Abstract
The aim of this study was to establish the therapeutic relevance of the CD33D2 isoform by developing novel antibodies targeting the IgC domain of CD33. Two novel IgC-targeting antibodies, HL2541 and 5C11-2, were developed, and CD33 isoforms were assessed using multiple assays in cells overexpressing either CD33FL or CD33D2 isoforms, unmodified acute myeloid leukemia (AML) cell lines and primary AML specimens representing different genotypes for the CD33 splicing single nucleotide polymorphism. CD33D2 was recognized on cells overexpressing CD33D2 and unmodified AML cell lines; however, minimal/no cell surface detection of CD33D2 was observed in primary AML specimens. Both isoforms were detected intracellularly using novel antibodies. Minimal cell surface expression of CD33D2 on primary AML/progenitor cells warrants further studies on anti-CD33D2 immunotherapeutics.
Keywords: acute myeloid leukemia, antibodies, CD33, immunotarget, immunotherapy
Plain language summary
CD33 is a small protein that is often present on tumor cells in acute myeloid leukemia and is therefore the focus of some research to develop targeted treatments for the disease. One such CD33-targeted treatment – gemtuzumab ozogamicin has shown promise in helping acute myeloid leukemia patients. However, some patients have a variation of this small protein, resulting in a shorter form called CD33D2, which makes the aforementioned treatment ineffective. In this study, the authors aimed to create two antibodies that would bind to both CD33 and CD33D2 so that we can try to develop a treatment that is effective for all patients with acute myeloid leukemia.
CD33 is a member of the sialic acid-binding immunoglobulin-like lectin family with restricted expression on myeloid cells [1,2]. Structurally, CD33 contains three domains: two Ig-like extracellular domains, the IgV domain and the IgC domain, and an intracellular immunoreceptor tyrosine-based inhibition motif domain [3]. In addition, CD33 has endocytic properties and internalizes in an immunoreceptor tyrosine-based inhibition motif-dependent manner when engaged by an appropriate sialylated ligand or antibody [4,5]. Recognition of this internalization mechanism and high expression on acute myeloid leukemia (AML) blasts in 85–90% of patients led to the development of gemtuzumab ozogamicin (GO), a CD33-directed immunoconjugate composed of a humanized IgV-targeting CD33 antibody, hP67.6, covalently linked to a cytotoxin, calicheamicin [6,7]. GO received accelerated approval as the first US FDA-approved antibody-drug conjugate in 2000; however, it was withdrawn from the market in 2010 because of high levels of mortality and no observed survival benefit in a post-approval Phase III study [8,9]. Subsequent follow-up randomized studies later demonstrated benefit with the addition of GO to AML treatment regimens, resulting in its reapproval in 2017 for treatment of de novo and relapsed/refractory AML [10,11]. The promising results from GO have paved the way for the development of several new CD33-directed immunotherapies, including monoclonal antibodies, novel antibody-drug conjugates, radio-immunconjugates, bi- and tri-specific antibodies for immune effector engagement and chimeric antigen receptor T-cell therapy, with efforts to develop more CD33-directed therapeutics underway [12–19].
The authors' group has had a long-standing interest in genetic and structural variation of CD33 and has established genetic variation in CD33 as a key predictive factor for leukemic cell surface CD33 levels and response to GO [20–25]. One of the most significant findings is a splicing single nucleotide polymorphism within CD33, rs12459419 (C >T; A14V), located four base pairs upstream from the junction of intron 1 and exon 2. The variant T allele of rs12459419 results in skipping of exon 2 and thus loss of the encoded IgV domain, leading to the production of the smaller isoform of CD33 (CD33D2) [26]. GO, which is the only currently approved CD33-directed therapy for AML, and P67.6, which is the antibody used for clinical immunophenotyping, recognize the IgV domain. Consistent with this fact, the authors' results from a randomized study comparing patients treated with standard chemotherapy with GO (GO arm) or without GO (No-GO arm) showed that AML patients with the variant T allele (CT and TT genotype groups) had low CD33 expression and derived no benefit from the inclusion of GO in their treatment regimen [26]. Of note are the heterozygous (CT) patients who, despite having intermediate levels of full-length CD33 (CD33FL) on the cell surface, did not show any benefit from the addition of GO to standard chemotherapy, suggesting that CD33D2 potentially has an impact on CD33FL function or expression. As expected, homozygous dominant (CC) patients showed significantly higher CD33 cell surface expression and significant improvement in outcome in the GO arm, resulting in approximately 50% lower risk of relapse compared with the No-GO arm [26].
Given that all currently available CD33-directed therapies and immunophenotyping diagnostics are based on recognition of the IgV domain and the observed genotype-dependent outcome using GO, there is a need to develop targeted therapeutics against CD33D2 as an alternative strategy for heterozygous and homozygous recessive (TT) patients. One of the challenges in pursuing studies to understand the biology and therapeutic potential of CD33D2 isoform is the lack of an antibody that specifically recognizes the IgC domain, which is common between the CD33FL and CD33D2 isoforms and is the only extracellular portion of CD33D2. Currently, HIM3-4 is the only available antibody known to recognize CD33D2; however, information on the epitope targeted by this antibody is lacking, and HIM3-4 has been shown to have compromised specificity in regards to CD33D2. This roadblock has limited the understanding regarding cell surface expression of CD33D2, its cellular localization patterns, its potential as a novel drug target and the therapeutic implications when it co-exists with CD33FL in heterozygous patients. In this study, the authors report the development and characterization of two novel CD33 antibodies directed to the IgC domain and use of these antibodies to evaluate CD33D2 expression in multiple AML cell lines and diagnostic AML bone marrow specimens of different rs12459419 genotypes.
Methods
Cell lines, antibodies & other reagents
The CD33- murine pro-B cell Ba/F3 cell line was cultured in Roswell Park Memorial Institute (RPMI-1640; Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS) (ATCC, VA, USA), 1% L-glutamine, 1% penicillin-streptomycin (Invitrogen, CA, USA) and 10 ng/ml of recombinant mouse IL-3 (Shenandoah, PA, USA). The human AML cell lines K562, Molm13, and MV4;11 (ATCC) were cultured in Iscove's modified Dulbecco's medium (IMDM; Gibco) or RPMI-1640 supplemented with 10% FBS. THP-1 (ATCC) cells were cultured in RPMI-1640 supplemented with 10% FBS and 0.05 mM ßME (Sigma-Aldrich, MO, USA). HL-60 and Kasumi-1 (ATCC) cells were cultured in IMDM or RPMI-1640 supplemented with 20% FBS. HEK293T17 and HEK293T cells (ATCC, VA, USA) used to generate viral titers for transduction were cultured in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% FBS and 1% penicillin-streptomycin. All antibodies used herein are listed in the Supplementary data. All other reagents used were molecular biology grade of the highest purity.
Generation of Ba/F3 cell lines stably overexpressing CD33 isoforms
Low passage HEK293T17 (ATCC) cells were thawed and grown to approximately 70% confluency. Afterward, cells were transfected with the following plasmids: pMXs-CD33FL, pMXs-CD33D2, pMXs-CD33FLGFP and pMXs-CD33D2GFP as well as the pEcoPak murine ecotropic retroviral helper plasmid (gifted by Julia Maxson, Oregon Health & Science University, OR, USA). Cells were returned to the incubator for 48 h, after which supernatants containing viral particles were collected and passed through 0.45-μm sterile filters (Corning, NY, USA). The filtered supernatants were then used for transduction at a ratio of 1 ml to 2 × 106 Ba/F3 cells to create the following CD33FL or CD33D2 overexpressing Ba/F3 cell lines: Ba/F3-CD33FL (pMXs-CD33FL), Ba/F3-CD33D2 (pMXs-CD33D2), Ba/F3-CD33FLGFP (pMXs-CD33FLGFP), Ba/F3-CD33D2GFP (pMXs-CD33D2GFP).
Immunoblotting
Cell pellets containing at least 5 × 106 cells (cell lines) or at least 7 × 105 cells (primary samples) were washed in triplicate in PBS and lysed in RIPA lysis buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS; Pierce Biotechnology Inc., IL, USA) supplemented with protease inhibitor cocktail (Roche, Basel, Switzerland) on ice according to the manufacturer's instructions. Lysates were cleared by centrifuging at approximately 18,000 × g for 15 min. A Bradford assay was used to estimate protein concentrations using diluted lysates. Where indicated, PNGase F (New England BioLabs, MA, USA) treatment was performed according to the manufacturer's instructions. Untreated and/or treated samples dissolved in NuPAGE LDS buffer were heated at 70°C for 10 min prior to separation by NuPAGE 10% Bis-Tris gel (Thermo Fisher Scientific, MA, USA). Proteins were transferred to polyvinylidene fluoride membranes using an iBlot dry blotting system (Invitrogen). Runtimes and voltages recommended by the manufacturers were used. Membranes were blocked in 4% bovine serum albumin (BSA) in 0.2% PBS-Tween and probed with specified antibodies overnight at 4°C. Stained membranes were washed in triplicate in 0.2% PBS-Tween and probed with IRDye 800CW goat anti-mouse and IRDye 680RD or IRDye 800 goat anti-rabbit fluorescent secondary antibodies (LI-COR Biosciences, NE, USA). Imaging was performed using the Odyssey CLx system (LI-COR Biosciences).
Flow cytometry
For cell staining, approximately 0.5–1 × 106 cells were harvested from culture and washed twice with staining buffer (2% PBS/FBS containing 0.05% NaN3, (Corning). Cells were then stained with LIVE/DEAD NIR viability dye (Thermo Fischer Scientific, MA, USA) for 30 min at room temperature (RT). After washing with staining buffer, cells were blocked for 5 min at RT using Human TruStain FcX and True-Stain Monocyte Blocker (BioLegend, CA, USA) and stained with the respective antibodies for 30 min at RT. Cells were then washed with the staining buffer and fixed using the Foxp3 transcription factor staining buffer set (eBioscience, CA, USA) according to the manufacturer's protocol before flow cytometry data acquisition and analysis.
Cryopreserved primary AML patient samples were first rapidly thawed at 37°C and then stained as described earlier. Data acquisition was performed using the Cytek Aurora™ (Cytek Biosciences, CA, USA) platform or a FACSCalibur instrument (BD Biosciences, NJ, USA), and data analysis was done using FCS Express 7 (De Novo Software, CA, USA) or the WinList (Verity Software House, ME, USA) software. Median fluorescence intensity (MFI) values were obtained using the aforementioned software. MFI ratios were calculated as the ratio between sample MFI/control MFI (indicated in bold). Percent positivity was calculated based on marker positivity, as shown in the gating strategies (Supplementary Figures 1A & 3A). Average percent positivity is reported as the average value ± standard error of the mean.
Immunofluorescence microscopy
Approximately 1 × 106 cells were harvested from culture and washed with 1% BSA/PBS (Fischer Bioreagents, PA, USA), fixed using a 3.7% formaldehyde solution and blocked with 1% BSA/PBS for 30 min at RT, followed by staining with the specified antibody overnight at 4°C. Afterward, the cells were washed with 1% BSA/PBS and stained with Alexa Fluor® 647-conjugated goat anti-mouse IgG H&L (Thermo Fischer Scientific) for 1 h at RT. Following secondary antibody incubation, cells were washed and incubated with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 5 min at RT in the dark, followed by a final wash using 1x PBS. Samples were then placed on poly-L-lysine-coated slides, mounted with coverslips using VECTASHIELD antifade mounting medium (Vector Laboratories, CA, USA) and examined using the Olympus IX81-DSU motorized spinning disk confocal microscope (Olympus, PS, USA). All images acquired represent several Z-stack optical sections obtained through a 20× objective at 0.02- to 0.03-mm intervals. The ImageJ platform (NIH, MD, USA) was used for analysis, and the RGB Profiler plug-in therein was used to observe the co-localization of color intensities. The antibody dilutions and amounts used in these experiments can be found in Supplementary Tables 1–3.
Primary patient specimens
Primary leukemic cells from patients of different rs12459419 genotypes enrolled in the AAML0531 clinical trial were used for this study. The details of this study, including study design, treatment regimen and clinical outcome, are published elsewhere [27]. Bone marrow aspirates were extracted at diagnosis, and peripheral blood mononuclear cells were isolated from the samples via Ficoll-Paque (Thermo Fischer Scientific). density gradient separation. Samples were subsequently cryopreserved in liquid nitrogen until use.
Regenerating bone marrow specimens from individuals with a history of leukemia were also obtained and used to profile regenerating monocytes for this study. The specimens were extracted, isolated and preserved as described earlier. All subjects provided written informed consent for the collection and use of their specimens for research purposes under protocols approved by the institutional review boards of all participating institutions, and the Children's Oncology Group Myeloid Disease Biology Committee approved this research.
Results
Novel IgC-specific CD33 antibodies recognize CD33D2 & not CD33FL on cell surface
Ba/F3 cells stably expressing CD33FL, CD33D2 and their respective GFP fusion proteins were utilized as a system to test the authors' new antibodies. To confirm the expression of CD33 in the correct confirmation, the authors performed a screen using flow cytometry for CD33 and GFP where applicable. As expected, Ba/F3-naive cells showed no GFP fluorescence signal, whereas Ba/F3-CD33FLGFP and Ba/F3-CD33D2GFP-transduced cells were positive for GFP, with MFI ratios >1000 each, confirming CD33 expression (Figure 1A). When stained with the IgV domain-directed P67.6 antibody, only Ba/F3-CD33FLGFP showed a positive signal, with an MFI ratio of 6.8, whereas Ba/F3-naive and Ba/F3-CD33D2GFP were negative, with MFI ratios of 1.0 each (Figure 1B). Screening of Ba/F3 cell lines engineered to express CD33FL and CD33D2 without GFP produced similar results, indicating no alteration in protein recognition resulting from overexpression of CD33 as a GFP fusion protein (Supplementary Figure 1).
Figure 1. . Generation and confirmation of IgC-specific CD33 antibody.
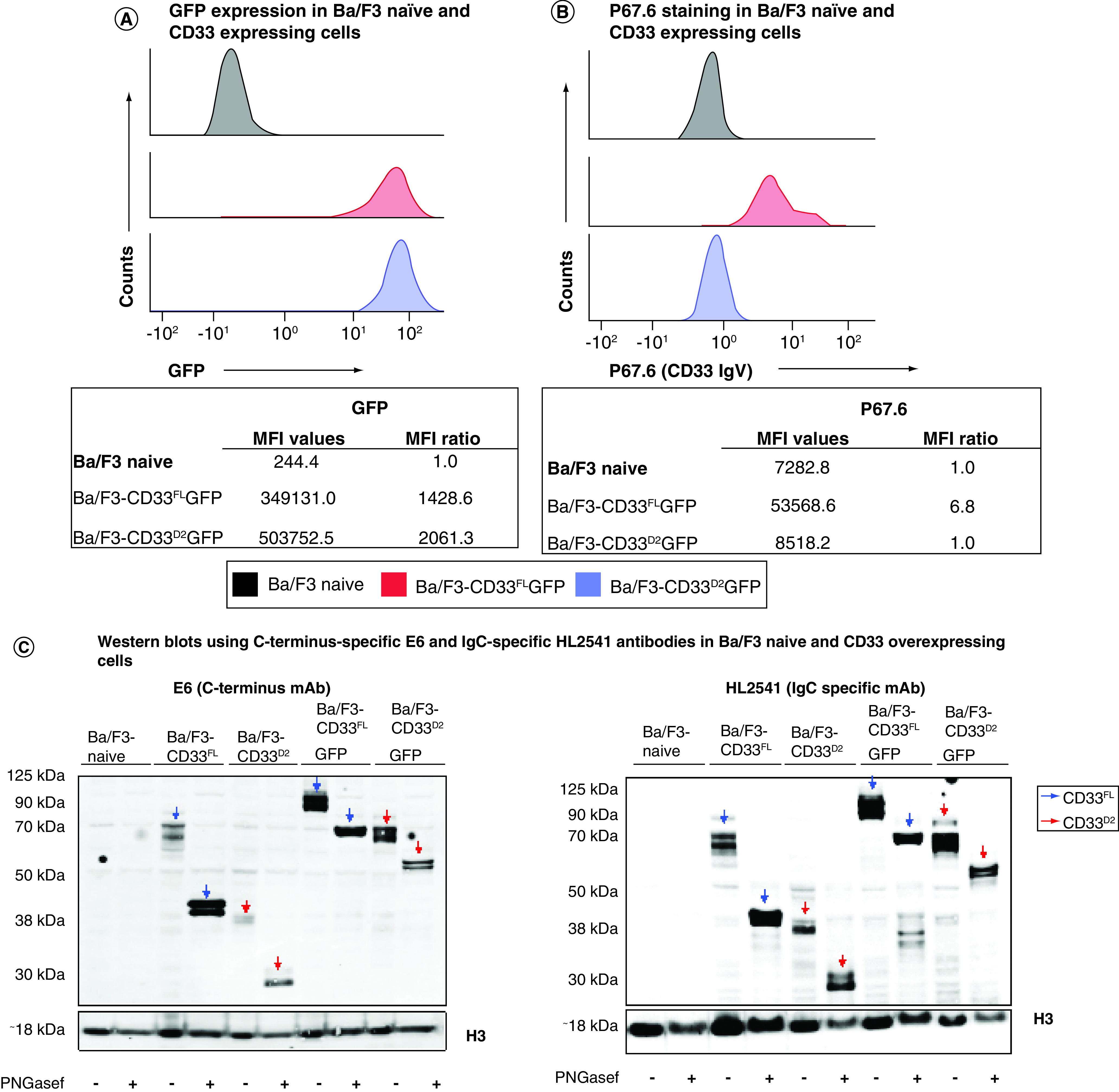
(A) Flow cytometry screen for GFP expression on naive and engineered Ba/F3 cells. (B) Flow cytometry screen for CD33FL by P67.6-PE (500 ng) on engineered Ba/F3 cells. (C) Recognition of CD33 is confirmed by western blotting analysis of engineered BaF3 cells using anti-CD33 C-terminus mAb E6 (ITIM-directed; 1:100), IgC-specific HL2541 (1:50) and anti-H3 antibody (1:1000) as a control. (D) Recognition of CD33 in flow cytometry assays using 5C11-2-PE (500 ng) on engineered Ba/F3 cells. (E) Recognition of CD33 in flow cytometry assays using HL2541-PE mAbs (500 ng) on engineered Ba/F3 cells. (F) Recognition of CD33 in confocal microscopic immunofluorescence assays using 5C11-2 and HL2541 mAbs (2 μg each) and engineered Ba/F3 cells. All images were acquired using the Olympus IX81-DSU motorized spinning disk confocal microscope and represent several Z-stack optical sections obtained through a ×20 objective at 0.02- to 0.03-mm intervals. All images were analyzed using the ImageJ platform. Flow cytometry data were collected on the Cytek Aurora platform and analyzed using FCS Express. MFI ratios were calculated as follows: (sample MFI/control MFI [indicated in bold]). All figures are representative of N ≥3 experiments.
ITIM: Immunoreceptor tyrosine-based inhibition motif; mAb: Monoclonal antibody; MFI: Median fluorescence intensity.
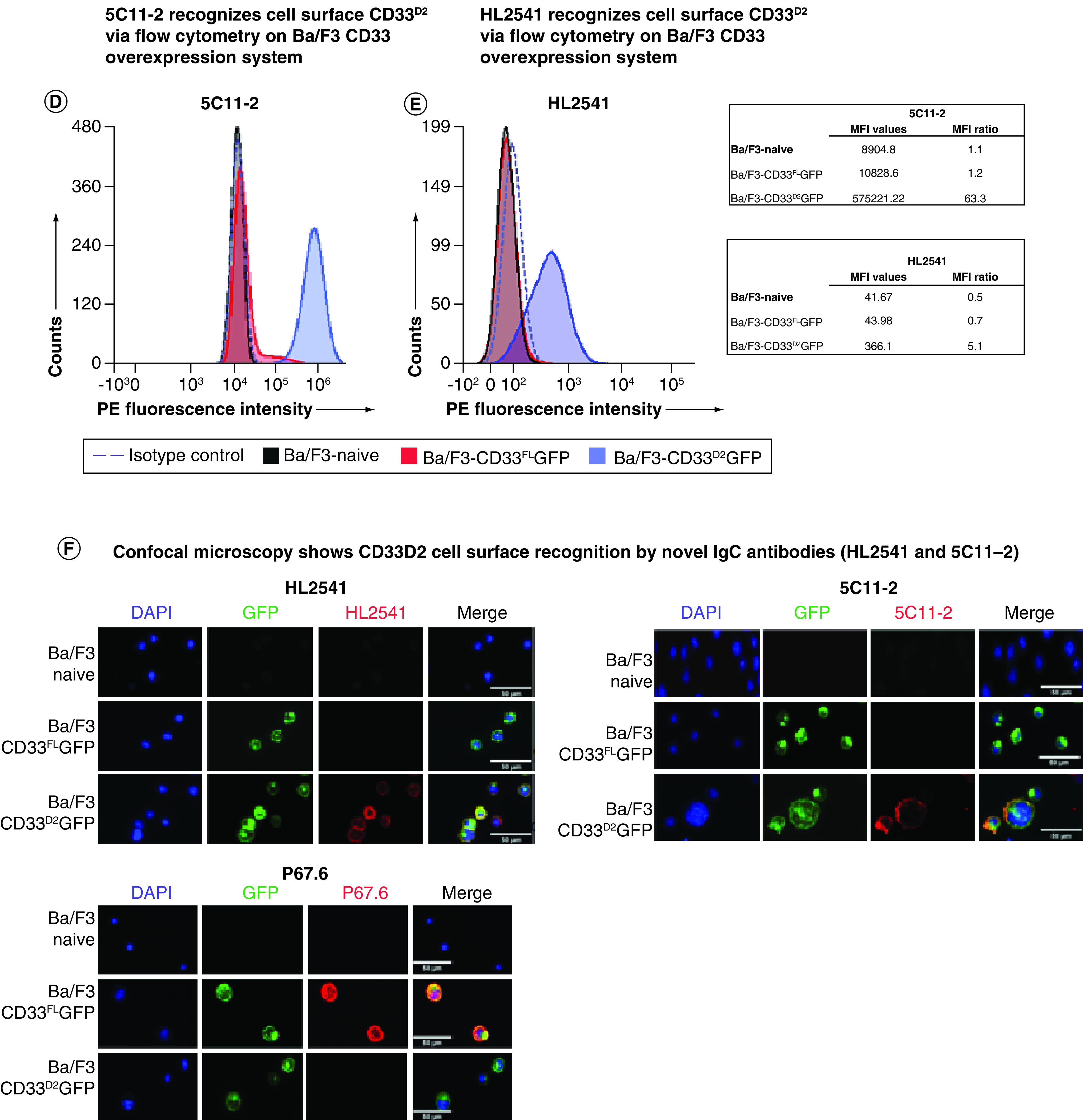
Subsequently, the two most promising hybridoma clonal populations, selected based on an initial screening of supernatants from the hybridoma using the same CD33 overexpression system (data not shown), were used to mass produce the new IgC-directed antibodies HL2541 and 5C11-2. The epitopes targeted by both antibodies are displayed in Supplementary Figure 2. Western blotting using Ba/F3 cell lines expressing either CD33FL or CD33D2 isoform confirmed recognition of both isoforms by HL2541 and 5C11-2. These results were concordant with the corresponding results using commercially available E6, an immunoreceptor tyrosine-based inhibition motif-directed anti-CD33 monoclonal antibody that recognizes both isoforms. Figure 1C shows protein bands corresponding to approximately 67 kDa and approximately 40–42 kDa for the glycosylated and unglycosylated forms of CD33FL and approximately 38 kDa and approximately 33 kDa for the glycosylated and unglycosylated forms of CD33D2 using the E6 and HL2541 monoclonal antibodies. CD33FLGFP and CD33D2GFP fusion proteins were similarly recognized with a corresponding increase of approximately 27 kDa, as expected (Figure 1C). Similar results were seen with 5C11-2, though some secondary bands were observed (Supplementary Figure 3).
Interestingly, in flow cytometry assays, both HL2541 and 5C11-2 recognized CD33D2 on the surface of Ba/F3-CD33D2GFP cells, with MFI ratios of 5.1 and 63.3 for each antibody, respectively, but displayed no recognition of CD33FL on the surface of Ba/F3-CD33FLGFP cells (Figure 1D & E). These results were confirmed by confocal microscopy, where HL2541 and 5C11-2 recognized CD33D2 only on the surface of Ba/F3-CD33D2GFP cells, whereas P67.6 recognized CD33FL only on the surface of Ba/F3-CD33FLGFP cells (Figure 1F). Co-localization of GFP and antibody signals was also confirmed using the RGB Profiler module in ImageJ (Supplementary Figure 4). Altogether, these results confirm suitability and specificity of 5C11-2 and HL2541 as IgC-directed novel CD33 monoclonal antibodies for detection of CD33D2 and CD33FL in western blotting and CD33D2 in immunofluorescence assays.
Evaluation of new antibodies for the screening of CD33FL & CD33D2 isoforms in AML cell lines
Endogenous CD33 cell surface expression was assessed in AML cell lines representative of different rs12459419 genotypes, including Molm-13 (CC), HL-60 (CC), MV4;11 (CT) and K562 (TT), using P67.6 and the authors' newly developed IgC domain-directed antibodies HL2541 and 5C11-2 (Figure 2A). As anticipated, P67.6 revealed higher cell surface expression of CD33FL in homozygous dominant (Molm-13: 99.2% positivity, MFI ratio = 17.6; HL-60: 94.5% positivity, MFI ratio = 8.4) and heterozygous (MV4;11: 98.2% positivity, MFI ratio = 9.4) cell lines in comparison to the homozygous recessive cell line (K562: 2.10% positivity, MFI ratio = 1.6). When staining with 5C11-2, CD33D2 was not recognized on the cell surface of any AML cell line irrespective of rs12459419 genotype, as all cell lines showed <1.0% positivity, with MFI ratios ≤1.3. However, when staining with HL2541, which is targeted to a different epitope within the IgC domain, a positive shift was observed for the MV4;11 (17.0% positivity, MFI ratio = 2.3) and K562 (94.4% positivity, MFI ratio = 68.9) cell lines. Percent positivity and MFI ratios for samples by genotype can be found in Tables 2 & 3, respectively.
Figure 2. . CD33 screening in AML cell lines.
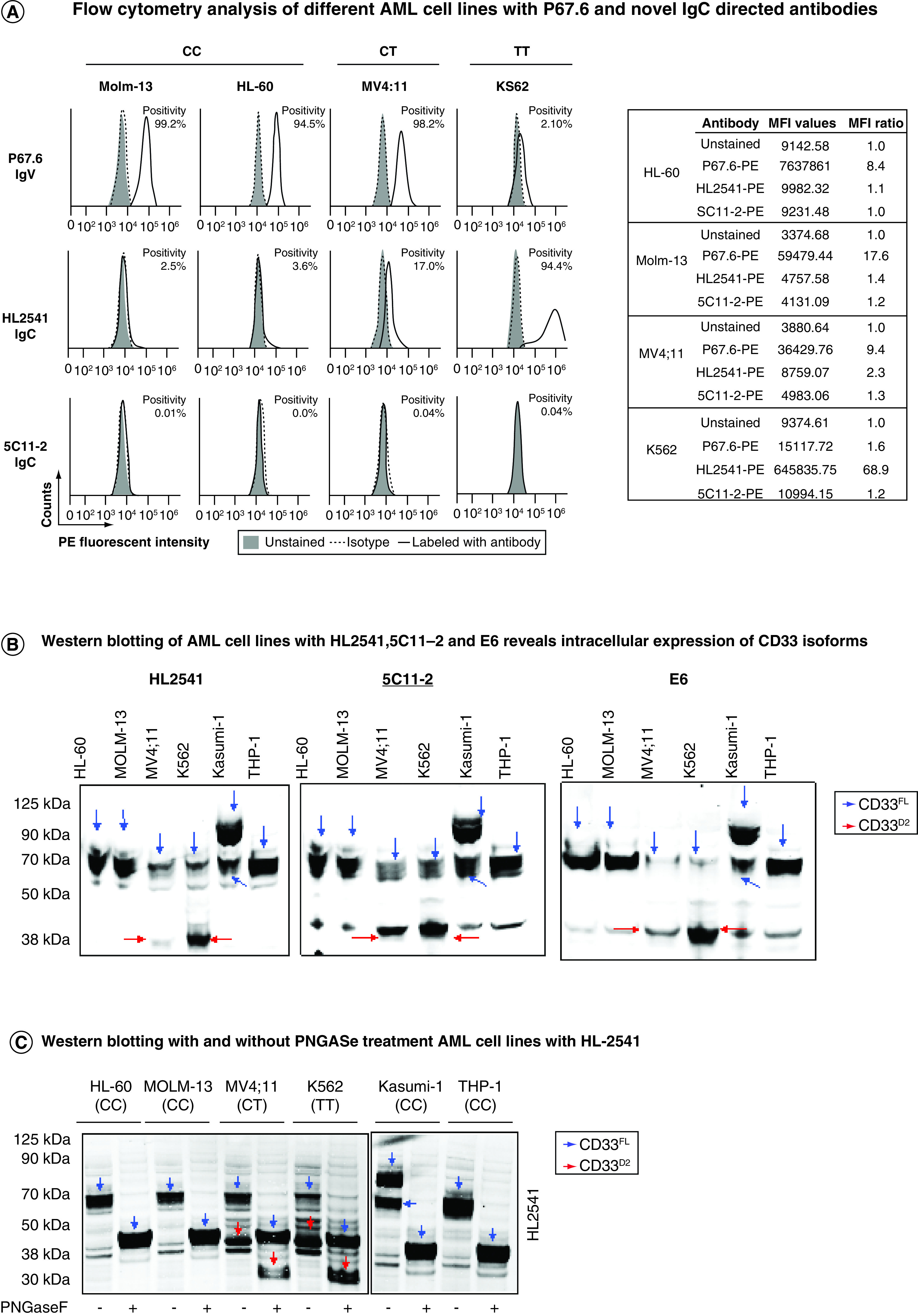
(A) AML cell lines of different rs12459419 genotypes stained with P67.6-PE, HL2541-PE or 5C11-2-PE antibodies (500 ng each). (B & C) Western blotting of AML cell lines using anti-CD33 C-terminus mAb E6 (1:100) and IgC-specific mAbs HL2541 (1:50) and 5C11-2 (1:250). (D) Immunophenotyping of AML cell lines revealed CD34, CD14 and CD45 cell surface expression. Flow cytometry data were collected using the Cytek Aurora platform and analyzed using FCS Express. MFI ratios were calculated as follows: (sample MFI/control MFI (indicated in bold)). Percent positivity was calculated based on marker positivity, as shown in the gating strategies (Supplementary Figures 1A & 3A). All figures are representative of N ≥3 experiments.
AML: Acute myeloid leukemia; mAb: Monoclonal antibody.
Table 2. . Percent positivity for cell lines and patient samples.
| Antibodies | Cell lines | Patient samples (by rs1259419 genotype) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HL-60 (CC) | Molm-13 (CC) | MV4;11 (CT) | K562 (TT) | CD14-CD45dimCD34+ AML progenitor cells | CD14+ monocytes | |||||
| CC (n = 5) | CT (n = 5) | TT (n = 4) | CC (n = 5) | CT (n = 5) | TT (n = 4) | |||||
| P67.6 | 94.5 | 99.2 | 98.2 | 2.1 | 44.1 | 35.0 | 30.2 | 75.8 | 80.5 | 30.1 |
| HL2541 | 3.6 | 2.5 | 17 | 94.4 | 23.0 | 14.3 | 35.9 | 67.7 | 80.1 | 64.5 |
| 5C11–2 | 0 | 0.01 | 0.04 | 0.04 | 7.5 | 4.4 | 19.1 | 10.7 | 20.0 | 15.6 |
AML: Acute myeloid leukemia; CC: Homozygous dominant; CT: Heterozygous; TT: Homozygous recessive.
Table 3. . Median fluorescence intensity ratios for cell lines and patient samples.
| Antibodies | Cell lines | Patient samples (by rs1259419 genotype) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HL-60 (CC) | Molm-13 (CC) | MV4;11 (CT) | K562 (TT) | CD14-CD45dimCD34+ AML progenitor cells | CD14+ monocytes | |||||
| CC (n = 5) | CT (n = 5) | TT (n = 4) | CC (n = 5) | CT (n = 5) | TT (n = 4) | |||||
| P67.6 | 8.4 | 17.6 | 9.4 | 1.6 | 2.9 | 2.0 | 2.2 | 9.3 | 6.0 | 3.7 |
| HL2541 | 1.1 | 1.4 | 2.3 | 68.9 | 2.3 | 1.3 | 2.4 | 16.3 | 14.4 | 18.2 |
| 5C11–2 | 1.0 | 1.2 | 1.3 | 1.2 | 1.2 | 1.2 | 1.5 | 3.3 | 2.0 | 2.3 |
AML: Acute myeloid leukemia; CC: Homozygous dominant; CT: Heterozygous; MFI: Median fluorescence intensity; TT: Homozygous recessive.
Western blotting of AML cell lines using the E6, HL2541 and 5C11-2 antibodies demonstrated robust recognition of CD33FL at approximately 67 kDa in all cell lines regardless of rs12459419 genotype. However, CD33D2 recognition at approximately 40 kDa was correlated with the rs12459419 genotype, whereas the Molm-13 and HL-60 cell lines displayed minimal expression of CD33D2, and higher levels were detected in the MV4;11 and K562 cell lines using all three antibodies (Figure 2B). Western blots of PNGase-treated and untreated AML cell lines using HL2541 also showed genotype-dependent expression of CD33 isoforms, with recognition of unglycosylated CD33FL and CD33D2 at approximately 40 kDa and approximately 33 kDa, as previously seen (Figure 2C). Of note, no additional protein bands were observed when probing with HL2541 in either the MV4;11 or K562 cell line, consistent with the results observed in flow cytometry assays using HL2541.
Evaluation of new antibodies in primary AML bone marrow specimens of different rs12459419 genotypes
Fourteen primary bone marrow specimens from AML patients with different rs12459419 genotypes were profiled for CD33D2 expression using P67.6 and new antibodies. Overall, no significant differences in patient demographics, including age, race and AML phenotypic characteristics, were observed across rs12459419 genotypes (Table 1).
Table 1. . Summary of patient characteristics for specimens used in the present study.
| Patient characteristics | rs12459419 genotype | |||||||
|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | Total | |||||
| (n = 5)† | (n = 5) | (n = 4)† | (N = 14) | |||||
| n | % | n | % | n | % | N | % | |
| Sex | ||||||||
| – Female | 2 | 50.0 | 2 | 40.0 | 3 | 60.0 | 7 | 50.0 |
| Age | ||||||||
| – Years, median (range) | 11.4 (8.7–16.0) |
13.6 (11.5–17.3) |
2.0 (1.7–16.2) |
11.8 (1.6–17.3) |
||||
| Race | ||||||||
| – White | 4 | 100.0 | 4 | 80.0 | 4 | 66.7 | 12 | 92.3 |
| – Unknown | 0 | 0.0 | 1 | 20.0 | 1 | 16.7 | 2 | 15.4 |
| Risk group | ||||||||
| – High | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| – Standard | 0 | 0.0 | 2 | 40.0 | 3 | 60.0 | 5 | 35.7 |
| – Low | 4 | 100.0 | 3 | 60.0 | 2 | 40.0 | 9 | 64.3 |
| Clinical features | ||||||||
| – BM blast (%), median (range) | 61 (36–90) |
65† (49–80) |
81 (25–90) |
80 (25–90) |
||||
One patient sample with unavailable demographic data.
BM: Bone marrow; CC: Homozygous dominant; CT: Heterozygous; TT: Homozygous recessive.
Primary cells were stained using CD34, CD14 and CD45 antibodies, allowing for gating on an immature CD14-CD45dimCD34+ population of AML cells and a more mature CD14+ monocytic population. Cells were also stained with P67.6, HL2541 or 5C11-2 antibodies, and samples were determined as positive based on the shift from fluorescence minus one control. All gating was done on live cells, and positivity was determined using negative gates (Figure 3A). In AML progenitor cells (CD14-CD45dimCD34+ cells), a genotype-dependent recognition of CD33FL was observed with P67.6. Specimens from homozygous dominant (CC genotype) patients displayed a higher percentage of CD33+ cells (average percent positivity: 44.1 ± 11.6%) in comparison to samples from heterozygous and homozygous recessive patients (CT and TT genotypes: 35.0 ± 10.9% and 30.2 ± 13.6%, respectively). HL2541 (average percent positivity: CC: 14.3 ± 2.3%; CT: 23.0 ± 14.0%; TT: 35.9 ± 17.6%) and 5C11-2 (average percent positivity: CC: 7.5 ± 5.4%; CT: 4.4 ± 3.5%; TT: 19.1 ± 9.3%) staining showed lower shifts in the histograms of stained samples compared with fluorescence minus one regardless of rs12459419 genotype, illustrating minimal/no CD33D2 cell surface recognition in CD14-CD45dimCD34+ progenitor cells. Representative samples corresponding to each genotype are shown in Figure 3B (Supplementary Figure 5 shows additional samples). When gating on CD14+ monocytes of the same patient samples, a shift using the HL2541 antibody in comparison to the fluorescence minus one control was observed in many patients regardless of genotype (average percent positivity: CC: 67.7 ± 11.6%; CT: 80.1 ± 8.1%; TT: 64.5 ± 16.8%) (Figure 3C; Supplementary Figure 6 shows additional samples), whereas minimal to no shift was observed using 5C11-2 (average percent positivity: CC: 10.7 ± 6.3%; CT: 20.0 ± 12.8%; TT: 15.6 ± 8.1%). Percent positivity and MFI ratios for samples by genotype and cell population can be found in Tables 2 & 3, respectively.
Figure 3. . Profiling of CD33D2 cell surface expression on primary AML cells obtained at diagnosis.
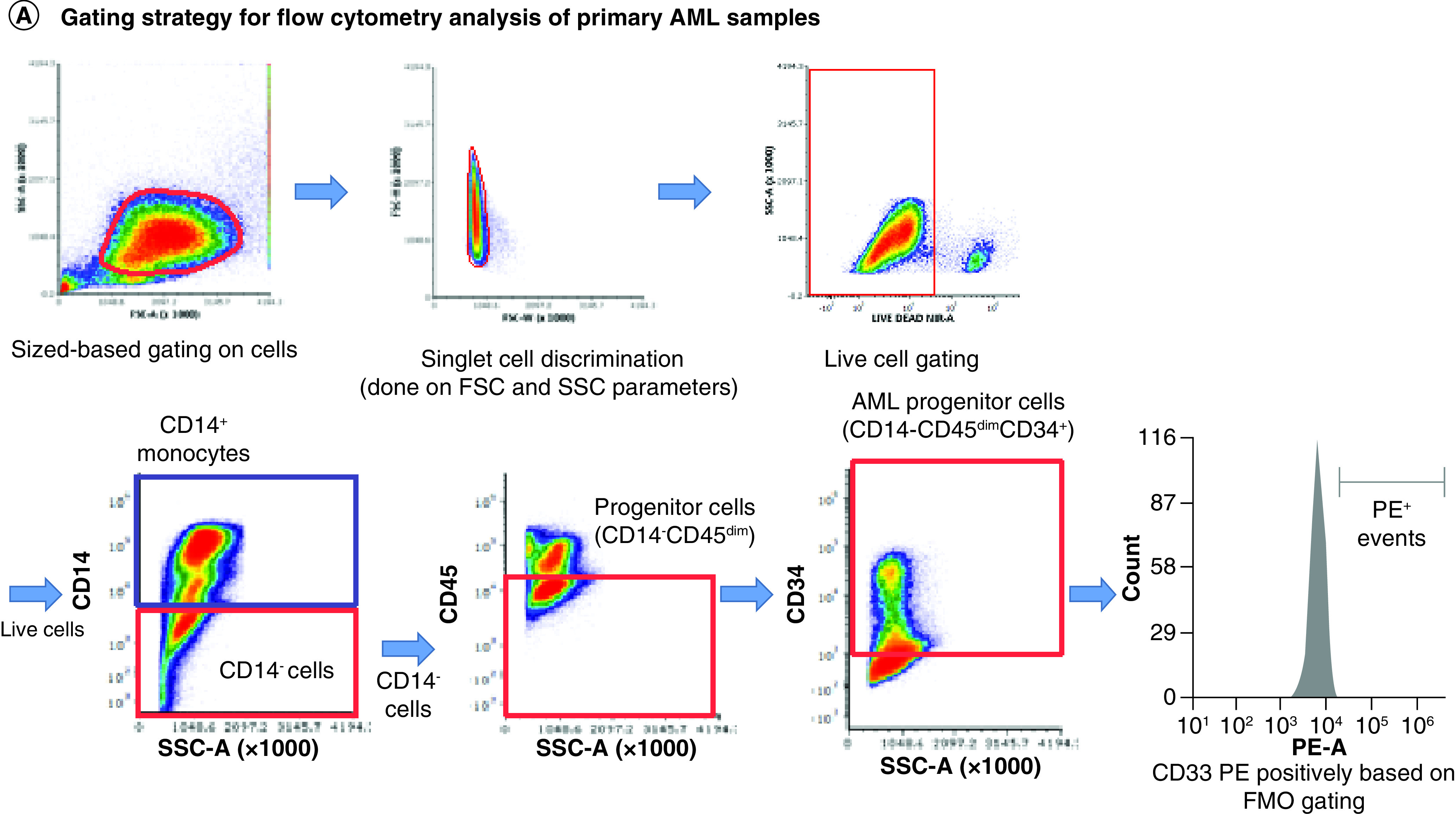
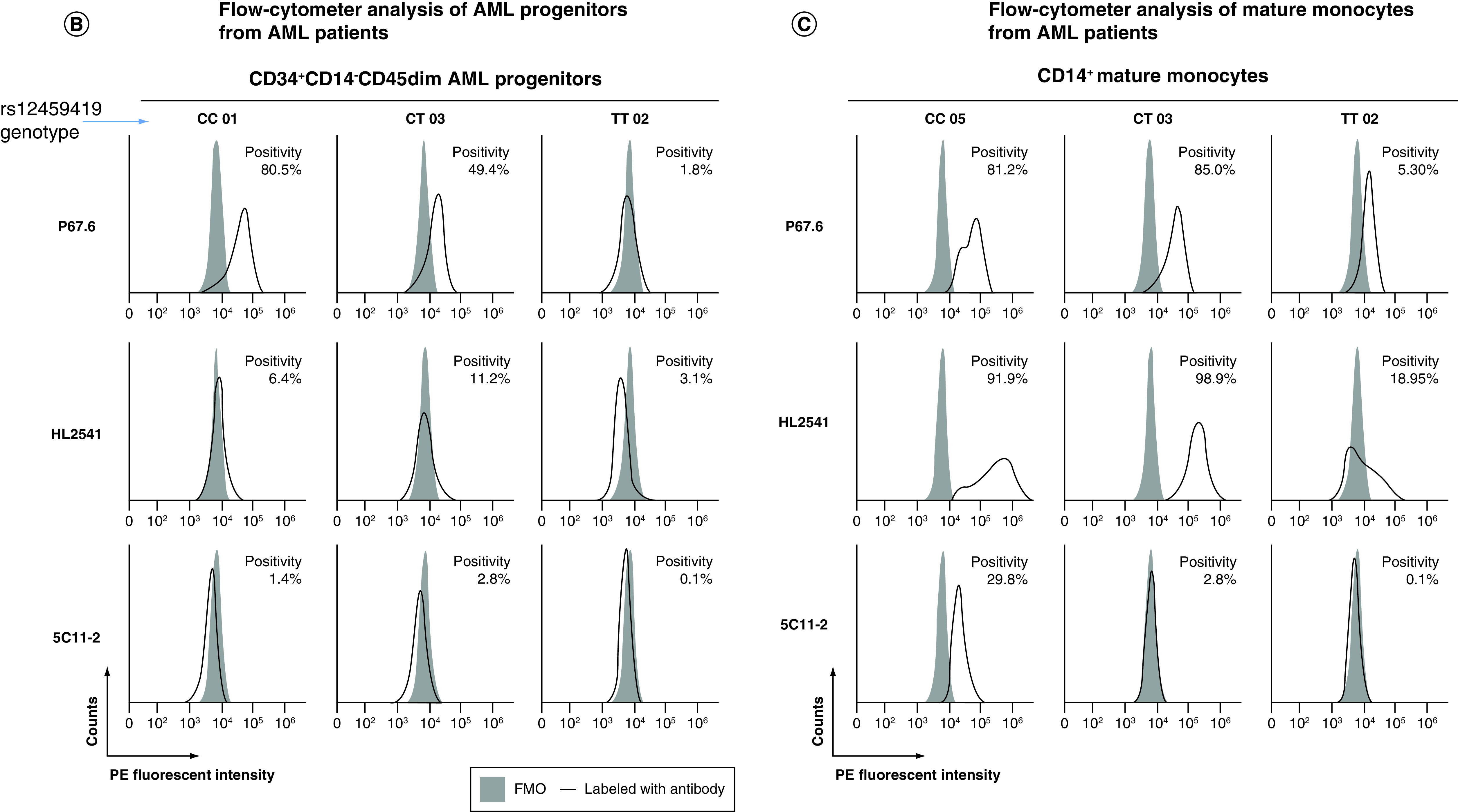
(A) Gating strategy for analysis of CD33D2 expression on CD34+CD14-CD45dim AML progenitors, CD14+ mature monocytes and other CD45high lymphocytes using CD34-APC (100 ng), CD14-pacific blue (800 ng) and CD45-super bright 600 antibodies (150 ng). Cells were first gated by size and viability. Subsequently, CD14- cells were gated, followed by a gate set to capture CD14-CD45dim and CD14-CD45high (other lymphocytes) and finally a gate to capture CD14-CD45dimCD34+ cells (AML progenitors). Following gating based on surface marker antibodies, a gate for positive fluorescence in the PE channel was set using FMO controls as negatives. All gates were set using unstained samples as a negative. (B) Staining for CD33D2 using HL2541-PE and 5C11-2-PE (500 ng each) on CD34+CD14-CD45dim AML progenitors from primary BM AML specimens of different rs12459419 genotypes. (C) Staining for CD33D2 using HL2541-PE and 5C11-2-PE on CD14+ mature monocytes from primary BM AML specimens of different rs12459419 genotypes. All flow cytometry data were collected on the Cytek Aurora platform and analyzed using FCS Express. MFI ratios were calculated as follows: (sample MFI/FMO MFI). Percent positivity was calculated based on marker positivity, as shown in the gating strategies (Supplementary Figures 1A & 3A). All figures are representative of N ≥3 experiments.
AML: Acute myeloid leukemia; BM: Bone marrow; FMO: Fluorescence minus one; MFI: Median fluorescence intensity; PE: Phycoerythrin.
Western blotting of primary AML cells from patients with different genotypes performed with and without PNGaseF treatment using HL2541 and E6 antibodies confirmed recognition and intracellular presence of CD33 isoforms (Supplementary Figure 7). In addition to primary AML bone marrow specimens, the authors also evaluated regenerating monocytes from bone marrow specimens obtained at remission from patients initially diagnosed with leukemia using P67.6, HL2541 and 5C11-2. The cells were stained for CD34, CD14 and CD45 as previously described, and individual cell populations, including CD14-CD45dimCD34+ progenitor cells and CD14+ mature monocytes, were gated out. In addition, lymphoblastic progenitors were separated from myeloblastic progenitor cells using CD34 and size based on right-angle scatter properties (Figure 4A). When staining with P67.6, both myeloblastic CD14-CD45dimCD34+ progenitor cells and CD14+ monocytes were positive for CD33FL cell surface expression, whereas lymphoblastic progenitor cells were negative for CD33FL cell surface expression, as expected. CD14-CD45dimCD34+ progenitor cells were negative for CD33D2 cell surface expression for HL2541 and 5C11-2, whereas CD14+ monocytes showed a positive signal for HL2541, consistent with the results from diagnostic AML samples (Figure 4B). Supplementary Figure 8 shows the results of the three additional bone marrow specimens stained with P67.6, HL2541 and/or 5C11-2.
Figure 4. . Profiling of CD33D2 cell surface expression on bone marrow specimens obtained at remission.
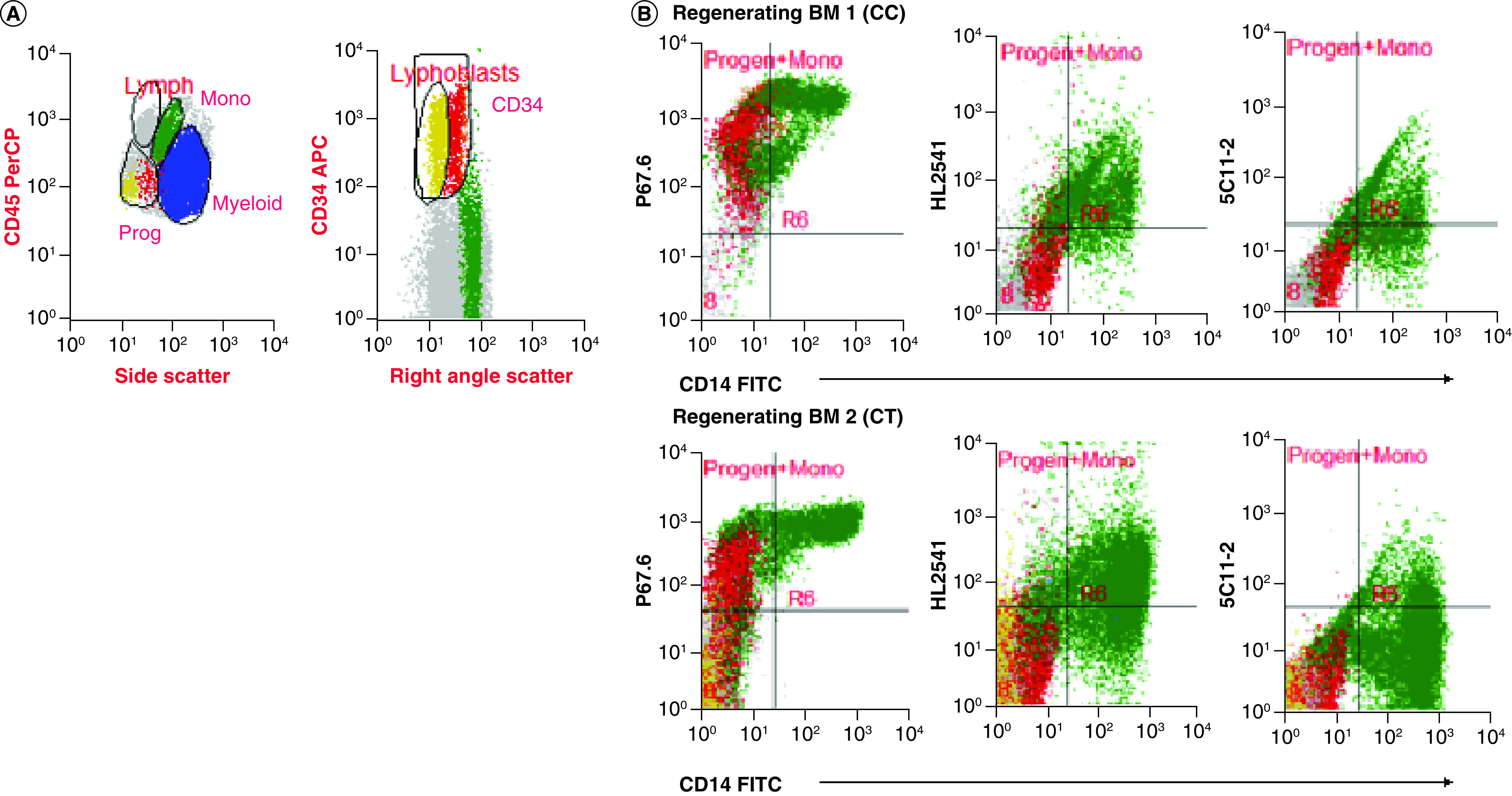
(A) Gating strategy for analysis of CD33D2 expression on CD34+CD14-CD45dim progenitors (both lymphoblastic and myeloblastic, as gated based on right-angle scatter properties) and CD14+ mature monocytes, as previously described, as well as other myeloid cells. All gates were set using unstained samples as a negative. Cells in red, yellow, green and blue represent CD34+CD14-CD45dim myeloblastic progenitors, CD34+CD14-CD45dim lymphoblastic progenitors, CD14+ monocytes and other myeloid cells, respectively (note: discretion of progenitor lineage was not possible for patient two). (B) Dot plot of overlay of progenitor cells and monocytes stained with P67.6 to measure CD33FL expression and HL2541 and 5C11-2 to measure CD33D2 expression. Figures are representative of data from N ≥2 sets of samples.
Discussion
Present in >85% of cases of AML, CD33 is a lucrative target for several immunotherapeutic strategies, with many clinical trials evaluating the efficacy of several anti-CD33 immunotherapeutics in the context of AML. The authors' group was the first to investigate genetic polymorphisms in CD33 and their effect on GO response and efficacy in AML, identifying rs12459419, a splicing single nucleotide polymorphism in CD33, as a significant prognostic factor. Since rs12459419 results in the skipping of exon 2 and thus loss of the encoded IgV domain, there is a need to investigate new strategies, such as the use of CD33D2-directed therapeutics, for patients with this splicing variant. Unfortunately, this endeavor has been hindered because of a lack of antibodies capable of detecting and characterizing CD33D2 expression in AML cells, which is needed as a first step to understand the biology and therapeutic potential of this isoform. Herein the authors report successful development of two novel antibodies, HL2541 and 5C11-2, directed to different epitopes of the IgC domain of CD33. Using the two new antibodies, CD33FL and CD33D2 were both detected by western blotting. However, by flow cytometry and confocal microscopy, both antibodies detected only CD33D2 on the surface of cells, suggesting that the proximity of the clustered extracellular Ig-like folds in CD33FL may create a steric hindrance that impacts recognition of CD33FL by the new IgC-directed antibodies [28].
While this study was being prepared for submission, a recent letter published by Godwin et al. reported the development of two antibodies directed to the IgC domain and the entire extracellular domain of CD33D2 [29]. Overall, the researchers concluded that their newly developed antibodies were unable to detect CD33D2 on AML cell lines and primary cells representing different genotypes. However, unlike a previous study, they were unable to detect CD33D2 in the intracellular spaces of AML cells or healthy donor myeloid cells even in immunoprecipitation and immunoblotting experiments, which raises questions regarding the ability of their antibodies to detect CD33D2 in AML cell lines or primary cells [30]. The HL2541 and 5C11-2 novel antibodies reported here detected cell surface expression of CD33D2 isoform in genetically modified cell lines, such as that seen using Ba/F3 cells modified to overexpress CD33D2 isoform; however, in unmodified AML cell lines, only HL2541 recognized CD33D2, and in primary AML cells there was minimal to no detection of surface expression using both antibodies. In the MV4;11 (CT) and K562 (TT) cell lines, HL2541, but not 5C11-2, demonstrated moderate positivity, whereas for diagnostic AML bone marrow specimens, an overwhelming majority of samples gated on a CD14-CD45dimCD34+ population of AML cells were negative for CD33D2 cell surface expression by 5C11-2 and HL2541. Moreover, the CD14+ monocyte cell population from the same patients showed moderate positivity with HL2541, suggesting a cell type-dependent expression pattern of CD33D2. Similar results were observed using regenerating cells from bone marrow specimens obtained from patients in remission, where progenitor cells, regardless of lineage, were negative for CD33D2 expression using both antibodies and monocytes were stained positive by HL2541. Unlike the results from the study by Godwin et al., in the authors' western blotting analysis using AML cell lines and the limited number of patient samples available, robust signals for CD33D2 were observed using both antibodies, suggesting intracellular presence of CD33D2 [29]. These results are consistent with previous studies in the context of Alzheimer's disease, which also reported primary expression of intracellular CD33D2 [28,30].
The observed differences in CD33D2 cell surface recognition may arise from differences in the epitopes targeted by each of the new antibodies (Supplementary Figure 2). As shown previously, recognition of the native confirmation of the IgC-like domain of CD33 can be impacted by glycosylation, which occurs in a cell type-dependent manner and may explain the cell type-specific differences in CD33D2 recognition using 5C11-2 and HL2541 [31–33]. This warrants in-depth studies focused on cell type-specific glycosylation and its influence on antibody recognition of CD33. Interestingly, a similar phenomenon has been reported with regard to CD300f, another myeloid-specific inhibitory transmembrane glycoprotein, where antibodies targeting different epitopes of CD300f displayed differential cell type-dependent recognition of its various isoforms [34].
A recent study published by Nair-Gupta et al. reported the development of a novel CD33-IgC domain-binding CD33xCD3 bispecific antibody (JNJ-67571244), which is currently in a Phase I trial for relapsed and refractory AML and high-risk myelodysplastic syndrome [35]. Binding of JNJ-67571244 to both CD33 isoforms was shown using cell lines representing different rs12459419 genotype. Interpatient variation in cytotoxicity and T-cell activation in response to JNJ-67571244 was observed, with half-maximal effective concentration ranging from 0.052 to 9.52 nM, and the lack of patient genotype information raises a few questions. Nevertheless, these results are in contrast to the previous work in Alzheimer's disease and the report by Godwin et al., in which cell surface CD33D2 expression was questioned [29,30]. In this study, the researchers claimed that the newly developed CD33xCD3 bispecific antibody could recognize and act against CD33D2 on the cell surface.
Although more in-depth investigations are required to establish the biological and therapeutic role of CD33D2 isoform, the authors' results using two novel IgC-targeting antibodies show poor cell surface expression of CD33D2 on AML and progenitor cells; however, in the context of other cell types and cell lines, the detection may be influenced by the epitope targeted. The authors' results using 5C11-2, which is directed to the N-terminus region of CD33D2, showed minimal or no cell surface recognition on AML cell lines and cells from primary patient samples, although there was robust detection of CD33D2 in overexpressing Ba/F3 cells. Using HL2541, CD33D2 was recognized on the surface of AML cell lines that are heterozygous and homozygous recessive for the rs12459419 splicing single nucleotide polymorphism as well as in mature monocytic cells, although poor cell surface expression and recognition of CD33D2 were seen on primary AML and healthy progenitor cells. Nonetheless, the authors saw significant intracellular detection of CD33D2 in AML and progenitor cells.
In light of these results and the results of the two recent studies, future mechanistic investigations exploring CD33D2 expression patterns and functionality are needed, especially with regard to different cell types and the heterozygous group of patients who do not benefit from GO despite having intermediate levels of CD33FL on the surface of their cells [26]. Given the established versatility of the authors' reported antibodies for use in flow cytometry, western blot and confocal microscopy, we anticipate future studies focused on characterizations of CD33D2, including the factors that govern its expression and specific intracellular localization and its impact on the expression and recycling of CD33FL.
Summary points.
CD33 is a highly sought-after target for development of novel immunotherapies for the treatment of acute myeloid leukemia (AML).
Gemtuzumab ozogamicin (GO) is the only anti-CD33 immunoconjugate approved by the US FDA for AML treatment, and the promising results with GO have paved the way for the development of several new CD33-directed immunotherapies.
Though CD33 immunotherapy such as GO has revolutionized AML treatment strategies, there is evidence of interpatient variation in therapeutic response. A splicing single nucleotide polymorphism, rs12459419 (C >T; A14V), in CD33 leads to loss of exon 2, resulting in production of CD33D2, a shorter isoform of CD33 that lacks the IgV domain. Since GO targets the IgV domain, the presence of the T variant allele impacts clinical response to GO, and thus development of new therapeutics targeting the IgC domain common to both full-length CD33 and CD33D2 isoforms is necessary.
Here the authors report development of novel IgC-directed CD33 antibodies that successfully detect CD33 isoforms by western blotting, flow cytometry and immunofluorescence microscopy in cells overexpressing CD33D2. The authors' novel antibody, HL2541, also successfully detected CD33D2 isoform in unmodified AML cell lines representing different genotypes for the splicing single nucleotide polymorphism.
In primary AML specimens, the authors observed minimal/no cell surface expression but detectable intracellular levels of CD33D2, consistent with two other reports, but in contrast to the study by Godwin et al., in which extracellular and intracellular expression of CD33D2 was undetectable [28–30].
Recent development of a novel CD33-IgC domain-binding CD33xCD3 bispecific antibody (JNJ-67571244) that is currently in a Phase I trial for relapsed/refractory AML and high-risk myelodysplastic syndrome implies the therapeutic relevance of the IgC domain and thus the potential for novel antibodies reported in this study [35].
Supplementary Material
Acknowledgments
The authors are grateful to the Antibody Generation Core at the Interdisciplinary Center for Biotechnology Research for the production of the HL2541 antibody. The authors also thank the Flow Cytometry Core at the Interdisciplinary Center for Biotechnology Research for their technical assistance and helpful discussions.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/fon-2020-0746
Author contributions
S Meshinchi and J K Lamba conceived of the project, contributed to project design and oversaw the project. M Gbadamosi, V Shastri, I Papageorgiou, T Hylkema and L Pardo designed and performed experiments, analyzed data and contributed to antibody generation efforts. L Pardo and M Loken contributed to experimental data and experimental design. A Doty contributed to experimental design and data analysis. All authors contributed to manuscript writing, reviewed and revised the manuscript, approved the final version and agreed to submit the manuscript for publication.
Financial & competing interests disclosure
L Pardo and M Loken are employed by Hematologics Inc., and M Loken holds a leadership position at and has stock/ownership in Hematologics Inc. This work was supported by the Leukemia & Lymphoma Society (no. 6610-20), St Baldrick's Foundation, the University of Florida Cancer Center and the University of Florida College of Pharmacy. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval, including from the Children's Oncology Group Myeloid Disease Biology Committee, or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Andrews RG, Torok-Storb B, Bernstein ID. Myeloid-associated differentiation antigens on stem cells and their progeny identified by monoclonal antibodies. Blood 62(1), 124–132 (1983). [PubMed] [Google Scholar]
- 2.Dinndorf PA, Andrews R, Benjamin D, Ridgway D, Wolff L, Bernstein I. Expression of normal myeloid-associated antigens by acute leukemia cells. Blood 67, 1048–1053 (1986). [PubMed] [Google Scholar]
- 3.McMillan SJ, Crocker PR. CD33-related sialic-acid-binding immunoglobulin-like lectins in health and disease. Carbohydr. Res. 343(12), 2050–2056 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Walter RB, Raden BW, Zeng R, Häusermann P, Bernstein ID, Cooper JA. ITIM-dependent endocytosis of CD33-related SIGLECs: role of intracellular domain, tyrosine phosphorylation, and the tyrosine phosphatases, Shp1 and Shp2. J. Leukoc. Biol. 83(1), 200–211 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Crocker PR, Paulson JC, Varki A. SIGLECs and their roles in the immune system. Nat. Rev. Immunol. 7(4), 255–266 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Ehninger A, Kramer M, Röllig C et al. Distribution and levels of cell surface expression of CD33 and CD123 in acute myeloid leukemia. Blood Cancer J. 4(6), e218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Velden VHJ, Te Marvelde JG, Hoogeveen PG et al. Targeting of the CD33-calicheamicin immunoconjugate Mylotarg (CMA-676) in acute myeloid leukemia: in vivo and in vitro saturation and internalization by leukemic and normal myeloid cells. Blood 97(10), 3197–3204 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Bross PF, Beitz J, Chen G et al. Approval summary: gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin. Cancer Res. 7(6), 1490–1496 (2001). [PubMed] [Google Scholar]
- 9.Petersdorf SH, Kopecky KJ, Slovak M et al. A Phase 3 study of gemtuzumab ozogamicin during induction and postconsolidation therapy in younger patients with acute myeloid leukemia. Blood 121(24), 4854–4860 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norsworthy KJ, Ko C, Lee JE et al. FDA approval summary: Mylotarg for treatment of patients with relapsed or refractory CD33-positive acute myeloid leukemia. Oncologist 23(9), 1103–1108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jen EY, Ko C-W, Lee JE et al. FDA approval: gemtuzumab ozogamicin for the treatment of adults with newly diagnosed CD33-positive acute myeloid leukemia. Clin. Cancer Res. 24(14), 3242–3246 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Lichtenegger FS, Krupka C, Haubner S, Köhnke T, Subklewe M. Recent developments in immunotherapy of acute myeloid leukemia. J. Hematol. Oncol. 10(1), 1–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutherland MSK, Walter RB, Jeffrey SC et al. SGN-CD33A: a novel CD33-targeting antibody–drug conjugate using a pyrrolobenzodiazepine dimer is active in models of drug-resistant AML. Blood 122(8), 1455–1463 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Jurcic JG, Rosenblat TL, McDevitt MR et al. Phase I trial of the targeted alpha-particle nano-generator actinium-225 (225Ac)-lintuzumab (anti-CD33; HuM195) in acute myeloid leukemia (AML). Blood 118(21), 768 (2011). [Google Scholar]
- 15.Jurcic JG, Levy MY, Park JH et al. Phase I trial of targeted alpha-particle therapy with actinium-225-lintuzumab and low-dose cytarabine (LDAC) in patients age 60 or older with untreated acute myeloid leukemia (AML). Blood 128(22), 4050 (2016). [Google Scholar]
- 16.Aigner M, Feulner J, Schaffer S et al. T lymphocytes can be effectively recruited for ex vivo and in vivo lysis of AML blasts by a novel CD33/CD3-bispecific BiTE antibody construct. Leukemia 27(5), 1107–1115 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Laszlo GS, Gudgeon CJ, Harrington KH et al. Cellular determinants for preclinical activity of a novel CD33/CD3 bispecific T-cell engager (BiTE) antibody, AMG 330, against human AML. Blood 123(4), 554–561 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenderian SS, Ruella M, Shestova O et al. CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia 29(8), 1637–1647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eksioglu EA, Chen X, Heider KH et al. Novel therapeutic approach to improve hematopoiesis in low risk MDS by targeting MDSCs with the Fc-engineered CD33 antibody BI 836858. Leukemia 31(10), 2172–2180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard JA, Alonzo TA, Loken M et al. Correlation of CD33 expression level with disease characteristics and response to gemtuzumab ozogamicin containing chemotherapy in childhood AML. Blood 119(16), 3705–3711 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan N, Hills RK, Virgo P et al. Expression of CD33 is a predictive factor for effect of gemtuzumab ozogamicin at different doses in adult acute myeloid leukaemia. Leukemia 31(5), 1059–1068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollard JA, Loken M, Gerbing RB et al. CD33 expression and its association with gemtuzumab ozogamicin response: results from the randomized Phase III children's oncology group trial AAML0531. J. Clin. Oncol. 34(7), 747–755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillaume O, Estelle G, Jullien G et al. The level of blast CD33 expression positively impacts the effect of gemtuzumab ozogamicin in patients with acute myeloid leukemia. Blood 127(17), 2155–2157 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Lamba JK, Pounds S, Cao X et al. Coding polymorphisms in CD33 and response to gemtuzumab ozogamicin in pediatric patients with AML. Leukemia 23(2), 402–404 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortland L, Alonzo TA, Walter RB et al. Clinical significance of CD33 nonsynonymous single-nucleotide polymorphisms in pediatric patients with acute myeloid leukemia treated with gemtuzumab-ozogamicin-containing chemotherapy. Clin. Cancer Res. 19(6), 1620–1627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamba JK, Chauhan L, Shin M et al. CD33 splicing polymorphism determines gemtuzumab ozogamicin response in de novo acute myeloid leukemia: report from randomized Phase III children's oncology group trial AAML0531. J. Clin. Oncol. 35(23), 674–2682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamis AS, Alonzo TA, Meshinchi S et al. Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized Phase iII children's oncology group trial AAML0531. J. Clin. Oncol. 32(27), 3021–3032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha S, Siddiqui SS, Khan N et al. Controversies about the subcellular localization and mechanisms of action of the Alzheimer's disease-protective CD33 splice variant. Acta Neuropathol. 138(4), 671–672 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Godwin CD, Laszlo GS, Wood BL et al. The CD33 splice isoform lacking exon 2 as therapeutic target in human acute myeloid leukemia. Leukemia 34(9), 2479–2483 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddiqui SS, Springer SA, Verhagen A et al. The Alzheimer's disease–protective CD33 splice variant mediates adaptive loss of function via diversion to an intracellular pool. J. Biol. Chem. 292(37), 15312–15320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pérez-Oliva AB, Martínez-Esparza M, Vicente-Fernández JJ, Corral-San Miguel R, García-Peñarrubia P, Hernández-Caselles T. Epitope mapping, expression and post-translational modifications of two isoforms of CD33 (CD33M and CD33m) on lymphoid and myeloid human cells. Glycobiology 21(6), 757–770 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Hernández-Caselles T, Martínez-Esparza M, Pérez-Oliva AB et al. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J. Leukoc. Biol. 79(1), 46–58 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12(4), 43R–56R (2002). [DOI] [PubMed] [Google Scholar]
- 34.Abadir E, Gasiorowski RE, Lai K et al. CD300f epitopes are specific targets for acute myeloid leukemia with monocytic differentiation. Mol. Oncol. 13(10), 2107–2120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair-Gupta P, Diem M, Reeves D et al. A novel C2 domain binding CD33xCD3 bispecific antibody with potent T-cell redirection activity against acute myeloid leukemia. Blood Adv. 4(5), 906–919 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


