Abstract
NOD1 and NOD2 sense small bacterial peptidoglycan fragments, often called muropeptides, that access the cytosol. These muropeptides include iE-DAP and MDP, the minimal agonists for NOD1 and NOD2, respectively. Here, we synthesized and validated alkyne-modified muropeptides, iE-DAP-Alk and MDP-Alk, for use in click-chemistry reactions. While it has long been known that many cell types respond to extracellular exposure to muropeptides, it is unclear how these innate immune activators access their cytosolic innate immune receptors, NOD1 and NOD2. The subcellular trafficking and transport mechanisms by which muropeptides access these cytosolic innate immune receptors are a major gap in our understanding of these critical host responses. The click-chemistry-enabled agonists developed here will be particularly powerful to decipher the underlying cell biology and biochemistry of NOD1 and NOD2 innate immune sensing.
Keywords: Click-chemistry, alkyne-modification, muropeptide, iE-DAP, MDP, NOD1, and NOD2
Introduction
Innate immune cells such as macrophages, neutrophils, NK cells, and dendritic cells harbor specific innate immune receptors on the cell surface, within intracellular compartments, or in the cytosol. Toll-like receptors (TLR) were the earliest discovered and most extensively studied cell surface and vesicular innate immune receptors. 1 However, cytosolic innate immune receptors play an equally critical role in host defense, especially in sensing and discriminating between pathogenic and commensal microbes. 1 This category of receptors includes several proteins capable of sensing nucleic acids, such as cGAS, RIG-I, NLRP1, and AIM2, recognizing bacterial products (NOD1/2, NAIPs/NLRC4) or other danger signals (NLRP1/3).2–5 Among these cytosolic receptors, NOD1 and NOD2 sense small bacterial cell wall fragments, often referred to as muropeptides, that access the intracellular environment. 6
Peptidoglycan (PGN), the major constituent of the bacterial cell wall, is a network of long carbohydrate chains cross-linked via short-stem peptides that provide the bacterium with mechanical strength and protection from internal and external stressors. 7 Muropeptides are often shed into the environment during cell wall turnover and biosynthesis, and trigger NOD1 or NOD2 upon gaining access to the cytosol. 8 For example, muramyl dipeptide (MDP) a major component of Freund's adjuvant, is the minimal molecular motif that activates NOD2 triggering an NF-κB inflammatory response. 9 Recent studies have shown that MDP must be first phosphorylated, by the host cytosolic enzyme N-acetylglucosamine kinase (NAGK), in order to activate NOD2. 10 Other muropeptides that contain meso-diaminopimelic acid (m-DAP), an amino acid unique to Gram-negative bacteria and Gram-positive bacilli peptidoglycan, activate NOD1. 11 Recently, it has been shown that mice lacking NOD1/2 receptors are more susceptible to intracellular and extracellular bacterial infections including H. pylori, Clostridium difficile, Listeria monocytogenes, Chlamydia pneumoniae, Neisseria gonorrhoeae, and Staphylococcus aureus.12–21 NOD2 is also strongly linked to inflammatory bowel disease 22 and several studies found that NOD2 is critical for the healthy gut epithelial structure and immunity. 6 Many gaps exist in our current knowledge of NOD1/2 pathways, such as if phosphorylation of all muropeptides is required for their NOD1/2 agonist activity, how different muropeptides access the cytosol to trigger NOD1 or NOD2, what other proteins are involved in binding and sensing these muropeptides, and if different agonists trigger distinct signaling pathways.
Click chemistry is a powerful tool in the fields of drug discovery, cell signaling, and pharmaceutical sciences. 23 This tool provides a rapid means to synthesize biomolecules useful in high throughput screening, examining biomolecular interactions, and fluorescent microscopic detection. 24 While several types of click reactions have been developed, including the copper-catalyzed azide-alkyne click reaction (CuAAC), strain-promoted alkyne-azide click reaction (SPAAC), and the inverse electron demand Diels-Alder reactions (IEDDA); notably the 2022 Noble prize in chemistry was awarded for this chemistry.25,26 Here we aimed to use the CuI-catalyzed Huisgen 1,3-dipolar cycloaddition of azides and terminal alkynes, which is the most widely implemented in the synthesis of natural products with increased efficiency and biologically compatible reagents.27,28 Using this approach, we have generated “click” modified d-isoGlutamate-meso-Diaminopimelic acid alkyne (iE-DAP-Alk) and 2-alkyne muramyl dipeptide (MDP-Alk), which represent the minimal NOD1 and NOD2 agonists respectively. 29 We note that others have made other PGN-related probes. Hang and co-workers used a similar approach in their synthesis of chemo-proteomic PGN probes, 30 combining photoactivatable cross-linkers with a clickable alkyne moiety; while others have used solid phase synthesis to incorporate click probes on PGNs. 31 A recent review from Tanner and co-workers succinctly highlights our and others’ synthetic efforts in this space. 32
Here, we report the chemical synthesis of iE-DAP-Alk and MDP-Alk, for use in click-chemistry reactions. This modification is smaller than those previously reported, and valuable in recent work where we probed the biological role of PGN transporters. 33 The modified iE-DAP and MDP showed similar potency as the native molecules in the NF-κB reporter assays in transfected cell lines, and in cytokine induction in IFNγ-primed bone marrow-derived macrophages (BMDM). Using these click-muropeptides and fluorescent confocal microscopy we have also confirmed the internalization of iE-DAP and MDP in WT BMDM and HCT116 cells. Below we describe the protocols for the preparation and utilization of these tools in visualizing cellular uptake of the minimal, synthetic bacterial cell wall immunostimulatory fragments.
Methods
Mice
All animal studies were performed in compliance with the federal regulations set forth in the Animal Welfare Act (AWA), the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and the guidelines of the UMass Chan Medical School Institutional Animal Use and Care Committee. All protocols used in this study were approved by the Institutional Animal Care and Use Committee at the UMass Chan Medical School (protocol 2056). All mice used in this study were maintained and bred in a specific pathogen-free environment. All mouse strains were backcrossed and maintained on a C57Bl/6J background. If not otherwise indicated, mice of the same sex and age were paired and assigned into experimental groups.
Nod1−/− and Nod2−/−mice were reported previously in.12,34
Cell culture
HCT116 cells were isolated from a male human colorectal carcinoma and originally acquired from American Type Culture Collection (ATCC, CCL-247). HCT116 cells were maintained in DMEM (Corning CellGro) supplemented with 10% heat-inactivated FBS at 37°C, and 5% CO2.
Dual-luciferase reporter assay
50,000 per well HCT-116 cells were incubated in 6-well plates for 24 h and then transfected with 0.3 μg NF-κB luciferase, 0.3 μg of pRL-TK (Promega), and 0.3 μg of pEF-Slc46 expression plasmid using GeneJuice (Millipore) for 24 h. In all cases, cells were then stimulated by adding indicated agonist directly to the media for 6 or 24 h and subject to dual-luciferase assay. The TK-renilla was used to normalize the values from firefly luciferase. The values from unstimulated controls were subtracted from those for muropeptide-stimulated conditions to display the signal over the background. Each transfection was similarly analyzed in two independently transfected wells in a single plate, and all experiments were repeated with at least four independent assays. 35
Mouse bone marrow-derived macrophages (BMDM)
Mouse BMDM were obtained from seven to twelve-week-old female mice maintained under specific pathogen-free conditions at the University of Massachusetts Medical School animal facilities, from C57BL/6J (WT), Nod1−/−, and Nod2−/− mice. Animals were sacrificed, femurs and tibia were dissected, and the bone marrow was flushed with PBS using a 30G needle connected to a 10 ml syringe. The cells were cultivated in RPMI (Corning) supplemented with 30% L929 cell-conditioned medium and 20% FBS (4 × 106 cells/10 cm plate) at 37°C and 5% CO2. Cultures were re-fed on day 3 with the same media and maintained in RPMI supplemented with 5% L929 cell-conditioned medium and 10% FBS. BMDMs were used in the experiments on days 7–10. 36
BMDM stimulation protocol
BMDMs were primed with murine IFNγ (5 ng/ml, R&D systems) for 16 h, followed by the medium being replaced with fresh IFNγ (5 ng/ml) containing media for 2 h before treatment with indicated stimulations: MDP (100 µM, Invivogen), iE-DAP (100 µM, Invivogen) and ultra-pure Lipopolysaccharide (LPS) from Escherichia coli 0111:B4 (50ng/ml, Invivogen). After 24h stimulation media were processed for ELISA (R&D systems) as per manufacturer protocol. 10
Microscopy
104 cells (HCT116 or BMDM) were seeded on a glass coverslip in 24 well plates overnight. Cells were challenged with iE-DAP-Alk (30 μM) or MDP-Alk (30 μM) at 37 ˚C for 1, 3 or 6 h, washed twice with 1xPBS to remove access of click-muropeptides and fixed with 4% paraformaldehyde in PBS at RT for 20 min. Cells were permeabilized with 0.1% Tween-20 in PBS for 10 min at RT and blocked with 1% BSA in PBS. These permeabilized cells were then incubated in click-reaction conditions (250 μM CuSO4, 35 μM BTAA, 60 μM sodium ascorbate) with 2.5 μM CalFluor 488 Azide at RT for 30 min. Cells were then washed with DPBS thrice and mounted on slides, with DAPI containing mounting media. Slides were imaged with a Leica SP8 confocal microscope.
Results
Synthesis of iE-DAP-Alk
Synthesis of iE-DAP-Alk follows an eight-step process (all of which have literature precedent).30,31,37–42 Overall chemical synthesis of iE-DAP-Alk is described below and presented in Figure 1. We note that the synthesis of intermediate 5 follows prior literature while transformation from 5 to 8 is supported by other related literature for the synthesis of similar molecules. All spectral shifts are referenced according to the chemical shift of the solvent. Solvent peaks are not reported but identified at the beginning of the line listing. In addition, green integration marks are included to indicate the area under the peak that references the protons we have identified to be significant in the compounds. The synthesis is detailed below:
Figure 1.
Schematic overview of synthesis of iE-DAP-Alk. The overall strategy for chemical synthesis of iE-DAP-alkyne with biologically active stereochemistry followed literature precedent; the protocols presented in this manuscript give critical details for this specific workflow, making it easier for the production of these important PGN probes.
Tert-butyl ((benzyloxy)carbonyl)-L-homoserinate (1): To a suspension of Z-L-Asp-ɑ-OtBu (2.04 g, 6.32 mmol, 1.0 eq.) and benzotriazol-1-yl-oxytris(dimethylamino)phosphonium hexafluorophosphate (BOP) (3.47 g, 7.84 mmol, 1.2 eq.) in anhydrous THF (50 ml) was added DIPEA (1.43 ml, 8.2 mmol, 1.3 eq.). The reaction mixture was stirred for 10 min at room temperature before cooling down to 0°C in an ice bath. Then, sodium borohydride (NaBH4) (312 mg, 8.2 mmol, 1.3 eq.) was added in two portions. The reaction was stirred overnight at room temperature. The reaction was then reduced in volume, diluted with ethyl acetate, and washed with 1N HCl (thrice), sat. sodium bicarbonate (thrice), brine (once), and dried over sodium sulfate. After concentration, the oily crude product was purified using flash chromatography (ethyl acetate/hexane - gradient from 5% to 20% ethyl acetate) to give 1 as a colorless oil, matching those previously reported in the literature. 43 (1.55 g, 86%). 1H NMR (600 MHz, MeOD) δ 7.41–7.23 (m, 5H), 5.13–5.02 (m, 2H), 4.20 (dd, J = 9.7, 4.7 Hz, 1H), 3.69–3.57 (m, 2H), 2.06–1.97 (m, 1H), 1.85–1.77 (m, 1H), 1.45 (s, 8H), 1.38 (s, 1H). 13C NMR (151 MHz, MeOD) δ 173.52, 158.67, 138.25, 129.45, 128.99, 128.84, 82.70, 67.91, 67.58, 59.21, 53.59, 53.49, 35.24, 28.21. LRMS (ESI) m/z: [M + Na]+ Calcd for C16H23NNaO5 332.14; Found 332.28. NMR spectra are shown in Figure S1A.
Tert-butyl (S)-2-(((benzyloxy)carbonyl)amino)but-3-enoate (2): a solution of 1 (2.0 g, 6.46 mmol, 1.0 eq.) in anhydrous THF (32 ml) was cooled down to 0°C in an ice bath. Tri-n-butylphosphine (TBP) (1.8 ml, 7.11 mmol, 1.1 eq.) was added dropwise followed by an addition of 2-nitrophenyl selenocyanate (1.61 g, 7.11 mmol, 1.1 eq.). The reaction was warmed to room temperature and stirred for 1 h. The reaction was cooled down to 0°C and 30% hydrogen peroxide solution (2.53 ml) was added. The reaction stirred overnight at room temperature. The reaction was then diluted into brine (60 ml) and extracted with ethyl acetate (thrice). Combined organics were washed with 1N HCl (thrice), washed with brine (once) and dried over sodium sulfate. After concentration with vacuum, the crude product was purified using flash chromatography (ethyl acetate/hexane - gradient from 0% to 30% ethyl acetate) to give 2 as a yellow oil, matching those previously reported in the literature. 42 (1.45 g, 77%). 1H NMR (400 MHz, MeOD) δ 7.41–7.25 (m, 5H), 5.93 (ddd, J = 16.7, 10.4, 5.9 Hz, 1H), 5.39–5.31 (m, 1H), 5.28–5.21 (m, 1H), 5.10 (t, J = 2.2 Hz, 2H), 4.69–4.62 (m, 1H), 1.45 (s, 8H), 1.38 (s, 1H). 13C NMR (101 MHz, MeOD) δ 171.35, 158.32, 138.19, 133.83, 129.46, 129.02, 128.86, 118.01, 83.13, 67.69, 58.72, 28.16. LRMS (ESI) m/z: [M + Na]+ Calcd for C16H21NNaO4 314.13; Found 314.43. NMR spectra shown in Figure S1B.
Tert-butyl (R)-2-((tert-butoxycarbonyl)amino)pent-4-enoate (3): To a solution of 2-allyl-N-Boc-D-glycine (0.932 g, 4.33 mmol, 1.0 eq.) in anhydrous DCM (4.33 ml) a solution of tert-butyl 22,2-trichloroacetimidate (1.9 g, 8.66 mmol, 2.0 eq.) in anhydrous cyclohexane (28 ml) was added dropwise. Then boron trifluoride diethyl etherate (100 µL) was added. The reaction was stirred at room temperature overnight. Upon reaction completion, monitored by TLC (30% ethyl acetate/hexane) and MS, solid NaHCO3 was added; the reaction was then filtered, and the reaction condensed to a yellow and white solid. The crude product was then purified using flash chromatography (ethyl acetate/hexane - gradient from 0% to 15% ethyl acetate) to give 3 as a clear oil (617 mg, 53%), matching those previously reported in the literature. 44 1H NMR (400 MHz, MeOD) δ 5.85–5.70 (m, 1H), 5.17–5.04 (m, 3H), 4.03 (dd, J = 7.9, 5.5 Hz, 1H), 2.53–2.45 (m, 1H), 2.43–2.34 (m, 1H), 1.46 (s, 9H), 1.44 (s, 11H). 13C NMR (101 MHz, MeOD) δ 172.91, 157.84, 134.61, 118.51, 82.67, 80.46, 56.50, 55.43, 37.20, 28.71, 28.29. LRMS (ESI) m/z: [M + H]+ Calcd for C14H26NO4 272.18; Found 272.20. NMR spectra shown in Figure S1C.
Di-tert-butyl(2S,6R,E)-2-(((benzyloxy)carbonyl)amino)-6-((tert-butoxycarbonyl) amino) hept-3-enedioate (4): 2 (458 mg, 1.57 mmol. 1.8 eq.) and 3 (237 mg, 0.87 mmol. 1.0 eq.) were dissolved in degassed anhydrous DCM (16 ml) and cannula transferred to an oven-dried round bottom flask charged with Grubbs’ catalyst 2nd generation (66.6 mg, 0.078 mmol. 0.05 eq) under nitrogen. We note that in order to prevent homodimer formation, equivalence of 2 and 3 were optimized. The reaction stirred 48 h monitored by MS. The reaction was quenched by removing solvent via rotovap, and purification of the residue by flash chromatography (ethyl acetate/hexane 0.01% Triethylamine - gradient from 0% to 12% ethyl acetate) to give 4 as a yellow oil (148 mg, 18%), matching those previously reported in the literature. 37 1H NMR (400 MHz, MeOD) δ 7.40–7.25 (m, 5H), 5.80–5.70 (m, 1H), 5.69–5.60 (m, 1H), 5.10 (s, 2H), 4.60 (d, J = 6.0 Hz, 1H), 4.04 (dd, J = 7.6, 5.3 Hz, 1H), 2.53–2.37 (m, 2H), 1.45 (s, 14H), 1.43 (s, 10H). 13C NMR (101 MHz, MeOD) δ 172.66, 171.51, 158.22, 157.84, 138.16, 130.05, 129.46, 129.02, 128.88, 83.15, 82.85, 80.61, 67.69, 57.94, 55.33, 49.85, 49.64, 49.43, 49.21, 49.00, 48.79, 48.57, 48.36, 35.50, 28.73, 28.29, 28.23.LRMS (ESI) m/z: [M + Na]+ Calcd for C28H42N2NaO8 557.28; Found 557.23. NMR spectra shown in Figure S1D.
Di-tert-butyl(2S,6R)-2-amino-6-((tert-butoxycarbonyl)amino)heptanedioate (5): 4 (453 mg, 0.84 mmol. 1.0 eq.) was dissolved in methanol (25 ml) and Pd/C catalyst (100 mg, 10% loading) was added. The reaction stirred under H2 (balloon) overnight. In order to quench the reaction, the reaction mixture was then filtered over a pad of Celite to remove the palladium, and the resulting filtrate was concentrated by rotovap to give 5 as a colorless oil (330 mg, 98%).1H NMR (600 MHz, MeOD) δ 3.94 (dd, J = 9.0, 5.2 Hz, 1H), 2.96 (dd, J = 9.0, 5.5 Hz, 1H), 2.33 (s, 6H), 1.80–1.67 (m, 2H), 1.67–1.56 (m, 2H), 1.49 (s, 9H), 1.46 (s, 9H), 1.44 (s, 11H). 13C NMR (151 MHz, MeOD) δ 173.70, 172.85, 82.58, 82.53, 80.42, 69.73, 55.62, 49.43, 49.28, 49.14, 49.00, 48.86, 48.72, 48.57, 42.09, 32.48, 30.40, 28.75, 28.52, 28.39, 28.27, 23.52. LRMS (ESI) m/z: [M + H]+ Calcd for C20H39N2O6 403.28; Found 403.27. NMR spectra shown in Figure S1E.
Tri-tert-butyl (5R,10S,14R)-1-(9H-fluoren-9-ylmethoxycarbonyl)-18,18 dimethyl3,8,16-trioxo-2,17-dioxa-4,9,15-triazanonadecane-5,10,14-tricarboxylate (6): 5 (30 mg, 0.074 mmol. 1.0 eq.) and Fmoc-D-Glu-ɑ-OtBu (31.7 mg, 0.075 mmol. 1.0 eq.) were dissolved in anhydrous DMF (1.5 ml). Then HBTU (31mg, 0.082 mmol, 1.1eq.) and HOBt (11 mg, 0.082 mmol, 1.1eq) were added and reaction stirred 5 min. DIPEA (52 µl, 0.29 mmol, 4 eq.) was added and reaction stirred overnight at room temperature. The reaction was quenched with water (2.5 ml) and extracted with ethyl acetate (thrice (5 ml)). The combined organics were then washed with 1N HCl (thrice), sat. sodium bicarbonate (thrice), brine (once) and dried over sodium sulfate. After concentration, the crude product was purified using flash chromatography (methanol/DCM/ 0.01% TEA gradient from 1% to 5% methanol) to give 6 as an off-white solid (51 mg, 85%). 1H NMR (400 MHz, MeOD) δ 7.80 (d, J = 7.5 Hz, 2H), 7.73–7.64 (m, 2H), 7.39 (t, J = 7.5 Hz, 2H), 7.36–7.27 (m, 3H), 4.45–4.28 (m, 2H), 4.23 (t, J = 6.9 Hz, 2H), 4.08 (dd, J = 9.6, 4.8 Hz, 1H), 3.94 (dd, J = 9.0, 5.0 Hz, 1H), 3.00 (d, J = 6.7 Hz, 1H), 2.93 (d, J = 4.4 Hz, 1H), 2.57–2.38 (m, 1H), 2.32 (t, J = 7.6 Hz, 1H), 2.18–2.09 (m, 1H), 1.96–1.87 (m, 1H), 1.83–1.72 (m, 1H), 1.68–1.57 (m, 1H), 1.46 (s, 11H), 1.44 (s, 7H), 1.43 (s, 7H). 13C NMR (101 MHz, MeOD) δ 172.65, 142.40, 128.59, 127.97, 126.04, 120.73, 82.69, 82.61, 82.37, 67.79, 49.43, 49.21, 49.00, 48.79, 48.57, 48.36, 48.15, 32.08, 28.54, 28.08, 28.06, 23.06. LRMS (ESI) m/z: [M + H]+ Calcd for C44H63N3O11 809.44; Found 810.44. NMR spectra shown in Figure S1F.
Tri-tert-butyl(6R,10S,15R)-2,2-dimethyl-4,12,17-trioxo-3-oxa-5,11,16-triazahenicos20-yne-6,10,15-tricarboxylate (7): 6 (57.8 mg, 0.071 mmol. 1.0 eq.) was dissolved in a solution of 20% piperidine in DMF (715 μL). The reaction stirred at room temperature for 4 h as monitored by TLC. The reaction was quenched with water (2 ml) and extracted with ethyl acetate (thrice). The combined organics were then washed with brine (once) and dried over sodium sulfate, concentrated and dried under vacuum overnight. The crude product was then dissolved in anhydrous DMF (1.42 ml). 4-pentynoic acid (11 mg, 0.106 mmol. 1.5 eq.) and EDC hydrochloride (34 mg, 0.177 mmol, 2.5 eq.) were added followed by DMAP (5.6 mg, 0.046 mmol, 0.65 eq.). The reaction stirred at room temperature overnight. The reaction was quenched with water (2 ml) and extracted with ethyl acetate (thrice). The combined organic solutions were then washed with 1N HCl (thrice), sat. sodium bicarbonate (thrice), brine (once) and dried over sodium sulfate. After concentration with vacuum, 7 a was obtained as an off-white solid (32.7 mg, 34.4%, over two steps). LRMS (ESI) m/z: [M + H]+ Calcd for C34H57N3O10 667. 40; found 668.40. Per literature precedent, further characterization is not performed. 30
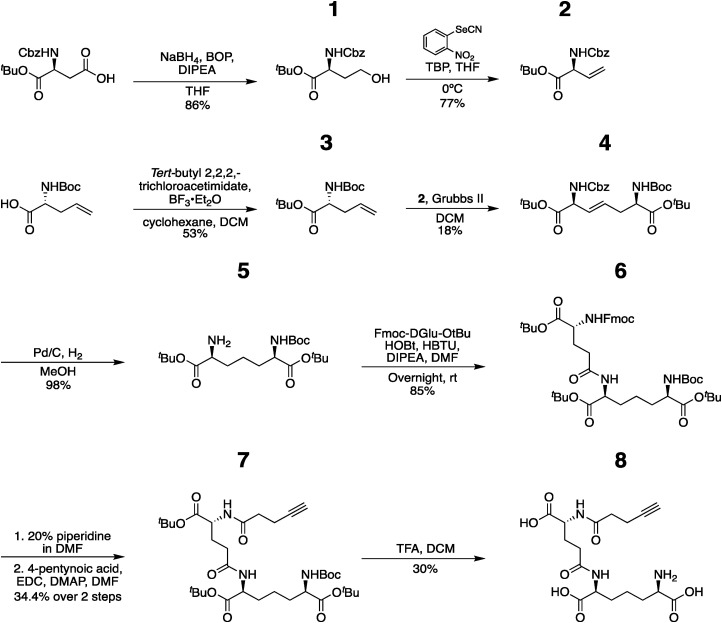
(6R,10S,15R)-2,2-dimethyl-4,12,17-trioxo-5,11,16-triazahenicos-20-yne-6,10,15- tricarboxylate (iE-DAP-Alk) (8): 7 (5 mg, 0.007 mmol, 1.0 eq.) was dissolved in 20% TFA in DCM (2.4mL) and the reaction stirred at room temperature for 6 h. We note that the reaction must be carefully monitored and checked by NMR to assure that all 3 tert-butyl and boc groups are removed. When NMR showed no more acetate peaks, the solvent was removed, resulting in a clear mixture, and the crude residue was triturated in cold diethyl ether, centrifuged, and purified with semi-prep C18 HPLC using a 0.1%TFA/ACN/H2O system (gradient 100% A for 4 min, followed by a gradient to 20%B over 6 min, 70%B to 30 min, 100% to 32 min, then 5 min at 100% B). The appropriate fractions were combined and lyophilized to give 8 as a white solid (1.0 mg, 30%). 1H NMR (600 MHz, D2O) δ 4.41 (dd, J = 8.8, 5.1 Hz, 1H), 4.36 (dd, J = 9.1, 5.3 Hz, 1H), 3.89 (t, J = 6.3 Hz, 1H), 2.56–2.50 (m, 2H), 2.45 (m, 2H), 2.40–2.37 (m, 1H), 2.24–2.16 (m, 1H), 2.08–2.02 (m, 2H), 2.01–1.87 (m, 3H), 1.80 (m, 2H), 1.58–1.46 (m, 2H).; 13C NMR (151 MHz, D2O) δ 175.47, 175.09, 174.99, 174.79, 172.25, 83.39, 70.11, 52.86, 52.37, 52.05, 34.09, 31.41, 29.87, 29.22, 26.49, 20.82, 14.34, 0.81 HRMS (ESI-Pos) m/z: [M + H]+ Calcd for C17H26N3O8 400.1714; Found 400.1744, see Figure 2 for proton and carbon NMR and high resolution Mass Spectra.
Figure 2.
Spectral data for iE-DAP-Alk (A) 1H NMR (B) 13C NMR (C) High resolution mass spec data top: experimental data; bottom: simulated data made through Xcaliber.
Synthesis of MDP-Alk
Synthesis of MDP-Alk follows an established synthesis of 2-amino-MDP 45 and instillation of the alkyne probe as the last step. 30 As shown in Figure 3:
Figure 3.
Schematic overview of the chemical synthesis of MDP-alkyne from the readily accessible 2-amino-MDP.
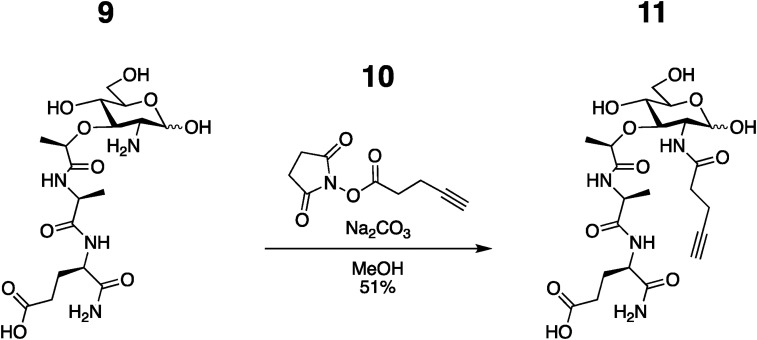
(R)-5-amino-4-((S)-2-((R)-2-(((3R,4R,5S,6R)-2,5-dihydroxy-6- (hydroxymethyl)-3-(pent-4-ynamido)tetrahydro-2H-pyran-4 yl) oxy) propanamido) propanamido)-5-oxopentanoic acid (11): 9 46 (0.2136 g, 0.5142 mmol) was dissolved in anhydrous MeOH (7.3 ml) and Na2CO3 (0.2736 g, 2.5814 mmol) and 10 47 (0.5016 g, 2.5701 mmol) was added in 5 portions over 1 h. Additional 10 (0.1009 g, 0.5170 mmol) was added at 24 h since the reaction was incomplete as determined by LC/MS. Once complete, the solvent was removed via rotovap and the residue was dissolved in water and IRA H + resin was added. The filtrate was then collected and concentrated. The off-white solid was purified with semi-prep C18 HPLC in a 0.1% TFA/ACN/H2O system (specifically: gradient 99% A for 1 min, followed by a gradient to 40%B over 30 min, 99%B to 35 min, then 5 min at 100% B). The appropriate fractions were combined and lyophilized to give 11 as a white solid (0.1386 g, 51% yield). 1H NMR (600 MHz, D2O): (Anomers 1.00α : 0.50β) δ 5.19 (d, J = 3.5 Hz, 1H), 4.71 (d, J = 8.4 Hz, 1H), 4.40 (dd, J = 9.7, 4.8 Hz, 1H), 4.34 (q, J = 6.8 Hz, 1H), 4.31–4.26 (m, 2H), 4.02 (dd, J = 10.5, 3.5 Hz, 1H), 3.97–3.64 (m, 5H), 3.63–3.53 (m, 3H), 3.53–3.47 (m, 1H), 2.53–2.42 (m, 6H), 2.39–2.34 (m, 1H), 2.26–2.18 (m, 1H), 2.08–1.89 (m, 1H), 1.49–1.43 (m, 3H), 1.41–1.35 (m, 3H); 13C NMR (151 MHz, D2O) δ 176.96, 175.91, 175.70, 175.15, 174.65, 94.90, 91.02, 83.27, 81.77, 78.95, 77.51, 77.30, 75.68, 71.50, 70.17, 70.11, 69.11, 68.80, 60.76, 60.56, 56.20, 53.64, 52.60, 49.81, 34.84, 34.49, 30.06, 26.13, 18.40, 18.30, 16.60, 14.41, 14.33; HRMS (ESI-Pos) m/z: [M + H]+ Calcd for C22H34N4O11 531.2297; Found 531.2299, see Figure 4 for NMR and high resolution Mass Spectra.
Figure 4.
Spectral analysis of MDP-Alk. (A) 1H NMR (B) 13C NMR (C) High-Resolution Mass Spec Data Top: experimental data; Bottom: Simulated data made through Xcaliber.
Validation of NF-κB stimulating activity
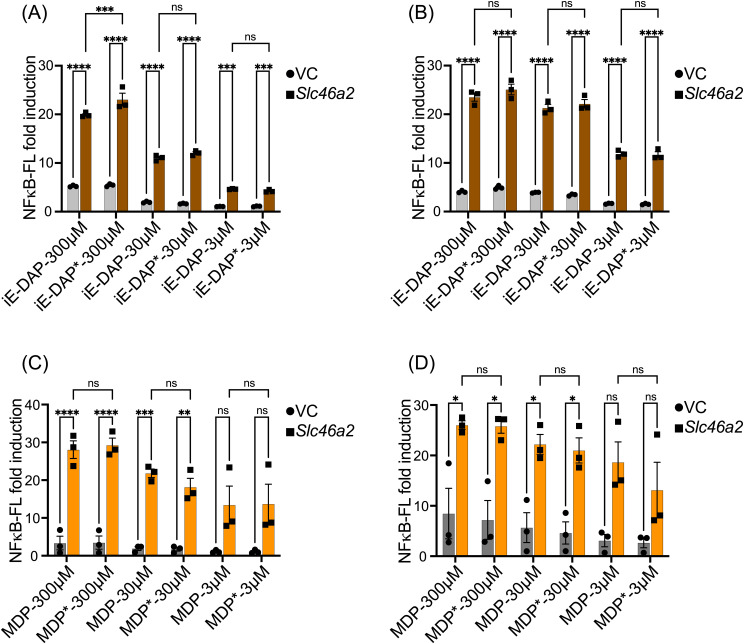
First, we checked the potency of iE-DAP-Alk and MDP-Alk using an established NF-κB luciferase assay. 35 We expressed by transient transfection human Slc46A2 in HCT116 colon carcinoma cells, which are known to display enhanced responses to NOD1/2 ligands when these SLC46s are expressed. 35 These cells were challenged with multiple doses of native iE-DAP, iE-DAP-Alk, native MDP, or MDP-Alk for 6hrs and 24hrs (Figure 5 A&B). iE-DAP and iE-DAP-Alk showed robust and similar induction of NF-κB reporter activity. MDP and MDP-Alk also showed an almost identical dose responses in NF-κB reporter activity (Fig. 5 C&D). Results from these experiments confirm that iE-DAP-Alk and MDP-Alk have similar potency as the native molecules in triggering the NOD1/NOD2 NF-kB pathway.
Figure 5.
Alkyne-modified iE-DAP and MDP showed similar potency in cell line reporter assays. NF-κB luciferase assays in HCT116 cells transiently expressing human Slc46a2 or empty vector (VC) showed that multiple doses of native iE-DAP/iE-DAP-Alk (iE-DAP*) and native MDP/MPD-Alk (MDP*) triggered indistinguishable NF-κB reporter activity at 6 or 24 h, A&C or B&D respectively. Values from unstimulated controls were subtracted from the values for muropeptide-stimulated conditions, to display the signal over the background. N = 4, two-way ANOVA with Tukey's multiple comparisons test to determine significance. **** P < 0.0001; *** P < 0.001; ** P < 0.01; * P < 0.05; ns, not significant.
BMDM cytokine stimulation assay
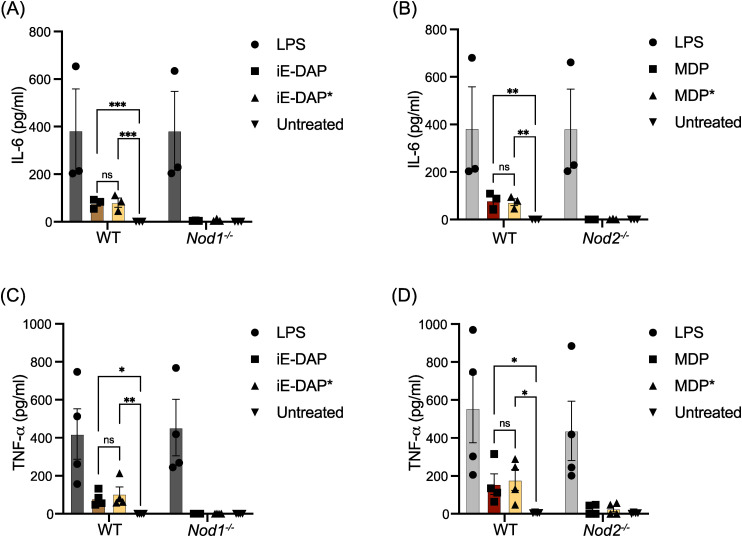
iE-DAP-Alk and MDP-Alk potency were further tested with IFNγ-primed bone marrow-derived macrophages (γ-BMDM) by measuring interleukin-6 (IL-6) and tumor necrosis factor (TNF) production. 10 BMDM from WT, Nod1−/− or Nod2−/− were challenged with native and iE-DAP-Alk and MDP-Alk (100 µM). Both native and alkyne-modified molecules similarly stimulated IL-6 and TNF production in WT γ-BMDM culture (Figure 6). Negligible cytokine production was observed in Nod1 or Nod2 deficient γ-BMDM challenged with iE-DAP or MDP, respectively (See Figure 6). These results demonstrate that the modified molecules have similar activity to the native NOD1/2 agonists. LPS was used as a control cytokine-inducing stimulus.
Figure 6.
Alkyne -and native iE-DAP and MDP stimulate similar levels of IL-6 and TNF-α in macrophages. (A and B) IFNγ-primed BMDMs with indicated genotypes were challenged with different agonists as labeled at 100 µM iE-DAP or MDP and 50 ng of LPS, and IL-6 was measured by ELISA. (C and D) TNFα was measured by ELISA after being challenged with different agonists as in Figures A and B. N ≥ 3, two-way ANOVA with Tukey's multiple comparisons test to determine significance. **** P < 0.0001; *** P < 0.001; ** P < 0.01; * P < 0.05; ns, not significant.
Internalization of modified ie-DAP and MDP
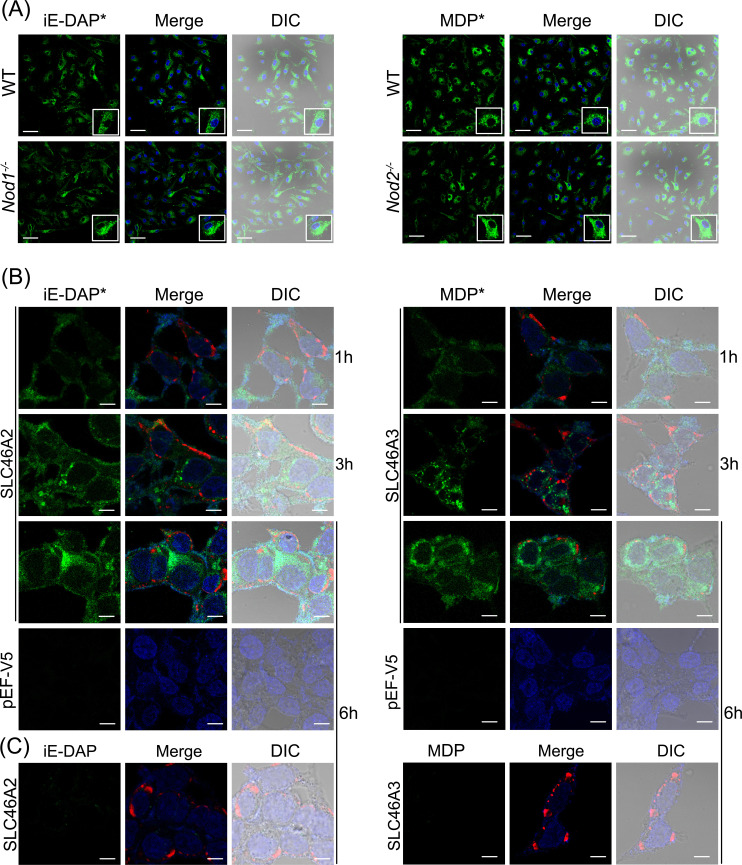
iE-DAP and MDP are recognized by intracellular receptors NOD1 and NOD2 respectively. Therefore, their delivery to the cytosol is critical for downstream signaling.10,11 To confirm the internalization of modified molecules into the cell, IFNγ-primed BMDMs were challenged with iE-DAP-Alk and MDP-Alk and visualized using click chemistry, with azide-linked Calflour-488 (AZDye™-488). Both iE-DAP-Alk and MDP-Alk were observed in intracellular compartments of WT as well as Nod1- and Nod2-deficient BMDMs after a 6h challenge, consistent with cytosolic delivery of modified iE-DAP and MDP into BMDMs. Moreover, it also shows that intracellular delivery of iE-DAP or MDP is not dependent on Nod1 or Nod2 (Figure 7A). We have further confirmed these results in HCT116 cells transiently expressing mouse Slc46a2 or Slc46a3. It was previously shown that HCT116 cells respond weakly to muropeptides at baseline, but Slc46A2 or Slc46A3 expression strongly enhanced responses to these NOD1/2 agonists ( 35 and Figure 5). HCT116 cells expressing Slc46a2 or Slc46a3 imported iE-DAP-Alk and MDP-Alk, respectively, using AZDye-488 and confocal microscopy for detection. With either probe, intracelluar signal was readily detectable as early as 1 h, exhibited a punctate pattern at 3 h, and increasingly diffuse staining throughout the cell by 6 h. On the other hand, control empty vector transfected cells did not show any sign of internalization of these modified muropeptides even at 6 h (Figure 7B). Additionally, native iE-DAP and MDP (without Alk-modification) did not react with AZDye-488 in these cells (Figure 7C). Results from these experiments confirm that alkyne modification did not affect the functionality or activity of iE-DAP and MDP, while these molecules are useful for cell biological analysis of muropeptide trafficking.
Figure 7.
Internalization of iE-DAP-Alk and MDP-Alk in BMDMs and HCT116 cells. (A) Using click-chemistry with AZDye-488 (green), the intracellular distribution of iE-DAP-Alk or MDP-Alk, independent of Nod1 or Nod2, was observed by confocal fluorescent microscopy in IFNγ-primed bone marrow-derived macrophages at 6hr. Nuclei stained with Hoechst (blue) (B) HCT116 cells transiently expressing mouse Slc46a2 or Slc46a3 similarly showed intracellular iE-DAP-Alk or MDP-Alk uptake (green) that increased with time from 1 to 6 h, whereas empty vector-transfected cells failed to import these NOD1/2 agonists. (C) Additionally, cells treated with native iE-DAP and MDP (without Alk-modification) did not react with AZDye-488. Throughout Panels B and C, the red stain is V5 tagged Slc46a2 or Slc46a3. Nuclei stained with Hoechst (blue). Images in panels are representative of three independent experiments. The scale bar in panel A is 50 µM and in panels B and C is 10 µM.
Discussion and conclusion
The microbiota is an essential component of animal anatomy and physiology and plays a critical role in human health. 48 Bacteria are the most abundant microorganism in the microbiota and play a significant role in maintaining homeostasis with innate and adaptive immune functions.49,50 Cytosolic innate immune receptors NOD1 and NOD2 play a major function in detecting bacteria or bacterial components, especially at barrier layers. 51 However, much still needs to be learned about how these receptors specifically recognize muropeptides and how these muropeptides gain access to these cytosolic receptors. iE-DAP and MDP have been identified as minimal agonists for NOD1 and NOD2, respectively. Here, we report the synthesis and validation of click reaction-supporting alkyne-linked iE-DAP and MDP. While the synthetic process can be extensive and time-consuming, synthesizing the fragments in-house allowed us to control the stereochemistry of the molecule.39–41 For example, in the synthesis of m-DAP we are able to selectively attain one enantiomer whereas commercially available iE-DAP is a mixture of both enantiomers. In the future, these “click-able” probes will be further modified, beyond the alkyne handle, to allow us to functionalize for further biological probing. In addition, these probes can be used to determine if other recently synthesized NOD1/2 ligands utilize the same transport system and establish inhibitory screens.52,53
Using an established NF-κB luciferase reporter assay, where expression of SLC46s in HCT116 cells supports robust NF-κB activity upon iE-DAP or MDP challenge, 35 we have compared the potency of iE-DAP-Alk and MDP-Alk with the native molecules. HCT116 cells express both NOD1 and NOD2, making them a suitable option to test both iE-DAP and MDP.54,55 These assays showed very similar activities for the alkyne-modified muropeptides, compared to their native counterparts, at multiple doses and at two different time points.
Further, we assessed the innate immune stimulatory activity of iE-DAP-Alk and MDP-Alk in IFNγ-primed mouse BMDMs and again found similar cytokine-promoting activity compared to their native counterparts. Modified molecules elicited the same level of IL-6 and TNF in BMDMs as the native NOD1/2 agonists. The same system (γBMDMs) as well as Slc46a2/a3 transfected HCT116 cells were utilized to establish visualization protocols and demonstrate the internalization of modified iE-DAP and MDP, taking advantage of the click handles. In the HCT116 cell system, the kinetics of muropeptide entry was also examined, showing diffuse intracellular staining by 6 h, which was preceded by a more punctate pattern at 3 h, suggestive of trafficking through an intracellular vesicular intermediate. Future studies are necessary to characterize the precise subcellular localization(s) of these probes through the trafficking process.
The investigations presented in these studies underscore the intrinsic worth of click-chemistry-enabled muropeptides in unraveling the cell biology underpinning immune activation induced by small molecules such as iE-DAP or MDP. This is achieved through the ingenious incorporation of an azide-linked fluor substrate in the presence of a copper catalyst. The exclusivity of the substrates for the click azide reaction ensures a highly specific response, further enhanced by the fact that the azide-dye reaction is selectively catalyzed only in the presence of copper. This precision is attributed to the absence of free alkyne or azide groups within biomolecules, thereby conferring a meticulous accuracy upon click reactions.56,57
In summary, these findings collectively emphasize the value of click-chemistry-facilitated muropeptides for detailed cell biological and immunological characterization of the NOD1 and NOD2 pathways.
Supplemental Material
Supplemental material, sj-pdf-1-ini-10.1177_17534259231207198 for Synthesis and validation of click-modified NOD1/2 agonists by Ravi Bharadwaj, Madison V. Anonick, Swati Jaiswal, Siavash Mashayekh, Ashley Brown, Kimberly A. Wodzanowski, Kendi Okuda, Neal Silverman and Catherine L. Grimes in Innate Immunity
Acknowledgments
The authors would like to acknowledge the NIH funding that supported this work: RO1AI060025 (to NS) and R01GM138599 (to CLG), and to thank the reviewers for their insightful critiques that improved the caliber of this work.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institute of General Medical Sciences, National Institute of Allergy and Infectious Diseases, (grant number R01GM138599, RO1AI060025).
ORCID iD: Neal Silverman https://orcid.org/0000-0002-4259-456X
Supplemental material: Supplemental material for this article is available online.
References
- 1.Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell 2020; 180: 1044–1066. 2020/03/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 2019; 19: 477–489. 2019/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okude H, Ori D, Kawai T. Signaling through nucleic acid sensors and their roles in inflammatory diseases. Front Immunol 2020; 11: 625833. 2021/02/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston EL, Heras B, Kufer TA, et al. Detection of bacterial membrane vesicles by NOD-like receptors. Int J Mol Sci 2021; 22: 1005. 2021/01/28. DOI: 10.3390/ijms22031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sundaram B, Kanneganti TD. Advances in understanding activation and function of the NLRC4 Inflammasome. Int J Mol Sci 2021; 22: 1048. DOI: 10.3390/ijms22031048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trindade BC, Chen GY. NOD1 And NOD2 in inflammatory and infectious diseases. Immunol Rev 2020; 297: 139–161. 2020/07/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan AJF, Errington J, Vollmer W. Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol 2020; 18: 446–460. 2020/05/20. [DOI] [PubMed] [Google Scholar]
- 8.Irazoki O, Hernandez SB, Cava F. Peptidoglycan muropeptides: release, perception, and functions as signaling molecules. Front Microbiol 2019; 10: 500. 2019/04/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dube JY, McIntosh F, Zarruk JG, et al. Synthetic mycobacterial molecular patterns partially complete Freund's adjuvant. Sci Rep 2020; 10: 5874. 2020/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stafford CA, Gassauer AM, de Oliveira Mann CC, et al. Phosphorylation of muramyl peptides by NAGK is required for NOD2 activation. Nature 2022; 609: 590–596. 2022/08/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masumoto J, Yang K, Varambally S, et al. Nod1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J Exp Med 2006; 203: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 2005; 307: 731–734. 2005/02/05. [DOI] [PubMed] [Google Scholar]
- 13.Hruz P, Zinkernagel AS, Jenikova G, et al. NOD2 Contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci U S A 2009; 106: 12873–12878. 2009/06/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosa A, Trumstedt C, Eriksson E, et al. Nonhematopoietic cells control the outcome of infection with Listeria monocytogenes in a nucleotide oligomerization domain 1-dependent manner. Infect Immun 2009; 77: 2908–2918. 2009/04/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimada K, Chen S, Dempsey PW, et al. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog 2009; 5: e1000379. 2009/04/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasegawa M, Yamazaki T, Kamada N, et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol 2011; 186: 4872–4880. 2011/03/18. [DOI] [PubMed] [Google Scholar]
- 17.Li E, Hamm CM, Gulati AS, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One 2012; 7: e26284. 2012/06/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabeling C, Scheer H, Schonrock SM, et al. Nucleotide oligomerization domain 1 ligation suppressed murine allergen-specific T-cell proliferation and airway hyperresponsiveness. Am J Respir Cell Mol Biol 2014; 50: 903–911. 2013/11/28. [DOI] [PubMed] [Google Scholar]
- 19.Askarian F, Wagner T, Johannessen M, et al. Staphylococcus aureus modulation of innate immune responses through toll-like (TLR), (NOD)-like (NLR) and C-type lectin (CLR) receptors. FEMS Microbiol Rev 2018; 42: 656–671. 2018/06/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minaga K, Watanabe T, Kamata K, et al. Nucleotide-binding oligomerization domain 1 and Helicobacter pylori infection: a review. World J Gastroenterol 2018; 24: 1725–1733. 2018/05/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavrogiorgos N, Mekasha S, Yang Y, et al. Activation of NOD receptors by Neisseria gonorrhoeae modulates the innate immune response. Innate Immun 2014; 20: 377–389. 2013/07/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macho Fernandez E, Valenti V, Rockel C, et al. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut 2011; 60: 1050–1059. 2011/04/08. [DOI] [PubMed] [Google Scholar]
- 23.Moses JE, Moorhouse AD. The growing applications of click chemistry. Chem Soc Rev 2007; 36: 1249–1262. [DOI] [PubMed] [Google Scholar]
- 24.Boyce M, Bertozzi CR. Bringing chemistry to life. Nat Methods 2011; 8: 638–642. 2011/07/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baskin JM, Prescher JA, Laughlin ST, et al. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci U S A 2007; 104: 16793–16797. 2007/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shieh P, Siegrist MS, Cullen AJ, et al. Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc Natl Acad Sci U S A 2014; 111: 5456–5461. 2014/04/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Appukkuttan P, Dehaen W, Fokin VV, et al. A microwave-assisted click chemistry synthesis of 1, 4-disubstituted 1, 2, 3-triazoles via a copper (I)-catalyzed three-component reaction. Org Lett 2004; 6: 4223–4225. [DOI] [PubMed] [Google Scholar]
- 28.Finn M, Fokin VV. Click chemistry: function follows form. Chem Soc Rev 2010; 39: 1231–1232. [DOI] [PubMed] [Google Scholar]
- 29.Traub S, von Aulock S, Hartung T, et al. Invited review: MDP and other muropeptides — direct and synergistic effects on the immune system. J Endotoxin Research 2006; 12: 69–85. [DOI] [PubMed] [Google Scholar]
- 30.Wang YC, Westcott NP, Griffin ME, et al. Peptidoglycan metabolite photoaffinity reporters reveal direct binding to intracellular pattern recognition receptors and arf GTPases. ACS Chem Biol 2019; 14: 405–414. 2019/02/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kekessie I, Goncharov T, Komuves LG, et al. A solid-phase approach for the synthesis of muramyl dipeptide conjugates for detection of NOD2. Bioorg Chem 2021; 116: 105360. 2021/09/26. [DOI] [PubMed] [Google Scholar]
- 32.Vacariu CM, Tanner ME. Recent advances in the synthesis and biological applications of peptidoglycan fragments. Chemistry 2022; 28: e202200788. 2022/05/14. [DOI] [PubMed] [Google Scholar]
- 33.Bharadwaj R, Lusi CF, Mashayekh S, et al. Methotrexate suppresses psoriatic skin inflammation by inhibiting muropeptide transporter SLC46A2 activity. Immunity 2023; 56: 998–1012. e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chamaillard M, Hashimoto M, Horie Y, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 2003; 4: 702–707. 2003/06/11. [DOI] [PubMed] [Google Scholar]
- 35.Paik D, Monahan A, Caffrey DR, et al. SLC46 Family transporters facilitate cytosolic innate immune recognition of monomeric peptidoglycans. J Immunol 2017; 199: 263–270. 2017/05/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okuda K, Silva Costa Franco MM, Yasunaga A, et al. Leishmania amazonensis sabotages host cell SUMOylation for intracellular survival. iScience 2022; 25: 104909. 2022/09/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy Chowdhury A, Boons G-J. The synthesis of diaminopimelic acid containing peptidoglycan fragments using metathesis cross coupling. Tetrahedron Lett 2005; 46: 1675–1678. [Google Scholar]
- 38.Girardin SE, Travassos LH, Herve M, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem 2003; 278: 41702–41708. 2003/07/23. [DOI] [PubMed] [Google Scholar]
- 39.Saito Y, Yoshimura Y, Wakamatsu H, et al. A facile synthesis of fully protected meso-diaminopimelic acid (DAP) and its application to the preparation of lipophilic N-acyl iE-DAP. Molecules 2013; 18: 1162–1173. 2013/01/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernandez N, Martin VS. General stereoselective synthesis of chemically differentiated alpha-diamino acids: synthesis of 2,6-diaminopimelic and 2,7-diaminosuberic acids. J Org Chem 2001; 66: 4934–4938. 2001/07/10. [DOI] [PubMed] [Google Scholar]
- 41.Kawasaki A, Karasudani Y, Otsuka Y, et al. Synthesis of diaminopimelic acid containing peptidoglycan fragments and tracheal cytotoxin (TCT) and investigation of their biological functions. Chemistry 2008; 14: 10318–10330. 2008/10/03. [DOI] [PubMed] [Google Scholar]
- 42.Grieco PA, Gilman S, Nishizawa M. Organoselenium chemistry. A facile one-step synthesis of alkyl aryl selenides from alcohols. J Org Chem 1976; 41: 1485–1486. [Google Scholar]
- 43.Gao Y, Lane-Bell P, Vederas JC. Stereoselective synthesis of meso-2,6-diaminopimelic acid and its selectively protected derivatives. J Org Chem 1998; 63: 2133–2143. [Google Scholar]
- 44.Guo Z-X, Schaeffer MJ, Taylor RJK. Synthesis of unsaturated α-amino acids using the ramberg–bäcklund reaction. J Chem Soc, Chem Commun 1993: 874–875. 10.1039/C39930000874. DOI: 10.1039/C39930000874. [DOI] [Google Scholar]
- 45.Melnyk JE, Mohanan V, Schaefer AK, et al. Peptidoglycan modifications tune the stability and function of the innate immune receptor Nod2. J Am Chem Soc 2015; 137: 6987–6990. 2015/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown AR, Wodzanowski KA, Santiago CC, et al. Protected N-acetyl muramic acid probes improve bacterial peptidoglycan incorporation via metabolic labeling. ACS Chem Biol 2021; 16: 1908–1916. 2021/09/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang H, DeMeester KE, Hou CW, et al. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat Commun 2017; 8: 15015. 2017/04/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altves S, Yildiz HK, Vural HC. Interaction of the microbiota with the human body in health and diseases. Biosci Microbiota Food Health 2020; 39: 23–32. 2020/04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 2013; 14: 676–684. 2013/06/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khosravi A, Mazmanian SK. Disruption of the gut microbiome as a risk factor for microbial infections. Curr Opin Microbiol 2013; 16: 221–227. 2013/04/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee T, Hovingh ES, Foerster EG, et al. NOD1 And NOD2 in inflammation, immunity and disease. Arch Biochem Biophys 2019; 670: 69–81. 2018/12/24. [DOI] [PubMed] [Google Scholar]
- 52.Griffin ME, Tsukidate T, Hang HC. N-Arylpyrazole NOD2 agonists promote immune checkpoint inhibitor therapy. ACS Chem Biol 2023; 18: 1368–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guzelj S, Bizjak Š, Jakopin Ž. Discovery of desmuramylpeptide NOD2 agonists with single-digit nanomolar potency. ACS Med Chem Lett 2022; 13: 1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krieg A, Correa RG, Garrison JB, et al. XIAP Mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci U S A 2009; 106: 14524–14529. 2009/08/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Correa RG, Khan PM, Askari N, et al. Discovery and characterization of 2-aminobenzimidazole derivatives as selective NOD1 inhibitors. Chem Biol 2011; 18: 825–832. 2011/08/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Presolski SI, Hong VP, Finn MG. Copper-Catalyzed azide-alkyne click chemistry for bioconjugation. Curr Protoc Chem Biol 2011; 3: 153–162. 2011/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lutz JF, Zarafshani Z. Efficient construction of therapeutics, bioconjugates, biomaterials and bioactive surfaces using azide-alkyne “click” chemistry. Adv Drug Deliv Rev 2008; 60: 958–970. 2008/04/15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ini-10.1177_17534259231207198 for Synthesis and validation of click-modified NOD1/2 agonists by Ravi Bharadwaj, Madison V. Anonick, Swati Jaiswal, Siavash Mashayekh, Ashley Brown, Kimberly A. Wodzanowski, Kendi Okuda, Neal Silverman and Catherine L. Grimes in Innate Immunity