Abstract
Random aliphatic-aromatic copolyesters synthesized from 1,4-butanediol, adipic acid, and terephthalic acid (BTA) have excellent thermal and mechanical properties and are biodegradable by mixed cultures (e.g., in compost). Over 20 BTA-degrading strains were isolated by using compost as a microbial source. Among these microorganisms, thermophilic actinomycetes obviously play an outstanding role and appear to dominate the initial degradation step. Two actinomycete strains exhibited about 20-fold higher BTA degradation rates than usually observed in a common compost test. These isolates were identified as Thermomonospora fusca strains. They appeared to be particularly suitable for establishment of rapid degradation tests and were used in comparative studies on the biodegradation of various polyesters.
Polymers designed to undergo controlled biological degradation are increasingly discussed as a favorable contribution to the solution of problems arising from plastic waste disposal, e.g., by recycling or land filling. The environmental safety of these novel materials has to be proven carefully (32). A number of standardized test methods using mixed cultures for evaluation of the biodegradability and compostability of plastics have been established recently (1, 5, 25, 30). However, for investigations of the degradation mechanism, it is advantageous to isolate individual strains which are able to degrade well-defined polymers.
For some polymers, including isoprene rubbers (7, 16, 34), polyvinyl alcohol (29), cellulose acetate (23), poly(ɛ-polycaprolactone) (24), and bacterial polyhydroxyalkanoates (6), a number of degrading microorganisms are described in the literature.
Recently, it has been shown that copolyesters containing adipic acid and terephthalic acid as aromatic acid components are also attacked by microorganisms (35). This group of copolyesters appears to be very promising with regard to widespread commercial applications (38). Studies have been performed with the particular goal of detecting the fate of the aromatic constituents and proving their biodegradation (36, 37).
However, in all previous investigations, inocula of undefined mixed cultures were used. Therefore, this study concentrated on the isolation of individual strains which are able to degrade random aliphatic-aromatic copolyesters. As composting is the most promising method of treating such biodegradable plastics, the microbial isolates were obtained from compost material. The isolates are appropriate candidates for use in the study of the mechanism of copolyester degradation and the establishment of improved and rapid test methods for evaluation of biodegradability.
MATERIALS AND METHODS
Polymers.
A copolyester made of 1,4-butanediol, terephthalic acid, and adipic acid (Fig. 1) was used to isolate degradative microorganisms. The origins, compositions, melting temperatures, and average molar masses of all of the polymers tested in the biodegradation studies are listed in Table 1. Polymer films 100 μm thick and 25 mm in diameter were prepared as described by Witt et al. (35). The films were washed in a 70% (vol/vol) ethanol solution for 30 min, dried at room temperature under a vacuum, and weighed to an accuracy of ±0.2 mg. For sterilization, polymer films were irradiated under a UV lamp (UVC 30; Hereaus, Hannover, Germany; 254 nm, 6 W/cm2 at a distance of 20 cm) at a lamp-to-film distance of 15 cm and an irradiation area of 38 by 18 cm for 15 min of exposure per side.
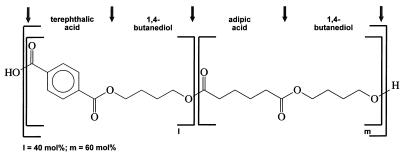
FIG. 1.
Formula of the aliphatic-aromatic copolyester BTA 40:60 used for the screening of microorganisms with regard to their degradation abilities. The copolyester consists of 1,4-butanediol, terephthalic acid, and adipic acid.
TABLE 1.
Compositions, melting temperatures, and average molar masses of tested polymers
| Polymera | Diol | Aromatic compound (mol%) | Aliphatic compound(s) (mol%) | Tm (°C)b | Mw (g/mol)c |
|---|---|---|---|---|---|
| BTA 40:60 | 1,4-Butanediol | Terephthalic acid (40) | Adipic acid (60) | 99 | 47,600 |
| BTA 60:40 | 1,4-Butanediol | Terephthalic acid (60) | Adipic acid (40) | 147 | 45,000 |
| SP313 | 1,3-Propanediol | Brassylic acid | 72 | 41,500 | |
| Bayer Tir 1874 | 1,4-Butanediol | Adipic acid (40), caprolactam (60) | 125 | NDd | |
| PHB | 3-Hydroxybutyrate | 175 | >800,000 | ||
| Bionolle | 1,4-Butanediol | Succinic acid | 113 | 66,000 |
Polymer sources: BTA 40:60 and BTA 60:40, Hüls AG, Marl, Germany; SP313, GBF, Braunschweig, Germany; Bayer Tir 1874, Bayer AG, Leverkusen, Germany; PHB, ICI, Billingham, United Kingdom; Bionolle, Showa Highpolymer, Tokyo, Japan.
Tm, melting temperature.
Mw, weight average molar mass (determined by gel permeation chromatography, based on polystyrene calibration).
ND, not determined.
Source of organisms.
Aerobic microorganisms were isolated from samples of approximately 6-month-old mature compost made from green waste (Compost Plant Watenbüttel, Watenbüttel, Germany).
Media and buffer.
The compositions of the media used are listed in Table 2 (3a, 4, 14). To retard drying of the agar media at high temperatures, petri dishes were incubated in sealed polyethylene bags (18). For preparation of suspensions and dilutions, a phosphate buffer (0.05 M Na2HPO4, 0.03 M KH2PO4, pH 7.0) was used.
TABLE 2.
Cultivation media
| Mediuma | Amt of component liter−1 (reference) |
|---|---|
| Mineral salt media | |
| MinB agar | NH4NO3, 1 g; MgSO · 7H2O, 0.5 g; NaCl, 0.5 g; FeSO4 · 7H2O, 0.01 g; K2HPO4, 1 g |
| MS agar | Mineral salt agar (4) |
| MSV agar | Optimized mineral salt medium (5) |
| Compost extract medium | CE agarb |
| Complex media | |
| SCK agar | Starch casein KNO3 agar (14) |
| TSB agar | Tryptic soy broth (Merck), 30 g |
| NB agar | Nutrient broth (Difco), 8 g |
| St-II-Soy agar | Standard-II-boullion (Merck), 15 g; peptone from soymeal, 5 g; yeast extract, 5 g |
| GYM agar | Streptomyces medium (3a) |
Twenty grams of agar per liter was added to all media. The pH was adjusted to 7.2 ± 0.2. Media were autoclaved for 15 min at 121°C. Mineral salt and compost extract media were used for organism enrichment and the degradation test. Complex media were used for isolation and cultivation of isolated strains.
To make 1 liter of compost extract, 100 g of mature compost (from green wastes) was added to 1 liter of demineralized water and stirred for 24 h at room temperature. The solution was separated from the particles by sieving and centrifugation. The extract had to be autoclaved (20 min, 121°C).
Isolation of BTA-degrading microorganisms.
An at least four-step procedure turned out to be the appropriate strategy for the isolation of BTA-degrading strains. Mixed populations were used as inocula. These were taken by either (i) scraping off and dissolving (2 ml of phosphate buffer) preadapted biofilms grown on BTA films which were incubated in compost reactors as described by Witt et al. (35) or (ii) dissolving 10 g of mature compost or compost harvested from reactors in 90 ml of phosphate buffer.
For enrichment of BTA-degrading microorganisms on agar plates, BTA films were inoculated with 100 μl of the inocula mentioned above. Different mineral salt and compost extract agar plates were used as enrichment media (Table 2). The enrichment cultures were incubated at 20, 40, and 55°C and examined daily for growth of colonies and visible disintegration of the polymer films.
For isolation, colonies were picked up from partly disintegrated areas of films and cultured on various complex agar media (Table 2) by using different inoculation techniques to provide favorable growth conditions for a wide range of microorganisms.
We could not produce homogeneous opaque top layer agar plates with granules of BTA (aggregation of particles due to the hydrophobic surfaces). Thus, direct selection of degrading microorganisms by a clear-zone method (2) was not possible. Because of these problems, the abilities of all isolates to degrade the copolyester BTA 40:60 had to be tested in a separate step.
The degradation test was carried out with polymer films on three mineral salt media and one compost extract medium (Table 2) on agar plates inoculated with actinospore or bacterial suspensions (≈107 microorganisms/ml) at the optimal growth temperature of the isolates. As an indicator for degradation, the weight loss of the polymer films was determined after 14 days of incubation. The results were compared with those obtained with noninoculated films (noninoculated controls) incubated in the respective media. The noninoculated controls showed no weight loss.
Cultivation and preservation of isolated strains.
The isolates were maintained on agar plates of complex media at temperatures satisfying the individual requirements for optimal growth (Table 3). Bacterial strains were preserved in 50% (vol/vol) glycerol at −20°C. For recultivation, 1 loopful of each glycerol suspension was streaked onto an NB agar plate (Table 2). After 48 h of incubation, single colonies were picked and suspended in 1 ml of phosphate buffer. These bacterial suspensions served as the inoculum for degradation tests.
TABLE 3.
Incubation temperatures and media for cultivation and testing of BTA-degrading isolates
| BTAdegrading isolate | Temp (°C)
|
Cultivation medium | Degradation of BTAb
|
||
|---|---|---|---|---|---|
| Rangea | Opti- mum | Medium or mediac | Capacity | ||
| K1a-1 | 45–65 | 55 | NB agar | MinB | + |
| K1a-2 | 45–65 | 55 | NB agar | MinB | + |
| K6a | 37–65 | 55 | SCK agar | MinB, MSA, CEA | ++ |
| K6b | NDd | 55 | SCK agar | CEA | +++ |
| K6c | ND | 55 | SCK agar | MinB, CEA | ++ |
| K7a-2 | 45–65 | 55 | St-II-Soy agar | MinB, MSV, CEA | ++ |
| K7a-3 | 37–65 | 55 | St-II-Soy agar | MSV | ++++ |
| K7e | ND | 55 | TSB agar | CEA | + |
| K7f | 45–60 | 55 | SCK agar | All media | ++ |
| K7h | 45–65 | 55 | SCK agar | MSV | +++ |
| K9a | 37–55 | 45 | SCK agar | MinB, MSA | ++ |
| K9b | 45–55 | 50 | St-II-Soy agar | All media | ++ |
| K10b | 20–50 | 40 | St-II-Soy agar | All media | ++ |
| K11a | 37–60 | 55 | SCK agar | All media | ++ |
| K11b | ND | 55 | St-II-Soy agar | MinB, CEA | +++ |
| K11d | 37–60 | 55 | St-II-Soy agar | MSV | +++ |
| K13a | ND | 55 | GYM agar | CEA | ++ |
| K13g | 37–65 | 55 | St-II-Soy agar | MSV | ++++ |
| K13j | ND | 55 | GYM agar | MSA, CEA | ++ |
| K13k | ND | 45 | GYM agar | All media | ++ |
| K13l | 20–50 | 40 | GYM agar | MSA, CEA | +++ |
Temperature range was tested on complex medium (TSB agar).
In milligrams per week per square centimeter: +, 0.15 to 0.3; ++, 0.3 to 1; +++, 1 to 2; ++++, >2.
Medium or media on which the isolate showed the highest BTA-degrading activity or activities.
ND, not determined.
For preservation of the isolated actinomycete strains, spores from well-sporulating actinomycete cultures were suspended in distilled water and kept at 4°C in screw-cap tubes. Spore suspensions routinely served as an inoculum source for degradation tests.
Determination of the optimum growth temperatures of the individual strains was carried out on sections of agar plates inoculated with 1 loopful of an actinospore or bacterial suspension and incubated at temperatures of 20 to 70°C. Aerial mycelium formation and/or colony growth was visually observed after 1 and 2 weeks. All further tests were carried out at the respective optimum temperature.
For chemotaxonomic analysis, actinomycete cells were grown in TSB shake flask cultures, harvested by centrifugation (20 min, 7,800 × g, 4°C), washed twice in phosphate buffer, and freeze-dried to provide cell preparations (exception: for analysis of fatty acids, wet cells were used).
Taxonomic studies.
For 16S ribosomal DNA (rDNA) sequencing, the genomic DNAs of K13g and K7a-3 were extracted and the 16S rRNA gene was amplified by PCR as described previously (27). Purified PCR products were directly sequenced by using the Taq Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Inc., Foster City, Calif.). Sequences were manually aligned with representatives of related sporoactinomycete taxa by using the ae2 editor available from the Ribosomal Database Project (17). The diagnostic regions of the 16S rDNA sequences were compared with those of the Thermomonospora type strains (28). Direct sequence similarities were calculated within the ae2 editor.
Analysis of cell wall amino acids and sugars.
Amino acid and sugar analysis of the whole-cell hydrolysate was done as described by Staneck and Roberts (31).
Extraction and analysis of isoprenoid quinones and polar lipids.
Isoprenoid quinones and polar lipids were extracted and purified by the small-scale integrated procedure of Minnikin et al. (21). The dried preparations of the quinone extracts were dissolved in 200 μl of isopropanol, and 1- to 10-μl amounts were separated by high-pressure liquid chromatography. The menaquinones were separated by high-pressure liquid chromatography on Lichrosorb RP-18 at 40°C using acetonitrile-isopropanol (65:35, vol/vol) as the solvent (10, 11). Polar lipid extracts were separated by two-dimensional thin-layer chromatography and identified by their Rf values and their reaction with diagnostic spray reagents (21).
Extraction and analysis of fatty acids.
Fatty acid methyl esters were obtained from 40 mg of wet cells by saponification, methylation, and extraction by using minor modifications (15) of the method of Miller (20). The fatty acid methyl ester mixtures were separated by using a 5898A Microbial Identification System (Microbial ID, Newark, Del.). Peaks were automatically integrated and fatty acids were identified by the Microbial Identification System Standard Software (Microbial ID).
RESULTS AND DISCUSSION
Screening and isolation of microorganisms.
Although it was expected that BTA-degrading microorganisms would be enriched in biofilms grown on BTA films during composting, there was no significant difference in the number of BTA-degrading strains isolated from compost eluates or preadapted biofilms.
Table 3 gives the isolates, their optimal growth temperatures, and the media on which the isolates revealed the highest BTA degradation. As expected, different isolates showed maximal activities on different media, none of which generally appeared to be the best. Further degradation tests with individual strains were then carried out with the appropriate media. We obtained the best degradation results with temperatures higher than 40°C; thus, we focused predominantly on thermophilic microorganisms.
It must be admitted that weight loss measurements only indicate disintegration of the polymer films likely caused by cleavage of the polymer chains. Complete metabolism of the polymer material has to be investigated in additional tests. However, for polyesters, the first step in attacking the polymer chain is often the step which determines the degradability of such materials.
Table 4 gives an overview of the number of isolated bacteria, actinomycetes, and fungi and their abilities to degrade the aliphatic-aromatic copolyester BTA 40:60. A total of 61 strains of thermophilic microorganisms were isolated from 13 different compost samples.
TABLE 4.
Numbers of microorganisms isolated and their abilities to degrade the aliphatic-aromatic copolyester BTA 40:60a
| BTA degradation (wt loss in mg/week · cm−2)b | No. of bacteria | No. of actinomycetes | No. of fungi |
|---|---|---|---|
| >2 | 2 | ||
| 1–2 | 5 | ||
| 0.3–1 | 11 | ||
| 0.15–0.3 | 2 | 1 | |
| 0.06–0.15c | 3 | 6 | |
| <0.06 | 25 | 4 | 2 |
The total number of isolates was 61.
Degradation abilities were measured on four different media. The best degradation results are indicated independently of the media used.
Weight loss of 0.06 mg/week · cm2 is the detectability limit.
Among 30 bacterial isolates, which were all aerobically growing rods mainly with endospore formation, only five strains were able to disintegrate BTA 40:60. Three of those strains exhibited only weak degradation activities (weight losses of 0.06 to 0.15 mg/week · cm2), just above the experimental error of the detection method used. Most fungi grow at temperatures of less than 50°C. This might be the reason why we isolated just two strains of fungi. Both strains were unable to disintegrate the copolyester BTA 40:60.
Within the group of thermophilic actinomycetes, only 4 of 29 isolates did not show significant degradation activities. Two of the most active actinomycetes, isolates K13g and K7a-3, were identified taxonomically and used for further investigations. They degraded films 100 μm thick up to 90% within 7 days.
More than 20 different microorganisms isolated from compost were able to depolymerize the synthetic polyester BTA 40:60. Probably due to the use of compost as the source of microorganisms, most isolates were thermophilic microorganisms. Especially, thermophilic actinomycetes play an outstanding role in degrading the BTA copolyester with regard to both the number of microorganisms isolated and their degradation rates. Actinomycetes are known to be involved in the degradation of several natural polymers, like chitin, celluloses, starch, agar, and lignocelluloses (3, 9, 18, 19). However, the role of this group of microorganisms in degrading synthetic polymers is rarely described in the literature (26).
Identification of selected strains.
The phenotypic and chemotaxonomic properties of strains K13g and K7a-3 are consistent with their classification in the genus Thermomonospora (13). The strains showed generation of a white aerial mycelium and individual spores which were formed in clusters on the tip of a short sporophore. Both strains have the same chemotaxonomic characteristics which are consistent with members of the Thermomonospora fusca taxon (13). The whole-cell hydrolysates of strain K13g and K7a-3 contained glucose and ribose as major sugars. Mesodiaminopimelic acid was the only diamino acid found in the cell walls. The polar lipids were composed of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, methylphosphatidylethanolamine, and some unspecified glycolipids. The fatty acid pattern of the strains contained mainly iso- and anteiso-branched fatty acids. Small amounts of 10-methyl branched and unbranched fatty acids were also found.
MK-11(H4), MK-11(H6), MK-12(H4), and MK-12(H6) were the predominant menaquinones of both strains. The combination of chemical markers found in K13g and K7a-3 is unique to the strains of T. fusca. Therefore, K13g and K7a-3 could be classified as T. fusca by chemotaxonomy. This was confirmed by the analysis of 16S rDNA sequences of both isolates, which showed that isolates K7a-3 and K13g belong to the family Thermomonosporaceae. The greatest similarities were found between the isolates and T. fusca DSM 43792T (K7a-3, 100%; K13g, 99.8%). Obviously, these two actinomycete isolates are representatives of this species.
BTA film degradation on agar plates by T. fusca K13g.
With regard to the application of isolated strains as test organisms in improved degradation tests, the time course of the degradation of BTA films on agar plates by T. fusca K13g was investigated. Usually, enzymatic degradation of plastics is a surface erosion process, because enzymes are not able to penetrate the bulk polymer. Thus, the rate of weight loss can be directly used to measure the enzymatic cleavage of the polymer chains.
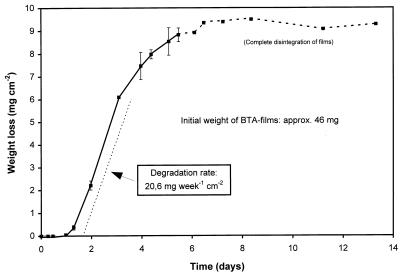
In Fig. 2, the weight loss of BTA films inoculated with a spore suspension of the actinomycete T. fusca K13g is plotted against the degradation time. A short period of only 7 days was sufficient to completely disintegrate the BTA films on the agar plates, resulting in a weight loss curve often observed for biological transformations. During a lag phase of 1 day, actinospores germinated; this was followed by a period of 3 days with constant weight loss of the exposed films. As the films started to fall into fragments, the degradation rate decreased gradually, and after 1 week, the BTA material disappeared totally. Although this high degree of degradation and the fact that the curve was obtained from films in parallel tests stopped at different times after incubation, the reproducibility of this experiment was very good.
FIG. 2.
Degradation of the aliphatic-aromatic copolyester BTA 40:60 on agar plates by the actinomycete T. fusca K13g. The test was carried out on MSV agar over a period of 13 days at 55°C with triplicate samples. From day 6 on, error bars are not shown because polymer films (diameter, 2.5 cm; thickness, 90 μm; initial weight, 46 mg; degradation area, 4.91 cm2) disappeared according to their initial weights (light films first) and error bars would represent the differences between the initial weights from that day on.
By calculating specific degradation rates from the linear part of the degradation curves in combination with the surface area of the films which could be attacked by the organisms, different experiments can be compared with regard to the degradation potential of the organisms involved.
Typical degradation rates of about 1 mg/week · cm−2 were obtained for BTA 40:60 films in a compost simulation test. Significantly higher degradation rates of up to 2.3 mg/week · cm−2 were found with preadapted mixed cultures from compost in an agar plate test. However, the increase in the degradation rate from compost to the preadapted mixed culture is not only related to the higher degradation potential of the organisms but also influenced by the optimized degradation conditions in the laboratory agar plate test. Remarkable degradation rates as high as 20 mg/week · cm−2 could be obtained with the actinomycete T. fusca K13g under defined laboratory conditions, which are about 10-fold higher than those achieved for the copolyester films incubated with adapted mixed cultures.
Compared to the fast degradation of T. fusca K13g, the degradation rate of bacterial strains K1a-1 and K1a-2 was quite slow. A continuous degradation of BTA films of only 0.1 mg/week · cm−2 within a test period of 6 weeks was observed.
The fast degradation of the BTA copolyester, in combination with the good reproducibility of the degradation rates in agar plate tests, predestine the actinomycete T. fusca K13g for use in rapid test methods and fundamental investigations of the degradation mechanism of the BTA copolyester and other polyesters as well.
Degradation of different kinds of polymers by two actinomycete isolates.
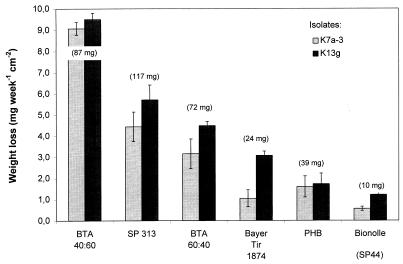
The aromatic-aliphatic copolyester BTA 40:60 used for the screening is a biodegradable polymer of high commercial interest but does not occur in nature. Thus, to determine the substrate specificity of the isolated microorganisms, we tested the degradation of other polyesters and one polyester amide (Fig. 3) by T. fusca K13g and K7a-3. The two strains exhibited similarly high BTA 40:60 degradation abilities.
FIG. 3.
Degradation of different polyesters and the polyester amide Bayer Tir 1874 on agar plates by T. fusca K7a-3 and K13g. Polymer films (diameter, 2.5 cm; degradation area, 4.91 cm2) were incubated at 55°C for 2 weeks on MSV agar. A second test series gave reproducible results. Each series was carried out with triplicate samples. The degradation rate was calculated with data from the linear part of the degradation curve.
In all cases, the degradation rates obtained with strain K7a-3 were somewhat lower than those of strain K13g. Of the polyester-based materials tested, the aliphatic-aromatic copolyester BTA 40:60 exhibited the highest degradation rate, while pure aliphatic materials like Bionolle, Bayer Tir 1874, or even the bacterial polyester poly(β-hydroxybutyrate) (PHB) were degraded much more slowly by both strains. This is surprising in that these polyesters are regarded to be easily biodegradable. Even a copolyester containing 60 mol% terephthalic acid disintegrates almost twice as fast as the natural material PHB. Polyesters with such a large aromatic compound fraction have been shown to degrade only very slowly in composting tests (35). The degradation behavior of SP313 is also unexpected. This aliphatic polyester with a long diacid component is not easily degraded in soil burial tests or in enzymatic degradation tests with lipases (22).
It can be anticipated that both thermophilic actinomycetes and their enzymatic systems prefer hydrophobic surfaces such as those offered by BTA copolyesters or the aliphatic polyester SP313. It will be challenging and illuminating to isolate and characterize the enzymatic system responsible for polymer chain cleavage and to compare its components with other polyester-degrading enzymes like lipases or PHB depolymerases (8, 33).
Despite the rapid depolymerization of the BTA copolyester, only poor growth of the actinomycetes could be observed on mineral salt agar with a polymer as the sole carbon source. This suggests that the actinomycetes are not able to completely metabolize the oligomers and monomers derived from the depolymerization of the polyester. As BTA copolyesters are abiotic synthetic materials, it is not likely that microorganisms have been specialized to this kind of carbon source. However, they are adapted to the use of other, similar polyester structures containing aromatic components like those in cutin or lignocelluloses.
Further investigations are in preparation to clarify the mechanism of BTA copolyester degradation by actinomycetes and to identify the enzymes involved.
ACKNOWLEDGMENTS
We thank B. Frerichs, G. Pötter, I. Kramer, and J. Swiderski for their skilled technical assistance.
REFERENCES
- 1.American Society for Testing and Materials. D 6002-96. Standard guide for assessing the compostability of environmentally degradable plastics. Washington, D.C: American Society for Testing and Materials; 1996. [Google Scholar]
- 2.Augusta, J., R.-J. Müller, and H. Widdecke. A rapid evaluation plate-test for the biodegradability of plastics. Appl. Microbiol. Biotechnol. 39:673–678.
- 3.Crawford D L, Sutherland J B. Isolation and characterization of lignocellulose-decomposing actinomycetes. In: Kirk T K, Higuchi T, Chang H, editors. Lignin biodegradation: microbiology, chemistry, and potential applications. II. Bota Raton, Fla: CRC Press, Inc.; 1980. pp. 95–101. [Google Scholar]
- 3a.Deutsche Sammlung von Mikroorganismen und Zellkulturen. DSM 65. DMS-catalogue of strains 1993, fifth ed. Braunschweig, Germany: Deutsche Sammlung von Mikroorganismen und Zellkulturen; 1993. p. 357. [Google Scholar]
- 4.Deutsches Institut für Normung e.V. DIN 53749. Lösungen und Nährmedien für die Prüfung mit Bakterien. Berlin, Germany: Beuth Verlag GmbH; 1984. p. 3. [Google Scholar]
- 5.Deutsches Institut für Normung e.V. DIN 54900–Entwurf. Prüfung der Kompostierbarkeit von polymeren Werkstoffen. Berlin, Germany: Beuth Verlag GmbH; 1997. pp. 1–7. [Google Scholar]
- 6.Doi Y. Microbial polyesters. New York, N.Y: VCH Publishers Inc.; 1990. [Google Scholar]
- 7.Heisey R M, Papadatos S. Isolation of microorganisms able to metabolize purified natural rubber. Appl Environ Microbiol. 1995;61:3092–3097. doi: 10.1128/aem.61.8.3092-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaeger K-E, Steinbüchel A, Jendrossek D. Substrate specificities of bacterial polyhydroxyalkanoate depolymerases and lipases: bacterial lipases hydrolyze poly(ω-hydroxyalkanoates) Appl Environ Microbiol. 1995;61:3113–3118. doi: 10.1128/aem.61.8.3113-3118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempf A, Kutzner H J. VDLUFA-Schriftenreihe 28. Oldenburg, Germany: Kongressband Teil II; 1988. Screening von biopolymerabbauenden Exoenzymen bei thermophilen Actinomyceten; pp. 979–989. [Google Scholar]
- 10.Kroppenstedt R M, Korn-Wendisch F, Fowler V J, Stackebrandt E. Biochemical and molecular genetic evidence for transfer of Actinoplanes armeniacus into the family Streptomycetaceae. Zentbl Bakteriol Hyg Abt Orig. 1981;C2:254–262. [Google Scholar]
- 11.Kroppenstedt R M. Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin D E, editors. Chemical methods in bacterial systematics. New York, N.Y: Academic Press, Inc.; 1985. pp. 173–199. [Google Scholar]
- 12.Kroppenstedt R M. The genus Nocardiopsis. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes, a handbook on the biology of bacteria; ecophysiology, isolation, identification, applications. 2nd ed. II. Berlin, Germany: Springer-Verlag KG; 1992. pp. 1139–1156. [Google Scholar]
- 13.Kroppenstedt R M, Goodfellow M. The family Thermomonosporaceae. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes, a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. II. Berlin, Germany: Springer-Verlaq KG; 1992. pp. 1085–1114. [Google Scholar]
- 14.Küster E, Williams S T. Selection of media for isolation of streptomycetes. Nature (London) 1964;202:928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- 15.Kuykendall L D, Roy M A, O’Neill J J, Devine T E. Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol. 1988;38:358–361. [Google Scholar]
- 16.Linos A, Steinbüchel A. Proceedings of the 10th International Biodeterioration and Biodegradation Symposium. Weinheim, Germany: VCH-Verlag; 1996. Investigations on the microbial breakdown of natural and synthetic rubber; pp. 211–219. [Google Scholar]
- 17.Maidak B L, Larsen N, McCoughey M J, Overbeek R, Olsen G J, Folgel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy A J, Cross T. A taxonomic study of Thermomonospora and other monosporic actinomycetes. J Gen Microbiol. 1984;130:5–25. [Google Scholar]
- 19.McCarthy A J. Lignocelllulose-degrading actinomycetes. FEMS Microbiol Rev. 1987;46:145–163. [Google Scholar]
- 20.Miller L T. A single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol. 1982;16:584–586. doi: 10.1128/jcm.16.3.584-586.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minnikin D E, O’Donnel A G, Goodfellow M, Alberton G, Ethel M, Scale A, Parlayed J H. An integrated procedure for the extraction of isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. [Google Scholar]
- 22.Müller R-J. Proceedings of the 10th International Biodeterioration and Biodegradation Symposium. Weinheim, Germany: VCH-Verlag; 1996. Mechanistic studies on the biodegradation of polyesters; pp. 211–219. [Google Scholar]
- 23.Nelson M, McCarthy S P, Gross R A. Isolation of a Pseudomonas paucimobilis capable of using insoluble cellulose acetate as a sole carbon source. Polym Mater Sci Eng. 1992;67:139–140. [Google Scholar]
- 24.Nishida H, Tokiwa Y. Distribution of poly(β-hydroxybutyrate) and poly(ɛ-caprolactone) degrading microorganisms and microbial degradation behaviour on plastic surfaces. Polym Mater Sci Eng. 1992;67:137–138. [Google Scholar]
- 25.Pagga, U. Biodegradability and compostability of polymeric materials in the context of the European packaging regulation. In Proceedings of the International Conference on Advanced Materials, in press.
- 26.Pommer E H. Synthetische organische Materialien. In: Brill H, editor. Mikrobielle Materialzerstörung und Materialschutz: Schädigungsmechanismen und Schutzmaßnahmen. Jena, Germany: Gustav Fischer Verlag; 1995. pp. 111–150. [Google Scholar]
- 27.Rainey F A, Dorsch M, Morgan H W, Stackebrandt E. 16S rDNA analysis of Spirochaeta thermophila: its phylogenetic position and implications for systematics of the order Spirochaetales. Syst Appl Microbiol. 1992;15:197–202. [Google Scholar]
- 28.Rainey F A, Ward-Rainey N, Kroppenstedt R M, Stackebrandt E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int J Syst Bacteriol. 1996;46:1088–1092. doi: 10.1099/00207713-46-4-1088. [DOI] [PubMed] [Google Scholar]
- 29.Sakai K, Hamada N, Watanabe Y. Degradation mechanism of poly(vinyl alcohol) by successive reactions of secondary alcohol oxidase and β-diketone hydrolase from Pseudomonas sp. Agric Biol Chem. 1986;50:989–996. [Google Scholar]
- 30.Sawada, H. ISO standard activities in standardization of biodegradable and/or compostable polymers; development of test methods, definitions. In Proceedings of the International Conference on Advanced Materials, in press.
- 31.Staneck J L, Roberts G D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–231. doi: 10.1128/am.28.2.226-231.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swift G. Biodegradable polymers in the environment: are they really biodegradable? Polym Mater Sci Eng. 1992;66:403–404. [Google Scholar]
- 33.Tokiwa Y, Ando T, Suzuki T, Takeda K. Biodegradation of synthetic polymers containing ester bonds. Am Chem Soc Symp Ser. 1990;433:136–148. [Google Scholar]
- 34.Tsuchii A, Takeda K. Rubber-degrading enzyme from a bacterial culture. Appl Environ Microbiol. 1990;56:1–269. doi: 10.1128/aem.56.1.269-274.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witt U, Müller R-J, Deckwer W-D. New biodegradable polyester-copolymers from commodity chemicals with favourable use properties. J Environ Polym Degrad. 1995;3:215–223. [Google Scholar]
- 36.Witt U, Müller R-J, Deckwer W-D. Evaluation of the biodegradability of copolyesters containing aromatic compounds by investigations of model oligomers. J Environ Polym Degrad. 1996;4:9–20. [Google Scholar]
- 37.Witt U, Müller R-J, Deckwer W-D. Studies on sequence distribution of aliphatic/aromatic copolyesters by high-resolution 13C nuclear magnetic resonance spectroscopy for evaluation of biodegradability. Makromol Chem Phys. 1996;197:1525–1535. [Google Scholar]
- 38.Witt U, Müller R-J, Deckwer W-D. Biodegradation behaviour and material properties of aliphatic/aromatic polyesters of commercial importance. J Environ Polym Degrad. 1997;5:81–89. [Google Scholar]