Abstract
Lately several naturally occurring peptides presenting antimicrobial activity have been described in the literature. However, snake venoms, which are an enormous source of peptides, have not been fully explored for searching such molecules. The aim of this work is to review the basis of antimicrobial mechanisms revealing snake venom as a feasible source for searching an antibiotic prototype. Therefore, it includes (i) a description of the constituents of the snake venoms involved in their main biological effects during the envenomation process; (ii) examples of snake venom molecules of commercial use; (iii) mechanisms of action of known antibiotics; and (iv) how the microorganisms can be resistant to antibiotics. This review also shows that snake venoms are not totally unexplored sources for antibiotics and complementary and alternative medicine (CAM).
Keywords: antibacterial, resistance, mechanism, peptide, reptiles
Introduction
Snake venoms contain a large number of biologically active proteins and peptides that are usually similar in structure but not identical to that of prey physiological systems. These molecules are produced by specialized glands, which are evolutionarily related to salivary glands, and are toxic to the prey (1). Interestingly, more than 100 million years ago, snakes evolved from lizards and since then, they independently evolved their own venom apparatus in ophidian evolution, at the base of the Colubroidea radiation1–3.
In an effort to show snake venoms as a promising source for antibiotics, this work briefly discusses the known biological activities of snake venoms, using snake venom molecules from the Viperidae family as examples, and concepts about antibiotics such as their mechanism and resistance. We also highlight the data about the antibacterial activity of some snake venoms described in the literature to date.
Snake Venom and its Constituents
All the known advanced snake species are venomous. Most of these snakes are found in the superfamily Colubroidea that also includes the families Elapidae (incl. Hydrophiidae; Cobras, Kraits, Coral Snakes, Sea Snakes) and Viperidae (Vipers and Pit Vipers)3. Their venoms are a wide mixture of proteins and peptides (90–95%), also including amino acids, nucleotides, free lipids, carbohydrates and metallic elements bound to proteins (5%) (2–5).
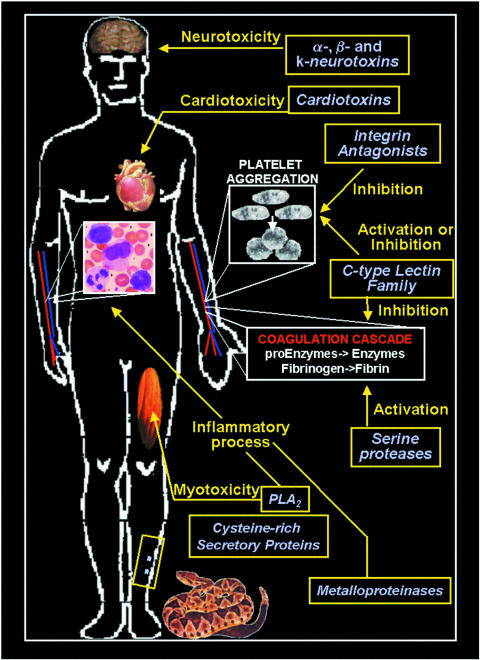
Snake venom protein constituents may present different biological activities that affect physiological processes such as neurotransmission, the complement system and homeostasis (6–8) (Fig. 1). These venoms can act in more than one system at the same time and they may present antigenic effects (8–10). Viperidae family venom molecules are good examples, such as in homeostasis, where they act as pro- and anticoagulant factors, and also as inducers and inhibitors of platelet aggregation (8,10–12).
Figure 1.
The biological effects of some snake venoms during the envenomation process.
Due to their diversity, the proteins from the Viperidae family members are classified (11) in the following categories: serine proteases, metalloproteinases, phospholipases A2 (PLA2), C-type lectins and disintegrins. Only the initial three groups display enzymatic activity (4,11).
Among the Viperidae serine proteases are (i) thrombin-like enzymes that convert fibrinogen into fibrin (13–15), such as batroxobin in Bothrops atrox (16), crotalase and gyroxin in Crotalus durissus terrificus (15,17), and LMTL in Lachesis muta venoms (18); (ii) factor X or factor V activators such as that described in Bothrops jararaca and B.atrox venoms (19,20); (iii) prothrombin activators present in bothropic venoms (21); and (iv) platelet activators such as that found in B.atrox and B.jararaca venoms (22,23) (Fig. 1).
Snake venom metalloproteinases are zinc-dependent enzymes that induce hemorrhaging by directly affecting capillary blood vessels and their interaction with endothelial cells (24). They cleave basement membranes, leading to blood extravasion that occurs through gaps formed in endothelial cells. Therefore, these metalloproteinases present a hemorrhagic effect such as BaH1 and BaP1 from the venom of the snake Bothrops asper (25). This ability also induces myonecrosis and plays a vital role in the significant local inflammatory response of the envenomation (21,24,26) (Fig. 1).
Phospholipases A2 (PLA2s) are enzymes that cleave phopholipids at the A2 position, and >150 have been identified in several snake venoms (27,28). They are described as responsible for some of the envenomation symptoms, which involve not only the hemostatic system, with an anticoagulant and an antiplatelet profile (29–32), but also inflammatory and myotoxic effects (33,34) (Fig. 1). Local inflammation and pain are important features of Viperidae and Elapidae snakebite envenomations that are rich in myotoxic nociceptive events induced by PLA2 (27,34,35). Interestingly, the elapid and viper PLA2 toxins belong to different groups (“pancreatic-type”-group I and “synovial-type”-group II, respectively). They represent independent use of PLA2 as toxins and are thus not homologous to each other as result from separate recruitment events. The snake presynaptic neurotoxins can also present PLA2 activity, which leads to the release of acetylcholine followed by impairment of synaptic functions. These neurotoxins are spread through several families of Colubroidea superfamily (8,36,37). β-bungarotoxin is a basic protein from Bungarus multicintus that partially paralyzed mouse hemi-diaphragm nerve–muscle preparations also due to the PLA2-mediated destruction of membrane phospholipids in motor nerve terminals (38) (Fig. 1).
The C-type lectin family from Viperidae is one of the most fully characterized lectin groups described in the literature (39,40). These calcium-dependent proteins are divided into two groups (I and II), those with a complete (I) or an incomplete (II) carbohydrate recognition domain (CRD) (39,41). The first group is involved in cell recognition such as adhesion, endocytosis and pathogen neutralization usually by using the CRD (42,43). Meanwhile, while conserving most of the primary structure, the incomplete CRD protein group displays different biological activities (40). These molecules are not able to bind carbohydrates but, by using different mechanisms, they induce or inhibit different steps of the same physiological system or even of different systems (39,40,44). These molecules can be found in several venoms such as botrocetin, a platelet-agglutinating protein (45), and bothrojaracin, a thrombin inhibitor, in B.jararaca venom (46,47); and convulxin, a pro-aggregating protein that binds to platelet GPVI receptor, in C.durissus terrificus venom (44,48,49) (Fig. 1).
Snake venoms also contain several peptides. They may vary from presenting neurotoxic (8,50,51) cardiotoxic (52,53) or even an inhibitory platelet profile (3,4,6,26,54,55). They also may present cytotoxic effects characterized by the cytolysins that present a cationic site flanked by a hydrophobic surface (56). In the group of peptides with inhibitory platelet activity, the disintegrins, also known as RGD peptides (molecules containing the Arg–Gly–Asp sequence), are integrin antagonists (Fig. 1). They act as potent inhibitors of platelet aggregation by binding specifically to integrins present on cell membranes of not only platelets (44,55,57) but also metastatic cells (54,58) (Fig. 1).
Finally, it is possible to observe the presence of other protein compounds with an enzymatic profile in snake venoms. Those include cysteine-rich secretory proteins, which inhibit smooth muscle contraction and cyclic nucleotide-gated ion channels (59) (Fig. 1). There are also phosphomonoesterases, phosphodiesterases, arginine esterases, hyaluronidases, L-aminooxidases, 5′ and NAD nucleotidases, and acetylcholinesterases in snake venoms (3,4,60,61). Interestingly, the concentration and distribution of all snake venom proteins and peptides vary from individual to individual, species to species, genus to genus and family to family, probably due to their features, feeding and environmental conditions (12,47,62–64).
Snake Venom Molecules of Commercial use
At the end of the last millennium, the development of therapeutic drugs made a significant improvement to the understanding of the mechanisms of action and structure–function relationship of important biological molecules (40,65,66). The broad spectrum of snake venom activities, including their biochemical, toxicological, physiological and pharmacological profiles, results from the action of their constituents. Therefore, snake venom are of biological interest as a potential source of active compounds. These molecules could act as (or be used as a prototype for) (i) therapeutic agents (67,68); (ii) research tools for use in the diagnosis of several diseases (68–70); and/or (iii) in basic research for understanding physiological and pathological processes (70–73). One of the most successful examples of using snake venom as a source for searching for drug prototypes also involved venom from the Viperidae family. In the 1960s, Ferreira, a PhD student at the time, and co-workers found a peptide presenting an angiotensin-converting enzyme inhibitory activity in B.jararaca venom. This molecule was named nonapeptide bradykinin potentiator (BPP9a) and was able to decrease arterial pressure using this mechanism (75,76). Based on BPP9a, several drugs were developed and protected by patents, now used by the international industry systems. These drugs, symbolized by captopril, represent a world market of billions of dollars annually. BPP9a is a good example where the use of a natural prototype found in a biological source can generate a medicine for worldwide use. In fact, there are many peptides from several natural sources, other than snake venom, described as potential prototypes for drug development. One of them is hirudin, a thrombin inhibitor from Hirudo medicinalis saliva, studied for its potential as an antithrombotic molecule (77–79).
Antimicrobial Peptides Versus Enzymes
Clearly snake venom peptides have the potential for practical and therapeutic use. However, enzymes and proteins are also very important as some of them are described as laboratory diagnosis reagents. Russel viper venom (RVV) X and V enzymes and ecarin from Echis carinatus venom are proteins used for factors X and V, and prothrombin determination in blood, respectively (19,20). Due to their characteristics, RVV enzymes have been used for the improvement of the detection of von Willebrand disease (6,15). Similarly, snake venom thrombin-like enzymes (SVTLEs) are very useful for blood measurements of several parameters of heparin-treated patients since they are not affected by heparin in the same way as thrombin, a key enzyme of the coagulation cascade (80). SVTLEs and snake venom proteases presenting fibrinolytic activity acting on coagulation contributed to the study of the treatment of vascular thrombosis. Included in this group are batroxobin (Defibrinase R), from B.atrox venom, and ancrod (Arvin R), from Calloselasma rhodostoma venom, currently used for controlled depletion of fibrinogen (80–82). They act as selective antithrombotic agents on deep vein thrombosis peripheral arterial diseases and on vascular surgery (69,80,82).
In the last decade, several snake venom compounds were used as important tools for the understanding of human physiological systems (83,84). Due to their similarity to physiological molecules, studies on myoblast fusion and fertilization, and matrix metalloproteinase (ADAMs)–cell interactions have been performed using the homologous snake venom metalloproteinases and peptide neurotoxins in order to characterize human cancers and small lung carcinoma. These studies are good examples of the use of snake venom molecules in basic research (25,26,70,83,84).
Antibiotics? What are they?
Antibiotics are a heterogeneous group of molecules produced by several organisms, including bacteria and fungi, presenting an antibacterial profile (85,86). At the present time, synthetic antimicrobials, known as chemotherapics, display different mechanisms of action and a broad antibacterial spectrum. The antimicrobials are produced by the international pharmaceutical industry and used worldwide. In fact, the control of the deleterious effects of microorganisms was significantly increased by the introduction of the sulfonamides (chemotherapics) and of penicillin (antibiotic) in 1936 and 1941, respectively (85–87). These drugs were crucial for the reduction of the incidence of several bacterial infections such as meningitis, endocarditis, pneumonia and gonorrhoea (85,86).
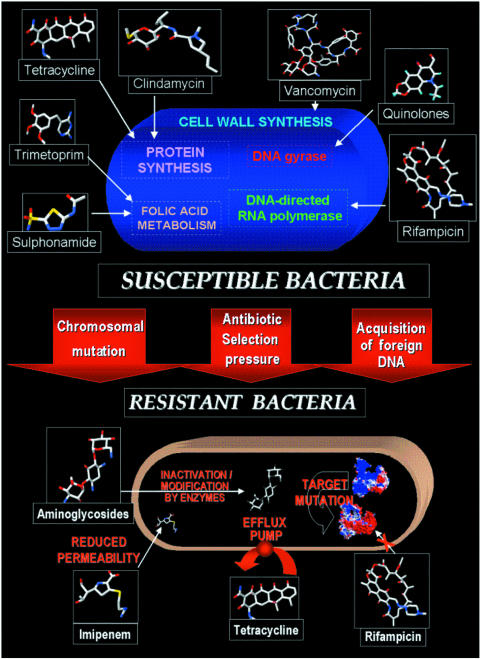
The main effects of antibiotics are: (i) inducing the death of the agent (bactericidal effect); and/or (ii) inhibition of bacterial growth (bacteriostatic effect). Their targets are the essential biosynthetic process or routes of these microorganisms (85,87). Among them, the inhibition of the synthesis of cell membrane, nucleotides and peptide bonds interferes directly with survival, chromosome replication and protein synthesis, respectively, of the bacteria (Fig. 2). They can also act by increasing cell permeability, or inhibiting through binding to ribosomes, which prevents nucleotide polymerization (85,87).
Figure 2.
Schematic representation of the emergence of resistant bacteria.
The main characteristic of antimicrobials (synthetic or natural) is their selective toxicity. This feature is based on the presence of the target only or mainly on the infectious agents, which allows their systemic administration without deleterious effects to the host cells (85,87).
Interestingly, antibiotics are usually produced by water- or soil-dwelling bacteria, where the absence or low concentrations of supplies turns the competition into an important issue for survival (88). The production of an antibiotic at the bacterial stationary phase probably reproduces the bacteria's behavior at a low nutrition environment, where these molecules are necessary for eliminating competitors and guarantee ‘food supplies’ (89). On the other hand, microorganisms that grow without food restriction, such as those of the intestinal flora (enterobacterias) or of an animal's oral cavity, generally produce bacteriocins, which are proteins presenting an antibacterial profile (90,91). These proteins are very different from antibiotics, clearly obvious by their chemical structure or non-metabolic characteristics, but mostly because they are produced during the exponential phase of Gram-positive and Gram-negative, pathogenic or non-pathogenic bacteria (e.g. Escherichia, Acetobacter, Actinobacillus, Bacillus, Clostridium, Lactobacillus, Streptococcus and Staphylococcus) (90,91). Colicin is a bacteriocin produced by E.coli against other homologous species. Similar to other bacteriocins, colicin's main effect is bactericidal (92). The inhibitory mechanism of these proteins is not fully characterized, but for colicin three steps are already confirmed: (i) binding of colicin to the receptor; (ii) its transport through the cell membrane; finally (iii) death of the agent (90–92).
The use of bacteriocins as therapeutic tools is very restricted since they can be destroyed due to their protein structure, and/or induce an immune response by the patient treated because of their antigenic profile (90,91). These proteins are mainly useful when present in food produced by using microorganisms such as yoghurt. In this specific case, these microorganisms, such as Lactobacillus, produce bacteriocins, which restrict the cell growth of other potential contaminants (93).
Still Searching for Antibiotics? What for?
Although extremely effective, antibiotics are able to induce resistance in bacteria. For >50 years, bacterial resistance has been the main factor responsible for the increase of morbidity, mortality and health care costs of bacterial infections (94). This bacterial defense mechanism is widely present in bacteria (e.g. Pseudomonas, Klebsiella, Enterobacter, Acinetobacter, Salmonella, Staphylococcus, Enterococcus and Streptococcus) and became a world health problem worsened by developments in human, animal and plant transportation (94–96). The airlines facilitated the rapid dissemination of resistant microorganisms through different countries and, as a consequence, the monitoring of those agents by the government became more and more difficult (94–96).
According to genetic studies, resistant bacteria are always present in a small number in any bacterial colony. This number of bacteria can increase by selective pressure induced by the presence of the drug used (94–96). The most common causes for the appearance of multiresistant bacteria are the inadequate or excessive use of antibiotics. In some countries, prescriptions are still determined empirically without previous identification of the pathogenic agents (94–96). In addition, inappropriate use such as (i) not following the intake schedule; (ii) giving up the treatment; (iii) the low quality of the medicines; (iv) self-medication; and (v) incorrect drug storage can lead to a selective pressure, which contributes to the selection of these multiresistant microorganisms. Other problems also include the amount of antimicrobials used in agribusiness and agriculture for protecting animal and plant growth. Environmental changes, and the increase of agro products and animal migration, contribute to the spreading of multiresistant agents (94–96).
The microorganisms can be resistant to antibiotics through an intrinsic resistance, which is determined by the original cell genes and is displayed by all individuals of the species (97). A good example is Lactobacillus that, similarly to mammalian cells, synthesizes tetrahydrofolate from p-aminobenzoic acid. Therefore, this microorganism is resistant to sulfas such as the mammal's cells (98).
The microorganisms can also become resistant through acquired resistance, which is represented by specific mutations on antibiotic targets acquired by plasmidial or transposon genes (94–97,99) (Fig. 2). This event leads to a new generation of insensitive cells. There are four known acquired resistance mechanisms.
Production of Enzymes or Isoenzymes
Enzymes and isoenzymes are usually produced by microorganisms as instruments for protection against antibiotics. A classical model is observed for streptomycin-resistant bacteria, which synthesize enzymes that phosphorylate, adenylate or acetylate hydroxyl or amino groups of amino glycosides, acquiring this ability through plasmids (94–96). The use of β-lactamases by resistant Gram-positive or Gram-negative bacteria in order to cleave the β-lactamic ring of penicillin. This cleavage leads to the formation of the penicillinoic acid that is devoid of antimicrobial activity. Using β-lactamases, these microorganisms are resistant not only penicillin to but also to, cephalosporin (100,101) (Fig. 2).
Because of this, the literature describes effort to synthesize new penicillin-like drugs by adding different chemical groups to the original penicillin, to modify the affinity between these drugs and this enzyme. However, these modifications also decrease absorption of these molecules and also induce a compensation system where the microorganisms increase the synthesis of the enzyme (1–2% of total proteins), which guarantees the resistance level (101).
Chloramphenicol acetyltransferase is another important enzyme directly involved in acquired resistance. This protein is able to inactivate chloramphenicol through the addition of an acetyl group from acetyl coenzyme-A, and its presence can be intrinsic or induced (102).
The synthesis of isoenzymes is also an induced resistant system. In sulfonamide resistance, the isoenzyme dihydropteroate synthase, acquired by plasmid genes, presents the same affinity for the substrate (p-aminobenzoate) but a 10 000 times lower Kd for the drug compared with the original enzyme (103).
Target Mutation
This acquired resistance is characterized by a specific mutation on the antibiotic target, which will result in drug-insensitive bacteria. In the case of streptomycin-resistant bacteria, mutation of the S12 protein prevents binding of the bacterial ribosome to this drug (104). Similarly, rifampicin-resistant bacteria present a mutation on the DNA polymerase β-subunit, which is sufficient to make this microorganism insensitive to rifampicin treatment (105–107) (Fig. 2).
Changes in Membrane Permeability
This acquired resistance can result from: (i) changes to the antibiotic structure, which make its passage difficult through bacterial permease, or to the cell membrane constitution, such as changing lipopolysaccharides; (ii) permease mutations, which decrease amino acid and antibiotic transport; and (iii) the efflux process that pumps the drug out of the cell (106–109) (Fig. 2).
Increase of Metabolic Molecules
Resistance can be acquired through an increase of a metabolic molecule when the drug's mechanism is in direct competition with this molecule (competitive antagonism), e.g. by increasing p-aminobenzoate production, sulfur-resistant bacteria are able to avoid antibiotic effects (110,111).
Are Snake Venoms Totally Unexplored Sources for Antibiotics? Not Really
More than 700 antimicrobial peptides have already been identified in all living species (112,113,114), including bacteria (86), fungi (115), amphibians (116), fish (117), insects (118) and mammals (119,120). These molecules are 5 kDa peptides with a high level of basic and hydrophobic amino acids. They present a broad antimicrobial spectrum against bacteria, fungi or parasites, by acting through insertion into the cell membrane or binding to receptors. These molecules are promising for development of antibiotics, especially for treatment of multiresistant microorganisms (112,113).
In the case of snake venoms, despite heavy snake oral and fang contamination with a wide variety of potentially pathogenic bacteria, envenomation is a process associated with a low incidence of bacterial infection (120,121). Therefore, this feature could indicate the presence of antibacterial molecules in the snake venoms that would protect the snakes during feeding. Some of the first reports about antibacterial activity in snake venoms were in 1948, and in 1968, involving Elapidae and Viperidae venoms (122,123). Viperidae were described as having antimicrobials against the Sarcina species, while in the Elapidae family, a lytic factor or cytotoxin composed of a basic, low molecular weight protein was found in Naja sp. and H.haemachatus. They were able to disrupt Staphylococcus aureus and E.coli phospholipid membranes respectively (122,123). Not only peptides but also enzymes were involved in the antimicrobial activity of snake venoms as described by Skarnes in 1970 (124). Crotalus adamanteus L-aminooxidase affects Gram-positive bacteria, while those from Agkistrodon halys pallas, Bothrops alternatus and Trimerusurus jerdoni have an inhibitory activity against E.coli, and S.aureus, Pseudomonas aeruginosa and Bacillus megaterium, respectively (124–127). Interestingly, LAO1, an L-aminooxidase from Pseudechis autralis, was 70 times more effective than tetracycline against Aeromonas (128).
Several antimicrobial studies involving many snake venoms have already been described in the literature. For example, Stocker and Traynor in 1986 wrote about the inhibitory effects of Naja naja soutratrix, Vipera russelli and C.adamanteus in E.coli (129); in 1991 Stiles described the antibacterial properties of 30 different snake venom where the Asian and African snakes (Naja sp.), Australian elapids (Notechis scutatus scutatus and Pseudechis australis) and North American snakes (Crotalus sp.) presented the highest activity and Talan and co-workers using Crotalid venoms against Gram-negative and Gram-positive bacteria (128). Recently, Blaylock studied Kwazulu Natal snake venoms in South Africa and showed that the eight venoms tested presented antibacterial activities. Adders showed most activity against aerobes, while cobras showed no distinct activity against aerobes or anaerobes (130). In this study, snake venoms from Causus rhombeatus, Bitis gabonica, Bitis arietans, Dendroaspis polylepsis, Dendroaspis augusticeps, Naja melanoleuca, Naja annulifera and Naja mossambica were detected presenting antibacterial activity against S.aureus, E.coli, P.aeruginosa, Bacterioides fragilis, Bacterioides intermedius, Clostridium sordellii and Clostridium perfringes (130). More recently, Xie and co-workers described peptides from Naja atra venom that act against multiresistant Mycobacterium tuberculosis in vitro (131).
Is There a Chance?
Currently certain bacterial infections are multidrug resistant. However, this worldwide problem may decrease if some attitudes can be adopted in a global perspective. Among them, the most important is still a reduction of the inappropriate and/or excessive use of antibiotics (132,133).
Despite reaching future positive statistics on antibiotic use, new antimicrobials will always be necessary to fight against multidrug-resistant microorganisms (132,133). Therefore, these drugs will be very important, particularly for treatment of the elderly, children and immune compromised patients (134–136). Thus, investment in antibiotic research and in finding new sources of new drugs or prototypes is of major interest to CAM.
This minireview does not intend to cover all data about snake venoms or antibiotics. Its main objective is to reinforce that both proteins and peptides from snake venoms can be good candidates for testing in antibiotic screening assays using multiresistant microorganisms. Compared with other snake venom biological activities, the antibacterial profile of these natural sources has not been fully investigated despite the positive results found to date. Although snake venom peptides and proteins have a direct therapeutic use limited by their antigenic and ‘digestible’ structure, their usefulness as prototypes has clear potential. These molecules could also be of interest for the food industry, since they can be easily degraded by the human digestive system and therefore could be useful to protect against contamination by food microorganism.
Acknowledgments
This study was supported by CNPq, FAPERJ, FUJB, CAPES and UFF (Brazil).
References
- 1.Kochva E. The origin of snakes and evolution of the venom apparatus. Toxicon. 1987;25:65–106. doi: 10.1016/0041-0101(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 2.Heise PJ, Maxson LR, Dowling HG, Hedges SB. Higher-level snake phylogeny inferred from mitochondrial DNA sequences of 12S rRNA genes. Mol Biol Evol. 1995;12:259–65. doi: 10.1093/oxfordjournals.molbev.a040202. [DOI] [PubMed] [Google Scholar]
- 3.Fry BG, Wuster W. Assembling an arsenal: origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol Biol Evol. 2004 May;21(5):870–83. doi: 10.1093/molbev/msh091. [DOI] [PubMed] [Google Scholar]
- 4.Russell FE. Venoms. In: Lippincott JB, Persol GM, editors. Snake Venom Poisoning. Philadelphia: Lippincott; 1980. pp. 139–234. [Google Scholar]
- 5.Tu AT. Venoms: Chemistry and Molecular Biology. New York: John Willey and Sons; 1988. Snake venoms: general background and composition; pp. 1–19. [Google Scholar]
- 6.Gold BS, Dart RC, Barish RA. Bites of venomous snakes. N Engl J Med. 2002;47:347–56. doi: 10.1056/NEJMra013477. [DOI] [PubMed] [Google Scholar]
- 7.Stocker K. Composition of snake venom. In: Stocker KF, editor. Medical Use of Snake Venom Proteins. Boca Raton: CRC Press; 1990. pp. 33–56. [Google Scholar]
- 8.Mion G, Olive F, Hernandez E, Martin YN, Vieillefosse AS, Goyffon M. Action of venoms on blood coagulation: diagnosis of hemorrhagic syndromes. Bull Soc Pathol Exot. 2002;95:132–8. [PubMed] [Google Scholar]
- 9.Lewis RL, Gutmann L. Snake venoms and the neuromuscular junction. Semin Neurol. 2004;24:175–9. doi: 10.1055/s-2004-830904. [DOI] [PubMed] [Google Scholar]
- 10.Gornitskaia OV, Platonova TN, Volkov GL. Enzymes of snake venoms. Ukr Biokhim Zh. 2003;75:22–32. [PubMed] [Google Scholar]
- 11.Markland FS., Jr Snake venoms. Drugs Suppl. 1997;54:31–40. doi: 10.2165/00003495-199700543-00003. [DOI] [PubMed] [Google Scholar]
- 12.Braud S, Bon C, Wisner A. Snake venom proteins acting on haemostasis. Biochimie. 2000;82:9–10. doi: 10.1016/s0300-9084(00)01178-0. [DOI] [PubMed] [Google Scholar]
- 13.Markland FS. Snake venoms and the haemostatic system. Toxicon. 1998;36:1749–800. doi: 10.1016/s0041-0101(98)00126-3. [DOI] [PubMed] [Google Scholar]
- 14.Eagle H. The coagulation of blood by snake venoms and physiologic significance. J Exp Med. 1937;65:613–75. doi: 10.1084/jem.65.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nahas L, Kamiguti AS, Barros MA. Thrombin-like and factor X-activator components of Bothrops snake venoms. Thromb Haemostasis. 1979;41:314–28. [PubMed] [Google Scholar]
- 16.Castro HC, Zingali RB, Albuquerque MG, Pujol-Luz M, Rodrigues CR. Snake venom thrombin-like enzymes: from reptilase to now. Cell Mol Life Sci. 2004;61:843–56. doi: 10.1007/s00018-003-3325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stocker K, Barlow GH. The coagulant enzyme from Bothrops atrox venom (batroxobin) Methods Enzymol. 1976;45:214–23. doi: 10.1016/s0076-6879(76)45021-8. [DOI] [PubMed] [Google Scholar]
- 18.Raw I, Rocha MC, Esteves MI, Kamiguti AS. Isolation and characterization of a thrombin-like enzyme from the venom of Crotalus durissus terrificus. Braz J Med Biol Res. 1986;19:333–8. [PubMed] [Google Scholar]
- 19.Magalhães A, de Oliveira GJ, Diniz CR. Purification and partial characterization of a thrombin-like enzyme from the venom of the bushmaster snake, Lachesis muta noctivaga. Toxicon. 1981;19:279–94. doi: 10.1016/0041-0101(81)90032-5. [DOI] [PubMed] [Google Scholar]
- 20.Rosing J, Govers-Riemslag JW, Yukelson L, Tans G. Factor V activation and inactivation by venom proteases. Haemostasis. 2001;31:241–6. doi: 10.1159/000048069. [DOI] [PubMed] [Google Scholar]
- 21.Tans G, Rosing J. Snake venom activators of factor X: an overview. Haemostasis. 2001;31:225–33. doi: 10.1159/000048067. [DOI] [PubMed] [Google Scholar]
- 22.Kini RM, Rao VS, Joseph JS. Procoagulant proteins from snake venoms. Haemostasis. 2001;31:218–24. doi: 10.1159/000048066. [DOI] [PubMed] [Google Scholar]
- 23.Niewiarowski S. Proteins secreted by the platelet. Thromb Haemostasis. 1977;38:924–38. [PubMed] [Google Scholar]
- 24.Serrano SM, Mentele R, Sampaio CA, Fink E. Purification, characterization, and amino acid sequence of a serine proteinase, PA-BJ, with platelet-aggregating activity from the venom of Bothrops jararaca. Biochemistry. 1995;34:7186–93. doi: 10.1021/bi00021a033. [DOI] [PubMed] [Google Scholar]
- 25.Kamiguti AS, Zuzel M, Theakston RD. Snake venom metalloproteinases and disintegrins: interactions with cells. Braz J Med Biol Res. 1998;31:853–62. doi: 10.1590/s0100-879x1998000700001. [DOI] [PubMed] [Google Scholar]
- 26.Rucavado A, Borkow G, Ovadia M, Gutierrez JM. Immunological studies on BaH1 and BaP1, two hemorrhagic metalloproteinases from the venom of the snake Bothrops asper. Toxicon. 1995;33:1103–6. doi: 10.1016/0041-0101(95)00039-o. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez JM, Rucavado A. Snake venom metalloproteinases: their role in the pathogenesis of local tissue damage. Biochimie. 2000;82:841–50. doi: 10.1016/s0300-9084(00)01163-9. [DOI] [PubMed] [Google Scholar]
- 28.Valentin E, Lambeau G. What can venom phospholipases A2 tell us about the functional diversity of mammalian secreted phospholipases A2? Biochimie. 2000;82:815–31. doi: 10.1016/s0300-9084(00)01168-8. [DOI] [PubMed] [Google Scholar]
- 29.Lizano S, Domont G, Perales J. Natural phospholipase A2 myotoxin inhibitor proteins from snakes, mammals and plants. Toxicon. 2003;42:963–77. doi: 10.1016/j.toxicon.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Lu QM, Jin Y, Wei JF, Wang WY, Xiong YL. Biochemical and biological properties of Trimeresurus jerdonii venom and characterization of a platelet aggregation-inhibiting acidic phospholipase A2. J Natural Toxins. 2002;11:25–33. [PubMed] [Google Scholar]
- 31.Roberto PG, Kashima S, Marcussi S, et al. Cloning and identification of a complete cDNA coding for a bactericidal and antitumoral acidic phospholipase A2 from Bothrops jararacussu venom. Protein J. 2004;23:273–85. doi: 10.1023/b:jopc.0000027852.92208.60. [DOI] [PubMed] [Google Scholar]
- 32.Fuly AL, Machado OL, Alves EW, Carlini CR. Mechanism of inhibitory action on platelet activation of a phospholipase A2 isolated from Lachesis muta (Bushmaster) snake venom. Thromb Haemostasis. 1997;78:1372–80. [PubMed] [Google Scholar]
- 33.Serrano SM, Reichl AP, Mentele R, et al. A novel phospholipase A2, BJ-PLA2, from the venom of the snake Bothrops jararaca: purification, primary structure analysis, and its characterization as a platelet-aggregation-inhibiting factor. Arch Biochem Biophys. 1999;367:26–32. doi: 10.1006/abbi.1999.1230. [DOI] [PubMed] [Google Scholar]
- 34.Teixeira CF, Landucci EC, Antunes E, Chacur M, Cury Y. Inflammatory effects of snake venom myotoxic phospholipases A2. Toxicon. 2003;42:947–62. doi: 10.1016/j.toxicon.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Fuly AL, Elias SC, Zingali RB, Guimarães JA, Melo PA. Myotoxic activity of an acid phospholipase A2 isolated from Lachesis muta (Bushmaster) snake venom. Toxicon. 2000;38:961–72. doi: 10.1016/s0041-0101(99)00208-1. [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez JM, Ownby CL. Skeletal muscle degeneration induced by venom phospholipases A2: insights into the mechanisms of local and systemic myotoxicity. Toxicon. 2003;42:915–31. doi: 10.1016/j.toxicon.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Hodgson WC, Wickramaratna JC. In vitro neuromuscular activity of snake venoms. Clin Exp Pharmacol Physiol. 2002;29:807–14. doi: 10.1046/j.1440-1681.2002.03740.x. [DOI] [PubMed] [Google Scholar]
- 38.Rossetto O, Rigoni M, Montecucco C. Different mechanism of blockade of neuroexocytosis by presynaptic neurotoxins. Toxicol Lett. 2004;149:91–101. doi: 10.1016/j.toxlet.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 39.Rowan EG. What does beta-bungarotoxin do at the neuromuscular junction? Toxicon. 2001;39:107–18. doi: 10.1016/s0041-0101(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 40.Clemetson KJ, Navdaev A, Dormann D, Du XY, Clemetson JM. Multifunctional snake C-type lectins affecting platelets. Haemostasis. 2001;31:148–54. doi: 10.1159/000048058. [DOI] [PubMed] [Google Scholar]
- 41.Morita T. Use of snake venom inhibitors in studies of the function and tertiary structure of coagulation factors. Int J Hematol. 2004;79:123–9. doi: 10.1532/ijh97.03172. [DOI] [PubMed] [Google Scholar]
- 42.Ozeki Y, Matsui T, Hamako J, et al. C-type galactoside-binding lectin from Bothrops jararaca venom: comparison of its structure and function with those of botrocetin. Arch Biochem Biophys. 1994;308:306–10. doi: 10.1006/abbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 43.Drickamer K, Taylor ME. Biology of animal lectins. Annu Rev Cell Biol. 1993;9:237–64. doi: 10.1146/annurev.cb.09.110193.001321. [DOI] [PubMed] [Google Scholar]
- 44.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 45.Andrews RK, Gardiner EE, Shen Y, Berndt MC. Structure–activity relationships of snake toxins targeting platelet receptors, glycoprotein Ib-IX-V and glycoprotein VI. Curr Med Chem Cardiovasc Hematol Agents. 2003;1:143–9. doi: 10.2174/1568016033477559. [DOI] [PubMed] [Google Scholar]
- 46.Brinkhous KM, Read MS, Fricke WA, Wagner RH. Botrocetin (venom coagglutinin) reaction with a broad spectrum of multimeric forms of factor VIII macromolecular complex. Proc Natl Acad Sci USA. 1983;80:1463–6. doi: 10.1073/pnas.80.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zingali RB, Jandrot-Perrus M, Guillin M, Bon C. Bothrojaracin, a new thrombin inhibitor isolated from Bothrops jararaca venom: characterization and mechanism of thrombin inhibition. Biochemistry. 1993;32:10794–802. doi: 10.1021/bi00091a034. [DOI] [PubMed] [Google Scholar]
- 48.Castro HC, Fernandes M, Zingali RB. Identification of bothrojaracin-like proteins in snakes venoms from Bothrops species and Lachesis muta. Toxicon. 1999;37:1403–16. doi: 10.1016/s0041-0101(99)00087-2. [DOI] [PubMed] [Google Scholar]
- 49.Prado-Franceschi J, Brazil OV. Convulxin, a new toxin from the venom of the South American rattlesnake Crotalus durissus terrificus. Toxicon. 1981;19:875–87. doi: 10.1016/0041-0101(81)90085-4. [DOI] [PubMed] [Google Scholar]
- 50.Francischetti IM, Saliou B, Leduc M, et al. Convulxin, a potent platelet-aggregating protein from Crotalus durissus terrificus venom, specifically binds to platelets. Toxicon. 1997;35:1217–28. doi: 10.1016/s0041-0101(97)00021-4. [DOI] [PubMed] [Google Scholar]
- 51.Harvey AL. Twenty years of dendrotoxins. Toxicon. 2001;39:15–26. doi: 10.1016/s0041-0101(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 52.Tsetlin VI, Hucho F. Snake and snail toxins acting on nicotinic acetylcholine receptors: fundamental aspects and medical applications. FEBS Lett. 2004;557:9–13. doi: 10.1016/s0014-5793(03)01454-6. [DOI] [PubMed] [Google Scholar]
- 53.Satora L, Morawska J, Targosz D. Cardiotoxicity of vertebrates venoms. Przegl Lek. 2003;60:199–201. [PubMed] [Google Scholar]
- 54.Ducancel F. The sarafotoxins. Toxicon. 2002;40:1541–5. doi: 10.1016/s0041-0101(02)00159-9. [DOI] [PubMed] [Google Scholar]
- 55.Niewiarowski S, Chambers AF, Groom AC. Effects of the disintegrin eristostatin on individual steps of hematogenous metastasis. Exp Cell Res. 1995;219:571–8. doi: 10.1006/excr.1995.1266. [DOI] [PubMed] [Google Scholar]
- 56.Lu X, Lu D, Scully MF, Kakkar VV. Modulation of integrin-binding selectivity by mutation within the RGD-loop of snake venom proteins: a novel drug development approach. Curr Med Chem Cardiovasc Hematol Agents. 2003;1:189–96. doi: 10.2174/1568016033477522. [DOI] [PubMed] [Google Scholar]
- 57.Kini RM, Evans HJ. A common cytolytic region in myotoxins, hemolysins, cardiotoxins and antibacterial peptides. Int J Peptide Protein Res. 1989;34:277–86. doi: 10.1111/j.1399-3011.1989.tb01575.x. [DOI] [PubMed] [Google Scholar]
- 58.McLane MA, Marcinkiewicz C, Vijay-Kumar S, Wierzbicka-Patynowski I, Niewiarowski S. Viper venom disintegrins and related molecules. Proc Soc Exp Biol Med. 1998;219:109–19. doi: 10.3181/00379727-219-44322. [DOI] [PubMed] [Google Scholar]
- 59.Morris VL, Schmidt EE, Koop S, et al. A study of the coagulant action of eight snake venoms. Thromb Diath Haemorrh. 1964;12:355–67. [PubMed] [Google Scholar]
- 60.Yamazaki Y, Morita T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon. 2004;44:227–31. doi: 10.1016/j.toxicon.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 61.Matsui T, Fujimura Y, Titani K. Snake venom proteases affecting hemostasis and thrombosis. Biochim Biophys Acta. 2000;1477:146–56. doi: 10.1016/s0167-4838(99)00268-x. [DOI] [PubMed] [Google Scholar]
- 62.Du XY, Clemetson KJ. Snake venom L-amino acid oxidases. Toxicon. 2002;40:659–665. doi: 10.1016/s0041-0101(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 63.Monteiro RQ, Carlini CR, Guimarães JA, Bon C, Zingali RB. Distinct bothrojaracin isoforms produced by individual jararaca (Bothrops jararaca) snakes. Toxicon. 1996;35:649–57. doi: 10.1016/s0041-0101(96)00176-6. [DOI] [PubMed] [Google Scholar]
- 64.Tsai IH, Wang YM, Chen YH, Tu AT. Geographic variations, cloning, and functional analyses of the venom acidic phospholipases A2 of Crotalus viridis viridis. Arch Biochem Biophys. 2003;411:289–96. doi: 10.1016/s0003-9861(02)00747-6. [DOI] [PubMed] [Google Scholar]
- 65.Monteiro RQ, Yamanouye N, Carlini CR, Guimarães JA, Bon C, Zingali RB. Variability of bothrojaracin isoforms and other venom principles in individual jararaca (Bothrops jararaca) snakes maintained under seasonally invariant conditions. Toxicon. 1998;36:153–63. doi: 10.1016/s0041-0101(97)00061-5. [DOI] [PubMed] [Google Scholar]
- 66.Shebuski RJ, Stabilito IJ, Sitko GR, Polokoff MH. Acceleration of recombinant tissue-type plasminogen activator-induced thrombolysis and prevention of reocclusion by the combination of heparin and the Arg–Gly–Asp-containing peptide bitistatin in a canine model of coronary thrombosis. Circulation. 1990;82:169–77. doi: 10.1161/01.cir.82.1.169. [DOI] [PubMed] [Google Scholar]
- 67.Volkers N. Venom anyone? Researchers tweak ‘bad’ compounds into good medicines. J Natl Cancer Inst. 1998;91:667–8. doi: 10.1093/jnci/91.8.667. [DOI] [PubMed] [Google Scholar]
- 68.Pal SK, Gomes A, Dasgupta SC, Gomes A. Snake venom as therapeutics agents: from toxin to drug development. Indian J Exp Biol. 2002;40:1353–8. [PubMed] [Google Scholar]
- 69.Bailey P, Wilce J. Venom as a source of useful biologically active molecules. Emerg Med (Fremantle) 2001;13:28–36. doi: 10.1046/j.1442-2026.2001.00174.x. [DOI] [PubMed] [Google Scholar]
- 70.Marsh NA. Diagnostic uses of snake venom. Haemostasis. 2001;31:211–7. doi: 10.1159/000048065. [DOI] [PubMed] [Google Scholar]
- 71.Andrews RK, Kamiguti AS, Berlanga O, Leduc M, Theakston RD, Watson SP. The use of snake venom toxins as tools to study platelet receptors for collagen and von Willebrand factor. Haemostasis. 2001;31:155–72. doi: 10.1159/000048059. [DOI] [PubMed] [Google Scholar]
- 72.Sher E, Giovannini F, Boot J, Lang B. Peptide neurotoxins, small-cell lung carcinoma and neurological paraneoplastic syndromes. Biochimie. 2000;82:927–36. doi: 10.1016/s0300-9084(00)01165-2. [DOI] [PubMed] [Google Scholar]
- 73.Wisner A, Braud S, Bon C. Snake venom proteinases as tools in hemostasis studies: structure–function relationship of a plasminogen activator purified from Trimeresurus stejnegeri venom. Haemostasis. 2001;31:133–40. doi: 10.1159/000048056. [DOI] [PubMed] [Google Scholar]
- 74.Rajendra W, Armugam A, Jeyaseelan K. Toxins in anti-nociception and anti-inflammation. Toxicon. 2004;44:1–17. doi: 10.1016/j.toxicon.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 75.Ferreira S. A bradykinin-potentiating factor (bpf) present in the venom of Bothrops jararaca. Br J Pharmacol. 1965;24:163–9. doi: 10.1111/j.1476-5381.1965.tb02091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferreira SH, Bartelt DC, Greene LJ. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry. 1970;9:2583–93. doi: 10.1021/bi00815a005. [DOI] [PubMed] [Google Scholar]
- 77.Cushman DW, Ondetti MA. History of the design of captopril and related inhibitors of angiotensin converting enzyme. Hypertension. 1991;17:589–92. doi: 10.1161/01.hyp.17.4.589. [DOI] [PubMed] [Google Scholar]
- 78.Markwardt F, Sturzebecher J, Glusa E. Antithrombin effects of native and recombinant hirudins. Biomed Biochim Acta. 1990;49:399–404. [PubMed] [Google Scholar]
- 79.Fareed J, Callas D, Hoppensteadt DA, Lewis BE, Bick RL, Walenga JM. Antithrombin agents as anticoagulants and antithrombotics: implications in drug development. Semin Hematol. 1999;36:42–56. [PubMed] [Google Scholar]
- 80.Markwardt F. Hirudin as alternative anticoagulant—a historical review. Semin Thromb Hemost. 2002;28:405–14. doi: 10.1055/s-2002-35292. [DOI] [PubMed] [Google Scholar]
- 81.Marsh NA, Fyffe TL. Practical applications of snake venom toxins in haemostasis. Boll Soc Ital Biol Sper. 1996;72:263–78. [PubMed] [Google Scholar]
- 82.Bell WR., Jr Defibrinogenating enzymes. Drugs. 1997;3:18–30. doi: 10.2165/00003495-199700543-00005. [DOI] [PubMed] [Google Scholar]
- 83.Meier J, Stocker K. Effects of snake venoms on hemostasis. Crit Rev Toxicol. 1991;21:171–82. doi: 10.3109/10408449109089878. [DOI] [PubMed] [Google Scholar]
- 84.Vogel CW, Fritzinger DC, Hew BE, Thorne M, Bammert H. Recombinant cobra venom factor. Mol Immunol. 2004;41:191–9. doi: 10.1016/j.molimm.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 85.Sagane K, Ohya Y, Hasegawa Y, Tanaka I. Metalloproteinase-like, disintegrin-like, cysteine-rich proteins MDC2 and MDC3: novel human cellular disintegrins highly expressed in the brain. Biochem J. 1998;334:93–8. doi: 10.1042/bj3340093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chopra I, Hesse L, O’Neill AJ. Exploiting current understanding of antibiotic action for discovery of new drugs. Symp Ser Soc Appl Microbiol. 2002;31:4S–15S. [PubMed] [Google Scholar]
- 87.Brown K. The history of penicillin from discovery to the drive to production. Pharm Hist (Lond) 2004;34:37–43. [PubMed] [Google Scholar]
- 88.Harris CR, Thorarensen A. Advances in the discovery of novel antibacterial agents during the year 2002. Curr Med Chem. 2004;11:2213–43. doi: 10.2174/0929867043364658. [DOI] [PubMed] [Google Scholar]
- 89.Bukharin OV, Usviatsov BIa, Khusnutdinova LM. Bacterial interactions. Zh Mikrobiol Epidemiol Immunobiol. 2003;4:3–8. (in Russian) [PubMed] [Google Scholar]
- 90.Wimpenny JW, Leistner L, Thomas LV, Mitchell AJ, Katsaras K, Peetz P. Submerged bacterial colonies within food and model systems: their growth, distribution and interactions. Int J Food Microbiol. 1995;28:299–315. doi: 10.1016/0168-1605(95)00065-8. [DOI] [PubMed] [Google Scholar]
- 91.Riley MA, Wertz JE. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 2002;84:357–64. doi: 10.1016/s0300-9084(02)01421-9. [DOI] [PubMed] [Google Scholar]
- 92.Cleveland J, Montville TJ, Nes IF, Chikindas ML. Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol. 2001;71:1–20. doi: 10.1016/s0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 93.Zakharov SD, Cramer WA. On the mechanism and pathway of colicin import across the E.coli outer membrane. Front Biosci. 2004;9:1311–7. doi: 10.2741/1334. [DOI] [PubMed] [Google Scholar]
- 94.Paraje MG, Albesa I, Eraso AJ. Conservation in probiotic preparations of Lactobacillus with inhibitory capacity on other species. N Microbiol. 2000;23:423–431. [PubMed] [Google Scholar]
- 95.Ang JY, Ezike E, Asmar BI. Antibacterial resistance. Symp Ser Soc Appl Microbiol. 2004;3:229–39. doi: 10.1007/BF02724275. [DOI] [PubMed] [Google Scholar]
- 96.Barbosa TM, Levy SB. The impact of antibiotic use on resistance development and persistence. Drug Resist Update. 2000;3:303–11. doi: 10.1054/drup.2000.0167. [DOI] [PubMed] [Google Scholar]
- 97.Jacobs MR, Anon J, Appelbaum PC. Mechanisms of resistance among respiratory tract pathogens. Clin Lab Med. 2004;24:419–453. doi: 10.1016/j.cll.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 98.McDermott PF, Walker RD, White DG. Antimicrobials: modes of action and mechanisms of resistance. Int J Toxicol. 2003;22:135–43. doi: 10.1080/10915810305089. [DOI] [PubMed] [Google Scholar]
- 99.Sybesma W, Starrenburg M, Tijsseling L, Hoefnagel MH, Hugenholtz J. Effects of cultivation conditions on folate production by lactic acid bacteria. Appl Environ Microbiol. 2003;69:4542–8. doi: 10.1128/AEM.69.8.4542-4548.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004;10:272–88. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 101.Majiduddin FK, Materon IC, Palzkill TG. Molecular analysis of beta-lactamase structure and function. Int J Med Microbiol. 2002;292:127–37. doi: 10.1078/1438-4221-00198. [DOI] [PubMed] [Google Scholar]
- 102.Buynak JD. The discovery and development of modified penicillin- and cephalosporin-derived beta-lactamase inhibitors. Curr Med Chem. 2004;11:1951–64. doi: 10.2174/0929867043364847. [DOI] [PubMed] [Google Scholar]
- 103.Potrykus J, Baranska S, Wegrzyn G. Inactivation of the acrA gene is partially responsible for chloramphenicol sensitivity of Escherichia coli CM2555 strain expressing the chloramphenicol acetyltransferase gene. Microb Drug Resist. 2002;8:179–85. doi: 10.1089/107662902760326887. [DOI] [PubMed] [Google Scholar]
- 104.Berglez J, Iliades P, Sirawaraporn W, Coloe P, Macreadie I. Analysis in Escherichia coli of Plasmodium falciparum dihydropteroate synthase (DHPS) alleles implicated in resistance to sulfadoxine. Int J Parasitol. 2004;34:95–100. doi: 10.1016/j.ijpara.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 105.Chumpolkulwong N, Hori-Takemoto C, Hosaka T, et al. Effects of Escherichia coli ribosomal protein S12 mutations on cell-free protein synthesis. Eur J Biochem. 2004;271:1127–34. doi: 10.1111/j.1432-1033.2004.04016.x. [DOI] [PubMed] [Google Scholar]
- 106.Musser JM. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hashimoto H. Acquisition of antibiotic-resistance in bacteria by alteration of molecular target, or by the decreased permeability. Nippon Rinsho. 1997;55:1167–72. [PubMed] [Google Scholar]
- 108.Webber MA, Piddock LJ. The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother. 2003;51:9–11. doi: 10.1093/jac/dkg050. [DOI] [PubMed] [Google Scholar]
- 109.Nikaido H. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin Cell Dev Biol. 2001;12:215–23. doi: 10.1006/scdb.2000.0247. [DOI] [PubMed] [Google Scholar]
- 110.Pages JM. Bacterial porin and antibiotic susceptibility. Med Sci (Paris) 2004;20:346–51. doi: 10.1051/medsci/2004203346. [DOI] [PubMed] [Google Scholar]
- 111.Patel OG, Mberu EK, Nzila AM, Macreadie IG. Sulfa drugs strike more than once. Trends Parasitol. 2004;20:1–3. doi: 10.1016/j.pt.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 112.Perez-Trallero E, Iglesias L. Tetracyclines, sulfonamides and metronidazole. Enferm Infecc Microbiol Clin. 2003;21:520–9. doi: 10.1016/s0213-005x(03)72999-1. [DOI] [PubMed] [Google Scholar]
- 113.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–20. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 114.Boman HG. Antibacterial peptides: basic facts and emerging concepts. J Intern Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 115.Nishikawa M, Ogawa K. Antimicrobial activity of a chelatable poly (arginyl-histidine) produced by the ergot fungus Verticillium kibiense. Antimicrob Agents Chemother. 2004;48:229–35. doi: 10.1128/AAC.48.1.229-235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hara T, Kodama H, Kondo M, et al. Efects of peptide dimerization on pore formation: antiparallel disulfide-dimerized magainin 2 analogue. Biopolymers. 2001;58:437–46. doi: 10.1002/1097-0282(20010405)58:4<437::AID-BIP1019>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 117.Noga EJ, Silphaduang U. Piscidins: a novel family of peptide antibiotics from fish. Drug News Perspect. 2003;16:87–92. doi: 10.1358/dnp.2003.16.2.829325. [DOI] [PubMed] [Google Scholar]
- 118.Gobbo M, Biondi L, Filira F, et al. Antimicrobial peptides: synthesis and antibacterial activity of linear and cyclic drosocin and apidaecin 1b analogues. J Med Chem. 2002;45:4494–504. doi: 10.1021/jm020861d. [DOI] [PubMed] [Google Scholar]
- 119.Rozek A, Friedrich CL, Hancock RE. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry. 2000;39:15765–74. [PubMed] [Google Scholar]
- 120.Trabi M, Schirra HJ, Craik DJ. Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from Rhesus macaque leukocytes. Biochemistry. 2001;40:4211–21. doi: 10.1021/bi002028t. [DOI] [PubMed] [Google Scholar]
- 121.Talan DA, Citron DM, Overturf GD, Singer B, Froman P, Goldstein EJ. Antibacterial activity of crotalid venoms against oral snake flora and other clinical bacteria. J Infect Dis. 1991;164:195–8. doi: 10.1093/infdis/164.1.195. [DOI] [PubMed] [Google Scholar]
- 122.White J. Bites and stings from venomous animals: a global overview. Ther Drug Monit. 2000;22:65–8. doi: 10.1097/00007691-200002000-00014. [DOI] [PubMed] [Google Scholar]
- 123.Glaser HRS. Bactericidal activity of Crotalus venom in vitro. Copeia. 1948;4:245–7. [Google Scholar]
- 124.Aloof-Hirsch S, de Vries A, Berger A. The direct lytic factor of cobra venom: purification and chemical characterization. Biochim Biophys Acta. 1968;22:53–60. doi: 10.1016/0005-2795(68)90257-2. [DOI] [PubMed] [Google Scholar]
- 125.Skarnes RC. L-Amino-acid oxidase, a bactericidal system. Nature. 1970;225:1072–3. doi: 10.1038/2251072a0. [DOI] [PubMed] [Google Scholar]
- 126.Yan XM, Zhang SQ, Chang Q, Liu P, Xu JS. Antibacterial and antifungal effects of Agkistrodon halys Pallas: purification of its antibacterial protein-LAO. Shi Yan Sheng Wu Xue Bao. 2000;33:309–16. (in Chinese) [PubMed] [Google Scholar]
- 127.Stabeli RG, Marcussi S, Carlos GB, et al. Platelet aggregation and antibacterial effects of an L-amino acid oxidase purified from Bothrops alternatus snake venom. Bioorg Med Chem. 2004;12:2881–6. doi: 10.1016/j.bmc.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 128.Lu QM, Wei Q, Jin Y, Wei JF, Wang WY, Xiong YL. L-Amino acid oxidase from Trimeresurus jerdonii snake venom: purification, characterization, platelet aggregation-inducing and antibacterial effects. J Natural Toxins. 2002;11:345–52. [PubMed] [Google Scholar]
- 129.Stiles BG, Sexton FW, Weinstein SA. Antibacterial effects of different snake venoms: purification and characterization of antibacterial proteins from Pseudechis australis (Australian king brown or mulga snake) venom. Toxicon. 1991;29:1129–41. doi: 10.1016/0041-0101(91)90210-i. [DOI] [PubMed] [Google Scholar]
- 130.Stocker JF, Traynor JR. The action of various venoms on Escherichia coli. J Appl Bacteriol. 1986;61:383–8. doi: 10.1111/j.1365-2672.1986.tb04300.x. [DOI] [PubMed] [Google Scholar]
- 131.Blaylock RS. Antibacterial properties of KwaZulu Natal snake venoms. Toxicon. 2000;38:1529–34. doi: 10.1016/s0041-0101(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 132.Xie JP, Yue J, Xiong YL, Wang WY, Yu SQ, Wang HH. In vitro activities of small peptides from snake venom against clinical isolates of drug-resistant Mycobacterium tuberculosis. Int J Antimicrob Agents. 2003;22:172–4. doi: 10.1016/s0924-8579(03)00110-9. [DOI] [PubMed] [Google Scholar]
- 133.Roos KL. Emerging antimicrobial-resistant infections. Arch Neurol. 2004;61:1512–4. doi: 10.1001/archneur.61.10.1512. [DOI] [PubMed] [Google Scholar]
- 134.Guardabassi L, Kruse H. Overlooked aspects concerning development and spread of antimicrobial resistance. Expert Rev Anti Infect Ther. 2003;1:359–62. doi: 10.1586/14787210.1.3.359. [DOI] [PubMed] [Google Scholar]
- 135.Hancock REW, Diamond G. The role of antimicrobial peptides in innate host defense. Trends Microbiol. 2000;8:402–10. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 136.Hujer AM, Bethel CR, Hujer KM, Bonomo RA. Antibiotic resistance in the institutionalized elderly. Clin Lab Med. 2004;24:343–61. doi: 10.1016/j.cll.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 137.Pirmohamed M, Park BK. HIV and drug allergy. Curr Opin Allergy Clin Immunol. 2001;1:311–6. doi: 10.1097/01.all.0000011032.69243.10. [DOI] [PubMed] [Google Scholar]