Abstract
Plasmids that can be used for controlled expression of the DnaK-DnaJ-GrpE and/or GroEL-GroES chaperone team were constructed in order to facilitate assessment of the effects of these chaperone teams on folding or assembly of recombinant proteins in Escherichia coli. A typical pACYC184-based plasmid which was obtained could express the major DnaK-DnaJ-GrpE and GroEL-GroES chaperone teams from separate promoters when l-arabinose and tetracycline, respectively, were added in a dose-dependent fashion. The model protein used to determine whether this system was useful was an allergen of Japanese cedar pollen, Cryj2, which was unstable when it was produced in E. coli K-12. The effects of chaperone coexpression on the folding, aggregation, and stability of Cryj2 were examined in the wild type and in several mutant bacteria. Coexpression of the DnaK-DnaJ-GrpE and/or GroEL-GroES chaperone team at appropriate levels resulted in marked stabilization and accumulation of Cryj2 without extensive aggregation. Experiments performed with mutants that lack each of the chaperone proteins (DnaK, DnaJ, GrpE, GroEL, and GroES) or heat shock transcription factor ς32 revealed that both chaperone teams are critically involved in Cryj2 folding but that they are involved in distinct ways. In addition, it was observed that the two chaperone teams have synergistic roles in preventing aggregation of Cryj2 in the absence of ς32 at certain temperatures.
Production of recombinant proteins in Escherichia coli often results in rapid degradation or aggregation of these proteins because of their inability to form native or correct tertiary structures. It is widely recognized that coexpression of molecular chaperones can assist protein folding and that in at least some cases this leads to increased production of active proteins. The most abundant and physiologically important chaperones in E. coli include DnaK, DnaJ, GrpE, GroEL, and GroES (2, 4), whose syntheses are under positive control of a minor ς factor (ς32) encoded by the rpoH gene (4, 18). Several lines of evidence have indicated that the two major chaperone teams, DnaK-DnaJ-GrpE and GroEL-GroES, play distinct but cooperative roles in protein folding in vivo (3, 5, 8). Extensive analyses of the effects of these chaperones on folding, aggregation, stability, and assembly of individual proteins, particularly in vitro, have provided numerous results that are consistent with these conclusions (2, 5).
Accordingly, much effort has gone into improving production of active recombinant proteins in E. coli by coexpressing one of the chaperone teams. In most cases the effects of GroE coexpression were examined, whereas the effects of DnaK-DnaJ coexpression often have been tested without simultaneous coexpression of GrpE. In general, these attempts have been only partially successful; the present state of our knowledge concerning protein folding seems to preclude rational predictions about which of the chaperones could be effective and useful for a given protein (16). In addition, the promoters used for expression of chaperones have often been the same promoters used for expression of the target protein, in part due to a lack of compatible plasmids that allow controlled expression of the chaperone teams. Systematic and well-controlled expression analyses are particularly important because overproduction of chaperones, as well as of recombinant proteins, can be deleterious to host cell growth. These and other considerations prompted us to construct a series of versatile expression plasmids that should permit expression of DnaK-DnaJ-GrpE and GroEL-GroES chaperone teams independently from each other and from target protein in the same strain, so that the effects of each chaperone team can be assessed easily and precisely.
The model target protein which we used was Cryj2, which was recently isolated as a major allergen of Japanese cedar (Cryptomeria japonica) pollen (15). The gene encoding this protein has been cloned and sequenced (11), and the mature 42-kDa protein product was shown to have weak polymethylgalacturonase activity (13). When this protein was expressed in E. coli, it was unstable and rapidly degraded. We used Cryj2 to examine the effects of each of the chaperone teams on the stability and aggregation of a protein under a variety of conditions. To do this, we used chaperone expression plasmids and several well-characterized mutants deficient in individual chaperones or the heat shock transcription factor ς32.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli K-12 strains MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR] (1) and MG1655 (prototroph) and their derivatives were used in all experiments. The temperature-sensitive chaperone mutants used were MC4100 ΔdnaK52 (10), MC4100 dnaJ259 (6), MC4100 grpE280 (6), MC4100 groEL44 (9), and MC4100 groES72 (7). Strain NK161 (= MG1655 ΔrpoH) was constructed by transducing ΔrpoH30::kan (19) into MG1655 with phage T4gt7. A Cryj2 expression plasmid, pKCJ2, which was derived from the pBR322-based plasmid pKK223-3 (Pharmacia), was kindly donated by M. Kurimoto. This plasmid contained the gene encoding Cryj2 mature protein (Arg-46–Ser-433), which was placed under the tac promoter. The lacIq repressor gene was then inserted upsteam of the tac promoter, and the resulting plasmid, pKCJ2l, was used for this study. pAR3 was a pACYC184-based arabinose-inducible expression plasmid compatible with ColE1-derived plasmids; it carried the araB promoter-operator, the araC activator-repressor gene, and the cat chloramphenicol acetyltransferase gene (14). Plasmid pUHE2Pzt-1 carrying the Pzt-1 promoter that can be induced by tetracycline was kindly provided by H. Bujard.
Construction of chaperone expression plasmids.
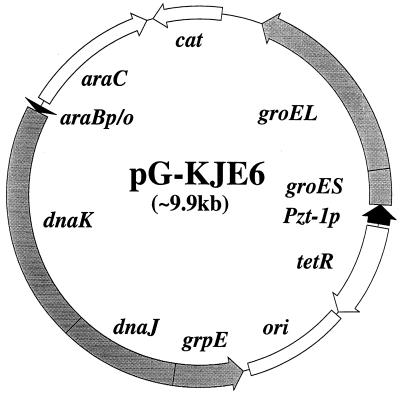
An artificial operon encoding the DnaK-DnaJ-GrpE chaperone team was constructed by inserting the grpE coding region (plus the Shine-Dalgarno sequence) into the dnaK-dnaJ operon (immediately after the dnaJ termination codon and upstream of the dnaJ transcription terminator. The resulting dnaK-dnaJ-grpE operon and the groES-groEL operon, each containing a Shine-Dalgarno sequence but lacking promoters, were placed under control of the araB promoter on a pACYC184-based plasmid (pAR3); the resulting plasmids were designated pKJE7 and pGro7, respectively. In a separate construction, the groES-groEL genes were placed downstream of the Pzt-1 promoter on plasmid pUHE2Pzt-1, and then the tetR repressor gene was inserted upsteam of the promoter in the opposite direction. The resulting tetR-Pzt-1p-groES-groEL segment was taken out and inserted into pKJE7, which yielded pG-KJE6 (Fig. 1).
FIG. 1.
Structure of chaperone coexpression plasmid pG-KJE6. ori, replication origin of pACYC184; cat, chloramphenicol acetyltransferase gene; Pzt-1p, Pzt-1 promoter; tetR, tetR repressor gene; araBp/o, araB promoter-operator; araC, araC repressor gene.
Media, chemicals, and culture conditions.
L broth with or without antibiotics was used for all experiments. Strains harboring pKCJ2l (expressing Cryj2) alone or together with a chaperone expression plasmid were grown to log phase in the presence of 50 μg of ampicillin per ml (plus 20 μg of chloramphenicol per ml when necessary) at 30°C unless otherwise indicated. To induce expression of chaperones, l-arabinose and/or tetracycline was added at various concentrations. Cryj2 was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). All chemicals were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
Protein extraction, detection, and analysis.
Culture samples were harvested and treated with trichloroacetic acid, and whole-cell proteins (from preparations having equivalent optical densities) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using a 10% gel as described previously (17). Protein contents were determined with a MicroBCA protein assay reagent kit (Pierce Chemical Co.). Chaperone proteins were detected by staining the gels with Coomassie brilliant blue and were quantified by immunoblotting with specific antisera. The Cryj2 protein was detected by immunoblotting with a mouse monoclonal antibody against Cryj2, N26 (kindly donated by Y. Taniguchi), and was quantified with an Intelligent Quantifier apparatus (BioImage Systems Co., Tokyo, Japan).
Analysis of protein aggregation and stability.
To examine the extent of aggregation of the Cryj2 produced, cells were disrupted by sonication (model XL2020 ultrasonic liquid processor; Heat Systems Inc.) for 30 s on ice, and then the preparations were centrifuged to remove debris. Subsequent centrifugation at 8,200 × g for 10 min separated the soluble and insoluble fractions, and fractions obtained from extracts having equivalent protein contents were analyzed by SDS-PAGE, followed by immunoblotting with specific antiserum. To determine the stability of Cryj2 in vivo, spectinomycin (500 μg/ml; Sigma-Aldrich) was added to log-phase cells to stop protein synthesis, samples were taken at intervals and treated with trichloroacetic acid, and whole-cell proteins were analyzed by SDS-PAGE and immunoblotting.
RESULTS
Plasmids that permit controlled expression of major chaperone teams.
We constructed a series of multicopy plasmids that permitted controlled expression of DnaK-DnaJ-GrpE and GroEL-GroES chaperone teams separately from each other and from the target protein. We used pACYC184-based chloramphenicol or kanamycin resistance plasmids that are compatible with the ColE1 type of plasmids widely used for production of recombinant proteins. One such plasmid, pG-KJE6, carried a newly constructed dnaK-dnaJ-grpE operon under control of the araB promoter-operator (araBp) and the groEL-groES operon under control of the Pzt-1 promoter (Pzt-1p) (Fig. 1). The expression of the DnaK-DnaJ-GrpE chaperone team and the expression of the GroEL-GroES chaperone team from this plasmid were quantitatively manipulated by adding l-arabinose (0.5 to 4 mg/ml) and tetracycline (1 to 10 ng/ml), respectively, at various times. Tetracycline at the low concentrations used had no effect on cell growth. Thus, the cellular level of each chaperone team could be increased separately or the levels of the two teams could be increased simultaneously up to levels which were approximately 10 times the respective wild-type levels. A number of chloramphenicol or kanamycin resistance plasmids that can express either or both of the chaperone teams from diverse promoters were constructed (Table 1), and some of these were used in the present study.
TABLE 1.
Chaperone expression plasmids constructed for this studya
| Plasmid | Promoter (chaperone genes)b | Regulatory gene | Drug resistancec |
|---|---|---|---|
| pKJE5 | trpp (dnaK-dnaJ-grpE) | trpR | Kmr |
| pKJE7 | araBp (dnaK-dnaJ-grpE) | araC | Cmr |
| pKJE8 | araBp (dnaK-dnaJ-grpE) | araC | Kmr |
| pGro6 | trpp (groES-groEL) | trpR | Kmr |
| pGro7 | araBp (groES-groEL) | araC | Cmr |
| pGro11 | Pzt-1p (groES-groEL) | tetR | Cmr |
| pGro12 | araBp (groES-groEL) | araC | Kmr |
| pG-KJE2d | trpp (dnaK-dnaJ-grpE) | trpR | Cmr |
| araBp (groES-groEL) | araC | ||
| pG-KJE3 | araBp (groES-groEL-dnaK-dnaJ-grpE) | araC | Cmr |
| pG-KJE6 | araBp (dnaK-dnaJ-grpE) | araC | Cmr |
| Pzt-1p (groES-groEL) | tetR | ||
| pG-KJE7 | araBp (groES-groEL-dnaK-dnaJ-grpE) | araC | Kmr |
All plasmids contained the replication origin of pACYC184 and genes encoding chaperones under control of the promoters indicated.
The chaperone genes (starting with the promoter-proximal gene) controlled by a given promoter are indicated in parentheses.
Kmr, kanamycin resistance; Cmr, chloramphenicol resistance.
The level of dnaK-dnaJ-grpE expression is low for unknown reasons.
Involvement of the chaperones in expression of Cryj2.
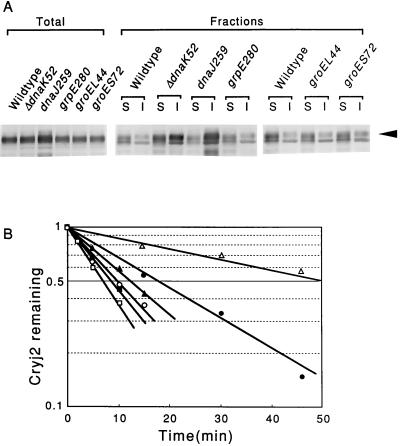
When E. coli cells carrying a Cryj2-expressing plasmid (pKCJ2l) were grown in L broth in the presence of 1 mM IPTG at 30°C, the Cryj2 that was produced was found to be rather unstable and to have a half-life of about 10 min. To examine the possible involvement of chaperones in expression and stability of Cryj2, plasmid pKCJ2l was transformed into a set of isogenic temperature-sensitive mutants, each deficient in an individual chaperone gene. The resulting strains were grown at 30°C (permissive temperature), Cryj2 was induced by adding IPTG, cells were harvested, and the amounts of Cryj2 produced in extracts were determined. When whole-cell proteins were analyzed by SDS-PAGE followed by immunoblotting with anti-Cryj2 serum, the Cryj2 level was markedly higher in the dnaJ or dnaK mutant than in the wild type, whereas the Cryj2 level was significantly lower in the groEL or groES mutant (Fig. 2A).
FIG. 2.
Expression and stability of Cryj2 in the wild type and in isogenic chaperone mutants harboring pKCJ2l. (A) Level of expression and fractionation of Cryj2. Cells were grown to the mid-log phase in L broth at 30°C, and Cryj2 was induced with IPTG (1 mM) for 2 h and analyzed by immunoblotting with whole-cell proteins (Total) or with the soluble (lanes S) or insoluble (lanes I) fractions obtained after centrifugation (Fractions), as described in the text. The arrowhead indicates the position of Cryj2. (B) Stability of Cryj2 in vivo. The fraction of Cryj2 remaining after incubation in the presence of spectinomycin (500 μg/ml) was plotted versus incubation time. Averages of data from three experiments are shown. Symbols: ○, MC4100 (wild type); •, ΔdnaK52 mutant; ▵, dnaJ259 mutant; ▴, grpE280 mutant; □, groEL44 mutant; ▪, groES72 mutant.
Fractionation of cell extracts by centrifugation revealed that there was marked aggregation of Cryj2, particularly in the dnaJ or dnaK mutant (Fig. 2A), indicating that the DnaK, DnaJ, and GrpE chaperones are required for efficient folding of Cryj2 and that marked misfolding occurs if these chaperones are not functional. Consistent with the increased levels of Cryj2 found in the dnaJ and dnaK mutant extracts, the stability of Cryj2 determined in the absence of protein synthesis (when spectinomycin was added) increased five- and twofold in the dnaJ and dnaK mutants, respectively (Fig. 2B). The grpE mutation had little or no effect on stabilization. In contrast, the groEL mutation (and possibly the groES mutation) destabilized Cryj2, suggesting that the GroEL and GroES chaperones also assist folding and tend to stabilize Cryj2 in wild-type E. coli. The latter results also seemed to account for the reduced level of Cryj2 detected in extracts of the groEL or groES mutant (Fig. 2A).
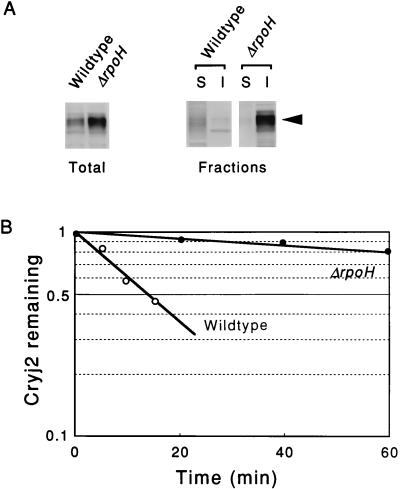
When Cryj2 was produced in NK161, the ΔrpoH mutant which lacks ς32 and grows only at a temperature of 20°C or less due to the absence of (or a severe reduction in the amount of) heat shock proteins, such as chaperones and ATP-dependent proteases (8, 19), strikingly large amounts of Cryj2 were obtained, mostly as insoluble aggregates at 20°C (Fig. 3A). Concomitant with the marked aggregation, drastic stabilization occurred under these conditions (ca. 15-fold increase) (Fig. 3B). These results are consistent with (and provide further support for) the results of the experiments described above performed with individual chaperone mutants. Moreover, a comparison of the stability data (Fig. 2B and 3B) suggested that reduced synthesis of heat shock proteins other than the major chaperones, such as ATP-dependent proteases (Lon, ClpP, FtsH/HflB, and HslVU/ClpQY), probably contributed to the extreme stabilization observed in the ΔrpoH mutant. Which of the heat shock proteins are responsible for the massive aggregation of Cryj2 is a question that is addressed below.
FIG. 3.
Expression and stability of Cryj2 in the wild type and in the isogenic ΔrpoH mutant harboring pKCJ2l. (A) Cells were grown in L broth at 20°C, Cryj2 was induced for 3 h, and cells were analyzed by immunoblotting before (left) and after (right) soluble (lanes S) and insoluble (lanes I) fractions were separated, essentially as described in the legend to Fig. 2. (B) Stability of Cryj2 in vivo, determined as described in the legend to Fig. 2. Averages of data from three experiments are shown. Symbols: ○, MG1655 (wild type); •, NK161 (ΔrpoH).
Coexpression of either chaperone team can stabilize Cryj2 in the wild-type host.
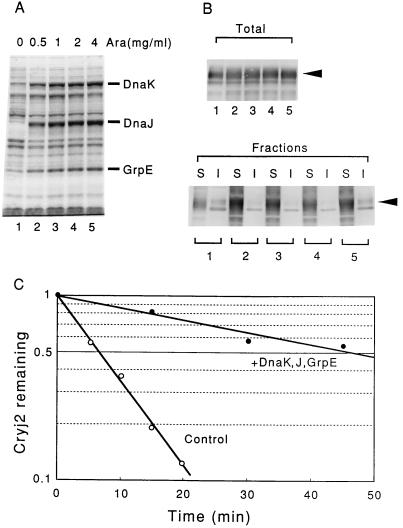
Next we examined the effects of coexpressing chaperones on the synthesis and stability of Cryj2 by using strain MG1655 harboring a pair of compatible plasmids, pKCJ2l (expressing Cryj2) and pG-KJE6 (expressing chaperones), at 30°C. The amounts of the DnaK-DnaJ-GrpE chaperone team increased with increasing concentrations of added inducer (l-arabinose) within the range tested (Fig. 4A), resulting in slight inhibition of cell growth (12 to 25% inhibition in the presence of 2 or 4 mg of arabinose per ml). The amounts of Cryj2 also increased with increasing chaperone coexpression (Fig. 4B). Concomitantly, Cryj2 was markedly stabilized; about fivefold stabilization was observed when a 10-fold excess chaperones was provided compared with the wild-type level in the absence of protein synthesis (Fig. 4C). In contrast to the results obtained with the dnaJ or dnaK mutant (Fig. 2A), in this case stabilization was not accompanied by aggregation, and most of the Cryj2 remained soluble (Fig. 4B).
FIG. 4.
Stabilization of Cryj2 by coexpression of the DnaK-DnaJ-GrpE chaperone team in the wild-type strain. Strain MG1655 harboring pKCJ2l and pG-KJE6 was grown in L broth containing various concentrations of l-arabinose (Ara) at 30°C for three or four generations, and Cryj2 was induced with IPTG (1 mM) for 2 h. (A) Expression of the DnaK-DnaJ-GrpE chaperone team as determined by SDS-PAGE followed by staining with Coomassie brilliant blue. l-Arabinose was added to lanes 1 through 5 as indicated. (B and C) Level of expression and fractionation of Cryj2 (B) and stability of Cryj2 in vivo (C) determined essentially as described in the legend to Fig. 2. Symbols: ○, no arabinose (control); •, 4 mg of l-arabinose per ml (chaperone coexpression). Lanes S, soluble fractions; lanes I, insoluble fractions.
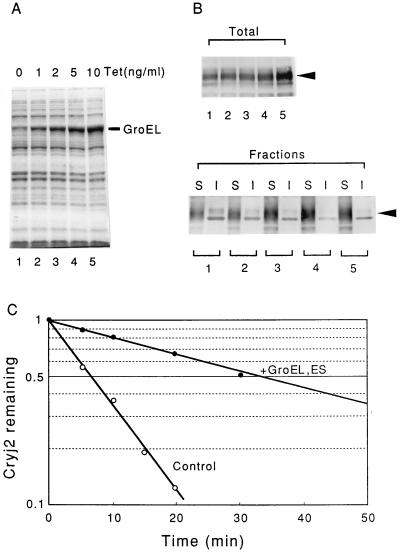
The effects of coexpressing the GroEL and GroES chaperones on Cryj2 were then examined by using the same strain; various concentrations of tetracycline were added to induce synthesis of the GroE chaperones to levels greater than the endogenous wild-type level (Fig. 5A). Again, the amount of Cryj2 produced increased with increasing levels of coexpressed GroE (Fig. 5B). The increase in the Cryj2 level was also accompanied by stabilization; about fourfold stabilization was observed when the GroE level increased 10-fold compared with the wild type, with no effect on cell growth (Fig. 5C). No appreciable aggregation was observed under these conditions (Fig. 5B). Thus, it was evident that excess synthesis of the DnaK-DnaJ-GrpE chaperone team and excess synthesis of the GroEL-GroES chaperone team can stabilize Cryj2 to comparable extents in wild-type bacteria, despite the distinctly different modes of action of these chaperone teams in protein folding. Also, no synergistic effect of coexpression of the DnaK-DnaJ-GrpE and GroEL-GroES chaperone teams was observed under the same conditions (data not shown).
FIG. 5.
Stabilization of Cryj2 by coexpression of the GroEL-GroES chaperone team in the wild-type strain. Strain MG1655 harboring pKCJ2l and pG-KJE6 was grown in L broth containing various concentrations of tetracycline (Tet) at 30°C, and Cryj2 was induced with IPTG. The GroE chaperones induced with tetracycline were monitored (A) and the level of expression and fractionation of Cryj2 (B) and the stability of Cryj2 in vivo (C) were determined essentially as described in the legends to Fig. 2 and 4. Overexpression of GroES was confirmed by SDS-PAGE and immunoblotting (data not shown). Symbols: ○, no tetracycline (control); •, 10 ng of tetracycline per ml (chaperone coexpression). Lanes S, soluble fractions; lanes I, insoluble fractions.
Synergistic roles of the two chaperone teams in preventing aggregation of Cryj2 in the ΔrpoH mutant.
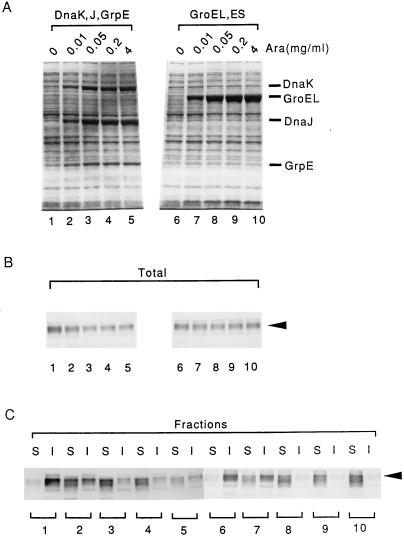
Having shown that a large amount of Cryj2 aggregate is produced in the ΔrpoH mutant (Fig. 3), we wanted to determine whether coexpression of either or both of the chaperone teams can prevent aggregation. In initial experiments, pKJE7 (expressing DnaK-DnaJ-GrpE) or pGro7 (expressing GroEL-GroES) instead of pG-KJE6 was used to avoid possible complications arising from basal expression of chaperones in the absence of inducers. When a pair of NK161 (ΔrpoH)-derived strains carrying pKCJ2l (expressing Cryj2) and pKJE7 or pGro7 were grown at 20°C and both Cryj2 and the chaperones were induced, Cryj2 was partially or fully recovered in supernatants (Fig. 6C). However, the effects observed differed markedly depending on which chaperones were employed. When DnaK, DnaJ, and GrpE were coexpressed, a modest excess (two to three times the wild-type level) was sufficient to prevent aggregation, and a greater excess resulted in a marked reduction in the amount of Cryj2 produced (Fig. 6B and C). In contrast, a greater excess (5 to 10 times the wild-type level) of the GroEL and GroES chaperones was required to prevent aggregation, and an even greater excess of these chaperones did not reduce the Cryj2 level.
FIG. 6.
Effect of chaperone coexpression on disaggregating Cryj2 in the ΔrpoH mutant at 20°C. A pair of NK161 (ΔrpoH) strains harboring pKCJ2l and pKJE7 or pGro7 were grown at 20°C, each chaperone team was induced with arabinose (Ara), and Cryj2 was induced with IPTG for 3 h. The levels of expression of chaperones (A) and the levels of expression of Cryj2 before (B) and after (C) fractionation were determined essentially as described in the legends to Fig. 2 and 4. Lanes S, soluble fractions; lanes I, insoluble fractions.
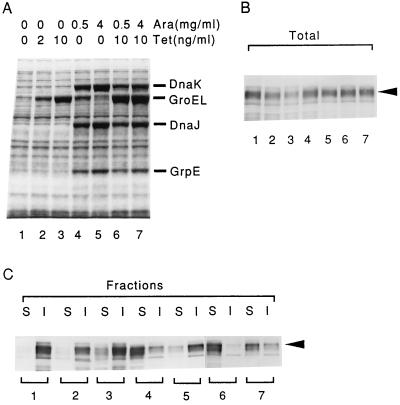
We then examined the effects of the chaperones at 30°C in the same mutant harboring pG-KJE6, which can express both chaperone teams, since the mutant containing this plasmid could grow at 30°C in the absence of arabinose or tetracycline. Apparently, the slight but significant inducer-independent expression of GroEL-GroES in this organism was sufficient to support growth at 30°C, which is consistent with previous findings obtained with other plasmids (8). In contrast to the results obtained at 20°C (Fig. 6C), an excess of GroEL-GroES alone (Fig. 7, lanes 2 and 3) or of DnaK-DnaJ-GrpE alone (lanes 5) did not prevent Cryj2 aggregation at 30°C. However, a modest excess of both chaperone teams was quite effective in preventing aggregation (Fig. 7, lanes 4 and 6) (see below for a discussion of the fortuitous coexpression of GroEL in lanes 4), although a greater excess of DnaK-DnaJ-GrpE markedly reduced the amounts of Cryj2 obtained (lanes 7). Taken together, these results indicate that the two chaperone teams play synergistic roles in preventing protein aggregation at 30°C in the ΔrpoH mutant, which is consistent with the previous results reported for the bulk E. coli proteins (3). In addition, both the level of expression and the ratio of the two chaperone teams appeared to be critical in obtaining the maximum yields of soluble Cryj2 under these conditions.
FIG. 7.
Synergistic effect of chaperones on preventing Cryj2 aggregation in the ΔrpoH mutant at 30°C. Strain NK161 (ΔrpoH) harboring pKCJ2l and pG-KJE6 was grown at 30°C, either or both chaperone teams were induced, and Cryj2 was induced for 2 h. The levels of chaperones expressed (A) and the levels of Cryj2 before (B) and after (C) fractionation were determined as described in the legends to Fig. 2 and 4. Lanes S, soluble fractions; lanes I, insoluble fractions. Ara, arabinose; Tet, tetracycline.
DISCUSSION
We constructed a series of plasmids that should be useful for systematic assessment of the effects of chaperone coexpression on the production of recombinant proteins in E. coli. One of these plasmids was the pACYC184-based plasmid pG-KJE6, which permits induction of the DnaK-DnaJ-GrpE and GroEL-GroES chaperone teams either simultaneously or separately from each other and from target recombinant proteins both in timing and expression levels. With this plasmid, it was possible to supply at least a 10-fold excess of either chaperone team compared with the normal wild-type level without appreciably affecting cell growth at physiological temperatures (20 to 42°C). We noted a minor difficulty in selectively coexpressing relatively small amounts of the DnaK-DnaJ-GrpE chaperone team with the plasmid used (pG-KJE6), because low concentrations (ca. 0.5 mg/ml) of l-arabinose somehow also induced modest amounts of GroEL-GroES for unknown reasons (Fig. 7, lanes 4); however, this problem could apparently be overcome by inserting an additional transcriptional terminator downstream of grpE (12).
Mutants of E. coli deficient in ATP-dependent proteases, such as Lon and Clp, have often been used to improve production of unstable recombinant proteins, such as Cryj2. However, this approach has had only limited success, since nonnative proteins that escape degradation often fail to refold properly and result in misfolding and aggregation. This is in part due to the fact that such protease mutants accumulate endogenous malfolded protein substrates that tend to reduce free chaperone pools that are normally available for assisting folding of recombinant proteins. Indeed, Cryj2 is highly stabilized in double mutants deficient in Lon and ClpP or in triple mutants deficient in Lon, ClpP, and HslVU, but they form mostly insoluble aggregates (12). In contrast, excess coexpression of either chaperone team from pG-KJE6 successfully stabilized Cryj2 without causing appreciable aggregation (Fig. 4 and 5), suggesting that high levels of chaperones can protect nascent Cryj2 polypeptides from proteolytic attack and facilitate production of native or quasinative protein.
It has been thought that the DnaK-DnaJ-GrpE chaperone team maintains nascent or other preexisting proteins in unfolded states, while the GroEL-GroES chaperone team can interact with partially folded polypeptides and assist in additional folding (5). The apparently similar effects of the two chaperone teams studied on Cryj2 stability observed in the wild type (Fig. 4 and 5) may indicate that these chaperone teams have synergistic and partially compensatory roles in assisting protein folding in such a way that an excess of one chaperone team can effectively reduce the requirement for the other and vice versa. On the other hand, the study performed with individual chaperone mutants showed that the dnaK and dnaJ mutations can stabilize Cryj2 in largely aggregated forms, while the groEL and groES mutations can destabilize the same protein with relatively little aggregation (Fig. 2). Furthermore, the ΔrpoH mutation, which should drastically reduce the levels of cytoplasmic chaperones and ATP-dependent proteases, caused drastic stabilization and aggregation of Cryj2 (Fig. 3).
Thus, the ΔrpoH mutant provided a unique opportunity to study the effects of both chaperones and proteases on protein folding, stability, and aggregation. We found that modest coexpression of either chaperone team alone in this mutant was effective in preventing aggregation of Cryj2 at a low temperature (20°C), at which relatively low levels of chaperones are needed for proper folding of proteins (Fig. 6). This agrees well with the previous observations that coexpression of GroE chaperones alone prevents the bulk of protein aggregation under similar conditions (3); little or no DnaK, DnaJ, and GrpE but significant amounts of GroE proteins are present in such mutants (8, 19). At a higher temperature (30°C), at which larger amounts of chaperones are normally required, however, both chaperone teams were needed for effective protection against protein aggregation (Fig. 7C). At both temperatures tested, a great excess of the DnaK-DnaJ-GrpE chaperone team was inhibitory since it resulted in reduced recovery of Cryj2 (Fig. 6 and 7), possibly because of enhanced proteolysis. In any event, the ΔrpoH mutant fortified by controlled coexpression of either or both chaperone teams may provide a useful system for future attempts to improve protein folding and production in E. coli or other related bacteria.
In conclusion, chaperone expression plasmids, such as those described here (Table 1), should help in assessing the effects of coexpression of chaperones on the synthesis, stability, and disaggregation of recombinant proteins in E. coli. In addition to Cryj2, which was examined in this study in some detail, we observed marked disaggregation effects of DnaK-DnaJ-GrpE (but relatively little effect of GroEL-GroES) on human prourokinase and ORP150 (an oxygen-regulated 150-kDa protein), which otherwise occur in large aggregates (12). The importance of systematic assessment of the effects of chaperone coexpression on protein production not only in wild-type bacteria but also in various mutant hosts, including the ΔrpoH strain, cannot be overemphasized.
ACKNOWLEDGMENTS
We are grateful to M. Kurimoto and Y. Taniguchi of Hayashibara Biochemical Laboratories (Okayama, Japan) for providing the Cryj2 expression plasmid and antibody, to H. Bujard (University of Heidelberg) for providing plasmids, and to M. Nakayama, M. Ueda, and H. Kanazawa for providing technical assistance.
REFERENCES
- 1.Casadaban M. Transposition and fusion of the lac gene to selected promoters in E. coli using bacteriophage lambda and mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 2.Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Morimoto R I, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- 3.Gragerov A, Nudler E, Komissarova N, Gaitanaris G A, Gottesman M E, Nikiforov V. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: AMS Press; 1996. pp. 1382–1399. [Google Scholar]
- 5.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 6.Ishiai M, Wada C, Kawasaki Y, Yura T. Mini-F plasmid mutants able to replicate in Escherichia coli deficient in the DnaJ heat shock protein. J Bacteriol. 1992;174:5597–5603. doi: 10.1128/jb.174.17.5597-5603.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanemori, M. Unpublished results.
- 8.Kusukawa N, Yura T. Heat shock protein GroE of Escherichia coli: key protective roles against thermal stress. Genes Dev. 1988;2:874–882. doi: 10.1101/gad.2.7.874. [DOI] [PubMed] [Google Scholar]
- 9.Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K. Effects of mutations in heat shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 1989;8:3517–3521. doi: 10.1002/j.1460-2075.1989.tb08517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagai H, Yuzawa H, Kanemori M, Yura T. A distinct segment of the ς32 polypeptide is involved in DnaK-mediated negative control of the heat shock response in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:10280–10284. doi: 10.1073/pnas.91.22.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Namba M, Kurose M, Torigoe K, Hino K, Taniguchi Y, Fukuda S, Usui M, Kurimoto M. Molecular cloning of the second major allergen, CryjII, from Japanese cedar pollen. FEBS Lett. 1994;353:124–128. doi: 10.1016/0014-5793(94)01022-6. [DOI] [PubMed] [Google Scholar]
- 12.Nishihara, K. Unpublished results.
- 13.Ohtsuki T, Taniguchi Y, Kohno K, Fukuda S, Usui M, Kurimoto M. Cryj2, a major allergen of Japanese cedar pollen, shows polymethylgalacturonase activity. Allergy. 1995;50:483–488. doi: 10.1111/j.1398-9995.1995.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 14.Pelez-Pelez J, Gutierrez J. An arabinose-inducible expression vector, pAR3, compatible with ColE1-derived plasmids. Gene. 1995;158:141–142. doi: 10.1016/0378-1119(95)00127-r. [DOI] [PubMed] [Google Scholar]
- 15.Sakaguchi M, Inouye S, Taniai M, Ando S, Usui M, Matuhasi T. Identification of the second major allergen of Japanese cedar pollen. Allergy. 1990;45:309–312. doi: 10.1111/j.1398-9995.1990.tb00501.x. [DOI] [PubMed] [Google Scholar]
- 16.Wall J G, Pluckthun A. Effects of overexpressing folding modulators on the in vivo folding of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:507–516. doi: 10.1016/0958-1669(95)80084-0. [DOI] [PubMed] [Google Scholar]
- 17.Yano R, Nagai H, Shiba K, Yura T. A mutation that enhances synthesis of ς32 and suppresses temperature-sensitive growth of the rpoH15 mutant of Escherichia coli. J Bacteriol. 1990;172:2124–2130. doi: 10.1128/jb.172.4.2124-2130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y-N, Kusukawa N, Erickson J W, Gross C A, Yura T. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor ς32. J Bacteriol. 1988;170:3640–3649. doi: 10.1128/jb.170.8.3640-3649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]