strains (data not shown) or genomic DNAs from several different xanthomonads, including the closely related pathogen X. oryzae pv. oryzicola. Although only a limited number of xanthomonads were tested and although they were tested for hybridization under high-stringency conditions, the findings suggest that the XorII R-M system did not arise recently from other xanthomonads. It is possible that the XorII system evolved after the xanthomonads were differentiated into species and pathovars or that it originated from other prokaryotes.
The genome organization around the region containing genes coding for the XorII R-M system of one Philippine race 1 strain, PXO61, was identical to that of this region in all of the Malaysian strains. This finding suggests either that PXO61 originated from Malaysia or that the Malaysian strains were derived from an introduction of the PXO61 type. Interpretation of these results is complicated by the fact that in a previous study, PXO61 was clustered with other Philippine strains by multicopy marker-mediated lineage analysis, and this cluster was distinct from the cluster containing the Malaysian strains (1). Perhaps the high degree of variability associated with multicopy markers masks differences in genome organization within the modification system. If so, a comparison of multicopy versus single-copy markers might be important for phylogenetic studies of these bacteria (13, 14, 21).
Analysis of the two modification systems in X. oryzae pv. oryzae provided additional insight into the geographic distribution of these systems in this pathogen throughout major rice-growing countries in Asia. After the distribution of a modification system, the particular modification lineages or clones apparently became fixed in the population and maintained their identity. All four phenotypes (XorI+ XorII+, XorI+ XorII−, XorI− XorII+, and XorI− XorII−) were found in Asia, and in some cases, mixed populations (more than one phenotype in a region or country) were observed. These mixed populations may have been derived from the introduction of new strains with different modification systems either by humans or by weather systems. This idea suggests that populations of X. oryzae pv. oryzae in different countries shared common genetic backgrounds from which loci related to fitness were further selected in a defined ecosystem. For example, the Korean strains with the XorII R-M system might have a genetic background similar to that of the Philippine strains besides race 6, and the Philippine race 6 strains might be similar to the Indian and XorI− XorII− Nepalese strains.
The genetic diversity of X. oryzae pv. oryzae populations detected by multicopy markers such as IS1112 (10, 29) or avrXa10 (7) in general was high throughout Asia, in spite of the presence or absence of different DNA modification systems. This finding suggests that the R-M systems may not play a role in reducing the genetic diversity of populations. However, the markers used in these studies to measure diversity were either themselves mutagenic (the mobile IS1112 element) or prone to selection (the avirulence gene family). Thus, these multicopy markers may produce an inflated measurement of diversity. Analysis of diversity with less variable components of the genome may provide more information on the effect of R-M systems on the genetic diversity of these bacteria.
ACKNOWLEDGMENTS
We thank T. Adhikari, Q. Zhang, M. Watabe, and M. R. White for preparation of some X. oryzae pv. oryzae genomic DNAs. We thank H. Leung, E. Ardales, and R. Nelson for valuable discussions and comments and D. Pavlisko for assistance in preparation of the manuscript.
S. H. Choi was supported by a graduate fellowship (P.3.29) from the Rockefeller Foundation.
Footnotes
Contribution no. 97-439-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Adhikari T B, Vera Cruz C M, Zhang Q, Nelson R J, Skinner D Z, Mew T W, Leach J E. Genetic diversity of Xanthomonas oryzae pv. oryzae in Asia. Appl Environ Microbiol. 1995;61:966–971. doi: 10.1128/aem.61.3.966-971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amábile-Cuevas C F, Chicurel M E. Horizontal gene transfer. Am Sci. 1993;81:332–341. [Google Scholar]
- 3.Ardales E Y, Leung H, Vera Cruz C M, Mew T W, Leach J E, Nelson R J. Hierarchical analysis of spatial variation of the rice bacterial blight pathogen across agroecosystems in the Philippines. Phytopathology. 1996;86:241–252. [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates-Wiley Interscience; 1991. [Google Scholar]
- 4a.Choi, S. H. Unpublished results.
- 5.Choi S H, Leach J E. Identification of the XorII methyltransferase gene and a vsr-homolog from Xanthomonas oryzae pv. oryzae. Mol Gen Genet. 1994;244:383–390. doi: 10.1007/BF00286690. [DOI] [PubMed] [Google Scholar]
- 6.George M L C, Bustamam M, Cruz W T, Leach J E, Nelson R J. Migration of Xanthomonas oryzae pv. oryzae in Southeast Asia detected using PCR-based DNA fingerprinting. Phytopathology. 1997;87:302–309. doi: 10.1094/PHYTO.1997.87.3.302. [DOI] [PubMed] [Google Scholar]
- 7.Hopkins C M, White F F, Choi S H, Guo A, Leach J E. Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol Plant-Microbe Interact. 1992;5:451–459. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- 8.Kiss A, Pósfai G, Keller C C, Venetianer P, Roberts R J. Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res. 1985;13:6403–6421. doi: 10.1093/nar/13.18.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krüger D H, Bickle T A. Bacteriophage survival: multiple mechanisms for avoiding deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983;47:345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leach J E, White F F, Rhoads M L, Leung H. A repetitive DNA sequence differentiates Xanthomonas campestris pv. oryzae from other pathovars of X. campestris. Mol Plant-Microbe Interact. 1990;3:238–246. [Google Scholar]
- 11.Leach J E, Rhoads M L, Vera Cruz C M, White F F, Mew T W, Leung H. Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae with a repetitive DNA element. Appl Environ Microbiol. 1992;58:2188–2195. doi: 10.1128/aem.58.7.2188-2195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leach J E, Leung H, Nelson R J, Mew T W. Population biology of Xanthomonas oryzae pv. oryzae and approaches to its control. Curr Opin Biotechnol. 1995;6:298–304. [Google Scholar]
- 13.Leung H, Nelson R J, Leach J E. Population structure of plant pathogenic fungi and bacteria. In: Andrews J H, Tommerup I C, editors. Advances in plant pathology. New York, N.Y: Academic Press, Inc.; 1993. pp. 157–205. [Google Scholar]
- 14.Levin B R. Restriction-modification immunity and the maintenance of genetic diversity in bacterial populations. In: Karlin S, Nevo E, editors. Evolutionary processes and theory. New York, N.Y: Academic Press, Inc.; 1986. pp. 669–688. [Google Scholar]
- 15.Mew T W. Current status and future prospects of research on bacterial blight of rice. Annu Rev Phytopathol. 1987;25:359–382. [Google Scholar]
- 16.Mew T W, Alvarez A M, Leach J E, Swings J. Focus on bacterial blight of rice. Plant Dis. 1993;77:5–12. [Google Scholar]
- 17.Mew T W, Vera Cruz C M, Medalla E S. Changes in race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Dis. 1992;76:1029–1032. [Google Scholar]
- 18.Morelle G. A plasmid extraction procedure on a miniprep scale. Focus. 1989;11:7–8. [Google Scholar]
- 19.Nei M. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. [Google Scholar]
- 20.Nei M, Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics. 1981;97:145–163. doi: 10.1093/genetics/97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson R J, Baraoidan M R, Vera Cruz C M, Yap I V, Leach J E, Mew T W, Leung H. Relationship between phylogeny and pathotype for the bacterial blight pathogen of rice. Appl Environ Microbiol. 1994;60:3275–3283. doi: 10.1128/aem.60.9.3275-3283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen R J, Borman P. A rapid biochemical method for purifying high molecular weight bacterial chromosomal DNA for restriction enzyme analysis. Nucleic Acids Res. 1987;15:3631. doi: 10.1093/nar/15.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pósfai J, Bhagwat A S, Pósfai G, Roberts R J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989;17:2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raymundo A K, Ardales E Y, Yap I V, Baroidan M R, Mew T W, Nelson R J. Analysis of genetic variation of Xanthomonas oryzae pv. oryzae using PstI digestion. Philipp J Biotechnol. 1993;4:9–27. [Google Scholar]
- 25.Sohail A, Lieb M, Dar M, Bhagwat A S. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methyltransferase gene. J Bacteriol. 1990;172:4214–4221. doi: 10.1128/jb.172.8.4214-4221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuchiya K, Mew T W, Wakimoto S. Bacteriological and pathological characteristics of wild types and induced mutants of Xanthomonas campestris pv. oryzae. Phytopathology. 1982;72:43–46. [Google Scholar]
- 27.Vera Cruz C M, Ardales E Y, Skinner D Z, Talag J, Nelson R J, Louws F J, Leung H, Mew T W, Leach J E. Measurement of haplotypic variation in Xanthomonas oryzae pv. oryzae within a single field by rep-PCR and RFLP analyses. Phytopathology. 1996;86:1352–1359. [Google Scholar]
- 28.Wang R Y H, Shedlarski J G, Farber M B, Kuebbing D, Ehrlich M. Two sequence specific endonucleases from Xanthomonas oryzae. Biochim Biophys Acta. 1980;606:371–385. doi: 10.1016/0005-2787(80)90047-7. [DOI] [PubMed] [Google Scholar]
- 29.Yun C H. Molecular characterization of a repetitive element of Xanthomonas oryzae pv. oryzae. Ph.D. thesis. Manhattan: Kansas State University; 1991. [Google Scholar]