Figure 6. Mesenchymal p16INK4A expression is required for epithelial regeneration in vivo.
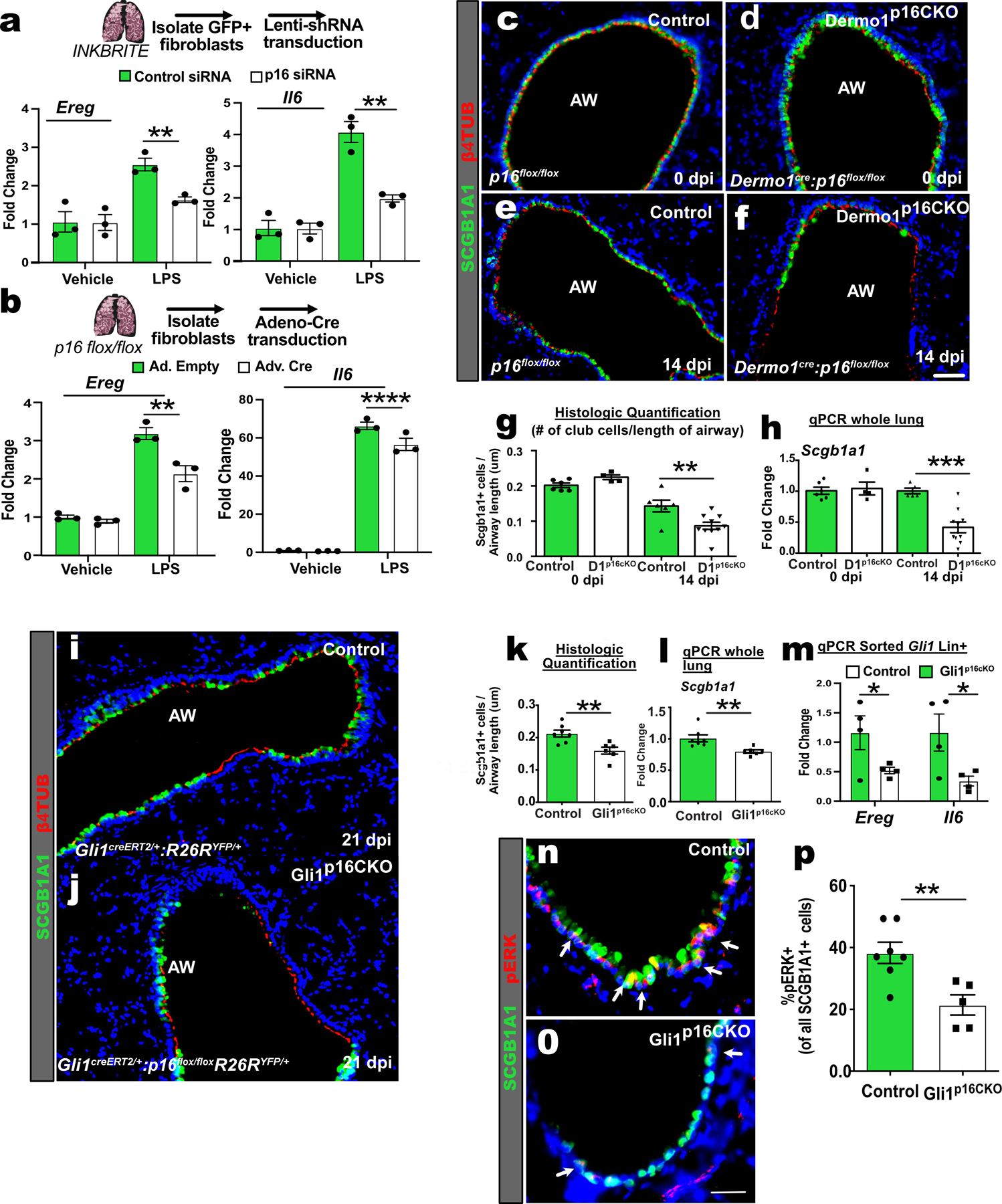
(A) qPCR of Ereg and Il6 in control or p16INK4A shRNA treated GFP+ fibroblasts after stimulation with vehicle or LPS for 6 hours (n=3 wells per conditions, 2 experiments). (B) qPCR of Ereg and Il6 in adeno-control or adeno-Cre treated p16flox/flox fibroblasts after stimulation with vehicle or LPS (n=3 wells per conditions, >2 experiments). (C to G) Images and quantification of the airways of control and Dermo1p16CKO lungs with and without naphthalene injury. (H) qPCR of Scgb1a1 of whole lung RNA (uninjured: n=6 Control, n=5 Dermo1p16CKO; injured: n=6 Control, n=10 Dermo1p16CKO,2 experiments ). (I to L) Images, histological quantification of SCGB1A1 club cells, and qPCR of Scgb1a1 on control and Gli1p16CKO lungs after naphthalene injury (n=7 Control, n=6 Gli1p16CKO). (M) qPCR of Ereg and Il6 of sorted Gli1 Lin+ cells after naphthalene (n=4 Control; n=4 Gli1p16CKO). (N to P) Images and quantification of pERK+ and SCGB1A1+ cells in control and Gli1p16CKO lungs after naphthalene injury (n=7 Control, n=6 Gli1p16CKO). AW=airway. Scale bars 100υm. Each point in graph represents one animal with mean ± s.e.m. All p values determined by one-tailed t-test and two-way ANOVA when applicable. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
