Abstract
There are numerous in vitro studies documenting the multiplication of Legionella species in free-living amoebae and other protozoa. It is believed that protozoa serve as host cells for the intracellular replication of certain Legionella species in a variety of environmental settings. This study describes the isolation and characterization of a bacterium initially observed within an amoeba taken from a soil sample. In the laboratory, the bacterium multiplied within and was highly pathogenic for Acanthamoeba polyphaga. Extracellular multiplication was observed on buffered charcoal yeast extract agar but not on a variety of conventional laboratory media. A 16S rRNA gene analysis placed the bacterium within the genus Legionella. Serological studies indicate that it is distinct from previously described species of the genus. This report also describes methods that should prove useful for the isolation and characterization of additional Legionella-like bacteria from free-living amoebae. In addition, the characterization of bacterial pathogens of amoebae has significant implications for understanding the ecology and identification of other unrecognized bacterial pathogens.
The number of species in the genus Legionella has increased dramatically since the original identification of Legionella pneumophila (9, 24). Members of this genus are widespread in natural settings as well as certain environments created as a result of human activity, such as cooling towers, various plumbing fixtures, and dental units (2, 5–8, 19). L. pneumophila and occasionally other legionellae found in these settings continue to be associated with sporadic episodes of respiratory illness in humans.
Efforts to understand the ecology of L. pneumophila and its distribution in the environment has led to an unexpected finding. In vitro studies demonstrated that L. pneumophila can use protozoa, such as free-living amoebae, as host cells for intracellular replication (1, 4, 14, 28, 34). Furthermore, some intracellular events following infection, such as the appearance of ribosomes and mitochondria in proximity to the membrane-enclosed bacteria, are common to both amoebae and human mononuclear phagocytes infected with L. pneumophila (1, 14, 26, 28). The host cells are subsequently lysed as a result of intracellular replication of the bacteria; therefore, L. pneumophila is a pathogen not only of human cells but also of amoebae. The multiplication of bacteria in amoebae, resulting in degeneration of the nuclei and lysis of the host cells, has been known for nearly 90 years (27). In recent years, however, interest in interactions between bacteria and free-living amoebae has increased. This is in part a reflection of studies of Legionella bacteria and amoebae.
Additional undescribed Legionella species that are pathogens of common free-living amoebae were reported in 1993 (31). As a group, they were originally described as Legionella-like amoebic pathogens (LLAPs) because they were capable of multiplying in the cytoplasms of amoebae, but they were difficult to cultivate on media designed to support the growth of Legionella species. These LLAPs, isolated in Europe, were taken from a variety of environmental sources, and one was a clinical isolate from an individual with persistent pneumonia (31). A recent phylogenetic analysis of their 16S rRNA genes (rDNAs) suggested that these isolates are members of the family Legionellaceae and that they may represent five new species in the genus Legionella (3). In a report by Drozanski in 1991, an obligate intracellular bacterial parasite of free-living amoebae was initially described and named Sarcobium lyticum (12). Subsequent analysis of the 16S rDNA of the bacterium suggested that it was a member of the genus Legionella but that it was different from previously described members of this genus (32). Additional studies showed that it could occasionally be cultured on buffered charcoal yeast extract (BCYE) agar (20) and that it exhibited a positive reaction with a Remel (Augusta, Ga.) Legionella Poly-ID kit, which consists of pooled immune sera to 22 species in the genus Legionella. Recently, it was proposed to transfer this bacterium to the genus Legionella as Legionella lytica comb. nov. (25).
There are numerous laboratory studies documenting the multiplication of Legionella in amoebae. Corroborating studies of amoebae naturally harboring Legionella from environmental sources have been lacking, with the exception of a report by Harf and Monteil where L. pneumophila was identified in culture lysates of amoebae originally isolated from river waters (23). Our study describes the isolation and characterization of a bacterium (LLAP-14) initially observed within an amoeba taken from a soil sample by light microscopy. 16S rDNA analysis of the bacterium indicates that it should be included within the genus Legionella. Serological studies indicate that it is distinct from previously described species of Legionella. Also described in this study are methods useful in the visualization and isolation of bacterial amoebic pathogens. These techniques should prove helpful in future studies directed toward bacteria that have parasitic or symbiotic relationships with amoebae.
MATERIALS AND METHODS
Initial isolation and cultivation of the bacteria.
A small moist soil sample of approximately 0.2 g was placed in the center of a nonnutrient agar (15 g of agar/liter of H2O) plate that had been streaked with heat-killed Escherichia coli in an X configuration, which promoted the multiplication and accumulation of amoebae in a defined area along the line of bacteria. The E. coli had been cultivated 24 h in Trypticase soy broth (TSB) and pelleted by centrifugation. After addition of the soil sample, the plate was sealed with parafilm and incubated at room temperature (25°C). After 48 h the plate was examined at magnifications of ×100 and ×400 for the presence of amoebae infected with bacteria. Several amoebae were distinguished by the presence of many motile intracellular bacteria within amoebic cytoplasms. A scalpel was used to remove a plug of agar (3 by 3 mm) containing an infected amoeba. The agar plug was transferred to a 25-cm2 tissue culture flask (Becton Dickinson, Franklin Lakes, N.J.) containing a monolayer of Acanthamoeba polyphaga (ATCC 30461) in spring water (Carolina Biological, Burlington, N.C.) that had been sterilized by autoclaving. A confluent monolayer had been formed by growing the amoebae in the flask containing TSB at 37°C for 48 h. Subsequently, the TSB was poured off and the adherent amoebae were washed with sterile spring water. Five milliliters of spring water was added prior to the addition of the agar plug with the infected amoeba. The replacement of the nutrient-rich TSB with nutrient-poor spring water served to greatly diminish the multiplication of contaminating bacteria before the amoeba pathogen was in pure culture. The preparation was incubated at room temperature for 72 h.
Obtaining a pure culture of the bacteria.
A plate of sterile nonnutrient agar was streaked with heat-killed E. coli in an X configuration. Then amoebae were concentrated by tapping a 25-cm2 tissue culture flask containing a monolayer of cells in TSB to dislodge the adherent cells, pelleting the cells by centrifugation, and then resuspending the cells in approximately 1 ml of the culture supernatant. Next, 0.1 ml of the concentrated suspension of amoebae (105 cells) was placed in the center of the X-shaped streak of E. coli and allowed to dry for 2 to 3 h at room temperature. One hundred microliters of the lysate (lysed amoebae resulting from intracellular replication of the bacterial pathogen) from a coculture which still contained other indigenous bacteria was placed directly on top of the amoebae and incubated for 2 to 3 days at room temperature. The infected amoebae were allowed to migrate across the sterile surface of the plate away from any contaminating bacteria. Within 48 to 72 h, infected amoebae could be observed in the peripheral areas of the plate away from contaminating bacteria. Small plugs of agar (3 by 3 mm) containing infected amoebae were cut out with sterilized tools under aseptic conditions and transferred to fresh monolayers of amoebae in spring water. This process not only promoted the initial isolation and culture of the amoebic pathogen (LLAP-14) but also allowed separation of the bacteria from contaminating bacteria in the original soil sample.
Giemsa stain procedure and electron microscopy.
Infected amoebae were Giemsa stained to determine if the bacteria initially accumulated within vacuoles or if the bacteria were free in the cytoplasms of the amoebae. Cytospins of amoebae cocultures were prepared with whole and lysed cells from 24-, 48-, and 72-h cocultures. The samples were spun at 700 × g for 6 min in a Shandon (Pittsburgh, Pa.) Cytospin-3. The samples were fixed in methanol for 3 min prior to staining. The Giemsa stain was prepared by adding two drops of Triton X-100 and 1 ml of stain (EM Diagnostic Systems, Gibbstown, N.J.) to 45 ml of deionized water. The slides were stained for 1 h and then rinsed with deionized water. For electron microscopy, aliquots of infected amoebae were fixed for 2 h in 0.1 M cacodylate buffer (pH 7.4) containing 3% glutaraldehyde and 1% osmium tetroxide. Cells were then washed in 0.1 M cacodylate buffer and dehydrated through a series of ethanols. Subsequently, preparations were embedded in epson-araldite resin and stained with 0.5% lead citrate and uranyl acetate (2%). Sections were examined with a Zeiss model 109 electron microscope.
Growth on laboratory media.
When the bacteria were in monoxenic coculture with A. polyphaga, we performed studies to assess the ability of the bacteria to grow independently of amoebae. Aliquots of the cocultures were plated onto Trypticase soy agar (TSA), blood agar, and BCYE differential and selective agars (Becton Dickinson) which are designed to support the growth of legionellae. The plates were incubated at room temperature (25°C), at 30°C, and at 37°C for up to 14 days.
Serological testing.
The bacteria were tested for reaction against Legionella-specific immune sera with a Remel Legionella Poly-ID kit. In addition, the bacteria were evaluated by slide agglutination with 17 pools of rabbit antiserum representing 41 species and 64 serogroups of Legionella at the Centers for Disease Control and Prevention, Atlanta, Ga. (CDC).
Amplification of 16S rDNA.
Genomic DNA was extracted from the bacteria cells with a Qiagen (Chatsworth, Calif.) blood kit. PCRs were carried out on the 16S rDNA with the eubacterial primers (obtained from the CDC) 8 forward (5′ AGTTTGATCCTGGCTCAG 3′) and 1510 reverse (5′ GGTTACCTTGTTACGACTT 3′). Each reaction mixture contained 2.0 μl of the DNA template, 0.25 μl (1.25 U) of Taq DNA polymerase (Boehringer Mannheim, Indianapolis, Ind.), 200 μM each deoxynucleoside triphosphate (Boehringer Mannheim), 5.0 μl of 10× Taq buffer (Boehringer Mannheim), and 1.0 μl each of the forward and reverse primers at a concentration of 50 pmol/μl. The reaction mixtures were then brought to a volume of 50 μl with sterile distilled water. Amplification was carried out with a Perkin-Elmer (Norwalk, Conn.) model 9600 thermal cycler. The cycling of the program involved an initial 2-min hold at 94°C followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s. Purification of the DNA fragments was accomplished by using the Wizard PCR Preps DNA purification system (Promega Corp., Madison, Wis.).
DNA sequence analysis.
The Taq Dye Deoxy Terminator cycle sequencing kit (Perkin-Elmer, Foster City, Calif.) was used to perform fluorescence-based dideoxy-chain termination sequencing reactions on the purified PCR products. The reaction mixtures contained 8.0 μl of Ready Mix (A Dye-C Dye-G Dye-T Dye Terminator, dITP, dATP, dTTP, dCTP, Tris-HCl [pH 9.0], MgCl2, thermal stable pyrophosphatase, AmpliTaq DNA polymerase), 1.0 μl (3 pmol/μl) of one of 35 different eubacterium- or Legionella 16S-protein-specific primers obtained from the CDC (Table 1), and 1.5 to 2.5 μl (0.2 μg/μl) of template and were brought to a volume of 20 μl with sterile deionized distilled water. Extension products of each sample were purified with a Centri-sep column (Princeton Separations, Inc., Adelphia, N.J.). The products of the sequencing reactions were separated with a 4.25% denaturing acrylamide gel in a model ABI-377 (Perkin-Elmer) automated DNA sequencer. The sequences were edited and assembled to give a contiguous sequence with the University of Wisconsin Genetics Computer Group sequence analysis program.
TABLE 1.
Eubacterium- and Legionella-specific 16S rDNA primer sequences
| Eubacterium- or Legionella-specific primera | Sequenceb |
|---|---|
| Eubacterium specific | |
| 8 f | AGT TTG ATC CTG GCT CAG |
| 104 f | GGA CGG GTG AGT AAC ACG TG |
| 357 f, r | TAC GGG AGG CAG CAG |
| 460 f | TAC CTG GGA AGA TAA TGA CGG |
| 530 f, r | CAG CAG CCG CGG TAA TAC |
| 652 f, r | GAT ATT CGG AGG AAC ACC AGT GGC |
| 690 f, r | GTG AAA TGC GTA GA |
| 734 f | TTA GAT ACC CTG GTA GTC CAC GCC |
| 790 f, r | ATT AGA TAC CCT GGT AG |
| 980 r | TTG CTT CGA ATT AAA CCA C |
| 981 f, r | CCC GCA ACG AGC GCA ACC C |
| 1100 f | CAA CGA CGC CAA CCC T |
| 1230 r | CAT TGT AGC ACG TGT GTA |
| 1390 r | CGG TGT GTA CAA CGC CC |
| 1510 r | GGT TAC CTT GTT ACG ACT T |
| Legionella specific | |
| 1 f | CAC ATG CAA GTC GAA CGG CAG |
| 300 f | AGA CAC GGT CCA GAC TCC TAC |
| 370 r | GTG CTT TAC AAC CCT CAG GCC |
| 500 f | AGC GTT AAT CGG AAT TAC TGG |
| 610 r | TCC ACT ACC CTC TCC CAT AC |
| 740 f | GAT TAG ATA CCC TGG TAG TC |
| 850 r | TTG AGT TTT AAT CTT GCG AC |
| 934 f | GCA CAA GCG GTG GAG CAT G |
| 969 f | AAC GCG AAG AAC CTA CCT AC |
| 1100 r | CGG CAG TCT CCT TAG AGT TG |
| 1283r | CTA GCG ATT CCG ACT TCA TG |
| 1321 r | TTG CAG ACT CCA ATC CGG AC |
| 1368 r | ACA TGC TGA TTC GCG ATT AC |
| 1400 r | AAC GTC CCC CCG AAG GTT AG |
f, forward; r, reverse; f, r, both forward and reverse.
Written 5′ to 3′.
Phylogenetic analysis.
A phenogram was established with 54 species of bacteria, including Legionella, LLAPs, and the outgroup Coxiella burnetii. The analysis was based on a 1,303-nucleotide region by the neighbor-joining method contained in the Phylogeny Inference Package (PHYLIP), version 3.5 (13). Homology values (percentages) between LLAP-14 and various species of Legionella and LLAPs were calculated as 1 − 3/4 [1 − e−4/3 (distance value)], where e is the base of the natural logarithm. The distance values were calculated with the Jukes-Cantor model in the DNADist program contained in PHYLIP.
Nucleotide sequence and American Type Culture Collection accession numbers.
A 1,474-bp nucleotide sequence of the 16S rDNA has been submitted to GenBank under accession no. U66104, and the bacterium was designated LLAP-14. The bacterium has been deposited at the American Type Culture Collection, Rockville, Md., under accession no. 700313.
RESULTS
Initial identification of the amoeba pathogen.
After 72 h of incubation at room temperature, amoebae in the soil sample had moved onto the E. coli and were actively multiplying. When viewed at magnifications of ×100 and ×400 amoebic cell structures such as the nucleus and vacuoles were clearly visible. Several amoebae were distinctly swollen and contained numerous bacteria that were highly motile. In addition, the host cell nucleus was not apparent and the vacuole containing the bacteria occupied most of the amoebic cytoplasmic space. Transferring an amoeba to a monolayer of A. polyphaga resulted in an increased number of the bacteria in the culture supernatant, although contaminating bacteria were still present. Subsequently, LLAP-14 was obtained in pure culture with amoebae.
Microscopic examination of the bacterial lytic cycle.
During the first 24 h of incubation of the amoeba coculture at room temperature, intracellular bacteria were not observed by Giemsa staining. Between 24 and 48 h, Giemsa staining revealed that the bacteria initially accumulated within vacuoles (Fig. 1). At this point in the lytic cycle amoebae retained their ability to adhere to the surface of the flask and retained internal morphological features, such as the nucleus. At 48 h most amoeba trophozoites contained intracellular bacteria, as determined by Giemsa staining and phase-contrast microscopy. The amoeba host cells frequently contained two or three vacuoles filled with the highly motile bacteria. An additional feature was the alignment of mitochondria in proximity to the vacuoles containing bacteria (Fig. 2). Seventy-two to 96 h after inoculation, the infected cells lacked defined organelle structures and the amoebae were no longer adherent to the surface of the flask. Typically, the bacteria remained motile through the end of the lytic cycle and only the cytoplasmic membrane of an amoeba host cell remained intact (Fig. 3). Following lysis of amoeba trophozoites, the bacteria remained viable for at least 7 days, as determined by reinfection of fresh monolayers of amoebae. Approximately 2.5 × 104 CFU per ml could be recovered on BCYE after completion of the lytic cycle in amoebae. No intracellular multiplication was observed at 37°C.
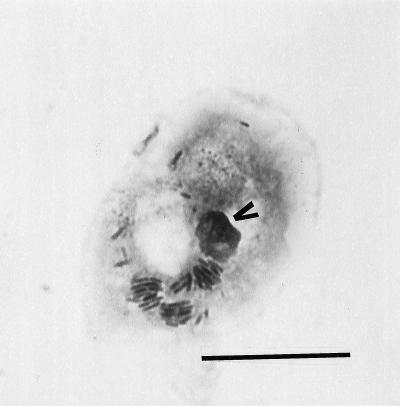
FIG. 1.
Giemsa stain showing the occurrence of bacteria in vacuoles after 24 and 48 h of incubation at 25°C. Characteristic morphological features of the amoeba host cell, such as the nucleus (arrowhead), were intact. Bar, 20 μm.
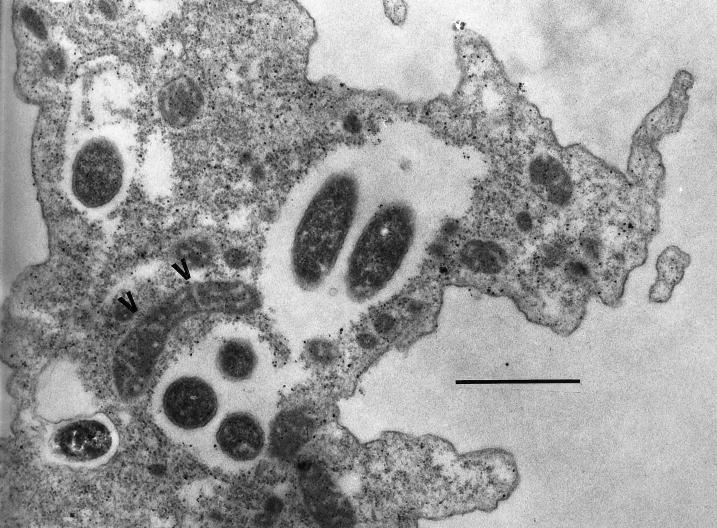
FIG. 2.
Alignment of mitochondria (arrowheads) to a vacuole containing bacteria during the early stages (24 to 48 h) of the lytic cycle. Bar, 1.0 μm.
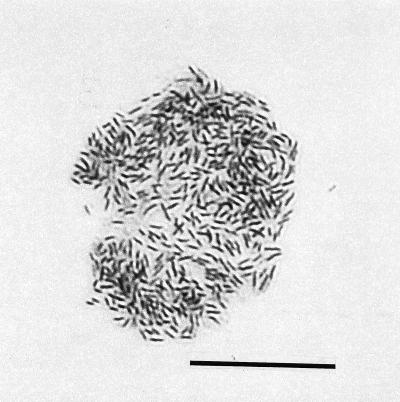
FIG. 3.
After 72 h, intracellular multiplication of bacteria was accompanied by loss of the amoeba host cell infrastructure, such as the nucleus and well-defined vacuoles. The bacteria remained bound by the amoeba cytoplasmic membrane. Subsequently, the membrane ruptured, which released the bacteria to infect adjacent amoebae, beginning a new lytic cycle. Bar, 20 μm.
Multiplication on bacteriological media.
The bacteria failed to multiply on TSA and blood agar. Growth was observed, however, on BCYE differential and selective agars at room temperature and 30°C. No growth was observed at 37°C. The colonies exhibited a gray, cut-glass appearance typical of other legionellae. Initial recovery of the bacteria from amoeba cocultures on BCYE differential and selective agars required 1 to 2 weeks at room temperature and 30°C. When agar-grown colonies were subcultured, the required incubation period decreased to 5 days after several passages. Substantial growth occurred on the BCYE differential medium, which does not contain antibiotics, while diminished growth occurred on the BCYE selective medium, which contains the antibiotics vancomycin and anisomycin. Confirmation that the agar-grown colonies were of the originally isolated bacterium (LLAP-14) was based on reinfectivity of the bacteria in A. polyphaga and the absence of growth on TSA and blood agar.
Serology.
No positive reactions were observed when cultures were tested with the Remel Legionella Poly ID kit and all 17 pools of Legionella antisera maintained at the CDC.
Phylogenetic analysis.
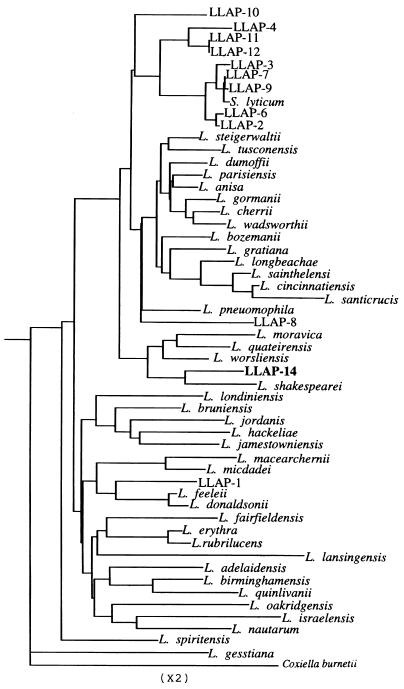
The phenogram based on 16S rDNA sequences clearly demonstrates that LLAP-14 merits inclusion within the genus Legionella, being most closely related to Legionella shakespearei (Fig. 4). The homology percentage comparisons showed that LLAP-14 was 97.0% homologous to L. shakespearei, 95.5 to 97% homologous to all four species within the same cluster, and 94.2 to 95.8% homologous to all LLAPs and S. lyticum (Table 2). Overall, LLAP-14 was between 92.0 and 97.0% homologous to all members of the genus Legionella.
FIG. 4.
16S rDNA-based phenogram reflecting the relationship between LLAP-14 and all other well-resolved species in the genus Legionella and in the outgroup C. burnetii. Analysis was based on a 1,303-bp region by the neighbor-joining method.
TABLE 2.
16S rDNA gene homology (%) between members of the genus Legionella and other LLAPs
| Legionella sp. or LLAP | % Homologya with:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LLAP-2 | LLAP-3 | LLAP-4 | LLAP-6 | LLAP-7 | LLAP-8 | LLAP-9 | LLAP-10 | LLAP-11 | LLAP-12 | LLAP-14 | S. lyticum | L. pneumophila | L. shakespearei | L. worsliensis | L. quateriensis | L. moravica | |
| LLAP-1 | 94.1 | 93.6 | 93.9 | 94.0 | 93.8 | 94.1 | 93.7 | 94.4 | 94.1 | 94.1 | 94.2 | 93.7 | 93.8 | 93.3 | 94.2 | 93.6 | 93.5 |
| LLAP-2 | 98.8 | 96.9 | 99.8 | 99.4 | 95.4 | 99.3 | 96.6 | 97.4 | 97.4 | 95.8 | 99.2 | 95.8 | 95.3 | 95.4 | 95.3 | 94.1 | |
| LLAP-3 | 96.9 | 99.0 | 99.5 | 94.9 | 99.5 | 96.3 | 97.1 | 97.1 | 95.4 | 99.4 | 95.8 | 94.9 | 95.1 | 94.9 | 93.9 | ||
| LLAP-4 | 97.0 | 97.2 | 95.1 | 97.1 | 96.1 | 98.5 | 98.5 | 95.7 | 97.1 | 95.5 | 95.4 | 95.3 | 94.7 | 93.9 | |||
| LLAP-6 | 99.3 | 95.1 | 99.2 | 96.8 | 97.3 | 97.3 | 95.7 | 99.2 | 95.7 | 95.1 | 95.2 | 95.1 | 93.9 | ||||
| LLAP-7 | 95.1 | 99.9 | 96.5 | 97.4 | 97.4 | 95.8 | 99.9 | 95.9 | 95.1 | 95.3 | 95.3 | 94.3 | |||||
| LLAP-8 | 94.9 | 95.0 | 95.7 | 95.7 | 94.3 | 94.9 | 96.1 | 94.0 | 94.6 | 94.0 | 93.2 | ||||||
| LLAP-9 | 96.4 | 97.3 | 97.3 | 95.7 | 99.8 | 95.8 | 95.0 | 95.2 | 95.2 | 94.2 | |||||||
| LLAP-10 | 96.4 | 96.4 | 95.7 | 96.4 | 96.7 | 94.9 | 96.8 | 96.5 | 95.0 | ||||||||
| LLAP-11 | 100.0 | 95.8 | 97.2 | 96.4 | 95.3 | 96.2 | 95.4 | 94.2 | |||||||||
| LLAP-12 | 95.8 | 97.2 | 96.4 | 95.3 | 96.2 | 95.4 | 94.2 | ||||||||||
| LLAP-14 | 95.7 | 95.2 | 97.0 | 96.0 | 96.3 | 95.5 | |||||||||||
| S. lyticum | 95.8 | 95.0 | 95.1 | 95.2 | 94.2 | ||||||||||||
| L. pneumophila | 95.1 | 96.6 | 96.5 | 95.3 | |||||||||||||
| L. Shakespearei | 95.9 | 96.4 | 95.3 | ||||||||||||||
| L. worsliensis | 97.8 | 96.5 | |||||||||||||||
| L. quateirensis | 97.0 | ||||||||||||||||
Percentages were rounded to the nearest 1/10. Boldface denotes LLAP-14 homology percentages.
DISCUSSION
To more completely define the genus Legionella, it may be pertinent to identify bacteria that occur naturally within free-living amoebae. The significance of the interaction between Legionella species and amoebae is also supported by the suggestion that the description for the genus Legionella in Bergey’s Manual of Systematic Bacteriology (26a) be amended to include the following statement: “some legionellae appear to be primary obligate intracellular parasites of amoebae and exhibit little or no growth on current laboratory media” (3). Documentation of naturally occurring bacteria within amoebae strengthens the assumption that certain members of the genus Legionella use amoebae for intracellular replication in natural settings. Additional studies of bacterial amoeba pathogens as described in this report will promote our understanding of the genus Legionella in both naturally occurring and human-made environments.
The bacterium described in this investigation is a facultative intracellular parasite of the free-living amoeba A. polyphaga and proliferates only on media designed to support Legionella species. During the early stages of amoebic infection, the alignment of host cell organelles such as mitochondria in proximity to bacteria in enclosed vacuoles is reminiscent of that observed with L. pneumophila in both amoebae and macrophages (1, 14, 26, 28). The multiplication and accumulation of the bacteria in amoebae and the corresponding loss of amoeba intracellular structures are features commonly observed in in vitro studies of Legionella-infected amoebae. The probability that this bacterium represents a member of the genus Legionella was confirmed by the phylogenetic analysis of its 16S rDNA. The 92.0 to 97.0% homology of LLAP-14 compares well with the 16S rDNA sequence identity (90.2 to 99.1%) that exists between the 39 validly described members of the genus (3). The 16S rDNA sequence over 1,303 bases was distinctive (maximum homology, 97.0%), and it may represent a new species in the genus Legionella. Sequence analysis of 16S rDNA was useful for establishing relationships; however, it should be used with caution to conclusively denote species identity (18). Species distinction of LLAP-14 and the LLAP-type bacteria would be best addressed by DNA-DNA hybridization studies of members of the genus Legionella (33). Based on serological analysis, the lack of reactivity with a battery of immune sera to previously described Legionella species additionally suggests that the bacterium LLAP-14 is an undescribed member of the genus Legionella.
The potential of Legionella bacteria that are also amoebic pathogens to cause disease in humans is best demonstrated with L. pneumophila. Infection of human cells with Legionella species was shown to be related to the ability of the bacteria to infect protozoa (15). It has been suggested that this ability may serve to enhance the virulence of Legionella and have importance in the pathogenic mechanism of the bacteria (10, 17, 21). For example, Legionella species such as L. parisiensis and L. jamestowniensis were originally not known to cause disease in humans. In vitro studies, however, demonstrated multiplication in macrophage-like cells (29). This result supported the concept that they could be human pathogens, which was recently confirmed when L. parisiensis was associated with pneumonic illness (30). One line of evidence indicates that multiplication of Legionella species in amoebae and the inability to multiply in human phagocytes does not preclude the possibility of causing human illness. For example, Legionella anisa readily multiplied in the amoeba Hartmannella vermiformis but not in human monocytes or U937 cells (16). However, L. anisa has been shown to cause Pontiac fever, which results from exposure to high levels of nonviable bacterial cells or bacteria unable to multiply in lung phagocytes (16). The inability of LLAP-14 to multiply at 37°C under laboratory conditions suggested that it would not be a highly virulent pathogen of humans or other endothermic animals, although the potential for exposure and disease cannot be discounted at this time. Bacteria in the genus Legionella infect a range of eukaryotic cells. This includes cells of the monocytic cell lineage, numerous amoeba species of different genera, and some protozoan ciliates as well (14, 26, 29). It is thus likely that the host range of LLAP-14 extends beyond the infected amoebae in the original soil sample and the laboratory cultures of A. polyphaga. Establishment of the host range of this bacterium and related Legionella-like amoebic pathogens would contribute to characterization and classification to species level. Multiplication in cells of monocytic lineage might also help establish any clinical relevance. These wide-ranging future studies would likely lead to descriptive amendments for the genus Legionella.
Finally, increased scientific awareness along with methods useful for the isolation and characterization of amoebic pathogens may serve to promote studies identifying any role these bacteria have on the distribution and occurrence of free-living amoebae in natural settings. Historically, the little information known about amoeba population dynamics has been based on observations of physical factors, food availability, and possible predation by micro- and macroinvertebrates. Protozoa such as amoebae are the significant predators of bacteria in both soil and aquatic environments (11, 22, 35). Amoebic consumption of prokaryotes is believed to play an important role in nutrient cycling and to be of practical importance for agriculture. The influence that these newly described amoebic pathogens may have on free-living amoebae in natural settings is presently open to speculation. The understanding of relative occurrence and host range of amoeba pathogens can provide insight into alternative means by which populations of amoebae are influenced by bacteria which invade and destroy their host cells.
ACKNOWLEDGMENTS
We thank Adenike Adeleke and Janet M. Pruckler of the CDC for their valuable assistance with the DNA sequencing and Ann Whitney for supplying the eubacterium primers and their sequences. We also thank Lisa Hill-Williams for her assistance with the electron microscopy.
REFERENCES
- 1.Abu Kwaik Y. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addiss D G, Davis J P, LaVenture M, Wand P J, Hutchinson M A, McKinney R M. Community-acquired Legionnaires’ disease associated with a cooling tower: evidence for longer-distance transport of Legionella pneumophila. Am J Epidemiol. 1989;130:557–568. doi: 10.1093/oxfordjournals.aje.a115370. [DOI] [PubMed] [Google Scholar]
- 3.Adeleke A, Pruckler J, Benson R, Rowbotham T, Halablab M, Fields B. Legionella-like amoebal pathogens: phylogenetic status and possible role in respiratory disease. Emerg Infect Dis. 1996;2:225–230. doi: 10.3201/eid0203.960311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand C M R, Skinner A, Malic A, Kurtz J B. Interaction of Legionella pneumophila and a free-living amoeba (Acanthamoeba palestinensis) J Hyg. 1983;91:167–178. doi: 10.1017/s0022172400060174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnow P M, Chou T, Shapiro E N, Kretzchmar C. Nosocomial Legionnaires’ disease caused by aerosolized tap water from respiratory devices. J Infect Dis. 1982;146:460–467. doi: 10.1093/infdis/146.4.460. [DOI] [PubMed] [Google Scholar]
- 6.Atlas R M, Williams J F, Huntington M K. Legionella contamination of dental-unit waters. Appl Environ Microbiol. 1995;61:1208–1213. doi: 10.1128/aem.61.4.1208-1213.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson R F, Thacker W L, Lanser J A, Sangster N, Mayberry W R, Brenner D J. Legionella adelaidensis, a new species isolated from cooling tower water. J Clin Microbiol. 1991;29:1004–1006. doi: 10.1128/jcm.29.5.1004-1006.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollin G E, Plouffe J F, Para M F, Hackman B. Aerosols containing Legionella pneumophila generated by shower heads and hot-water faucets. Appl Environ Microbiol. 1985;50:1128–1131. doi: 10.1128/aem.50.5.1128-1131.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner D J, Steigerwalt A G, McDade J E. Classification of the Legionnaires’ disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann Intern Med. 1979;90:656–658. doi: 10.7326/0003-4819-90-4-656. [DOI] [PubMed] [Google Scholar]
- 10.Brieland J, McClain M, Heath L, Chrisp C, Huffnagle G, LeGendre M, Hurley M, Fantone J, Engleberg C. Coinoculation with Hartmannella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires’ disease. Infect Immun. 1996;64:2449–2456. doi: 10.1128/iai.64.7.2449-2456.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarholm M. Heterotrophic, free-living protozoa: neglected microorganisms with an important task in regulating bacterial populations. In: Klug M J, Reddy C A, editors. Current perspectives in microbial ecology. Washington, D.C: American Society for Microbiology; 1984. pp. 321–326. [Google Scholar]
- 12.Drozanski W J. Sarcobium lyticum gen. nov., sp. nov., an obligate intracellular bacterial parasite of small free-living amoebae. Int J Syst Bacteriol. 1991;41:82–87. [Google Scholar]
- 13.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 14.Fields B S. Legionella and protozoa: interaction of a pathogen and its natural host. In: Barbaree J M, Breiman R F, Dufour A P, editors. Legionella: current status and emerging perspectives. Washington, D.C: American Society for Microbiology; 1993. pp. 129–136. [Google Scholar]
- 15.Fields B S, Barbaree J M, Shotts E B, Jr, Feeley J C, Morrill W E, Sanden G N, Dykstra M J. Comparison of guinea pig and protozoan models for determining virulence of Legionella species. Infect Immun. 1986;53:553–559. doi: 10.1128/iai.53.3.553-559.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields B S, Barbaree J M, Sanden G N, Morrill W E. Virulence of a Legionella anisa strain associated with Pontiac fever: an evaluation using protozoan, cell culture, and guinea pig models. Infect Immun. 1990;58:3139–3142. doi: 10.1128/iai.58.9.3139-3142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fliermans C B. Ecology of Legionella: from data to knowledge with a little wisdom. Microb Ecol. 1996;32:203–228. doi: 10.1007/BF00185888. [DOI] [PubMed] [Google Scholar]
- 18.Fox G E, Wisotzkey J D, Jurtshuk P J R. How close is close; 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 19.Garbe P L, Davis B J, Weisfeld J S, Markowitz L, Miner P, Garrity F, Barbaree J M, Reingold A L. Nosocomial Legionnaires’ disease: epidemiologic demonstration of cooling towers as a source. JAMA. 1985;254:521–524. doi: 10.1001/jama.254.4.521. [DOI] [PubMed] [Google Scholar]
- 20.Giles D L, Fields B S, Newsome A L, Drozanski W J. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. Cultivation of Sarcobium lyticum on artificial medium, abstr. Q-447; p. 478. [Google Scholar]
- 21.Glavin F L, Winn W C, Jr, Craighead J E. Ultrastructure of lung in Legionnaires’ disease. Observations of three biopsies done during the Vermont epidemic. Ann Intern Med. 1978;90:555–558. doi: 10.7326/0003-4819-90-4-555. [DOI] [PubMed] [Google Scholar]
- 22.Gonzales J M, Iriberri J, Egea L, Barcina I. Differential rates of digestion of bacteria by freshwater and marine phagotropic protozoa. Appl Environ Microbiol. 1990;56:1851–1857. doi: 10.1128/aem.56.6.1851-1857.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harf C, Monteil H. Interactions between free-living amoebae and Legionella in the environment. Water Sci Technol. 1988;20:235–239. [Google Scholar]
- 24.Harrison T G, Saunders N A. Taxonomy and typing of legionellae. Rev Med Microbiol. 1994;5:79–90. [Google Scholar]
- 25.Hookey J V, Saunders N A, Fry N K, Birtles R J, Harrison T G. Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int J Syst Bacteriol. 1996;46:526–531. [Google Scholar]
- 26.Horwitz M A. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: Williams & Wilkins; 1984. [Google Scholar]
- 27.Nagler K. Fakultativ parasitische Micrococcen in Amöben. Arch Protistenkd. 1910;19:246. [Google Scholar]
- 28.Newsome A L, Baker R L, Miller R D, Arnold R R. Interactions between Naegleria fowleri and Legionella pneumophila. Infect Immun. 1985;50:449–452. doi: 10.1128/iai.50.2.449-452.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connell W A, Dhand L, Cianciotto N P. Infection of macrophage-like cells by Legionella species that have not been associated with disease. Infect Immun. 1996;64:4381–4384. doi: 10.1128/iai.64.10.4381-4384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Presti F L, Riffard S, Vandenesch F, Reyrolle M, Ronco E, Ichai P, Etienne J. The first clinical isolate of Legionella parisiensis, from a liver transplant patient with pneumonia. J Clin Microbiol. 1997;35:1706–1709. doi: 10.1128/jcm.35.7.1706-1709.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowbotham T J. Legionella-like amoebal pathogens. In: Barbaree J M, Breiman R F, Dufour A P, editors. Legionella: current status and emerging perspectives. Washington, D.C: American Society for Microbiology; 1993. pp. 137–140. [Google Scholar]
- 32.Springer N, Ludwig W, Drozanski W, Amann R, Schleifer K H. The phylogenetic status of Sarcobium lyticum, an obligate intracellular bacterial parasite of small amoebae. FEMS Microbiol Lett. 1992;96:199–202. doi: 10.1016/0378-1097(92)90403-b. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 34.Tyndall R L, Dominique E L. Cocultivation of Legionella pneumophila and free-living amoebae. Appl Environ Microbiol. 1982;44:954–959. doi: 10.1128/aem.44.4.954-959.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weekers P H H, Bodelier P L E, Wijen J P H, Vogels G D. Effects of grazing by the free-living soil amoebae Acanthamoeba castellanii, Acanthamoeba polyphaga, and Hartmannella vermiformis on various bacteria. Appl Environ Microbiol. 1993;59:2317–2319. doi: 10.1128/aem.59.7.2317-2319.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]