Abstract
So-called sulfur-turf microbial mats, which are macroscopic white filaments or bundles consisting of large sausage-shaped bacteria and elemental sulfur particles, occur in sulfide-containing hot springs in Japan. However, no thermophiles from sulfur-turf mats have yet been isolated as cultivable strains. This study was undertaken to determine the phylogenetic positions of the sausage-shaped bacteria in sulfur-turf mats by direct cloning and sequencing of 16S rRNA genes amplified from the bulk DNAs of the mats. Common clones with 16S rDNA sequences with similarity levels of 94.8 to 99% were isolated from sulfur-turf mat samples from two geographically remote hot springs. Phylogenetic analysis showed that the phylotypes of the common clones formed a major cluster with members of the Aquifex-Hydrogenobacter complex, which represents the most deeply branching lineage of the domain bacteria. Furthermore, the bacteria of the sulfur-turf mat phylotypes formed a clade distinguishable from that of other members of the Aquifex-Hydrogenobacter complex at the order or subclass level. In situ hybridization with clone-specific probes for 16S rRNA revealed that the common phylotype of sulfur-turf mat bacteria is that of the predominant sausage-shaped bacteria.
Microbial mats develop in a wide variety of aquatic environments, including geothermal hot springs and hydrothermal vents. There are several types of thermophilic microbial mats, e.g., those of cyanobacteria, anoxygenic phototrophic bacteria, and chemotrophic sulfur bacteria, which differ according to the physical and chemical conditions they favor and other environmental factors (10, 38). These microbial mats in thermal habitats have been studied extensively as a peculiar microbial community of the ecosystem, in relation to the phylogeny and evolution of thermophilic prokaryotes, or as a source of new functional enzymes.
So-called sulfur-turf microbial mats are macroscopic bundles of white filaments consisting of colorless sulfur bacteria and elemental sulfur particles that form in shallow streams of sulfide-containing high-temperature hot springs. Since first reported by Miyoshi in 1897 (33), this kind of microbial mat has been recorded for several geographically remote hot springs in Japan, although there have been only scattered reports of sulfur-turf microbial mats or chemotrophic sulfur streamers in geothermal springs in other countries (9, 13, 14). The sulfur-turf mats generally develop within a temperature range of 45 to 73°C, within a pH range of 6 to 9, and at discrete sulfide-oxygen interfaces in geothermal springs. These characteristics suggest that the major constituents of the sulfur-turf prokaryotic community are (hyper)thermophilic, neutrophilic, microaerophilic, and chemolithotrophic bacteria. Early studies of these sulfur-turf mats distinguished microscopically three morphotypes of bacteria, two of which were tentatively named Thiovibrio miyoshi and Thiothrix miyoshi (15). Moreover, in situ ecophysiological and microscopic studies have shown that one of these bacteria, the large sausage-shaped “Thiovibrio miyoshi,” predominates in sulfur-turf mats and oxidizes environmental sulfide to elemental sulfur and then to sulfate via thiosulfate (27–31). So far, however, it has not been possible to isolate and cultivate any thermophilic prokaryotes from the sulfur-turf mats predominated by these sausage-shaped bacteria with artificial media, and no attempt has been made to clarify their taxonomic and phylogenetic positions.
Determination of 16S rRNA genes is a useful research strategy for identifying uncultivated prokaryotes and is now commonly performed in ecological studies. This technique, involving PCR amplification of 16S rRNA genes or synthesis of cDNAs from bulk 16S rRNAs of natural mixed microbial populations, has been used successfully for the phylogenetic characterization of prokaryotes in hydrothermal environments (6, 7, 34, 40, 41, 47, 48). In the present study, this approach was applied to characterize the sausage-shaped bacteria in sulfur-turf mats without isolating and cultivating them. Here we report that sulfur-turf mats contain novel thermophilic bacteria belonging to the earliest-branching lineage of the domain bacteria.
MATERIALS AND METHODS
Mat and water samples.
The sulfur-turf mats were collected from two hot-springs: Nakanoyu (36°11′N, 137°37′E) in Nagano Prefecture and Ganiba (39°47′N, 140°48′E) in Akita Prefecture, Japan. A portion of the sulfur-turf mat was fixed with 70% ethanol and examined immediately upon return to the laboratory. The physicochemical characteristics of the hot spring water in situ were investigated with portable analytical equipment, including a model pH81 meter (Yokogawa Co. Ltd., Tokyo, Japan) for measurement of pH, a model DO-14P analyzer (Toa Co. Ltd., Tokyo, Japan) for measurement of dissolved oxygen, and a model CM-14P meter (Toa) for measurement of electrical conductivity. The dissolved sulfide concentration (S2−) was measured with a lead acetate colorimetric detection tube kit for sulfide ion (catalog no. 211L; GASTEC Co., Tokyo, Japan). Dissolved organic carbon in the hot spring water was determined with a total organic carbon analyzer (model TOC-5000; Shimadzu Co. Ltd., Kyoto, Japan).
Catalase test.
A small portion of the sulfur-turf mat was dropped into 3% (vol/vol) hydrogen peroxide solution and examined for development of gas bubbles.
DNA extraction.
Mat samples (ca. 3 g [wet weight]) in ethanol were collected by centrifugation, washed twice with TE buffer (10 mM Tris-HCl, 1 mM Na2-EDTA [pH 7.8]), and resuspended in sucrose lysis buffer (10% sucrose, 0.7 M NaCl, 40 mM Na2-EDTA, 50 mM Tris-HCl [pH 8.5]) to a total volume of 10 ml, to which lysozyme (1 mg/ml; Wako Pure Chemicals Co., Tokyo, Japan) and achromopeptidase (50 μg/ml; Wako) were then added. This suspension was subjected to three cycles of disruption in a French pressure cell at 20,000 lb/in2. The disrupted sample was treated with proteinase K (50 μg/ml; Wako) at 55°C for 30 min and then with 1% sodium dodecyl sulfate (SDS) at 55°C for 60 min. The slurry was further treated with 1% hexadecyltrimethyl ammonium bromide at 55°C for 30 min to remove polysaccharides and residual proteins as precipitates. The digested sample was treated with phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]) with shaking and then centrifuged. The crude DNA in the resulting aqueous layer was obtained by ethanol precipitation and centrifugation and subjected to a standard purification procedure consisting of RNase digestion, chloroform-isoamyl alcohol treatment, and ethanol precipitation. The DNA sample was further purified by cesium chloride gradient ultracentrifugation as described previously (42). A simplified procedure for DNA extraction was also used as follows. The mat samples (ca. 0.1 g [wet weight]) were washed twice with 1 ml of TE buffer, resuspended in 100 μl of TE buffer containing lysozyme (50 μg/ml) and achromopeptidase (50 μg/ml), and incubated at 37°C for 60 min. Proteinase K (100 μg/ml) was then added, and the mixture was incubated at 55°C for 60 min and finally kept in boiling water for 5 min to terminate the enzyme activity. The resulting crude lysate was measured for absorbance at 260 nm and diluted with TE buffer to give an A260 of 3.0. A 5-μl portion of the diluted sample was subjected to PCR amplification.
PCR amplification, cloning, and sequencing of 16S rDNA.
PCR was performed with a commercial Taq polymerase kit and a pair of universal primers for 16S rRNA gene (rDNA) sequences (25); forward primer 5′-AGA GTT TGA TCA TGG CTC-3′ (S-D-Bact-0027-a-S-18) or 5′-CTG GTT GAT CCT GCC AG-3′ (S-D-Arch-0025-a-S-17) and reverse primer 5′-GTA TTA CCG CGG CTG CTG G-3′ (S-*-Univ-0518-a-A-19), 5′-CTA GCG ATT CCG ACT TCA-3′ (S-D-Bact-1327-a-A-18), or 5′-GGC TAC CTT GTT ACG ACT T-3′ (S-*-Proc-1492-a-A-19) (2). The numbers refer to positions in the Escherichia coli 16S rRNA (11). The thermal conditions were as follows: 40 cycles of denaturation at 95°C for 15 s, primer annealing at 50°C for 60 s, and extension at 70°C for 60 s in a thermal cycler (Perkin-Elmer ABI Japan, Tokyo, Japan). Amplified DNAs were purified by the spin column method with S-400 MicroSpin columns (Pharmacia Biotech Inc., Uppsala, Sweden). The purified DNAs were cloned directly by the TA cloning method (32) with a pGEM-T vector kit (Promega Co., Madison, Wis.) and a DNA ligation kit (version 2; Takara Shuzo Co., Kyoto, Japan). Clone libraries were constructed by transformation of E. coli JM109 or GIFU 12484 (same as XL1-Blue strain). DNAs were sequenced by linear PCR sequencing with either a SequiTherm Long-Read cycle sequencing kit (Epicentre Technologies, Madison, Wis.) or a DyeTerminator cycle sequencing kit (Perkin-Elmer ABI) and analyzed with a Pharmacia ALF DNA sequencer or a Perkin-Elmer ABI 373A DNA sequencer, respectively.
Phylogenetic analysis.
Sequence data were compiled with the GENETYX-MAC program (Software Developing Co., Tokyo, Japan) and examined for sequence homology with the 16S rDNA sequences deposited in the databases employing the BLAST search program (3). Possible chimera artifacts of the sequences were determined by the CHECK CHIMERA program of the Ribosomal Database Project (RDP) server (http://rdp.life.uiuc.edu) (26). Other sequences to be compared were obtained from the small-subunit rRNA database (release 5.0) of the RDP server. Multiple alignment of sequences and calculation of nucleotide substitution rates by Kimura’s two-parameter model (23) were performed with the CLUSTAL W program (46) and the SeqPup program (16). A phylogenetic tree was constructed by using the maximum-likelihood algorithm (36) and illustrated with the TreeView drawing program (37). RNA secondary structures were predicted by using the free-energy minimization algorithm with the MFOLD program, version 2.0 (21, 51). Signatures of 16S rRNA sequences were examined based on the definitions of the domains archaea and bacteria (49).
Oligonucleotides probes.
Oligonucleotide probes complementary to the NAK and GANI clone-specific regions of 16S rRNA sequences were designed and tested for their specificities against sequences in the rRNA database of the RDP and the DNA database of the DDBJ. From among the sequences tested, the following probe for positions 89 to 106 of the 16S rRNA was selected: 5′-GTC GCC AGC ACT ATT ACC-3′ (S-*-ST-0089-a-A-18). The rhodamine-labeled oligonucleotide was synthesized and purified by Takara Shuzo Co.
In situ hybridization.
Samples of sulfur-turf mat were fixed in 4% paraformaldehyde in 3× phosphate-buffered saline (PBS) for 6 to 12 h, washed three times in 1× PBS, and then stored in 50% (vol/vol) ethanol in 1× PBS at −20°C until further use (4, 40). Small portions of the fixed samples were smeared onto gelatin-coated glass slides and air dried. The slides were rinsed in a series of ethanol (50, 80, and 99%). Buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.4], 0.01% SDS, 20% formamide) containing 5 ng of fluorescently labeled oligonucleotide probe per μl was mounted on the slides and hybridization was done at 46°C for 1.5 h in a humidified box. The slides were washed with washing buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.4], 0.01% SDS), submerged in the buffer at 48°C for 15 min, stained with 4′, 6-diamidino-2-phenylindole (DAPI; 0.1 μg/ml in distilled water), and rinsed with distilled water. Cells of E. coli and Bacillus subtilis were employed as negative controls of hybridization. The slides were observed with an Olympus fluorescence microscope, model BX 40.
Nucleotide sequence accession numbers.
The 16S rDNA sequences determined in this study have been deposited in the DDBJ nucleotide sequence database under the accession no. AB005735 to AB005738.
RESULTS
Traits of sulfur-turf microbial mats.
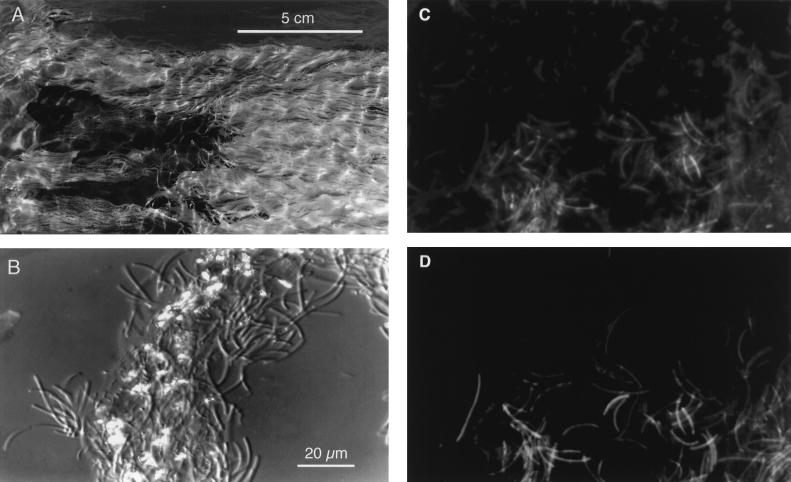
The sulfur-turf microbial mats of the Nakanoyu and Ganiba hot springs showed a typical appearance: white ruffled fur or turf-like massive filamentous streamers (Fig. 1A). These mats grow in shallow hot spring streams and sometimes spread out for several square meters. The waters of both hot springs were at 52 to 72°C, pH 7.2 to 8.0, had an electrical conductivity of 100 mS/m, and contained 3 to 6 mg of dissolved sulfide, less than 1 mg of dissolved oxygen, and 0.41 to 0.72 mg of dissolved organic carbon per liter. In both of the sulfur-turf mats, large sausage-shaped bacteria (1 μm wide and 5 to 20 μm long) predominated and many elemental sulfur particles adhered to the surfaces of the bundles (Fig. 1B). When a portion of the sulfur-turf mat was dropped into H2O2 solution, it exhibited no bubble production, indicating that the predominant sulfur-turf bacteria had no catalase activity.
FIG. 1.
Photographs of a sulfur-turf microbial mat. (A) Ruffled fur or turf-like appearance of a mat in a shallow hot spring stream; (B) Nomarski interference contrast micrograph of the sulfur-turf mat consisting of bundles of large sausage-shaped bacteria and glittering elemental sulfur particles; (C) epifluorescence microscopic image of the sulfur-turf mat stained with DAPI; (D) image of fluorescently labeled probe hybridized to large sausage-shaped cells in the same microscopic field shown in panel C. The microscopic images were obtained with a charge-coupled device camera (model C5910; Hamamatsu Photonics K.K., Hamamatsu, Japan).
Amplification efficiencies with different primers and template preparations.
We performed a preliminary examination using a sample from the Ganiba hot spring to examine the PCR amplification efficiency of 16S rRNA genes with different primer pairs and with two kinds of template DNA prepared by different methods. One method involved bulk DNA purified by ultracentrifugation, and the other method involved crude lysate prepared by simplified extraction by enzymatic digestion only. In both tests, positive PCR signals of the expected molecular sizes were detected by agarose gel electrophoresis, but the signal intensities differed markedly according to the PCR primer used. The primer pairs (S-D-Bact-0027-a-S-18 with S-*-Univ-0518-a-A-19 or S-*-Univ-1327-a-A-19) gave pronounced single bands (0.5- and 1.3-kb fragments, respectively) upon electrophoresis, whereas the remaining pair of bacterial universal primers, which were expected to give a 1.5-kb fragment, generated only a weak PCR band or smeared signals. The amount of this 1.5-kb fragment amplified was too small to be cloned with high efficiency for transformation. PCR assays with a primer pair for archaeal 16S rRNA (S-D-Arch-0025-a-S-17 and a reverse primer) did not produce any pronounced signals (data not shown). These results indicated that the major populations of the sulfur-turf microbial mats were of the domain bacteria and that archaeal members were few or nonexistant.
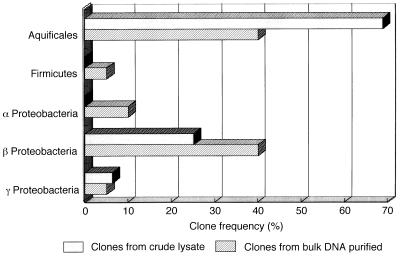
We constructed two 16S rDNA clone libraries using the 0.5-kb PCR products amplified from the purified DNA and the crude lysate of the Ganiba mat and sequenced 16 to 20 positive clones of these libraries for comparison. The homology search with the BLAST system showed that the sulfur-turf clones could be classified into three major phylotypes: the Aquifex-Hydrogenobacter complex, Firmicutes spp. (gram-positive bacteria), and Proteobacteria. Figure 1 shows the distribution of different phylotypes in the two 16S rDNA libraries we constructed. In both libraries, the phylotypes related to the Aquifex-Hydrogenobacter complex predominated (40 to 68% of all the clones examined). A preliminary examination indicated that the crude lysate prepared by enzymatic digestion was usable as a source of DNA templates for PCR amplification of the 16S rDNA of sulfur-turf mat samples.
Phylogenetic analysis of sulfur-turf mat bacteria.
Based on the above-described results, we used the crude lysate only as the PCR template in further experiments. In order to obtain more-detailed sequence information, we constructed DNA libraries using the 1.3-kb PCR fragments amplified directly from the crude lysates of the two mat samples. Then, a total of 25 positive clones, 15 from Nakanoyu and 10 from Ganiba, were sequenced. When the 5′-terminal regions of the 16S rDNA clones were first sequenced, the sequences of 2 clones from Nakanoyu and all 10 clones from Ganiba were identical between positions 8 and 314 and these sequences corresponded to the Aquifex-Hydrogenobacter complex. The remaining Nakanoyu clones were assigned to either Firmicutes or Proteobacteria but were not more than 93% similar to any of the Ganiba clones of the same phylogenetic groups. Only the clones belonging to the Aquifex-Hydrogenobacter complex were of the common phylotype. Thus, the results suggested that these common clones in samples from both hot springs represent the predominant sausage-shaped bacteria of the sulfur-turf mats.
Of the major clones isolated, two each of the Nakanoyu (NAK-9 and -14) and Ganiba (GANI-3 and -4) clones were chosen and analyzed for their full sequences (a nucleotide stretch from positions 8 to 1345). All of the four clones from the sulfur-turf mat specimens were closely related, with similarity levels of 99.4 to 94.8% (evolutionary distances, 0.005 to 0.043) (Table 1). However, there seemed to be some local variations between the sequences of the GANI and NAK clones.
TABLE 1.
Levels of sequence similarity and evolutionary distances for 16S rRNAs
| No. | Name of species or clone | Sequence similarity (%) (lower triangle) or evolutionary distance (Knuc) (upper triangle) of species or clone no.:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| 1 | Haloferax volcanii | 0.23 | 0.29 | 0.27 | 0.37 | 0.371 | 0.367 | 0.370 | 0.369 | 0.397 | 0.384 | 0.385 | |
| 2 | Thermococcus celer | 67.6 | 0.21 | 0.16 | 0.32 | 0.323 | 0.312 | 0.313 | 0.306 | 0.367 | 0.327 | 0.324 | |
| 3 | Sulfolobus acidocaldarius | 59.8 | 72.1 | 0.12 | 0.35 | 0.349 | 0.340 | 0.342 | 0.333 | 0.374 | 0.345 | 0.338 | |
| 4 | Pyrodictium occultum | 63.8 | 78.7 | 85.7 | 0.33 | 0.336 | 0.326 | 0.329 | 0.310 | 0.370 | 0.326 | 0.323 | |
| 5 | GANI-3 | 42.9 | 51.1 | 46.5 | 48.9 | 0.005 | 0.040 | 0.041 | 0.199 | 0.210 | 0.195 | 0.187 | |
| 6 | GANI-4 | 42.7 | 51.0 | 46.3 | 48.7 | 99.4 | 0.041 | 0.043 | 0.200 | 0.210 | 0.196 | 0.186 | |
| 7 | NAK-9 | 43.8 | 52.5 | 47.3 | 50.2 | 94.8 | 94.8 | 0.012 | 0.186 | 0.203 | 0.183 | 0.175 | |
| 8 | NAK-14 | 43.4 | 52.5 | 47.1 | 50.0 | 94.8 | 94.8 | 98.7 | 0.185 | 0.204 | 0.183 | 0.174 | |
| 9 | Aquifex pyrophilus | 37.6 | 49.2 | 47.0 | 50.2 | 71.8 | 71.8 | 73.2 | 73.5 | 0.190 | 0.113 | 0.109 | |
| 10 | Hydrogenobacter acidophilus | 34.6 | 39.1 | 36.6 | 39.2 | 67.3 | 67.4 | 69.2 | 69.3 | 73.5 | 0.174 | 0.159 | |
| 11 | EM17 phylotype of Yellowstone hot spring | 35.3 | 43.6 | 41.5 | 46.8 | 68.4 | 68.4 | 70.2 | 70.4 | 82.7 | 79.8 | 0.062 | |
| 12 | Calderobacterium hydrogenophilum | 37.8 | 46.9 | 44.0 | 48.1 | 70.4 | 70.6 | 72.6 | 72.9 | 83.6 | 81.8 | 91.9 | |
| 13 | Hydrogenobacter thermophilus | 38.4 | 47.8 | 45.3 | 49.8 | 70.3 | 70.6 | 72.9 | 73.1 | 83.7 | 82.2 | 91.5 | 97.9 |
| 14 | EM19 phylotype of Yellowstone hot spring | 36.1 | 44.4 | 40.8 | 46.5 | 60.1 | 60.1 | 62.7 | 62.6 | 67.8 | 62.2 | 66.6 | 67.4 |
| 15 | Thermosipho africanus | 37.5 | 45.9 | 39.5 | 45.3 | 64.9 | 65.0 | 67.5 | 67.5 | 68.5 | 62.6 | 64.9 | 66.0 |
| 16 | Thermotoga maritima | 42.3 | 55.6 | 47.4 | 52.4 | 66.6 | 66.6 | 68.8 | 68.6 | 72.7 | 64.4 | 67.1 | 70.8 |
| 17 | EM3 phylotype of Yellowstone hot spring | 38.8 | 48.4 | 42.4 | 47.1 | 58.3 | 58.3 | 60.4 | 60.2 | 61.0 | 59.0 | 63.9 | 65.3 |
| 18 | Thermus aquaticus | 39.9 | 46.1 | 43.8 | 48.4 | 62.1 | 62.0 | 64.5 | 64.7 | 66.5 | 63.0 | 67.4 | 69.4 |
| 19 | Thermomimcrobium roseum | 41.1 | 49.2 | 45.5 | 50.5 | 60.4 | 60.4 | 63.9 | 63.8 | 64.4 | 60.5 | 63.0 | 65.1 |
| 20 | Chloroflexus aurantiacus | 38.4 | 42.8 | 39.7 | 42.6 | 59.1 | 59.1 | 60.7 | 60.5 | 59.1 | 59.7 | 63.4 | 64.8 |
| 21 | CLostridium thermoaceticum | 38.3 | 43.6 | 42.3 | 46.0 | 64.4 | 64.4 | 66.8 | 66.9 | 65.9 | 62.7 | 65.5 | 65.4 |
| 22 | Bacillus subtilis | 37.8 | 44.7 | 40.3 | 44.0 | 60.3 | 60.4 | 62.8 | 63.0 | 64.8 | 63.0 | 60.9 | 64.1 |
| 23 | Agrobacterium tumefaciens | 43.2 | 46.8 | 41.6 | 45.5 | 64.2 | 64.2 | 66.8 | 66.8 | 58.7 | 61.1 | 57.4 | 61.2 |
| 24 | Anabaena “cylindrica” | 22.0 | 25.7 | 22.2 | 24.0 | 41.9 | 42.1 | 44.1 | 44.3 | 34.3 | 43.1 | 40.0 | 42.6 |
| 25 | Oscillatoria williamsii | 31.1 | 36.6 | 32.5 | 34.4 | 54.2 | 54.2 | 56.7 | 56.5 | 50.4 | 51.1 | 49.5 | 54.0 |
| 26 | Alcaligenes xylosoxidans | 34.3 | 38.2 | 34.4 | 38.2 | 59.7 | 59.6 | 62.7 | 62.9 | 63.3 | 62.7 | 60.0 | 62.6 |
| 27 | Thiobacillus hydrothermalis | 37.7 | 41.7 | 37.0 | 40.7 | 59.8 | 59.7 | 63.5 | 63.3 | 64.4 | 61.7 | 60.8 | 63.2 |
| 28 | Escherichia coli | 33.6 | 38.1 | 35.9 | 37.9 | 58.0 | 58.0 | 61.6 | 61.9 | 62.5 | 61.4 | 58.8 | 61.1 |
| Sequence similarity (%) (lower triangle) or evolutionary distance (Knuc) (upper triangle) of species or clone no.:
|
Accession no. | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |
| 0.381 | 0.382 | 0.376 | 0.363 | 0.388 | 0.383 | 0.382 | 0.398 | 0.380 | 0.387 | 0.385 | 0.394 | 0.385 | 0.401 | 0.378 | 0.398 | K00421 |
| 0.319 | 0.329 | 0.328 | 0.282 | 0.323 | 0.328 | 0.331 | 0.357 | 0.345 | 0.345 | 0.353 | 0.362 | 0.343 | 0.372 | 0.350 | 0.368 | M21529 |
| 0.334 | 0.355 | 0.361 | 0.327 | 0.344 | 0.330 | 0.346 | 0.360 | 0.345 | 0.362 | 0.366 | 0.363 | 0.348 | 0.388 | 0.369 | 0.377 | D14876 |
| 0.317 | 0.330 | 0.331 | 0.298 | 0.327 | 0.317 | 0.321 | 0.356 | 0.329 | 0.344 | 0.357 | 0.355 | 0.343 | 0.373 | 0.353 | 0.370 | M21087 |
| 0.190 | 0.265 | 0.246 | 0.237 | 0.268 | 0.254 | 0.280 | 0.279 | 0.243 | 0.261 | 0.260 | 0.272 | 0.254 | 0.273 | 0.271 | 0.277 | AB005735 |
| 0.189 | 0.266 | 0.246 | 0.238 | 0.269 | 0.256 | 0.281 | 0.280 | 0.244 | 0.261 | 0.261 | 0.272 | 0.255 | 0.275 | 0.273 | 0.278 | AB005736 |
| 0.175 | 0.253 | 0.233 | 0.222 | 0.253 | 0.239 | 0.258 | 0.271 | 0.230 | 0.247 | 0.244 | 0.261 | 0.240 | 0.262 | 0.253 | 0.261 | AB005737 |
| 0.175 | 0.255 | 0.234 | 0.225 | 0.255 | 0.240 | 0.260 | 0.273 | 0.231 | 0.247 | 0.245 | 0.260 | 0.242 | 0.261 | 0.256 | 0.260 | AB005738 |
| 0.111 | 0.235 | 0.238 | 0.211 | 0.241 | 0.230 | 0.250 | 0.248 | 0.254 | 0.257 | 0.268 | 0.292 | 0.248 | 0.263 | 0.251 | 0.265 | M83548 |
| 0.156 | 0.285 | 0.274 | 0.262 | 0.292 | 0.279 | 0.277 | 0.282 | 0.268 | 0.274 | 0.260 | 0.268 | 0.249 | 0.270 | 0.277 | 0.276 | D16296 |
| 0.069 | 0.249 | 0.246 | 0.229 | 0.252 | 0.238 | 0.246 | 0.247 | 0.239 | 0.276 | 0.275 | 0.282 | 0.248 | 0.278 | 0.270 | 0.281 | U05661 |
| 0.019 | 0.247 | 0.240 | 0.213 | 0.242 | 0.231 | 0.240 | 0.243 | 0.241 | 0.259 | 0.257 | 0.280 | 0.233 | 0.269 | 0.259 | 0.272 | Z30242 |
| 0.248 | 0.243 | 0.217 | 0.243 | 0.233 | 0.239 | 0.244 | 0.239 | 0.264 | 0.258 | 0.276 | 0.237 | 0.270 | 0.263 | 0.272 | Z30214 | |
| 67.4 | 0.256 | 0.242 | 0.252 | 0.238 | 0.247 | 0.253 | 0.255 | 0.263 | 0.290 | 0.278 | 0.273 | 0.297 | 0.262 | 0.291 | U05662 | |
| 65.7 | 63.9 | 0.114 | 0.220 | 0.223 | 0.236 | 0.260 | 0.221 | 0.254 | 0.244 | 0.264 | 0.223 | 0.261 | 0.259 | 0.275 | M83140 | |
| 70.7 | 66.5 | 85.8 | 0.195 | 0.204 | 0.213 | 0.247 | 0.220 | 0.237 | 0.247 | 0.271 | 0.231 | 0.243 | 0.219 | 0.254 | M21774 | |
| 65.4 | 63.3 | 66.5 | 69.7 | 0.251 | 0.247 | 0.267 | 0.259 | 0.289 | 0.293 | 0.291 | 0.257 | 0.278 | 0.266 | 0.285 | U05660 | |
| 69.4 | 67.2 | 69.6 | 71.9 | 65.8 | 0.200 | 0.238 | 0.223 | 0.246 | 0.248 | 0.277 | 0.231 | 0.260 | 0.237 | 0.241 | L09663 | |
| 65.4 | 63.0 | 67.1 | 71.0 | 65.0 | 73.3 | 0.221 | 0.226 | 0.244 | 0.264 | 0.273 | 0.238 | 0.270 | 0.248 | 0.264 | M34115 | |
| 64.7 | 61.4 | 60.0 | 61.7 | 63.1 | 68.3 | 71.3 | 0.257 | 0.261 | 0.268 | 0.286 | 0.269 | 0.275 | 0.255 | 0.261 | D38365 | |
| 65.9 | 63.3 | 73.2 | 72.3 | 61.5 | 69.5 | 69.6 | 62.1 | 0.169 | 0.201 | 0.223 | 0.192 | 0.228 | 0.199 | 0.217 | M59121 | |
| 63.6 | 61.3 | 66.9 | 69.5 | 56.7 | 66.8 | 67.8 | 61.9 | 79.6 | 0.201 | 0.234 | 0.211 | 0.244 | 0.202 | 0.228 | X60646 | |
| 61.3 | 55.7 | 64.1 | 64.4 | 57.4 | 65.8 | 64.9 | 66.0 | 71.1 | 71.2 | 0.242 | 0.224 | 0.203 | 0.186 | 0.205 | M11223 | |
| 42.5 | 40.9 | 39.6 | 40.3 | 40.6 | 44.2 | 43.3 | 48.2 | 47.5 | 47.4 | 53.3 | 0.130 | 0.246 | 0.227 | 0.252 | See RDP | |
| 53.4 | 46.0 | 55.4 | 55.3 | 49.9 | 56.8 | 57.9 | 54.6 | 61.3 | 59.9 | 63.4 | 63.5 | 0.225 | 0.210 | 0.233 | X58359, X58360, X58361 | |
| 62.6 | 56.5 | 65.1 | 67.5 | 55.5 | 63.9 | 63.8 | 62.3 | 70.5 | 68.1 | 73.4 | 47.2 | 59.5 | 0.180 | 0.186 | M22509 | |
| 62.8 | 60.2 | 65.3 | 69.1 | 58.2 | 66.6 | 65.0 | 63.1 | 74.2 | 74.0 | 74.9 | 49.2 | 61.0 | 78.6 | 0.159 | M90662 | |
| 61.2 | 56.1 | 62.8 | 64.0 | 55.6 | 65.7 | 62.5 | 62.1 | 71.5 | 70.5 | 72.7 | 45.3 | 57.2 | 77.3 | 82.6 | J01695 | |
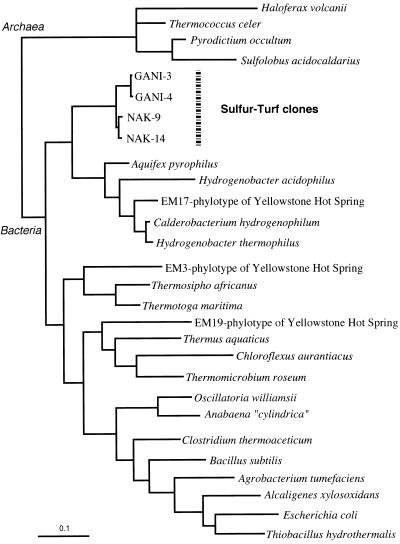
A phylogenetic tree was constructed by using the maximum-likelihood algorithm based on the distance matrix data shown in Table 1. The four clones from the sulfur-turf mat formed a novel phylogenetic clade that branched deeply off the order Aquificales, which includes the genera Aquifex, Hydrogenobacter, and Calderobacterium (Fig. 2). Thus, the predominant phylotypes of the sulfur-turf mats belonged to the phylum of the Aquifex-Hydrogenobacter complex but were distinguishable at the order or subclass level from previously known members of this phylum.
FIG. 2.
Frequencies of 16S rDNA clones derived from the sulfur-turf microbial mat of the Ganiba hot spring.
Signatures and secondary structures of 16S rRNA.
The 16S rRNAs of the GANI and NAK phylotypes deduced from the primary sequences of their 16S rDNAs show the nucleotide signatures specific to members of the domain bacteria with few exceptions (Table 2). The base at position 558 (A) was idiosyncratic to that of the Aquifex-Hydrogenobacter phylum, which includes the GANI and NAK phylotypes. The signatures of the base pairs at positions 340 and 349 of the GANI and NAK clones, as well as those of other members of the Aquifex-Hydrogenobacter phylum, were of the archaeal type. The sulfur-turf mat phylotypes showed characteristic bacterial signatures at base pair positions 367 and 393 and 684 and 706 and at position 923, whereas other members of the Aquifex-Hydrogenobacter phylum showed different signatures at these positions.
TABLE 2.
Compositions of 16S rRNA sequences for the signature positions
| Position(s) of base or base pair | Amino acid(s)a for:
|
||||
|---|---|---|---|---|---|
| Bacteria | Members of sulfur-turf mat phylotypes | H. acidophilus | A. pyrophilus | Archaea | |
| 8 | A | A | A | U | |
| 9, 25 | G, C | G, C | G, C | C, G | |
| 10, 24 | R(A), U | A, U | A, U | Y, R | |
| 33, 551 | A, U | A, U | A, U | A, U | Y(C), R(G) |
| 52, 359 | Y, R | C, G | C, G | C, G | G, C |
| 53, 358 | A, U | A, U | A, U | A, U | C, G |
| 113, 314 | G, C | G, C | G, C | G, C | C, G |
| 307 | H | U | U | C | G |
| 338 | A | A | A | A | G |
| 339, 350 | C, G | C, G | C, G | C, G | G, Y(C) |
| 340, 349 | U, A | M, G | C, G | C, G | C, G |
| 361 | R(G) | G | G | G | C |
| 365 | U | U | U | U | A |
| 367, 393 | U, A | U, A | G, C | G, C | C, G |
| 377, 386 | R(G), Y(C) | G, Y | G, C | G, C | Y(C), G |
| 514, 537 | Y(C), R(G) | C, G | C, G | C, G | G, C |
| 549 | C | C | C | C | U |
| 558 | G | A | A | A | Y |
| 585, 756 | R(G), Y(C) | G, C | C, G | C, G | C, G |
| 675 | A | A | A | A | U |
| 684, 706 | U, A | U, A | U, A | G, C | G, Y(C) |
| 716 | A | A | A | A | C |
| 867 | R(G) | G | G | G | Y(C) |
| 923 | A | A | G | G | G |
| 930 | Y(C) | C | C | C | A |
| 931 | C | C | C | C | G |
| 933 | G | G | G | G | A |
| 962 | C | C | C | C | G |
| 966 | G | G | G | G | U |
| 974 | M | A | A | A | G |
| 1098 | Y(C) | C | C | C | G |
| 1109 | C | C | C | C | A |
| 1110 | A | A | A | A | G |
| 1194 | U | U | U | U | R(G) |
| 1211 | U | U | U | U | G |
| 1212 | U | U | U | U | A |
R, purine (A/G); Y, pyrimidine (U/C); H, A, C, or U; M, A or C. Boldface type indicates a possible plesiomorphic signature retained in the Aquifex-Hydrogenobacter complex.
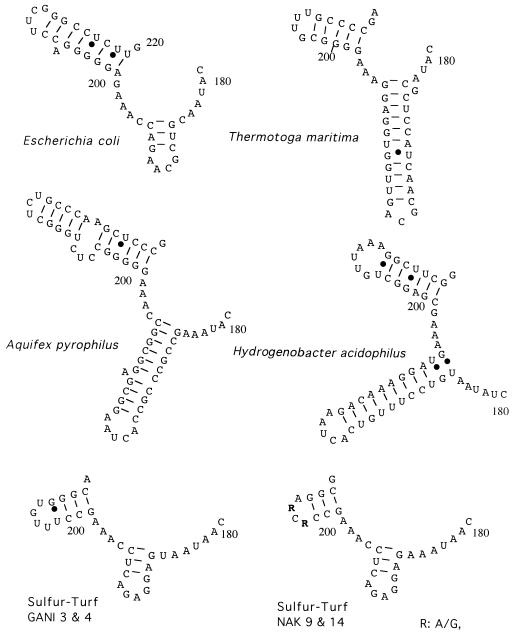
Comparison of the 16S rRNA secondary structures showed that the NAK and GANI clones had unique structural features at positions 180 to 220 (Fig. 3). In the helix at positions 180 to 195 characterized by the capping loop (GAGA) motif, bacteria of the sulfur-turf mat phylotypes were more similar to the proteobacterium E. coli than to the phylogenetically inherent hyperthermophilic bacteria Aquifex pyrophilus, Hydrogenobacter acidophilus, and Thermotoga maritima. In the 16S rRNAs of the bacteria of the sulfur-turf mat phylotypes, another helix at positions 199 to 220 consisted of a shorter length of nucleotides (10 bases) and the capping loop sequences of the bacteria of the two local phylotypes of Nakanoyu and Ganiba varied.
FIG. 3.
Comparative secondary structures of the 16S rRNAs of bacteria of the sulfur-turf mat phylotypes and some other eubacteria at positions 179 to 220 (E. coli numbering) estimated by the free-energy minimization algorithm.
In situ hybridization of sulfur-turf mat bacteria.
In order to determine whether the common phylotypes (NAK and GANI) of bacteria from both hot springs correspond to that of the predominant sausage-shaped bacteria in the sulfur-turf mat, fluorescent in situ hybridization with an oligonucleotide probe specific for 16S rRNA was carried out. Fluorescent microscopic observation by DAPI staining showed that the sulfur-turf mat of the Nakanoyu hot spring stream at 53°C consists of several morphotypes (Fig. 1C). In the same microscopic field, the large sausage-shaped bacteria emitted fluorescence only with the rhodamine of the probe specific for sequences of the NAK and GANI phylotypes (Fig. 1D). None of the specific probe hybridized to the negative-control bacteria (data not shown).
DISCUSSION
The hot spring sulfur-turf microbial mats of Japan have long been of interest because of their conspicuous appearance. However, the community structure of the mats has yet to be investigated in detail mainly because of the difficulty in cultivating mat-building thermophilic prokaryotes. In this study, we adopted a 16S rDNA-based molecular approach to characterize the sulfur-turf bacteria without isolation and cultivation and found common phylotypes in the hot spring sulfur-turf mats from both Ganiba and Nakanoyu. The phylogenetic tree we obtained shows that the clade of novel sulfur-turf mat phylotypes belongs to the phylum of the Aquifex-Hydrogenobacter complex and is distinguishable at the order or subclass level from previously recognized members of this phylum. The unique phylogenetic positions of the sulfur-turf phylotypes are supported by the signatures and secondary structures of their rRNAs. Fluorescent in situ hybridization with a specific probe demonstrated that the clones of NAK and GANI are of a phylotype identical to that of sausage-shaped bacteria, which are the microscopically predominant morphotype in the sulfur-turf mats.
The Aquifex-Hydrogenobacter complex consists of hyperthermophilic, aerobic, obligate, chemolithotrophic bacteria capable of growth with molecular hydrogen or sulfur as an energy source (5, 18, 22, 24, 43) and represents one of the earliest-branching lineages of the domain bacteria (12, 39, 44). These bacteria are divided into two groups with respect to their habitats: members of the genus Aquifex inhabit marine hydrothermal environments, whereas those of the genera Hydrogenobacter and its relatives inhabit hot springs. Hydrogenobacter species are also reported to be unusual in that they fix CO2 through the reductive tricarboxylic acid cycle and contain a sulfur-containing naphthoquinone, methionaquinone, in their respiratory chains (20, 22). The results of this study and previous research on sulfur-turf mats (27–31) show that there is phenotypic and ecological resemblance between the predominant bacteria of sulfur-turf mats and Hydrogenobacter. The sausage-shaped bacteria inhabit sulfide-oxygen interfaces in geothermal hot springs and have a tendency to show microaerophilic chemolithotrophy, with sulfide as the electron donor (28, 29). Concurrent biomarker studies of hot spring microbial mats have also revealed that a methionaquinone homolog is the most abundant quinone in the sulfur-turf mats (17). Moreover, like A. pyrophilus (18) and Hydrogenobacter thermophilus (19), the sulfur-turf bacteria are strongly resistant to bacteriolytic enzymatic treatment with lysozyme or achromopeptidase. Although the sausage-shaped bacteria have not yet been isolated as a cultivable strain, present results strongly suggest that they are (hyper)thermophilic, chemolithotrophic, microaerophilic, and methionaquinone-producing respiratory bacteria.
Since the sulfur-turf mats are conspicuously white bundles of sausage-shaped bacteria with elemental sulfur particles, it is easy to find them in hot spring streams. Similar filamentous and mat-building bacteria have been reported at Octopus Springs of Yellowstone National Park and characterized phylogenetically (40). However, their phylotypes (EM phylotypes) were isolated from pink-colored microbial mats and the similarity of their 16S rRNA gene sequences to those of bacteria of the sulfur-turf mat phylotypes we defined were quite low (Table 1 and Fig. 4). Also, members of the genus Thermothrix, chemolithotrophic sulfur-oxidizing thermophilic bacteria, form filamentous mats in geothermal hot springs (9, 13, 35) but their phylogenetic positions based on 16S rDNA sequences are in the beta subclass of the Proteobacteria (35), indicating that they are very distant from the Aquifex-Hydrogenobacter lineage and from the sulfur-turf mat phylotypes.
FIG. 4.
Phylogenetic tree deduced from 16S rRNA gene sequences of the sulfur-turf mat clones and of representative members of the domains bacteria and archaea. Scale, 10% nucleotide substitution.
The sausage-shaped bacteria of the sulfur-turf mats from the Ganiba and Nakanoyu hot springs exhibited common morphotypes and phylotypes. Sulfur-turf mats created by sausage-shaped bacteria in hot spring streams may offer a habitat or microniche to other microorganisms. However, other clones, members of Proteobacteria or Firmicutes, isolated in this study varied greatly among the mat samples. The population diversity of the sulfur-turf mats seems to be affected by environmental factors of hot springs, e.g., water temperature, sulfide concentration, and human alteration. Geothermal and hydrothermal environments harbor a wide variety of hyperthermophilic and thermophilic microbes belonging to various phylogenetic positions, e.g., those showing chemolithotrophy, photolithotrophy, and chemoorganotrophy. Further study of hot spring biomats is required to analyze not only a mechanism of population dynamics but also the evolutionary development of a pristine ecosystem that existed on early Earth (1, 8, 50).
ACKNOWLEDGMENTS
We thank T. Umezawa and Y. Ueda for their technical assistance in the PCR experiments. We are also grateful to H. Ogawa of the Ocean Research Institute, University of Tokyo, for the determination of organic carbon.
This study was supported in part by the Decoding the Earth Evolution Program of the Intensified Study Area Program of the Ministry of Culture, Science, Sports and Education, Tokyo, Japan (grant 259, 1955–1997).
REFERENCES
- 1.Achenbach-Richter L, Gupta R, Stetter K O, Woese C R. Were the original eubacteria thermophiles? Syst Appl Microbiol. 1987;9:34–39. doi: 10.1016/s0723-2020(87)80053-x. [DOI] [PubMed] [Google Scholar]
- 2.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aragno M. Thermophilic aerobic hydrogen-oxidizing (Knallagas) bacteria. In: Ballos A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 3917–3933. [Google Scholar]
- 6.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspective on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bengtson S, editor. Early life on Earth. Nobel Symposium no. 84. New York, N.Y: Columbia University Press; 1994. [Google Scholar]
- 9.Brannan D K, Cladwell D E. Ecology and metabolism of Thermothrix thiopara. Adv Appl Microbiol. 1986;31:233–270. [Google Scholar]
- 10.Brock T D. Thermophilic microorganisms and life at high temperatures. New York, N.Y: Springer-Verlag; 1978. [Google Scholar]
- 11.Brosius J, Palmer J L, Kennedy J P, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burggraf S, Olsen G J, Woese C R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- 13.Caldwell D E, Caldwell S J, Laycock J P. Thermothrix thioparus gen. et sp. nov., a facultatively anaerobic facultative chemolithoautotroph living at neutral pH and high temperature. Can J Microbiol. 1976;22:1509–1517. doi: 10.1139/m76-223. [DOI] [PubMed] [Google Scholar]
- 14.Castenholtz R W. The effect of sulfide on the blue-green algae in hot springs. I. New Zealand and Iceland. J Phycol. 1976;12:54–68. [Google Scholar]
- 15.Emoto Y, Hirose H. Studien über die Thermal flora von Japan. XIX. Thermal Bakterien und Algen aus thermal Quellen von Hatimantai und Yakeyama. Bot Mag Tokyo. 1942;56:332–343. [Google Scholar]
- 16.Gilbert D G. SeqPup biological sequence editor and analysis program, version 0.6. Bloomington: Biology Department, Indiana University; 1996. [Google Scholar]
- 17.Hiraishi, A., T. Umezawa, H. Yamamoto, K. Kato, and Y. Maki. Unpublished data. [DOI] [PMC free article] [PubMed]
- 18.Huber R T, Huber W D, Trincone A, Burggraf S, Konig H, Rachel R, Rockinger I, Fricke H, Stetter K O. Aquifex pyrophilus gen. nov., sp. nov. represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst Appl Microbiol. 1992;15:340–351. [Google Scholar]
- 19.Ishii, M. Personal communication.
- 20.Ishii M, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. 2-Methylthio-1,4-naphthoquinone, a unique sulfur-containing quinone from a thermophilic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. J Bacteriol. 1987;169:2380–2384. doi: 10.1128/jb.169.6.2380-2384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;56:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasumi T, Igarashi Y, Kodama T, Minoda Y. Hydrogenobacter thermophilus gen. nov., sp. nov., an extremely thermophilic, aerobic, hydrogen-oxidizing bacterium. Int J Syst Bacteriol. 1984;34:5–10. [Google Scholar]
- 23.Kimura M. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 24.Kryukov V R, Savel’eva N D, Pusheva M A. Calderobacterium hydrogenophilum gen. et sp. nov., an extremely thermophilic hydrogen bacterium and its hydrogenase activity. Mikrobiologiya. 1983;52:611–618. [Google Scholar]
- 25.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 26.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maki Y. Factors in habitat preference in situ of sulfur-turfs growing in hot spring effluents: dissolved oxygen and current velocities. J Gen Appl Microbiol. 1986;32:203–313. [Google Scholar]
- 28.Maki Y. Biological oxidation of sulfide and elemental sulfur by A-type sulfur-turf growing in hot springs effluents. J Gen Appl Microbiol. 1987;33:123–134. [Google Scholar]
- 29.Maki Y. Effects of dissolved oxygen concentration on the biological oxidation of sulfide and elemental sulfur by the A-type sulfur-turf growing in hot spring effluents. J Gen Appl Microbiol. 1987;33:391–400. [Google Scholar]
- 30.Maki Y. Study of the sulfur-turf: a community of colorless sulfur bacteria growing in hot spring effluent. Bull Jpn Soc Microb Ecol. 1991;6:33–43. [Google Scholar]
- 31.Maki Y. Rapid biological sulfide oxidation in the effluent of a hot spring. Bull Jpn Soc Microb Ecol. 1993;8:175–179. [Google Scholar]
- 32.Marchuk D, Drumm M, Saulino A, Collins F C. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi M. Studien über die Schwefelrasenbildung und die Schwefelbacterien der Thermen von Yumoto bei Nikko. J Coll Sci Imp Univ Tokyo. 1997;10:143–173. [Google Scholar]
- 34.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odintsova E V, Jannasch H W H, Mamone J A, Langworthy T A. Thermothrix azorensis sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing, thermophilic bacterium. Int J Syst Bacteriol. 1996;46:422–428. doi: 10.1099/00207713-46-2-422. [DOI] [PubMed] [Google Scholar]
- 36.Olsen G J, Matsuda H, Hagstron R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 37.Page R D M. TreeView, version 1.4. Glasgow, United Kingdom: University of Glasgow; 1997. [Google Scholar]
- 38.Pierson B K. Modern mat-building microbial communities: a key to the interpretation of Proterozoic stromatolitic communities. In: Schoph J W, Klein C, editors. The Proterozoic biosphere. A multidisciplinary study. London, United Kingdom: Cambridge University Press; 1994. pp. 247–251. [Google Scholar]
- 39.Pitulle C, Yang Y, Marchiani M, Moore E R B, Siefert J L, Arano M, Jurtshuk P, Jr, Fox G E. Phylogenetic position of the genus Hydrogenobacter. Int J Syst Bacteriol. 1994;44:620–626. doi: 10.1099/00207713-44-4-620. [DOI] [PubMed] [Google Scholar]
- 40.Reysenbach A-L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruff-Roberts A L, Kuenen J G, Ward D M. Distribution of cultivated and uncultivated cyanobacteria and Chloroflexus-like bacteria in hot spring microbial mats. Appl Environ Microbiol. 1994;60:697–704. doi: 10.1128/aem.60.2.697-704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shima S, Suzuki K-I. Hydrogenobacter acidophilus sp. nov., a thermoacidophilic, aerobic, hydrogen-oxidizing bacterium requiring elemental sulfur for growth. Int J Syst Bacteriol. 1993;43:703–708. [Google Scholar]
- 44.Shima S, Yanagi M, Saiki H. The phylogenetic position of Hydrogenobacter acidophilus based on 16S rRNA sequence analysis. FEMS Microbiol Lett. 1994;119:119–122. doi: 10.1111/j.1574-6968.1994.tb06877.x. [DOI] [PubMed] [Google Scholar]
- 45.Stetter K O. Hyperthermophilic procaryotes. FEMS Microbiol Rev. 1996;18:149–158. [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weller R, Bateson M M, Heimbuch B K, Kopczynski E D, Ward D M. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and spirochete-like inhabitants of a hot spring microbial mat. Appl Environ Microbiol. 1992;58:3964–3969. doi: 10.1128/aem.58.12.3964-3969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weller R, Weller J W, Ward D M. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl Environ Microbiol. 1991;57:1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winker S, Woese C R. A definition of the domains Archaea, Bacteria, Eucarya in terms of small subunit ribosomal RNA characteristics. Syst Appl Microbiol. 1991;14:305–310. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- 50.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuker M. On folding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]