Abstract
Two toluene-degrading strains, T103 and T104, were isolated from rock surface biomass in a freshwater stream contaminated with toluene. The strains exhibit different capacities for degradation of toluene and other aromatic compounds and have characteristics of the genus Mycobacterium. Both are aerobic, rod-shaped, gram-positive, nonmotile, and acid-alcohol fast and produce yellow pigments. They have mainly straight-chain saturated and monounsaturated fatty acids with 10 to 20 carbon atoms and large amounts of tuberculostearic acid that are typical of mycobacteria. Fatty acid analyses indicate that T103 and T104 are different mycobacterial strains that are related at the subspecies level. Their identical 16S rDNA sequences are most similar to Mycobacterium aurum and Mycobacterium komossense, and they constitute a new species of fast-growing mycobacteria. Ecological studies reveal that toluene contamination has enriched for toluene-degrading bacteria in the epilithic microbial community. Strains T103 and T104 play only a small role in toluene degradation in the stream, although they are present in the habitat and can degrade toluene. Other microorganisms are consequently implicated in the biodegradation.
In the United States, toluene is ranked 27th among the top 50 chemical products by production volume, with 931 million gallons (3.5 × 109 liters) of toluene manufactured in 1994 (22). Industry uses toluene in refining gasoline; chemical manufacturing; the manufacture of paints, lacquers, and adhesives; and some printing and leather-tanning processes. Toluene is usually disposed of as a used solvent. Toluene is listed as a priority pollutant (46), because contamination of drinking water can pose a potential health hazard. In a 1988 study, toluene was detected in groundwater, surface water, or soil at 29% of the hazardous waste sites surveyed; the average amounts detected were 0.2 μM in groundwater, 0.1 μM in surface water, and 77 μg/kg in soil (46).
Since toluene is ubiquitous in the environment, it is not surprising that microorganisms capable of degrading toluene have been isolated from a variety of environments, including polluted topsoils and oil tanker ballast waters (4, 5, 10, 29, 38, 48, 50). However, little is known about the role these isolates may play in the conversion of toluene in the environment. Therefore, studies that address the response of stream bacteria to effects of anthropogenic chemical impact are important in enhancing our understanding of the ecology of contaminant degradation.
This study describes the isolation and characterization of two closely related toluene-degrading strains of a novel Mycobacterium species from rock surface biofilms in a toluene-contaminated reach of a small freshwater stream. Ecological data are also presented to assess the role of these Mycobacterium strains in degrading toluene in the rock surface biofilms and to determine the impact of toluene contamination on the stream’s microbial community. Earlier studies showed biodegradation to be the most significant sink for toluene in this stream (20), and microorganisms attached to stream sediments and rock surfaces accounted for most of the biodegradative activity (7).
MATERIALS AND METHODS
Study site.
The East Drainage Ditch is a small stream in an industrial area of Wilmington and Woburn, Mass. (7, 20), and forms part of the Aberjona Watershed, a 90-km2 area 20 km north of Boston. Toluene in the stream arises from subsurface contamination below an 80-m-long culvert (11) located 1,600 m upstream from the confluence of the East Drainage Ditch with Halls Brook. Toluene levels typically range from 0.6 to 4.2 μM in the streamwater near the source (20). Sampling stations are upstream 50 m (U50) and 100 m (U100) and downstream 5 m (D5) and 50 m (D50).
Isolation of bacterial strains.
Rocks were collected in July 1992 from the streambed at station D5. Rock biomass was scraped with sterile spatulas, serially diluted, and plated onto a mineral salts (MS) agar medium (35) supplemented with trace elements (49). The plates were incubated in a desiccator with a beaker of water (15 ml) previously equilibrated with toluene. The water was replaced every 2 to 3 days. Toluene was supplied by diffusion from the beaker in the desiccator that resulted in theoretical concentrations of approximately 110 μM in the agar based on Henry’s law (36), although the actual concentrations might be affected by diffusion kinetics and hydrophobic interactions between toluene and organic constituents of the agar. Plates were monitored over 7 weeks, during which colonies were picked and restreaked on fresh plates. Several colonies grew in the presence of toluene, and four strains, numbered sequentially starting from T101, were selected for further characterization. Strains T103 and T104 possessed traits typical of mycobacteria and are described in this paper.
Phenotypic characterization.
Tests for Gram stain, oxidase activity, catalase activity, carbon source utilization, nitrate reduction, and acid fastness (Ziehl-Neelsen method) were performed as previously described (27, 37). Tests for growth on aromatic compounds were conducted in the same manner as with toluene. Substrates were supplied by diffusion from a reservoir in the desiccator resulting in theoretical concentrations in the agar of 10 mg of benzene, o-xylene, m-xylene, p-xylene, phenol, or chlorobenzene per liter. Plates were monitored for 4 weeks, and visible colonies were picked and restreaked on fresh plates that were further incubated in either the presence or absence of the relevant substrate.
Toluene biodegradation kinetics.
Rates of toluene degradation were determined by a headspace gas chromatography method (7). Cells were grown in 1,100-ml Teflon-stoppered glass bottles with 600 ml of MS medium initially containing 110 μM toluene. Toluene levels were monitored daily and replenished when depleted. After every five toluene feedings, the headspace was flushed with air to replenish the oxygen. Cells were harvested by centrifugation during the exponential phase and resuspended in 100 ml of fresh medium. Kinetic experiments were performed with 60-ml Teflon-stoppered serum bottles (The West Co., Phoenixville, Pa.) with 19 ml of medium and 0.5 ml of concentrated cell suspension (230 μg of protein/ml for strain T103 and 185 μg of protein/ml for strain T104). Toluene was injected at approximately 1.1, 2.2, 5.4, and 10.9 μM (aqueous concentration). Control experiments were performed with autoclaved cell suspensions. Bottles were shaken in the dark at 20°C on a rotary shaker at 150 rpm and assayed hourly for toluene. Initial rates of toluene disappearance were determined to establish Michaelis-Menten kinetic parameters. Protein was measured with the bicinchoninic acid protein assay (Pierce, Rockford, Ill.).
Fatty acid analyses.
Cells were grown on Middlebrook 7H10 agar (9) for 7 days at 28°C, harvested, and saponified to prepare fatty acid methyl esters (33). The analysis was performed at Microbial ID, Inc. (MIDI, Newark, Del.), by using the MIDI Microbial Identification System software for identification of fatty acids. Strains T103 and T104, together with the M. komossense type strain (ATCC 33013), were analyzed in duplicate and compared with profiles from the Microbial Identification System (MIS) library (34).
16S rDNA sequencing.
Strains T103 and T104 were cultured in nutrient broth (Difco Laboratories, Detroit, Mich.) in Teflon-stoppered serum bottles by shaking at 20°C on a rotary shaker at 150 rpm. Cells were harvested by centrifugation, and genomic DNA was extracted with a miniprep (1). The nearly full-length 16S rRNA gene was amplified from genomic DNA by PCR with forward primer Eubac27F and reverse primer Universal 1492R (23). All reactions were run in triplicate under PCR conditions as described elsewhere (8), and PCR products were purified with the Wizard PCR Prep (Promega Corp., Madison, Wis.) for automated dye-dideoxy terminator sequencing at the Michigan State University Sequencing Facility with a 373A DNA sequencing system (Applied Biosystems, Foster City, Calif.). Sequences for oligonucleotides complementary to the conserved regions of the eubacterial 16S rRNA gene were kindly provided by Debra J. Lonergan (United States Geological Survey, Reston, Va.); these oligonucleotides were chosen to prime the sequencing reactions. Sequencing reaction mixes consisted of 12 pmol of sequencing primer and 50 to 250 ng of PCR template in a total volume of 20 μl of sterile H2O. The sequences of approximately 1,425 nucleotide bases, corresponding to the Escherichia coli 16S rDNA sequence from nucleotides 55 to 1501, were obtained in both directions for the two strains.
Phylogenetic analyses.
The 16S rDNA secondary structures of strains T103 and T104 were constructed manually with templates published in the Ribosomal Database Project (RDP) (25) to aid in the identification of homologous sequence positions. Sequence alignments were performed manually in the Genetic Data Environment (39). All reference sequences and the basic alignment were obtained from the RDP. Only homologous sites at which the 16S rDNA sequences of strains T103 and T104 could be aligned unambiguously with the reference sequences were included in a final data set of 1,165 nucleotides for further analyses. Distance, parsimony, and maximum likelihood analyses were performed with PHYLIP 3.5 (13), PAUP 3.1 (42), and fastDNAml (12, 28), respectively, as described elsewhere (31).
Bacterial counts.
Rock samples were collected from stations U100, U50, D5, and D50 along the East Drainage Ditch on 7 September 1993. Biomass was scraped with sterile spatulas and suspended in 10 ml of MS medium, and the resulting cell suspension was successively diluted to obtain dilutions of 10−2 to 10−7 g of biomass/ml. Petri dishes containing 1% PTYG agar (3) were inoculated in duplicate with 100 μl of each dilution and incubated at 20°C for 3 weeks. Heterotrophic colonies were counted on the plates every 3 days, until no new colonies were observed. Since counts were based on the highest-dilution plates, colony overgrowth on the plates was not a problem.
Toluene-degrading bacteria were enumerated with MS agar medium inoculated in duplicate with 100 μl of each dilution, with toluene supplied as described earlier. Colonies growing on the highest-dilution plates were picked, transferred to new plates, and incubated in both the presence and absence of toluene to confirm the abilities of these isolates to grow with toluene as an energy source.
Toluene levels in the stream.
Duplicate water samples were collected at stations U100, U50, D5, and D50 on 19 September 1993 with 40-ml vials provided with hollow screw caps and Teflon-coated silicone septa. Mercuric chloride was added to the samples to a final concentration of 15 mg/liter. Toluene was measured as described previously (7).
Nucleotide sequence accession number.
The sequences for strains T103 and T104 have been deposited in the GenBank database under accession no. U62889 and U62890. The GenBank accession numbers of the other sequences used in the analyses are as follows: Mycobacterium aurum, X55595; Mycobacterium chelonae subsp. abscessus L948, M29559; Mycobacterium chitae, X55603; Mycobacterium chlorophenolicus PCP-I, X79094; Mycobacterium diernhoferi SN 1418, X55593; Mycobacterium fortuitum subsp. fortuitum, X52933; Mycobacterium gilvum, X55599; Mycobacterium komossense Ko2, X55591; Mycobacterium neoaurum, M29564; Mycobacterium sphagni Sph29, X55590; Mycobacterium thermoresistible, X55602; Mycobacterium vaccae, X55601; Mycobacterium asiaticum N61H, X55604; Mycobacterium avium serovar 1, M29573; Mycobacterium haemophilum, L24800; Mycobacterium tuberculosis H37/Rv, X52917; Mycobacterium xenopi, X52929; Corynebacterium xerosis, M59058; Gordona terrae, X79286; Nocardia otitidiscaviarum, M59056; and Rhodococcus equi, M29574. Primary literature references for these sequences are available from the RDP (25).
RESULTS
Enrichment and isolation.
Two mycobacterial toluene-degrading strains, T103 and T104, were independently isolated from two yellow colonies, 1 to 2 mm in diameter, that appeared between 3 and 8 days on toluene-incubated MS plates inoculated with 10−6 g of biomass, indicating that these strains were present in the rock surface biomass at a density of 106 cells/g of biomass. Biomass scrapings averaged 0.018 g (fresh weight)/cm2 of rock surface. Both strains grew slowly (relative to other nonmycobacterial toluene-degrading strains similarly isolated) on solid media when incubated with toluene. Mycobacterial colonies were not detected on plates inoculated with higher dilutions of biomass.
Morphological and phenotypic characteristics.
Strains T103 and T104 had morphologically similar rod-shaped cells when grown on solid media. Cells were nonmotile, and flagella were not observed under scanning electron microscopy (data not shown). The strains were aerobic, gram positive, and acid-alcohol fast (Table 1), which is characteristic of the mycobacteria (47). Strain T104 differed physiologically from strain T103 in that T104 could grow on xylenes (Table 1).
TABLE 1.
Characteristics of toluene-degrading strains T103 and T104
| Characteristic | Result for straina
|
|
|---|---|---|
| T103 | T104 | |
| Morphology | Rod | Rod |
| Flagella | − | − |
| Pigmentation | Yellow | Yellow |
| Gram stain | + | + |
| Acid fast | + | + |
| Nitrate reduction | − | − |
| Catalase | + | + |
| Oxidase | − | − |
| Citrate | + | + |
| Growth on sugars | ||
| Glucose | + | + |
| Lactose | − | − |
| Maltose | + | + |
| Sucrose | − | − |
| Growth on aromatic compoundsb | ||
| Toluene | + | + |
| Benzene | − | − |
| Chlorobenzene | − | − |
| o-Xylene | − | ++ |
| m-Xylene | − | ++ |
| p-Xylene | − | ++ |
| Phenol | − | − |
+, good growth or activity; −, poor or no growth or activity.
For growth on aromatic compounds, cultures were streaked onto agar plates containing a minimal salts media. Plates were then incubated in desiccators with substrate concentrations of approximately 10 mg/liter. The incubation temperature was 20°C. Each compound was tested twice. ++, very good growth within 3 days; +, good growth after 3 days; −, poor or no growth.
Toluene biodegradation kinetics.
Strains T103 and T104 had maximal toluene consumption rates (Vmax) of 1.0 ± 0.1 and 6.0 ± 1.3 μmol of toluene/mg of protein per h, respectively; their half-saturation constants (Ks) were 0.6 ± 0.4 and 3.8 ± 1.9 μM, respectively.
Fatty acid analyses.
Strains T103 and T104 were made up of mainly straight-chain saturated and monounsaturated fatty acids, as well as substantial amounts of tuberculostearic acid (Table 2) that are typical of mycobacteria (16). Fatty acid analyses (34) indicated that T103 and T104 were different strains of a novel Mycobacterium sp. that was most closely related to M. aurum (similarity indices of 0.31 and 0.24 with strains T103 and T104, respectively).
TABLE 2.
Whole-cell fatty acid compositions of strains T103, T104, M. komossense, and M. aurum
| Fatty acid | % of total (coefficient of variation [n = 2]) in:
|
|||
|---|---|---|---|---|
| Strain T103 | Strain T104 | M. komossense (ATCC 33013) | M. auruma | |
| Decanoic acid | 1.98 (1.1) | 1.49 (3.8) | NDb | ND |
| Dodecanoic acid | 2.11 (1.0) | 1.64 (5.1) | 0.25 (2.9) | ND |
| Tetradecanoic acid | 6.58 (0.4) | 7.25 (4.1) | 8.18 (2.4) | 5.93 |
| Pentadecanoic acid | 0.79 (5.4) | 0.45 (1.6) | 0.34 (8.3) | ND |
| cis-7-Hexadecenoic acid | 0.70 (1.0) | 0.76 (15.9) | 0.95 (9.7) | 2.19 |
| cis-9-Hexadecenoic acid | 2.63 (0.0) | 2.36 (2.7) | 2.34 (1.2) | ND |
| cis-10-Hexadecenoic acid | 4.70 (4.1) | 5.09 (10.4) | 8.11 (0.5) | 5.58 |
| Hexadecanoic acid | 22.93 (0.0) | 23.02 (0.5) | 27.2 (0.6) | 30.12 |
| Summed feature 1c | 0.78 (12.7) | 1.14 (3.1) | 0.85 (2.5) | ND |
| Summed feature 2d | 14.68 (3.2) | 19.69 (0.1) | 12.15 (6.1) | 10.01 |
| Heptadecanoic acid | ND | ND | 0.24 (9.0) | ND |
| cis-9,12-Octadecadienoic acid | 0.85 (5.0) | 0.89 (14.3) | 1.00 (2.1) | ND |
| cis-9-Octadecenoic acid | 20.93 (0.6) | 17.98 (5.9) | 16.0 (3.8) | 27.35 |
| cis-11-Octadecenoic acid | 0.61 (3.5) | 0.25 (141.4) | 0.54 (6.6) | ND |
| Octadecanoic acid | 0.87 (12.3) | 1.35 (6.3) | 3.46 (0.6) | 2.66 |
| 10-Methyloctadecanoic acid (tuberculostearic acid) | 10.96 (2.2) | 12.11 (1.7) | 14.98 (1.9) | 9.09 |
| Summed feature 3e | 7.23 (1.0) | 4.05 (3.5) | 3.18 (8.4) | 3.74 |
| Eicosanoic acid | 0.72 (18.8) | 0.51 (8.3) | 0.28 (5.1) | 2.23 |
From the Microbial Identification System library.
ND, not detected.
Summed feature 1 contains 8-methylhexadecanoic acid and 10-methylhexadecanoic acid.
Summed feature 2 contains cis-10-heptadecenoic acid, cis-11-heptadecenoic acid, ωcyclopropane 7-8 hexadecanoic acid, and 2-octadecanol.
Summed feature 3 contains ωcyclopropane 8-9 octadecanoic acid, ωcyclopropane 10-11 octadecanoic acid, and 2-eicosanol.
16S rDNA sequence analyses.
Strains T103 and T104 possessed identical 16S rDNA sequences. The secondary structures of the sequences of T103 and T104 and other fast-growing mycobacteria were identical, and they contained the shortened stem structure bounded by positions 455 to 477 (E. coli numbering) that typically distinguishes the fast-growing from the slow-growing mycobacteria (41).
The levels of identity between the T103/T104 sequence and the sequences of the fast-growing mycobacteria, slow-growing mycobacteria, and other nonmycobacterial nocardioform bacteria ranged from 96.9 to 99.0%, 96.1 to 97.3%, and 93.0 to 95.7%, respectively. The T103/T104 sequence was most identical to the sequences of M. aurum (identity, 99.0%) and M. komossense (identity, 98.9%).
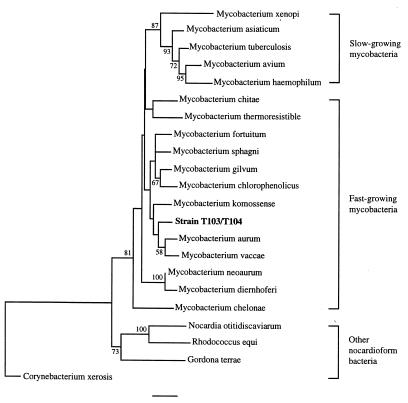
Distance and bootstrap analyses confirmed that strains T103 and T104 clustered with fast-growing mycobacteria (Fig. 1). The mycobacteria fell into a closely related, coherent group, distinct from the other high-G+C gram-positive bacteria examined. Within the genus, the slow-growing mycobacteria defined a distinct line of evolutionary descent, while the precise relationships among the fast-growing species remained unresolved because of low bootstrap values. Parsimony and maximum likelihood analyses also showed that strains T103 and T104 belonged with the fast-growing mycobacteria (data not shown).
FIG. 1.
Unrooted evolutionary distance tree based on the 16S rDNA sequences of strains T103 and T104, representative members of the genus Mycobacterium, and other high-G+C gram-positive bacteria. Bootstrap values greater than 50% are shown at the nodes. Bar = 0.01 nucleotide difference per sequence position.
Bacterial counts and toluene levels.
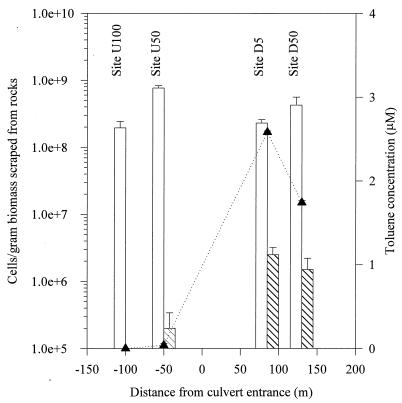
No toluene was detected at station U100, although very low levels of toluene (0.04 μM) were found at station U50. Downstream stations D5 and D50 had high toluene concentrations of 2.6 and 1.7 μM, respectively, in September 1993 (Fig. 2). The total heterotrophic bacterial plate counts at these stations ranged from 2 × 108 to 7.7 × 108 cells/g of biomass. Toluene-degrading bacteria were not detected for station U100, where no toluene was present. The counts of toluene-degrading bacteria for the other three stations increased with increasing toluene concentration and comprised 0.03, 1.09, and 0.35% of the total heterotrophic plate counts at stations U50, D5, and D50, respectively.
FIG. 2.
Bacterial counts (sampled on 7 September 1993) and toluene levels (sampled on 19 September 1993) along the East Drainage Ditch. Error bars represent standard deviations of duplicate samples. □, viable count of heterotrophic bacteria on 1% PTYG plates incubated at 20°C for up to 4 weeks in the dark. ▧, plate count of toluene-degrading bacteria on minimal salts agar with 110 μM toluene. Incubations were performed at 20°C for up to 4 weeks in the dark. ▴, toluene concentration.
DISCUSSION
Taxonomy and phylogeny.
Although strains T103 and T104 both degrade toluene and have major characteristics of the genus Mycobacterium, they exhibit some differences in physiology. Unlike strain T103, strain T104 is able to grow on the xylenes. Also, their fatty acid profiles are sufficiently different for them to be considered different strains of a novel species of fast-growing mycobacteria. Compared to the slow-growing mycobacteria, many of which are human and animal pathogens, most of the fast-growing mycobacteria are common saprophytes in natural habitats (18). They have been isolated from a diverse array of habitats, are able to survive and multiply under a wide range of environmental conditions, and can biotransform a variety of xenobiotic compounds and pollutants, including polycyclic aromatic hydrocarbons (15) and groundwater-pollutant mixtures (4). There is a long history of isolation of mycobacteria that have the capacity to degrade the aromatic fraction of the oil in contaminated soils (44, 45).
The closest known relative of strains T103 and T104, M. aurum, is a fast-growing species commonly isolated from soils (47) that is able to metabolize morpholine (6, 26) and vinyl chloride (17). The other closely related species, M. komossense, is a fast-growing nonpathogenic species isolated from Sphagnum vegetation of moors in south Sweden and the Atlantic coastal area of Norway (19). It is unable to utilize benzoate and benzamide; it is not known if it possesses the ability to utilize other aromatic hydrocarbons or xenobiotic compounds. Other closely related species have been found to degrade a range of aromatic hydrocarbons. M. vaccae is the only other Mycobacterium species known to grow on toluene. It also grows on acetone and can degrade acetone, cyclohexane, styrene, benzene, ethylbenzene, propylbenzene, dioxane, and 1,2-dichloroethylene (4). A Mycobacterium sp. that was closely related to M. gilvum (14) was isolated from the soil of a former coal gasification site and is able to degrade the polycyclic aromatic hydrocarbons phenanthrene, pyrene, and fluoranthene (2). Overall, their physiological versatility suggests that the fast-growing mycobacteria should be important in pollutant biodegradation, and our results point to the possibility that mycobacteria may be useful in the bioremediation of contaminated environments.
Biodegradation kinetics.
Toluene contamination of the East Drainage Ditch appears to selectively enrich for toluene-degrading bacteria within the epilithic microbial community. Maximal velocities are somewhat higher than those reported for other aerobic toluene-degrading bacteria; strain T104 also has a larger Ks (3.8 μM) than those reported for other toluene-degrading bacterial strains. Pseudomonas sp. strain T2 had a maximal velocity of 0.30 μmol of toluene/mg of protein per h (assuming that 50% of a typical cell’s dry weight is protein) and a Ks of 0.48 μM (32). Corresponding values for a terrestrial strain of Pseudomonas putida, PpF1, were 0.43 μmol of toluene/mg of protein per h and 0.68 μM, respectively (32). That Ks values for strains T103 and T104 lie within the range of toluene concentrations observed in the stream suggests that these strains are adapted to the ambient level of toluene contamination in the stream. Noncarbon nutrients are unlikely to be limiting in this case, because these are present in the stream at high levels (43).
In order to assess the role that strains T103 and T104 may play in toluene biodegradation in intact biofilms, toluene biodegradation rates for the pure cultures were compared with rates obtained in the laboratory for rock biofilms from a previous study (7). For East Drainage Ditch rocks with their natural biofilms under summer conditions, the rate was observed to be first order for toluene concentrations up to 2.2 μM and approached zero order for concentrations greater than 4.3 μM; the Vmax was 2.0 nmol/cm2 of rock surface per h (7). Assuming that mass transport is not limiting, the potential contributions of strains T103 and T104 to toluene biodegradation on the rock biofilm were estimated according to their cell densities (CD) on the rock surfaces and their individual kinetic parameters. The CD of 1.8 × 104 cells/cm2 of rock surface was estimated from plate counts of the mycobacterial isolates (106 cells/g of biomass) and biomass density (0.018 g of biomass/cm2 of rock surface). Since the biofilm data were most reliable for toluene concentrations greater than 4.3 μM (7), comparisons were performed for toluene concentrations greater than or equal to 4.3 μM. Assuming a typical cell protein weight of 0.2 pg (24) and a uniform distribution of mycobacteria in the biofilm, the relative contributions of strains T103 and T104 to toluene biodegradation by the biofilm can be estimated as follows:
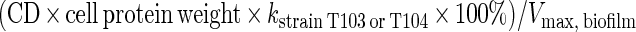 |
where
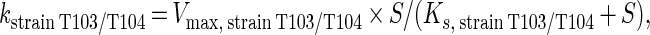 |
and S is the toluene concentration.
At a toluene concentration of 4.3 μM, strains T103 and T104 are estimated to account for 0.2 and 0.6%, respectively, of the toluene biodegradation that occurs on the rock surfaces. These numbers should be interpreted as upper bounds on the relative contributions of the pure cultures to toluene degradation in the biofilms; to the extent that diffusion limitation occurs, these numbers may be lower, especially in the case of T104. Strain T103, having a Ks much lower than 4.3 μM, would be less affected. The low relative contributions suggest that other bacterial species play a larger role than strains T103 and T104 in toluene biodegradation on the rock surfaces. Cohen et al. (7) showed that constantly shaken flasks provided a reasonable microcosm simulation of the fast-flowing stream. In addition, mass transport of the substrate to the biofilm surface is not calculated to be a limiting factor in the turbulent flow regime of this stream, although biofilms themselves are not always free of transport limitation (21). We therefore expect that the results from laboratory microcosms offer a reasonable estimate of the importance of T103 and T104 to toluene degradation in the East Drainage Ditch itself.
Microbial ecology.
Toluene levels in the stream do not appear to have a significant effect on total heterotrophic bacterial counts. On the other hand, counts of toluene-degrading bacteria in samples from the contaminated stations are about an order of magnitude higher than those from pristine stations, which suggests that toluene in the contaminated reaches of the stream has caused the epilithic bacterial communities to adapt by selectively enriching for toluene-degrading bacteria. Other studies of freshwater environments (30, 40) also reported viable heterotrophic counts to be unaffected by the presence of a xenobiotic contaminant, while the numbers of organisms capable of degrading the contaminant of interest increased by between 2 and 3 orders of magnitude. The increased counts in contaminated reaches versus uncontaminated reaches of the East Drainage Ditch are not as high as observed in these other studies, possibly because of greatly reduced microbial activity at lower temperatures (30). The East Drainage Ditch study was conducted in the fall when stream temperatures averaged 11°C. In the other studies, temperatures averaged 18°C (40) or ranged from 19 to 30°C (30).
Although strains T103 and T104 were isolated from rock biofilms collected in July 1992, a 16S rDNA clone with a sequence that is identical to that of T103 and T104 was recovered independently from DNA from rock biofilms in July 1993 (43). This detection of the presence of the Mycobacterium sp. in the stream at different times suggests that this species is a permanent member of the stream’s microbial community. Attempts to isolate the Mycobacterium sp. from the pristine stations were unsuccessful. This, coupled with the relative ease with which the Mycobacterium sp. strains were isolated from station D5, suggests a greater abundance of this species in the contaminated reaches of the stream, perhaps as a result of selection by toluene.
In summary, we describe two closely related toluene-degrading mycobacterial strains isolated from rock surface biofilms from a toluene-contaminated freshwater stream. These strains constitute a novel fast-growing Mycobacterium species and extend our knowledge of the list of Mycobacterium species that can grow on toluene. Fast-growing mycobacteria are seen to play an important role in the environment, as evidenced by increasing discoveries of members within this group that can degrade a wide range of xenobiotic compounds. Our ecological experiments show a stream response to the presence of low levels of toluene. This community response appears to be more complex than the stimulation of a single indigenous toluene-degrading species; other microorganisms that are involved in toluene degradation must also be present.
ACKNOWLEDGMENTS
This work was supported by NIEHS Superfund Basic Research Program grant 5P42ES04675-06 and Office of Naval Research grant N00014-91-J-1489.
We thank Michael Collins for carrying out the kinetics experiment, Karen Dohrman for performing the fatty acid analysis, Sue Lootens for performing 16S rDNA sequencing, and Debra Lonergan for advising on aspects of 16S rDNA sequencing and analyses.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 2.Boldrin B, Tiehm A, Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1993;59:1927–1930. doi: 10.1128/aem.59.6.1927-1930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bone T L, Balkwill D L. Morphological and cultural comparison of microorganisms in surface soil and subsurface sediments at a pristine study site in Oklahoma. Microbiol Ecol. 1988;16:49–64. doi: 10.1007/BF02097404. [DOI] [PubMed] [Google Scholar]
- 4.Burback B L, Perry J J. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. Appl Environ Microbiol. 1993;59:1025–1029. doi: 10.1128/aem.59.4.1025-1029.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Button D K, Robertson B R, Craig K S. Dissolved hydrocarbons and related microflora in a fjordal seaport: sources, sinks, concentrations, and kinetics. Appl Environ Microbiol. 1981;42:708–719. doi: 10.1128/aem.42.4.708-719.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Čech J S, Hartman P, Šlosárek M, Chudoba J. Isolation and identification of a morpholine-degrading bacterium. Appl Environ Microbiol. 1988;54:619–621. doi: 10.1128/aem.54.2.619-621.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen B A, Krumholz L R, Kim H, Hemond H F. In-situ biodegradation of toluene in a contaminated stream. 2. Laboratory studies. Environ Sci Technol. 1995;29:117–125. doi: 10.1021/es00001a015. [DOI] [PubMed] [Google Scholar]
- 8.DeLong E. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Difco Laboratories. Difco manual: dehydrated culture media and reagents for microbiology. Detroit, Mich: Difco Laboratories; 1985. [Google Scholar]
- 10.Duetz W A, de Jong C, Williams P A, van Andel J G. Competition in chemostat culture between Pseudomonas strains that use different pathways for the degradation of toluene. Appl Environ Microbiol. 1994;60:2858–2863. doi: 10.1128/aem.60.8.2858-2863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durant J L. M.S. thesis. Cambridge: Massachusetts Institute of Technology; 1991. [Google Scholar]
- 12.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 14.Fritzsche C. Degradation of pyrene at low defined oxygen concentrations by a Mycobacterium species. Appl Environ Microbiol. 1994;60:1687–1689. doi: 10.1128/aem.60.5.1687-1689.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerin W F, Jones G E. Mineralization of phenanthrene by a Mycobacterium sp. Appl Environ Microbiol. 1988;54:937–944. doi: 10.1128/aem.54.4.937-944.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Häggblom M M, Nohynek L J, Palleroni N J, Kronqvist K, Nurmiaho-Lassila E-L, Salkinoja-Salonen M S, Klatte S, Kroppenstedt R M. Transfer of polychlorophenol-degrading Rhodococcus chlorophenolicus (Apajalahti et al. 1986) to the genus Mycobacterium as Mycobacterium chlorophenolicum comb. nov. Int J Syst Bacteriol. 1994;44:485–493. doi: 10.1099/00207713-44-3-485. [DOI] [PubMed] [Google Scholar]
- 17.Hartmans S, de Bont J A M. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl Environ Microbiol. 1992;58:1220–1226. doi: 10.1128/aem.58.4.1220-1226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmans S, de Bont J A M. The genus Mycobacterium—nonmedical. In: Balows A, Truper H, Dworkin M, Harder L, Schleifer K H, editors. The prokaryotes. N.Y: Springer-Verlag; 1992. pp. 1214–1237. [Google Scholar]
- 19.Kazda J, Müller K. Mycobacterium komossense sp. nov. Int J Syst Bacteriol. 1979;29:361–365. [Google Scholar]
- 20.Kim H, Hemond H F, Krumholz L R, Cohen B A. In-situ biodegradation of toluene in a contaminated stream. 1. Field studies. Environ Sci Technol. 1995;29:108–116. doi: 10.1021/es00001a014. [DOI] [PubMed] [Google Scholar]
- 21.Kim H. Ph.D. thesis. Cambridge: Massachusetts Institute of Technology; 1995. [Google Scholar]
- 22.Kirschner E M. Production of top 50 chemicals increased substantially in 1994. Chem Eng News. 1995;73(15):16–22. [Google Scholar]
- 23.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 24.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular cell biology. 3rd ed. New York, N.Y: Scientific American; 1995. [Google Scholar]
- 25.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazure N, Truffaut N. Degradation of morpholine by Mycobacterium aurum MO1. Can J Microbiol. 1994;40:761–765. doi: 10.1139/m94-120. [DOI] [PubMed] [Google Scholar]
- 27.Murray R G E, Doetsch R N, Robinow C F. Determinative and cytological light microscopy. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 21–41. [Google Scholar]
- 28.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 29.Ooyama J, Foster J W. Bacterial oxidation of cycloparaffinic hydrocarbons. Antonie Leeuweenhoek. 1965;31:45–65. doi: 10.1007/BF02045875. [DOI] [PubMed] [Google Scholar]
- 30.Pignatello J J, Johnson L K, Martinson M M, Carlson R E, Crawford R L. Response of the microflora in outdoor experimental streams to pentachlorophenol: environmental factors. Can J Microbiol. 1986;32:38–46. doi: 10.1139/m86-008. [DOI] [PubMed] [Google Scholar]
- 31.Polz M F, Odintsova E V, Cavanaugh C M. Phylogenetic relationships of the filamentous sulfur bacterium Thiothrix ramosa based on 16S rRNA sequence analysis. Int J Syst Bacteriol. 1996;46:94–97. doi: 10.1099/00207713-46-1-94. [DOI] [PubMed] [Google Scholar]
- 32.Robertson B R, Button D K. Toluene induction and uptake kinetics and their inclusion in the specific-affinity relationship for describing rates of hydrocarbon metabolism. Appl Environ Microbiol. 1987;53:2193–2205. doi: 10.1128/aem.53.9.2193-2205.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. Newark, Del: Microbial ID, Inc.; 1990. [Google Scholar]
- 34.Sasser M. “Tracking” a strain using the microbial identification system. MIDI technical note 102. Newark, Del: Microbial ID, Inc.; 1990. [Google Scholar]
- 35.Schraa G, Boone M L, Jetten M S M, van Neerven A R W, Colberg P J, Zehnder A J B. Degradation of 1,4-dichlorobenzene by Alcaligenes sp. strain A175. Appl Environ Microbiol. 1986;52:1374–1381. doi: 10.1128/aem.52.6.1374-1381.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarzenbach R P, Gschwend P M, Imboden D M. Environmental organic chemistry. New York, N.Y: John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 37.Smibert R M, Krieg N R. Phenotypic characterization. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 607–654. [Google Scholar]
- 38.Smith M R. The biodegradation of aromatic hydrocarbons by bacteria. Biodegradation. 1990;1:191–206. doi: 10.1007/BF00058836. [DOI] [PubMed] [Google Scholar]
- 39.Smith S W, Overbeek R, Olsen G, Woese C, Gillevet P M, Gilbert W. Genome mapping and sequencing. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. p. 190. [Google Scholar]
- 40.Spain J C, Van Veld P A, Pritchard P H, Cripe C R. Comparison of p-nitrophenol biodegradation in field and laboratory test systems. Appl Environ Microbiol. 1984;48:944–950. doi: 10.1128/aem.48.5.944-950.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahl D A, Urbance J W. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J Bacteriol. 1990;172:116–124. doi: 10.1128/jb.172.1.116-124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swofford D L. PAUP: phylogenetic analysis using parsimony, version 3.1. Champaign, Ill: Illinois Natural History Survey; 1991. [Google Scholar]
- 43.Tay S T-L. Ph.D. thesis. Cambridge: Massachusetts Institute of Technology; 1998. [Google Scholar]
- 44.Traxler R W, Proteau P R, Traxler R N. Action of microorganisms on bituminous materials. I. Effect of bacteria on asphalt viscosity. Appl Microbiol. 1965;13:838–841. doi: 10.1128/am.13.6.838-841.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traxler R W, Robinson J A, Wetmore D E, Traxler R N. Action of microorganisms on bituminous materials. II. Composition of low molecular weight asphaltic fractions determined by microbial action and infra-red analyses. J Appl Chem. 1966;16:266–271. [Google Scholar]
- 46.U.S. Public Health Service. Toxicological profile for toluene. Publication ATSDR/TP-89/23. Atlanta, Ga: Agency for Toxic Substances and Disease Registry, U.S. Public Health Service; 1989. [Google Scholar]
- 47.Wayne L G, Kubica G P. Genus Mycobacterium Lehmann and Neumann 1896, 363. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams and Wilkins Co.; 1986. pp. 1436–1457. [Google Scholar]
- 48.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zehnder A J B, Huser B A, Brock T D, Wuhrmann K. Characterization of an acetate-decarboxylating non-hydrogen-oxidizing methane bacterium. Arch Microbiol. 1980;124:1–11. doi: 10.1007/BF00407022. [DOI] [PubMed] [Google Scholar]
- 50.Zeyer J, Kuhn E P, Schwarzenbach R P. Rapid microbial mineralization of toluene and 1,3-dimethylbenzene in the absence of molecular oxygen. Appl Environ Microbiol. 1986;52:944–947. doi: 10.1128/aem.52.4.944-947.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]