Abstract
Seven blue nucleic acid dyes from Molecular Probes Inc. (SYTO-9, SYTO-11, SYTO-13, SYTO-16, SYTO-BC, SYBR-I and SYBR-II) were compared with the DAPI (4′,6-diamidino-2-phenylindole) method for flow cytometric enumeration of live and fixed bacteria in aquatic systems. It was shown that SYBR-II and SYTO-9 are the most appropriate dyes for bacterial enumeration in nonsaline waters and can be applied to both live and dead bacteria. The fluorescence signal/noise ratio was improved when SYTO-9 was used to stain living bacteria in nonsaline waters. Inversely, SYBR-II is more appropriate than SYTO dyes for bacterial enumeration of unfixed and fixed seawater samples.
Quantification of total bacterial numbers is a basic and essential task in several areas of microbiology, including public health, biotechnology, the food, water, and pharmaceutical industries, and natural environments. During the last two decades, total direct counting methods that use fluorochrome stains and epifluorescence microscopy have become increasingly popular because most naturally occurring communities cannot be enumerated accurately as CFU by culturing on various agar media (16, 18). Advances in the field of fluorescent dye technology and flow cytometry now allow the application of this rapid, automated technique to such studies. Flow cytometry has become increasingly popular because it offers the advantage over microscopy of rapid, easy, and accurate enumeration (6, 12, 14, 15, 19). This increasing popularity is also due to the recent development of low-cost compact flow cytometers.
The laser-based flow cytometers that are currently commercially available are equipped with argon lasers. Air-cooled argon lasers are not tunable, running at only one wavelength, namely 488 nm. For this reason, an increasing number of blue-light-excited nucleic acid dyes have been developed. Although some of these dyes have been successfully applied to natural bacterial communities, comparisons of the performance of these dyes in different types of aquatic environments have never been made and should be useful for new investigators.
An expanding list of blue-excitable dyes such as TOTO, TO-PRO, YOYO, YO-PRO, PicoGreen, SYTO-13, and SYBR Green I (referred to hereafter as SYBR-I) have been developed for the quantification of microorganisms in aquatic systems (4, 12, 14, 15). Among these, SYTO-13 and SYBR-I have been shown to be well suited for the enumeration of bacterioplankton in natural aquatic ecosystems (4, 15). Recently, new nucleic acid-specific dyes with different quantum yields on DNA and RNA have been developed. For instance, the quantum yield of SYTO-16 for DNA is much higher than those of SYTO-13 and SYTO-11, which have higher binding affinities for RNA. SYTO-9, which is not commercially available alone but is provided in the LIVE/DEAD bacterial viability kit (Molecular Probes Inc., Eugene, Oreg.), yields bright fluorescence when applied to living bacteria. A new specific bacterial counting kit from the same company has been marketed based on the use of the SYTO-BC stain (Molecular Probes Inc.). SYBR-II has been developed for DNA and RNA staining in gels and has never been compared to SYBR-I for bacterial enumeration. The SYTO dyes penetrate intact live cells, while others are generally applied to fixed samples. Staining procedures that work with live cells are of great interest in situations which require (i) rapid assessment of total bacterial counts and (ii) total cell enumeration after a first staining step of living cells with viability or activity dyes. It is also important because fixation results in cell shrinkage and induces biased light-scattering measurements and a decrease in fluorescence emission for SYTO dyes (4).
This study was undertaken (i) to compare the staining efficiencies of different dyes, including those most recently developed, and (ii) to estimate total bacterial abundances in live and fixed samples from different aquatic systems, including fresh and saline waters.
MATERIALS AND METHODS
Natural samples.
Bottled mineral waters were purchased in 1-liter plastic bottles. River water samples were collected in the Tech River (along the Mediterranean coast of France) at three stations (Pas-du-Loup, Saint-Paul, and Pont d’Elne) in February 1997. Surface seawater samples were collected with Niskin bottles at three different stations in the Mediterranean Sea near Banyuls-sur-Mer (France) in January and February 1997. SYTO and SYBR bacterial counts were determined by flow cytometry, and DAPI (4′,6-diamidino-2-phenylindole) counts were determined by epifluorescence microscopy. We counted bacteria in fresh samples within 2 h of sampling and in fixed samples within 2 days. Fixed samples were stored at 4°C in the dark.
Cultures.
Salmonella typhimurium CIP 60.62T (Collection Institut Pasteur, Paris, France) was grown at 37°C on Trypticase soy agar (TSA) or broth (TSB) medium (bioMérieux). For growth experiments, 100 ml of fresh TSB medium prewarmed at 37°C was inoculated with 1% (vol/vol) of an overnight culture (at 37°C under continuous shaking). Growth was monitored spectrophotometrically at 600 nm, and 30-ml subsamples were taken and fixed at different stages of the growth curve for RNA analysis (see below). An aliquot of 10 ml was used for live cell analysis, and two aliquots of 10 ml were fixed as described below.
Fixation.
An aliquot (10 ml) of each sample was fixed for at least 15 min with 2% (vol/vol) formaldehyde (final concentration) and stored in the dark at 4°C until analysis. For fluorescent in situ hybridizations (FISH), a second aliquot (10 ml) was fixed with 3 volumes of 4% paraformaldehyde in phosphate-buffered saline (PBS) (130 mM sodium chloride, 10 mM sodium phosphate buffer [pH 7.2]) and incubated at 4°C for 4 h. Then, cells were pelleted by centrifugation in an Eppendorf microcentrifuge (8,000 × g, 2 min), washed with PBS, and resuspended in 1 volume of PBS and 1 volume of absolute ethanol at a concentration of 108 to 109 cells per ml. The fixed cells were stored at −20°C.
Whole-cell hybridizations.
The rRNA contents of individual cells were determined by whole-cell hybridizations using EUB338 and non-EUB338 (negative control) oligonucleotide probes (20). Cells were hybridized at 46°C for 2 h in 50 μl of hybridization buffer (0.9 M sodium chloride, 20 mM Tris-HCl, 100 μg of polyadenylic acid per ml, 10% of a Denhardt solution [100×; Sigma], 0.01% sodium dodecyl sulfate [pH 7.2]) containing 1 ng of probe per μl in a 1.6-ml Eppendorf tube. The hybridization was stopped by adding 500 μl of cold PBS solution (pH 8.4, 0°C). Samples were stored on ice (0.5 to 1 h) and in the dark until analysis. Both electrophoretically purified unlabeled probes and fluorescence-labeled oligonucleotide probes were obtained from Eurogentec (Seraing, Belgium). An amino group was attached to the 5′ end of the oligonucleotide in the last stage of synthesis. Labeling was performed by linking fluorescein isothiocyanate to the 5′ end of the oligonucleotide via a six-carbon spacer arm. Fluorescence was analyzed by flow cytometry (see below).
Dyes and staining conditions.
SYBR-I, SYBR-II, SYTO-9, SYTO-11, SYTO-13, and SYTO-16 are high-affinity nucleic acid stains (Molecular Probes Inc.). The spectral characteristics and quantum yields on DNA and RNA of these dyes are reported in Table 1. SYTO-9 is not commercially available alone but is provided in the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes Inc.). The bacterial counting kit from the same company is based on the use of a SYTO-BC stain, but no further information concerning the molecule is provided by the manufacturer. All dyes were delivered in dimethyl sulfoxide, and commercial stock solutions were stored at −20°C.
TABLE 1.
Absorption, fluorescence maxima, and fluorescence quantum yields for DNA and RNA of the dyes tested in this studya
| Dye | Abs | Em | QY
|
|
|---|---|---|---|---|
| DNA | RNA | |||
| SYTO-9 | 480 | 500 | 0.60*b | 0.20* |
| SYTO-11 | 508–510 | 527–530 | 0.49 | 0.39 |
| SYTO-13 | 488–491 | 509–514 | 0.4 | 0.4 |
| SYTO-16 | 488 | 518–525 | 0.65 | 0.24 |
| SYBR-I | 494 | 521 | 0.80 | 0.40* |
| SYBR-II | 494 | 521 | 0.36 | 0.54 |
| Counting kit | 488 | 530 | —c | — |
Abbreviations: Abs, absorption; Em, fluorescence maximum; QY, quantum yield.
∗, estimation provided by Molecular Probes Inc.
—, unknown (information not provided by the manufacturer).
The staining procedures for new dyes were optimized for both dead and live cells. For SYTO-9, -11, -13, and -16 dyes and SYBR-II, dye concentrations and incubation times were optimized on fixed S. typhimurium cells and on natural waters by testing three concentrations (2.5, 5, and 10 μM) and recording the intensities of cellular fluorescence at time intervals. Then, for optimization of the staining solution, the effects of both sodium citrate and potassium citrate were tested (14).
For all live and fixed samples and after optimization of staining procedures, sodium citrate (pH 7.4, 50 mM final concentration) and potassium citrate (pH 7.4, 30 mM final concentration) were added when staining with SYTO and SYBR dyes, respectively. For cultures, live and fixed cells were directly diluted in the saline solution. SYBR-I and SYBR-II dyes were added at a final concentration of 10−4 of the stock solution and incubated for 15 min in the dark. The optimized staining procedure for SYBR-II was similar to that used for SYBR-I. For the counting kit (SYTO-BC dye), the conditions were those recommended by the manufacturer.
Evaluations of both DNA and RNA staining efficiency were done for SYTO-9 and SYBR-II. In this case, optimized staining procedures were applied after the fixed cells had been incubated (37°C, 60 min) in the presence of RNase A (type 1A, Sigma R-4875) at 400 Kunitz units ml−1 and RNase B (Sigma R-5750) at 500 Kunitz units ml−1 (14) or not incubated. Ribonucleases were rendered free of DNase (13). Nonspecific staining was tested by combining DNA and RNA digestions. DNA digestion was performed with DNase I (type 4, Sigma D-5025) at 2,000 Kunitz units ml−1 (37°C, 60 min). Pronase E (Sigma P6911) was used alone or combined with nucleases at 50 μg ml−1 to test for the possibility of dye-restricting nucleoproteins (12).
Flow cytometry.
All experiments were performed with a FACS-Calibur flow cytometer (Becton Dickinson) equipped with an air-cooled laser providing 15 mW at 488 nm and the standard filter setup. All parameters were collected as logarithmic signals. Green fluorescence was collected in the FL1 channel (530 ± 15 nm). For marine samples, the presence of phytoplanktonic cells in natural samples, mainly Prochlorococcus spp., was checked in the FL3 channel (>630 nm).
For cell enumeration, two procedures were tested because quantification of the analyzed volume is not available on the FACS-Calibur flow cytometer: (i) a 1-ml sample was put in a 12- by 75-mm plastic tube and was weighed before and after analysis in order to determine the analyzed volume, and (ii) cells were enumerated during a fixed time (generally 1 min) at a given flow rate which was calibrated at the beginning and end of each analysis session. The second procedure was finally used for routine work but both procedures yielded similar results. Yellow-green fluorescent microspheres (0.95-μm-diameter fluorescent size-standard beads; Polysciences Inc., Warrington, Pa.) were systematically added to each sample as an internal reference. This internal standard allows the normalization of cell fluorescences, which were expressed in bead fluorescence units (19). All microscopic counts were performed by DAPI staining, as previously reported, with 47-mm-diameter filters (10).
RESULTS AND DISCUSSION
Staining procedures.
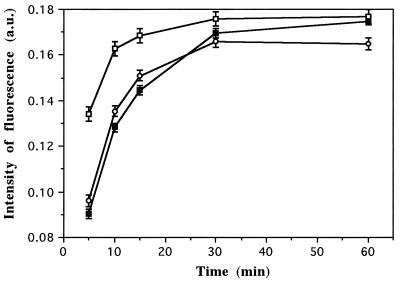
The staining kinetics of SYTO dyes were determined by flow cytometry of live and fixed S. typhimurium cells from a stationary-phase culture. Three dye working concentrations (2.5, 5, and 10 μM) were chosen after preliminary experiments (data not shown). Figure 1 shows the results obtained for fixed S. typhimurium cells stained with SYTO-9. The green fluorescence intensity increased rapidly during the first 15 min, and equilibrium was reached by 30 min. Similar results were obtained for all SYTO dyes (data not shown). Although staining for 30 min at 2.5 μM was not significantly different from staining at 5 μM, the reproducibility of results including those from natural samples was better at the higher concentration. Thus, for further tests with SYTO-type dyes, the staining concentration was 5 μM and incubations were performed at room temperature in the dark for 30 min. When applied to natural samples, the kinetics of staining of SYTO dyes were similar. Fluorescence of stained cells peaked after 15 min for both SYBR-I and -II dyes (data not shown). When applied to living cells, equilibrium was reached after 30 and 40 min for SYBR and SYTO dyes, respectively (data not shown).
FIG. 1.
Staining kinetics of fixed S. typhimurium cells stained with SYTO-9 at three final concentrations: 2.5 μM (closed squares), 5 μM (circles), and 10 μM (open squares). Standard deviations were determined with triplicate samples. a.u., arbitrary units.
Both the concentrations and incubation times found for SYTO dyes were higher than those reported by del Giorgio et al. (4), who worked with lake waters stained with SYTO-13. As stated by those authors, we also found that the fluorescence of fixed cells was lower than that of live cells; this difference was much greater for marine samples (see below). The staining conditions found for SYBR-I and SYBR-II dyes were in general agreement with those reported for SYBR-I by Marie et al. (15).
Effects of salts.
The effect of salt addition on the fluorescence signal was also tested for each dye (Table 2). The addition of 50 mM sodium citrate improved staining with all SYTO dyes, whereas the fluorescence of SYBR (I and II)-stained cells was higher when incubated with 30 mM of potassium citrate. These results are similar to those reported with SYBR-I for natural seawater samples (15). For all other experiments, the protocol used for staining cultures and natural samples consisted of staining with SYTO dyes at 5 μM (final concentration) in the presence of 30 mM sodium citrate and an incubation of 30 and 45 min for fixed and live cells, respectively. For SYBR-I and SYBR-II, cells were stained with a 10−4 concentration of stock solution in the presence of 30 mM potassium citrate and incubated for 15 and 30 min for fixed and unfixed bacteria, respectively. All incubations were at room temperature in the dark.
TABLE 2.
Relative fluorescence intensities of fixed bacterial cells from mineral water and seawater samples with various dyes
| Sample and dye | Mean relative fluorescence intensity (SE) (101)a
|
||
|---|---|---|---|
| Normal | Sodium citrate | Potassium citrate | |
| Mineral water | |||
| SYTO-9 | 0.27 (<0.01) | 0.38 (<0.01) | 0.38 (<0.01) |
| SYTO-11 | 0.03 (<0.01) | 0.06 (<0.01) | 0.06 (<0.01) |
| SYTO-13 | 0.22 (0.02) | 0.29 (0.05) | 0.28 (0.04) |
| SYTO-16 | 0.24 (<0.01) | 0.35 (<0.01) | 0.33 (<0.01) |
| SYBR-I | 0.26 (<0.01) | 0.27 (0.02) | 0.31 (<0.01) |
| SYBR-II | 0.27 (<0.01) | 0.27 (<0.01) | 0.33 (<0.01) |
| Seawater | |||
| SYTO-9 | 0.12 (0.03) | 0.17 (0.02) | 0.06 (<0.01) |
| SYTO-11 | 0.08 (<0.01) | 0.09 (<0.01) | 0.08 (<0.01) |
| SYTO-13 | 0.08 (<0.01) | 0.10 (<0.01) | 0.04 (<0.01) |
| SYTO-16 | 0.07 (<0.01) | 0.10 (<0.01) | 0.08 (<0.01) |
| SYBR-I | 0.16 (<0.01) | 0.08 (<0.01) | 0.19 (<0.01) |
| SYBR-II | 0.15 (<0.01) | 0.10 (<0.01) | 0.19 (0.01) |
Samples were labeled with each of seven dyes: SYTO-9, -11, -13, -16, SYBR-I, and SYBR-II. For labeling, samples were not supplemented (normal) or supplemented with Tris-EDTA with buffer containing sodium citrate (50 mM, final concentration) or potassium citrate (30 mM, final concentration). Data are from three separate experiments.
Comparisons of the different dyes.
Table 3 shows comparisons of relative green fluorescence of fixed bacterial cells from a culture and from mineral, river, and seawater samples stained with the different dyes. For each water type, the mean fluorescence intensities vary greatly between the different dyes. For low-salinity samples (culture, mineral, and river water samples), the fluorescence intensity was high with SYTO-9, SYTO-16, SYBR-I and SYBR-II, whereas SYTO-13, SYTO-11, and the counting kit yielded lower signals. The highest mean intensity of green fluorescence was obtained with SYTO-9. Among SYBR-type dyes, the higher fluorescence signals were obtained with SYBR-II. Conversely, for marine samples the fluorescence intensity was higher with SYBR (I and II) dyes. Surprisingly, the fluorescence intensities of cells stained with both SYBR-I and SYBR-II dyes revealed little difference, whereas the quantum yields of the two stains for DNA and RNA are very different (Table 1). These results suggest that quantum yields should be used with caution in assessing the apparent DNA and RNA contents of individual cells.
TABLE 3.
Relative fluorescence intensities of fixed bacterial cells from culture, mineral water, river water, and seawater samples with various dyes
| Sample | Relative fluorescence intensity (101)a
|
||||||
|---|---|---|---|---|---|---|---|
| SYTO-9 | SYTO-11 | SYTO-13 | SYTO-16 | SYBR-I | SYBR-II | Kitb | |
| Culture | |||||||
| S. typhimurium | 1.52 | 0.80 | 1.21 | 1.37 | 1.34 | 1.48 | 1.20 |
| Mineral water | |||||||
| Type 1 | 0.43 | 0.13 | 0.27 | 0.35 | 0.30 | 0.38 | 0.25 |
| Type 2 | 0.43 | 0.08 | 0.25 | 0.42 | 0.31 | 0.37 | 0.23 |
| Type 3 | 0.38 | 0.06 | 0.29 | 0.30 | 0.27 | 0.32 | 0.27 |
| River water | |||||||
| Station 1 | 0.65 | 0.09 | 0.34 | 0.62 | 0.40 | 0.46 | 0.35 |
| Station 2 | 0.71 | 0.04 | 0.32 | 0.67 | 0.37 | 0.52 | 0.30 |
| Station 3 | 0.43 | 0.10 | 0.28 | 0.41 | 0.38 | 0.35 | 0.29 |
| Seawater | |||||||
| Station 1 | 0.68 | 0.25 | 0.36 | 0.16 | 0.69 | 0.69 | 0.49 |
| Station 2 | 0.17 | 0.09 | 0.10 | 0.10 | 0.19 | 0.18 | 0.12 |
| Station 3 | 0.21 | 0.09 | 0.11 | 0.07 | 0.23 | 0.27 | 0.12 |
The standard error determined for triplicate samples was less than 0.002 in each case.
Counting kit, with SYTO-BC dye.
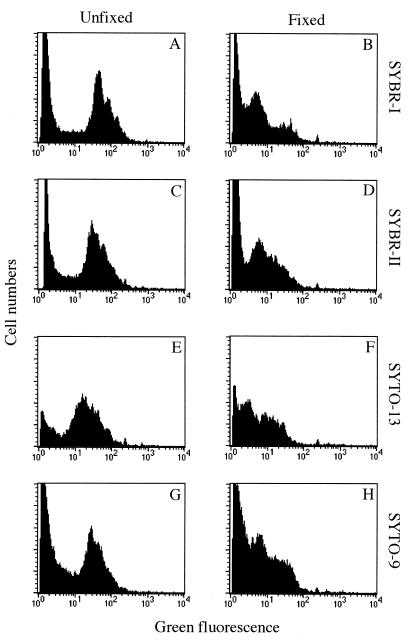
For all SYTO and SYBR dyes, fixation of the cells before staining resulted in a decrease in the fluorescence of fixed relative to unfixed bacteria. Table 4 shows the effect of formaldehyde fixation on the green fluorescence emission of cells stained with SYTO-9 and SYBR-II. We noted a strong decrease in the mean fluorescence emissions of stained cells after formaldehyde fixation. The effect of fixation on stationary-phase cells from a pure culture was lower. This decrease resulted in lower fluorescence signal/noise ratios, which were due to the lower fluorescence signals of stained bacteria (Fig. 2). Although this lower fluorescence ratio had no effect on cell counting in nonsaline samples because bacteria remained clearly discriminable, it was more problematic for marine samples (Fig. 2). In marine waters, the discrimination of bacterial cells was easier with live cells since the fluorescence distribution of fixed cells was closer to the origin (Fig. 2). With fixed seawater samples, the fluorescence noise, which most likely results from the presence of fluorescent organic particles naturally occurring in seawater, interfered with the cytometric signals of stained bacteria. These interferences, which were more important with SYTO dyes, were due to a variable overlapping of both signal and noise fluorescence distributions and resulted in biased estimations (generally an overestimation) of bacterial counts (data not shown). The higher signal/noise ratio was obtained with SYBR-II (Fig. 2).
TABLE 4.
Relative fluorescence intensities of live and dead (fixed) S. typhimurium log-phase cells from mineral and marine waters stained with SYTO-9 and SYBR-II
| Sample | Mean relative fluorescence intensity (SE) (101)a
|
|||
|---|---|---|---|---|
| SYTO-9
|
SYBR-II
|
|||
| Live | Dead | Live | Dead | |
| Culture | ||||
| S. typhimurium | 11.17 (0.52) | 11.52 (0.89) | 11.14 (0.35) | 11.48 (0.40) |
| Mineral water | ||||
| Type 1 | 0.52 (0.01) | 0.43 (<0.01) | NDb | ND |
| Type 2 | 0.48 (<0.01) | 0.43 (0.01) | ND | ND |
| Type 3 | 0.56 (0.01) | 0.38 (0.01) | 0.42 (0.01) | 0.32 (0.01) |
| Type 4 | 0.61 (0.01) | 0.43 (0.01) | 0.54 (0.01) | 0.35 (0.01) |
| Seawater | ||||
| Station 1 | 0.77 (<0.01) | 0.68 (<0.01) | 0.81 (0.03) | 0.69 (0.02) |
| Station 2 | 0.24 (0.01) | 0.17 (0.01) | 0.38 (0.01) | 0.30 (0.01) |
| Station 3 | 0.23 (0.01) | 0.21 (0.01) | ND | ND |
| Station 4 | 0.22 (0.01) | 0.10 (0.01) | 0.52 (0.02) | 0.43 (0.01) |
| Station 5 | 0.23 (0.02) | 0.16 (<0.01) | 0.45 (0.02) | 0.33 (0.01) |
| Station 6 | 0.25 (0.01) | 0.18 (<0.01) | 0.42 (0.02) | 0.31 (0.01) |
Data are from three separate experiments.
ND, not determined.
FIG. 2.
Fluorescence histograms of bacteria from unfixed and fixed seawater samples stained with SYBR-I, SYBR-II, SYTO-9, and SYTO-13.
This effect of formaldehyde fixation on fluorescence intensity was reported previously by del Giorgio et al. (4) for SYTO-13. Those authors suggested that it could be explained by morphological changes after fixation and/or by decreased permeability of formaldehyde-fixed cells to the dyes. We suggest that the positive charges of SYTO (cyanines) and SYBR-type dyes (at neutral pH) facilitate their penetration into living cells, which have a membrane potential, as opposed to formaldehyde-fixed cells with compromised membranes. Permeabilization by formaldehyde fixation was not sufficient to allow passive entrance of the dyes. Inversely, when natural samples and cultures were fixed by heat treatment to permeabilize the cells, the fluorescence emission was higher than that of live cells, suggesting better permeabilization of membranes to the dyes (data not shown). These results suggest that when applied to natural samples, the fluorescence intensities of SYTO- and SYBR-stained bacteria are improved with live bacteria having polarized membranes. Moreover, the presence of cells having damaged membranes within natural communities may result in lower fluorescence signals due to less-efficient penetration of the dyes, as observed with formaldehyde-fixed cells.
Comparison of counts.
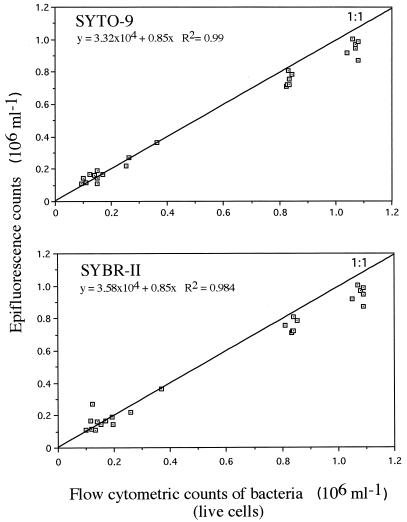
For nonsaline samples, counts made by epifluorescence microscopy of DAPI-stained cells were well correlated with those of live cells stained with SYTO-9 (r2 = 0.99, n = 23) and SYBR-II (r2 = 0.984, n = 23) and counts made by flow cytometry (Fig. 3). The correlation between counts obtained with SYTO-9 and DAPI samples was high for both fixed (r2 = 0.978) and live (r2 = 0.959) cells. A similar strong relationship was found for SYBR-II for live (r2 = 0.99, n = 23) and fixed (r2 = 0.985, n = 23) cells. For unfixed seawater samples, the bacterial counts obtained for samples stained with SYTO- and SYBR-type dyes were well correlated (Table 5). However, the correlations reported between counts made with these dyes and those obtained by DAPI staining and epifluorescence microscopy were slightly lower. These differences may be due to less-accurate enumerations with epifluorescence microscopy, since natural seawater samples contain many more particles than mineral waters and pure cultures. The presence of organic and mineral particles and the small sizes of most marine bacteria may result in lower bacterial discrimination when cells are counted by microscopic examination. Moreover, although this is common to both fresh and marine samples, microscopic enumeration is less accurate than flow cytometric enumeration because (i) a low number of cells is counted (400 to 500), (ii) cells are not always homogeneously distributed on the membrane filter, and (iii) the determination of the microscope factor (ratio between the surface of the microscopic field and that of the filters on which cells are distributed) suffers from some inaccuracies (10).
FIG. 3.
Relationship between epifluorescence bacterial counts with DAPI staining and cytometric counts with SYTO-9 and SYBR-II staining of S. typhimurium cells in culture, mineral, and river waters (n = 23). The straight line corresponds to a 1:1 relationship.
TABLE 5.
Correlations between bacterial counts determined from unfixed seawater samples (n = 28) stained with different dyes
| Dyea | Correlation with:
|
||||
|---|---|---|---|---|---|
| SYTO-9 | SYTO-13 | SYBR-I | SYBR-II | DAPI | |
| SYTO-9 | 1.000 | 0.989 | 0.960 | 0.956 | 0.850 |
| SYTO-13 | 1.000 | 0.971 | 0.965 | 0.871 | |
| SYBR-I | 1.000 | 0.997 | 0.898 | ||
| SYBR-II | 1.000 | 0.897 | |||
| DAPI | 1.000 | ||||
Bacteria stained with SYTO-9, SYTO-13, SYBR-I, and SYBR-II were enumerated by flow cytometry; DAPI-stained bacteria were counted by epifluorescence microscopy.
Specificity of the nucleic acid stains SYTO-9 and SYBR-II for Salmonella.
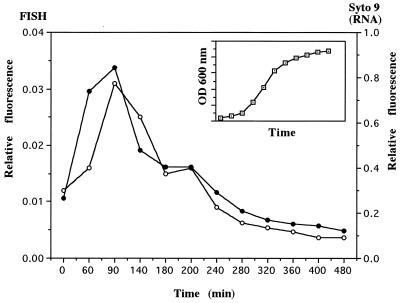
Initially, we were interested in testing dyes with high binding affinities for both DNA and RNA to simultaneously determine both total counts and active cells in natural marine waters. It has been suggested that a cell containing rRNA in quantities detectable by 16S rRNA hybridization probably contains the essential ingredients to be viable (2, 7–9). However, the rRNA content of most bacteria in oligotrophic environments is often undetectable by in situ hybridization with rRNA fluorescent probes without amplification techniques (1, 11, 17). Inversely, other workers have suggested that DNA, RNA, and proteins are not good indicators of activity in the environment due to low metabolic activity and heterogeneous composition (5). The sensitivity of SYTO-9 and SYBR-II to the rRNA content of individual cells of S. typhimurium was estimated by comparing FISH hybridization with SYTO-9 and SYBR-II staining. Pronase E combined with nucleases had no effect on the fluorescence signals, suggesting the absence of dye-restricting nucleoproteins. DNase treatment resulted in an important background of fluorescence and could not be applied alone or combined with RNase prior staining. A good correlation (r2 = 0.89, n = 12) between the fluorescence values of cells hybridized by FISH techniques and those obtained by the difference between SYTO-9-stained cells treated and not treated with RNases was found for growing cells (Fig. 4). The fluorescence signal of S. typhimurium cells stained with SYTO-9 was much higher than that of cells hybridized with the rRNA fluorescent probe (Fig. 4). A similar correlation was obtained with SYBR-II (r2 = 0.91, n = 12), but the fluorescence intensity relative to the RNA content of the cells, as estimated by the difference between SYBR-II-stained cells treated and not treated with RNases, was similar to that of SYTO-9-stained cells. Although these dyes have very different quantum yields when they bind to RNA (Table 1), the fluorescence intensities of stained cells with high RNA contents were not higher with SYBR-II (data not shown). Thus, the application of dyes, such as SYTO-9 and SYBR-II, to the discrimination of active and/or growing cells containing RNA within natural communities remains unclear since (i) DNase treatment induces an important background of fluorescence and cannot be applied prior to staining, (ii) the quantum yield of the dyes may be different when cells rather than double-stranded RNA and double-stranded DNA in solution are stained, and (iii) the quantitative relationship between RNA and activity may be highly species dependent. Alternatively, the use of such dyes without any nuclease treatment, to discriminate subpopulations having different nucleic acid contents and membrane polarization within natural communities, seems promising but requires further experiments.
FIG. 4.
Evolution of RNA contents of S. typhimurium cells during successive phases of growth. RNA contents were determined by whole-cell hybridization (closed circles) and by SYTO-9 staining (open circles). The insert shows the positions of sampling points on the growth curve. OD, optical density.
In this paper, we have presented data that suggest that SYTO-9 and SYBR-II are the best-suited blue-light-excited fluorescent dyes for bacterial enumeration in aquatic systems and that both can be used to stain living cells. A major advantage of staining living cells is that it prevents cell shrinkage and reduces the time of staining. However, SYTO-9 yields higher fluorescence signals in freshwater samples, whereas SYBR-II dye is more efficient in saline waters.
Within the range of SYTO dyes, SYTO-9 is much more efficient than SYTO-13, SYTO-11, and the counting kit for bacterial enumeration. However, one limitation to further use of this dye is that it is not commercially available at present as a single dye but only in combination with another dye in the LIVE/DEAD BacLight kit from Molecular Probes Inc. We hope that it will be available soon as a single dye, due to its excellent discrimination of bacterial populations when it is applied to different types of nonsaline environments. At the moment, SYBR-II is the best candidate for bacterial cell counting in water samples.
ACKNOWLEDGMENTS
This work was funded in part by Chemunex (Maisons-Alfort, France) and by contract ELOISE PL950439 from the European Community. The FACS-Calibur flow cytometer was funded by CNRS-INSU and by contract ELOISE PL950439.
We are grateful to Hendrik Schäfer for language improvements, and we thank the anonymous reviewers for valuable comments.
REFERENCES
- 1.Alfreider A, Pernthaler J, Amann R, Sattler B, Glöckner F-O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boye M, Ahl T, Molin S. Application of a strain-specific rRNA oligonucleotide probe targeting Pseudomonas fluorescens Ag1 in a mesocosm study of bacterial release into the environment. Appl Environ Microbiol. 1995;61:1384–1390. doi: 10.1128/aem.61.4.1384-1390.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1527–1542. [Google Scholar]
- 4.del Giorgio P A, Bird D F, Prairie Y T, Planas D. Flow cytometric determination of bacterial abundance in lake plankton with the green nucleic acid stain SYTO 13. Limnol Oceanogr. 1996;41:783–789. [Google Scholar]
- 5.Jeffrey W H, Haven R V, Hoch M P, Coffin R B. Bacterioplankton RNA, DNA, protein content and relationships to rates of thymidine and leucine incorporation. Aquat Microb Ecol. 1996;10:87–95. [Google Scholar]
- 6.Joux F, Lebaron P, Troussellier M. Succession of cellular states in a Salmonella typhimurium population submitted to starvation in artificial seawater microcosms. FEMS Microbiol Ecol. 1997;22:65–76. [Google Scholar]
- 7.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemp P F, Lee S, LaRoche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer J G, Singleton F L. Measurement of rRNA variations in natural communities of microorganisms on the southeastern U.S. continental shelf. Appl Environ Microbiol. 1993;59:2430–2436. doi: 10.1128/aem.59.8.2430-2436.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebaron P, Troussellier M, Got P. Accuracy and precision of epifluorescence microscopy count for direct estimates of bacterial numbers. J Microbiol Methods. 1993;19:89–94. [Google Scholar]
- 11.Lebaron P, Catala P, Fajon C, Joux F, Baudart J, Bernard L. A new sensitive, whole-cell hybridization technique for detection of bacteria involving a biotinylated oligonucleotide probe targeting rRNA and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3274–3278. doi: 10.1128/aem.63.8.3274-3278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W K W, Jellett J F, Dickie P M. DNA distributions in planktonic bacteria stained with TOTO or TO-PRO. Limnol Oceanogr. 1995;40:1485–1495. [Google Scholar]
- 13.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 14.Marie D, Vaulot D, Partensky F. Application of the novel nucleic acid dyes YOYO-1, YO-PRO-1, and PicoGreen for flow cytometric analysis of marine prokaryotes. Appl Environ Microbiol. 1996;62:1649–1655. doi: 10.1128/aem.62.5.1649-1655.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marie D, Partensky F, Jacquet S, Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McFeters G A, Yu F P, Pyle B H, Stewart P S. Physiological assessment of bacteria using fluorochromes. J Microbiol Methods. 1995;21:1–13. doi: 10.1016/0167-7012(94)00027-5. [DOI] [PubMed] [Google Scholar]
- 17.Ouverney C, Furhman J A. Increase in fluorescence intensity of 16S rRNA in situ hybridization in natural samples treated with chloramphenicol. Appl Environ Microbiol. 1997;63:2735–2740. doi: 10.1128/aem.63.7.2735-2740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter J, Deere D, Pickup R, Edwards C. Fluorescent probes and flow cytometry: new insights into environmental bacteriology. Cytometry. 1996;23:91–96. doi: 10.1002/(SICI)1097-0320(19960201)23:2<91::AID-CYTO1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 19.Troussellier M, Courties C, Vaquer A. Recent applications of flow cytometry in aquatic microbial ecology. Biol Cell. 1993;78:111–121. doi: 10.1016/0248-4900(93)90121-t. [DOI] [PubMed] [Google Scholar]
- 20.Wallner G, Amann R I, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]