Abstract
We recently described a reverse transcription-PCR (RT-PCR) for detecting low numbers of viable Cryptosporidium parvum oocysts spiked into clarified environmental water concentrates. We have now modified the assay for direct analysis of primary sample concentrates with simultaneous detection of viable C. parvum oocysts, Giardia cysts, and a novel type of internal positive control (IPC). The IPC was designed to assess both efficiency of mRNA isolation and potential RT-PCR inhibition. Sensitivity testing showed that low numbers of organisms, in the range of a single viable cyst and oocyst, could be detected when spiked into 100-μl packed pellet volumes of concentrates from creek and river water samples. The RT-PCR was compared with an immunofluorescence (IF) assay by analyzing 29 nonspiked environmental water samples. Sample volumes of 20 to 1,500 liters were concentrated with a wound fiberglass cartridge filter. Frequency of detection for viable Giardia cysts increased from 24% by IF microscopy to 69% by RT-PCR. Viable C. parvum oocysts were detected only once by RT-PCR (3%) in contrast to detection of viable Cryptosporidium spp. in four samples by IF microscopy (14%), suggesting that Cryptosporidium species other than C. parvum were present in the water. This combination of the large-volume sampling method with RT-PCR represents a significant advance in terms of protozoan pathogen monitoring and in the wider application of PCR technology to this field of microbiology.
The ability of Giardia intestinalis and Cryptosporidium parvum to cause waterborne disease is well documented (12, 19, 22, 38). However, analytical methods widely used to detect the presence of these organisms in water do not provide the quality of data required to assess health risk and effectively manage this problem (15, 21, 27, 42). Considerable effort is being made worldwide to improve detection methodologies through the application of techniques such as flow cytometry (45), laser scanning (5), immunomagnetic separation (7), and PCR. In particular, PCR is an attractive diagnostic procedure as it is rapid, sensitive, and pathogen specific. While many PCR methods have been described for both Giardia and Cryptosporidium detection (1, 11, 18, 20, 23, 24, 26, 29, 39, 47), this technology is only slowly emerging as a practical method for pathogen assessment of water quality. This is largely due to the low numbers of cysts and oocysts in water and the requirement for significant sample concentration factors to reduce large sample volumes to the microliter quantities that are compatible with PCR. Concentration of microorganisms in water samples, or their nucleic acids, leads to the coconcentration of substances such as humic acids which can severely reduce PCR efficiency and reliability (41, 44, 48). One approach that is becoming widely used to overcome these problems is to use paramagnetic beads to purify target nucleic acids (14, 25, 39). Magnetic bead technology is a powerful tool for water microbiology as it is simple to use, does not require expensive equipment, and can effectively separate and concentrate a range of rare molecules from inhibitory sample matrices. We have previously reported the use of this approach to successfully capture heat shock protein (HSP) mRNA for reverse transcription-PCR (RT-PCR) detection of low numbers of viable Cryptosporidium parvum oocysts in water (39).
While sensitive detection methods are important, equal consideration must be given to sample concentration methodology. Required are concentration techniques that are compatible with viability detection systems and suitable for rapid processing of large sample volumes. Sample volumes for analyses of surface water and drinking water of 100 and ≥1,000 liters, respectively, have been recommended elsewhere (16), and the most commonly used method for concentration of Giardia cysts and Cryptosporidium oocysts from water is the American Society for Testing and Materials yarn-wound cartridge filter method (16). However, parasite recovery with this technique is generally poor (10). Vortex flow filtration, membrane filtration, and capsule filters have been proposed for concentration of 10-liter volumes in the U.S. Environmental Protection Agency draft Cryptosporidium method (6). However, each method is limited to some degree by either small sample volumes, interference by high sample turbidity, high equipment costs, long processing times, or laboratory-based equipment, necessitating the transportation of water to the laboratory. Chemical flocculation, while an effective procedure (35, 37, 46), may inactivate viable organisms (9) and is generally restricted to volumes of less than 20 liters. One technique that offers many advantages is a filtration-elution procedure which uses cheap, negatively charged fiberglass filters in a simple entrapment-backwash elution format (28). High efficiencies of recovery for Giardia cysts in spiked 20-liter samples have been reported elsewhere (28).
The aim of this study was to develop an RT-PCR assay for the simultaneous detection of viable Giardia and Cryptosporidium in large-volume water samples. Our approach was to concentrate cysts and oocysts from water samples with the Diamond filter (28), then selectively capture mRNA with oligo(dT)25 magnetic beads, and detect viable organisms by RT-PCR. An RNA internal control that hybridized to the magnetic beads was used to monitor the entire detection process from mRNA capture through RT-PCR for every sample.
MATERIALS AND METHODS
Cyst and oocyst stock preparation.
Viable Giardia intestinalis cysts and C. parvum oocysts used throughout this study were obtained from several sources. Viable G. intestinalis cysts and C. parvum oocysts, cultured and harvested from Mongolian gerbils, were purchased (DyNAgenics, Neosho, Mont.) as crude, partially purified, fecal suspensions. C. parvum oocysts harvested from an endemically infected dairy herd were supplied by Peter Cox (AWT Ensight, Sydney, Australia), and C. parvum oocysts were also obtained from calf fecal specimens supplied by Michael O’Callaghan (Department of Primary Industries, Adelaide, South Australia, Australia). Additional G. intestinalis cysts from human fecal specimens were supplied by Gribbles Pathology (Melbourne, Australia). All fecal samples were emulsified in phosphate-buffered saline (PBS), semipurified by Percoll-sucrose density gradient centrifugation, and stored at 4°C. The concentrations of cyst and oocyst stocks were determined in triplicate with a hemocytometer.
Dilution series of cysts and oocysts used in spiking experiments were enumerated microscopically after staining them in suspension with the Hydrofluor-Combo indirect immunofluorescence detection kit (Meridian Diagnostics). Viability of C. parvum oocysts was assessed by concurrent staining with 4′,6-diamidino-2-phenylindole (DAPI) and propidium iodide (PI). Giardia cysts were considered viable when PI was excluded (33, 34). Oocysts were considered viable when inclusion of DAPI in sporozoite nuclei was observed and PI was excluded from the oocyst (8, 17). Epifluorescence was observed with an Axioskop microscope (Zeiss, Oberkochen, Germany).
Water samples.
Volumes of water between 20 and 1,500 liters were concentrated by a modification of a method described by Payment et al. (28). This involved filtration through a wound fiberglass depth cartridge filter with a nominal pore size of 1 μm (Memtec Limited, Timonium, Md.) and elution with a beef extract eluent (1.5% beef extract, 0.5% Tween 80, pH 9.75). The flow rate of water through the filters varied between 4 and 15 liters min−1. Either samples were filtered on-site and the filter was transported to the laboratory on ice, or volumes of water were collected in sterile 20-liter plastic containers which were transported to the laboratory and filtered within 4 h of collection. Additional 500-ml samples were collected from each site to determine the physicochemical quality of each sample as described in Standard Methods for the Examination of Water and Wastewater, 18th ed. (3). Cysts and oocysts were eluted off the filter within 24 h of sample collection by backflushing with 1.8 liters of eluent. The eluate was immediately adjusted to pH 7.2 ± 0.2 with the addition of 1 N HCl. Backflushings were concentrated by centrifugation at 3,500 × g for 20 min, and the final pellet was resuspended in PBS to a volume between 3 and 10 ml. Equal volumes of concentrates were analyzed by both immunofluorescence (IF) microscopy and RT-PCR, but sample portions for IF-DAPI-PI microscopy first required clarification by Percoll-sucrose density gradient centrifugation (4). Cysts and oocysts were enumerated, and viability was determined as described above. Density gradient centrifugation was not required for RT-PCR analysis of sample concentrates.
Preparation of spiked samples for Diamond filters.
Ten spiking experiments were performed on 50-liter volumes of tap water with a turbidity range of 3 to 5 nephelometric turbidity units (NTU). The water was seeded with 500 to 2,000 cysts and oocysts, which were 8 to 12 weeks old. The seeded samples were filtered (flow rates, 7 to 15 liters min−1) and recovered by backflushing and centrifugation as described above. Recovered cysts and oocysts were enumerated, and the percentage of recovery was determined.
Preparation of spiked samples for RT-PCR.
Sample concentrates were heated to 60°C for 30 min to inactivate any indigenous viable Giardia cysts or Cryptosporidium oocysts. Fresh dilutions of viable cysts and oocysts were prepared in PBS. The G. intestinalis and C. parvum stocks were approximately 75 and 90% viable, respectively. Fifty-microliter volumes containing approximately 500, 100, 50, 10, 5, and 0 viable cysts and oocysts were inoculated into 100-μl packed pellet volumes (volume of material remaining after centrifugation at 5,000 × g and removal of supernatant) of heat-inactivated sample concentrates or directly into InstaGene (Bio-Rad). Sterile water was inoculated into sample concentrates or InstaGene for negative controls. A 50-μl volume of each dilution was also stained and enumerated as described above to obtain a count of each inoculum.
RNA internal positive control (IPC).
An RNA fragment was added to each sample at the stage of oocyst and cyst lysis to monitor the efficiency of both mRNA capture by the magnetic beads and the subsequent RT and PCR. The RNA IPC was constructed with the use of modifying primers to amplify a 290-bp sequence of nontarget DNA by PCR (39). The 5′ end of the upstream primer was modified to contain the T7 phage promoter sequence followed by an additional 6 nucleotides (13, 40). The downstream primer (5′ modified with 15 thymidine bases) was designed to add a 3′ tail of 15 adenosine bases to the IPC. The PCR product was transcribed with the riboprobe T7 in vitro transcription system (Promega). After in vitro transcription, the DNA was digested with RQ1 DNase (Promega) and the RNA was purified by phenol-chloroform extraction followed by precipitation with ethanol (32). The resultant RNA IPC containing a 15-nucleotide-long polyadenylated tail was resuspended in diethylpyrocarbonate-treated water (DEPC-H2O), diluted to a concentration of approximately 5 fg μl−1, and stored in 100-μl aliquots at −70°C. PCR was performed on the preparation to confirm the absence of DNA.
Heat shock and mRNA extraction.
Both spiked and nonspiked Diamond filter concentrates were pelleted by centrifugation at 5,000 × g for 3 min. Pellets containing cysts and oocysts were resuspended in 200 μl of InstaGene and incubated at 45°C for 20 min to maximize HSP mRNA production. Following heat shock, 200 μl of lysis-binding buffer (100 mM Tris-HCl [pH 8.0], 500 mM LiCl, 10 mM EDTA [pH 8.0], 1% lithium dodecyl sulfate, 5 mM dithiothreitol) was added to each sample, and this mixture was then subjected to freeze-thaw treatment to rupture cysts and oocysts and release nucleic acids. This involved five cycles of freezing the samples in liquid nitrogen for 30 s and thawing them at 65°C for 1 min. Extraction and isolation of mRNA were achieved with 40 μg of oligo(dT)25 magnetic beads (Dynal, Oslo, Norway) per sample, as previously described (39). Immediately after freeze-thaw lysis, debris was pelleted by centrifugation at 17,000 × g for 3 min and the supernatant containing the mRNA was added to prepared beads. At this stage, 2 μl of RNA IPC was added to assess the ability of the beads to capture the mRNA, ensure that RNases did not destroy the RNA prior to addition to the RT, and also ensure that negative results were not due to inhibition of the RT or PCR. Hybridization was performed with gentle mixing by rolling at 30°C for 30 min, and then the beads were washed once with 400 μl of wash buffer (10 mM Tris-HCl [pH 8.0], 0.15 M LiCl, 1.0 mM EDTA, 0.1% lithium dodecyl sulfate) and once with 400 μl of 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl) with a magnetic concentrator to retain the beads. The bound mRNA and beads were finally resuspended in 5 μl of DEPC-H2O and added to the RT reaction mixture. mRNA for use as controls was extracted from approximately 103 G. intestinalis cysts and C. parvum oocysts, resuspended in 100 μl of DEPC-H2O, stored at −70°C, and used neat or diluted as required.
RNase and DNase treatment of Giardia mRNA.
Experiments were performed to confirm that RNA was being specifically isolated from Giardia with the oligo(dT)25 magnetic beads, following the protocol previously described (39).
Inactivation of indigenous Giardia cysts in water concentrates.
In order to confirm the viability of Giardia detected in nonspiked water concentrates, concentrates positive for Giardia were treated with either heat or formaldehyde to inactivate the cysts. Heat treatment involved incubation of the water concentrate at 60°C in a water bath for 30 min. Formaldehyde treatment was performed by adding sufficient formaldehyde to the sample concentrate to obtain a final concentration of 10% (vol/vol). After incubation at room temperature for 2 h, the formaldehyde was removed by centrifugation at 5,000 × g for 3 min. The supernatant containing the formaldehyde was discarded, and the pellet was washed once in PBS. Both heat-inactivated and formaldehyde-inactivated concentrates were then concentrated by centrifugation, and the pellets were resuspended in InstaGene and treated as described above within 2 h of treatment.
PCR primers and probes.
The sequences and references for all PCR primers and oligonucleotide probes used in this study are presented in Table 1. For Giardia detection, we initially tried the G. intestinalis-specific GHSP1-GHSP2 primer set (1); however, neither a PCR nor an RT-PCR product could be reliably detected with these primers. The Giardia spp. primer set GGL-GGR (23, 24) was subsequently used, as it demonstrated reliable and sensitive Giardia detection. Primers for multiplex PCR (Giardia spp., C. parvum, and IPC) were each modified by the 5′ addition of a 20-mer universal primer sequence (UPS; 5′-GCGGTCCCAAAAGGGTCAGT-3′) (36).
TABLE 1.
PCR primers and oligonucleotide probes used for IPC construction and detection of Giardia, Cryptosporidium, and IPC
| Oligonucleotide
|
Target | Sequence | PCR product size (bp) | Reference(s) | |
|---|---|---|---|---|---|
| Primer pair or probe | Name | ||||
| Primer pair 1 | GGL | Giardia giardin gene | 5′-AAGTGCGTCAACGAGCAGCT-3′ | 171 | 23, 24 |
| GGR | 5′-TTAGTGCTTTGTGACCATCGA-3′ | ||||
| Primer pair 2 | CHSP1 | C. parvum hsp70 gene | 5′-AGCAATCCTCTGCCGTACAGG-3′ | 590 | 39 |
| CHSP4 | 5′-AAGAGCATCCTTGATCTTCT-3′ | ||||
| Primer pair 3 | IPCF | IPC | 5′-ATGACAGCCACTCCT-3′ | 290 | This study |
| IPCR | 5′-ATGTCAGTTGTGACCACGAA-3′ | ||||
| Primer pair 4 | GHSP1 | Giardia heat shock gene | 5′-AGGGCTCCGGCATAACTTTCC-3′ | 163 | 6 |
| GHSP2 | 5′-GTATCTGTGACCCGTCCGAG-3′ | ||||
| Primer pair 5 | CPHSP2423 | Cryptosporidium spp. hsp70 gene | 5′-AAATGGTGAGCAATCCTCTG-3′ | 307 | 29 |
| CPHSP2764 | 5′-CTTGCTGCTCTTACCAGTAC-3′ | ||||
| Primer pair 6 | T7 promoter-CHSP1 link | IPC | 5′-GGATCCTAATACGACTCACTATAATGACAGCCACTCCT-3′ | 328 | This study |
| Poly(T)15-CHSP4 link | 5′-TTTTTTTTTTTTTTTATGTCAGTTGTGACCACGAA-3′ | ||||
| Probe 1 | GGP | GGL-GGR PCR product | 5′-TCGAGGACGTCGTCTCGAAGATCCAG-3′ | 23, 24 | |
| Probe 2 | CPHSP2475 | CPHSP2423-CPHSP2764 PCR product | 5′CCATTATCACTCGGTTTAGA-3′ | 29 | |
RT-PCR.
RT-PCR was performed with the GeneAmp RNA PCR kit (Perkin-Elmer); however, Amplitaq Gold DNA polymerase (Perkin-Elmer) was used in place of the Amplitaq DNA polymerase. The captured mRNA was reverse transcribed with oligo(dT)16 primers (Perkin-Elmer). Each RT reaction mixture (30 μl) contained 1× PCR buffer II (10× PCR buffer II contains 500 mM KCl and 100 mM Tris-HCl [pH 8.3]), 5 mM MgCl2, 1 mM deoxynucleoside triphosphates (1 mM [each] dATP, dTTP, dCTP, and dGTP), 1.7 μM oligo(dT)16 primer, 20 U of RNase inhibitor, 50 U of murine leukemia virus reverse transcriptase, and 5 μl of DEPC-H2O containing mRNA-bead complex. RT was performed in an FTS-960 thermal sequencer (Corbett Research, Sydney, Australia) at 42°C for 30 min. Following RT, the samples were heated to 95°C for 5 min to inactivate the reverse transcriptase and were then held at 4°C until aliquots were added to the PCR mixture. A 10-μl aliquot of the RT reaction mixture was added directly to separate PCR mixtures. Each PCR mixture contained 1× PCR buffer II, 2.5 mM MgCl2, 0.5 μM (each) primer, and 1.5 U of Amplitaq Gold DNA polymerase. Amplification was performed in the FTS-960 thermal sequencer with the following protocol: activation of the Amplitaq Gold DNA polymerase at 95°C for 10 min; then 40 cycles of 95°C for 20 s, 60°C for 45 s, and 72°C for 45 s; and a final extension step at 72°C for 5 min. The PCR products were held at 4°C until analyzed. The annealing temperature used for the GHSP primer set was 55°C. PCR products were detected by polyacrylamide gel electrophoresis with silver staining (39).
RESULTS
Detection of Giardia and C. parvum by RT-PCR.
Our intention was to develop a multiplex RT-PCR for the simultaneous detection of Giardia spp., C. parvum, and IPC. The primers used for detection of each target were modified by the 5′ addition of a 20-mer UPS, making the PCR products 40 bp larger. This modification has been reported to eliminate most of the optimization steps required for multiplex PCR (36). The UPS modification of primers facilitated successful multiplex PCR (Fig. 1); however, the sensitivity of the CHSP1-CHSP4 PCR was decreased 10-fold (data not shown). Optimum detection sensitivity was achieved with one RT step, and aliquots of the resultant cDNA pool were added to two separate PCRs, one for the detection of Giardia and the IPC with the UPS-modified primers and one for the detection of C. parvum with the unmodified CHSP1-CHSP4 primer pair. This represented a compromise between detection sensitivity and ease of use.
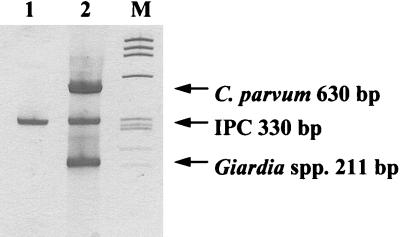
FIG. 1.
Silver-stained polyacrylamide gel showing multiplex RT-PCR with UPS-modified primers for detection of viable Giardia spp., viable C. parvum, and IPC. Lane 1, negative control; lane 2, 500 C. parvum oocysts, 500 Giardia cysts, and 10 fg of IPC; lane M, molecular weight markers (φX174 HaeIII) (Promega).
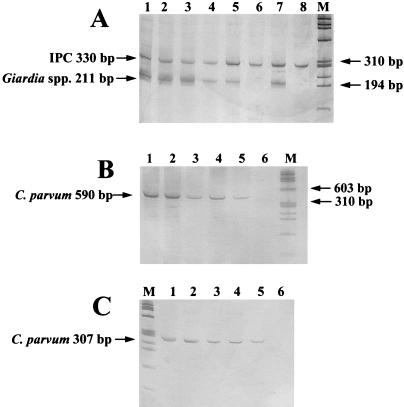
Dilutions of enumerated cysts and oocysts were spiked into equal aliquots of sample concentrate to determine the sensitivity of the RT-PCR. Each aliquot had a final packed pellet volume of approximately 100 μl and was equivalent to approximately 50 liters of creek and river water. These experiments demonstrated detection of low numbers of Giardia cysts and C. parvum oocysts in the range of a single viable cyst and oocyst. The results are summarized in Table 2, and an example from one spiking experiment is given in Fig. 2. No difference in detection sensitivity was observed between the two sets of C. parvum HSP primers (Fig. 2B and C) (29, 39). A comparison of the intensity of the IPC RT-PCR product from the spiked samples with that of the positive and negative controls in Fig. 2 indicates efficient mRNA capture and no observable RNA degradation or inhibition of the RT-PCR.
TABLE 2.
Sensitivity of the RT-PCR assay for detection of G. intestinalis cysts and C. parvum oocysts spiked into 100-μl packed pellet concentrates from river and creek samples as determined by silver-stained polyacrylamide gel electrophoresis
| Spike dosea
|
RT-PCR resultc for:
|
|||
|---|---|---|---|---|
| G. intestinalis | C. parvum | Giardia | C. parvum | IPCb |
| 340 | 210 | + | + | + |
| 100 | 62 | + | + | + |
| 31 | 12 | + | + | + |
| 8 | 2 | + | + | + |
| 2 | 1 | + | + | + |
| 0 | 0 | − | − | + |
Cyst and oocyst concentrations (numbers of cysts or oocysts per sample) representing the averages from 10 spiking experiments.
Ten femtograms of IPC added to each reaction mixture.
+, detected; −, not detected.
FIG. 2.
Sensitivity of RT-PCR for simultaneous detection of Giardia and IPC (A), C. parvum with CHSP1 and CHSP4 primers (B), and C. parvum PCR with CPHSP2423 and CPHSP2764 primers (C) as determined by silver-stained polyacrylamide gel electrophoresis. Lanes 1 to 6 in all panels represent 50 liters of river water (100-μl packed pellet) spiked with approximately 1,000, 100, 50, 10, 1, and 0 cysts or oocysts; lanes 7 and 8 in panel A show positive and negative RT-PCR controls, respectively. Lanes M, DNA molecular weight markers (φX174 HaeIII). IPC was spiked at a concentration of 10 fg per sample.
We previously reported that mRNA and not DNA was specifically captured from C. parvum by oligo(dT)25 beads with up to 103 oocysts (39). To ensure that RNA was specifically captured from Giardia, mRNA was extracted with paramagnetic beads from 2 × 103 cysts and subjected to RNase and DNase treatments. No product was detected by RT-PCR when portions were subjected to RNase treatment alone or when portions were both RNase and DNase treated. The expected product of 171 bp was detected when portions were subjected to DNase treatment or when no treatment was applied. These results confirmed that RNA was specifically isolated from the cysts and that DNA was not being captured and carried over to the PCR.
Water sample analyses for Giardia and C. parvum.
Wound fiberglass depth filter cartridges (Diamond filters) were used to concentrate Giardia cysts and Cryptosporidium oocysts from water. Ten spiking experiments were conducted in 50-liter volumes of tap water to evaluate the performance of the Diamond filter. These gave recoveries for Giardia cysts and Cryptosporidium oocysts of 45 to 88% (average, 74%) and 43 to 84% (average, 54%), respectively. DAPI-PI staining indicated that the C. parvum oocyst stock was 75 to 79% viable while the viability of oocysts in sample concentrates was 73 to 75%, indicating that there was no loss of viability throughout the concentration process.
To compare the performance of the RT-PCR method with that of IF-DAPI-PI microscopy, 29 water samples were collected with the Diamond filters. Sample volumes concentrated were between 20 and 1,500 liters. Several physicochemical and microbiological water quality parameters were tested for these samples and are shown in Table 3, demonstrating the range of water types that are compatible with this method. Separate volumes of the water concentrates were tested by RT-PCR and IF-DAPI-PI microscopy. mRNA was extracted directly from the water concentrates. However, due to interfering debris in the concentrates, a clarification step was required prior to fluorescent staining and microscopy. Table 4 shows the results of RT-PCR detection of viable Giardia and C. parvum compared to those of Giardia and Cryptosporidium detection by IF-DAPI-PI microscopy. Overall, viable Giardia cysts were detected in 20 (69%) samples by RT-PCR and only 7 (24%) samples by IF-DAPI-PI microscopy. In the river and creek samples alone, the detection of viable Giardia was fourfold higher by RT-PCR than by microscopy. An example of results obtained from analysis of creek samples is given in Fig. 3. Viable Cryptosporidium oocysts were detected in four (14%) samples by IF-DAPI-PI microscopy. However, viable C. parvum oocysts were detected only once by RT-PCR (Table 4). Viable Giardia cysts and C. parvum oocysts were not detected in drinking water samples by either method. Comparable results were obtained by both detection methods for treated sewage samples. Interestingly, while Giardia cysts were present, no C. parvum oocysts were detected in these samples by either assay.
TABLE 3.
Microbiological and physicochemical parameters of water samples concentrated for Giardia and Cryptosporidium analyses
| Sample | Value for parameter:
|
|||||
|---|---|---|---|---|---|---|
| 37°C total plate count (no. of organisms/ml) | pH | Electric conductivity (μS/cm at 25°C) | Color (Pt-Co units) | Turbidity (NTU) | Total organic carbon (mg/liter) | |
| Treated sewage discharge | NDa | 6.6–6.9 | 780–800 | 20–50 | 0.8–1.0 | ND |
| River or creek water | 380–2,900 | 7.0–7.9 | 65–780 | 52–160 | 3.5–120 | 4.0–9.0 |
| Drinking water | 0–600 | 6.5–9.0 | 48–60 | 2–15 | <0.5–2 | 1–2 |
ND, not determined.
TABLE 4.
Comparison of IF-DAPI-PI microscopy with RT-PCR for detection of viable Giardia and Cryptosporidium in nonspiked water sample concentrates
| Sample | n | Vol analyzed (liters) | No. of samples positive by:
|
|||
|---|---|---|---|---|---|---|
| IF microscopya
|
RT-PCR
|
|||||
| Giardia spp. | Cryptosporidium | Giardia spp. | C. parvum | |||
| Treated sewage discharge | 6 | 10 | 3 (1–2) | 0 | 4 | 0 |
| River or creek water | 19 | 2–110 | 4 (1) | 4 (1–3) | 16 | 1 |
| Treated drinking water | 4 | 32–300 | 0 | 0 | 0 | 0 |
Numbers in parentheses are the count ranges per volumes analyzed.
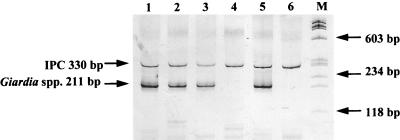
FIG. 3.
Detection of Giardia in water samples by RT-PCR as determined by silver-stained polyacrylamide gel electrophoresis. Lanes 1 to 3, river and creek samples positive for Giardia spp.; lane 4, river sample negative for Giardia spp.; lane 5, RT-PCR positive control; lane 6, RT-PCR negative control; lane M, DNA molecular weight markers (φX174 HaeIII). Sample turbidities were 120 (lane 1), 6.5 (lane 2), 17 (lane 3), and 3.5 (lane 4) NTU. Sample volumes concentrated by Diamond filters were 300 liters; sample volumes processed by RT-PCR were 100 liters.
The low rate of isolation of C. parvum from river, creek, and treated sewage concentrates was surprising. The recently reported Cryptosporidium genus primers, CPHSP2423 and CPHSP2764 (29), were tested in order to directly compare IF-DAPI-PI microscopy to RT-PCR. These primers produced RT-PCR-positive results in all water samples tested (n = 16) including drinking water samples. Southern blot analysis with the C. parvum probe CPHSP2475 indicated the absence of C. parvum (data not shown). An analysis of the Cryptosporidium primers was conducted. Homology searches of GenBank with the BLASTN algorithm (2) showed broad primer cross-reactivity with other heat shock genes from several organisms, including amoeboid slime molds and yeasts which were predicted to produce PCR products of the same size as those of Cryptosporidium.
Further testing was performed on 6 of the 16 sample concentrates which were positive for Giardia by RT-PCR to ensure that the detection of the mRNA correlated with the presence of viable organisms in the samples. Sample concentrates were treated with either formaldehyde or heat to inactivate cysts. No RT-PCR product was detected in any sample after treatment, confirming that detection of mRNA correlated with cyst viability.
DISCUSSION
We have developed an assay which utilizes RT coupled with two different PCRs for the simultaneous detection of viable Giardia spp. and C. parvum. This is a reliable and economic test which is more sensitive than a widely used IF microscopy procedure. In addition, this method specifically detects the human pathogen C. parvum. We attempted to use the GHSP PCR primers (1) in our assay to detect the human pathogen G. intestinalis, but to date we have found this RT-PCR to be unreliable.
Initially, we designed this test as a multiplex PCR for single-tube detection of C. parvum, Giardia, and IPC, but a 10-fold loss in detection sensitivity was observed for the C. parvum component. During the course of this study, another laboratory reported successful multiplex PCR for G. intestinalis and C. parvum with HSP primers (29). This multiplex was not used in an RT-PCR format, and while detection of cysts and oocysts in environmental water concentrates was possible, the detection limit was not specified. In our study, isolation and purification of mRNA with magnetic beads coupled to a single RT with a two-tube PCR permitted sensitive detection of low numbers of cysts and oocysts from sample concentrates. A brief comparison of two reported C. parvum HSP primer sets (29, 39) showed no difference in detection sensitivity (Fig. 2B and C), despite a reported significant theoretical difference in melting temperatures between CHSP1 and CHSP4 primers (29).
Wound fiberglass cartridge filters (Diamond filters) were used for concentrating sample volumes of up to 420 liters of river and creek water and 1,500 liters of drinking water. These filters have been reported elsewhere for concentration of 1,000 to 2,000 liters of drinking water for Giardia detection (28). Assessment of filter performance with 50-liter spiked samples during this study demonstrated an average recovery of 74 and 54% for Giardia and Cryptosporidium, respectively. In addition, the viability of seeded C. parvum oocysts was not altered during collection and processing of samples. Based on these results, concentration of Giardia and Cryptosporidium with the Diamond filter is an efficient method, convenient for processing large-volume water samples. Furthermore, even when sample turbidity was high (6.5 to 120 NTU), sample volumes up to 420 liters could be easily filtered and positive results could be obtained by RT-PCR for viable Giardia (Table 3; Fig. 3). We are currently evaluating the performance of these filters under high-turbidity conditions as measured by spiking experiments.
Clarification of samples by Percoll-sucrose density gradient centrifugation contributes significantly to losses of Giardia and Cryptosporidium (21), compromising the sensitivity of any subsequent detection method. mRNA capture with oligo(dT)25 beads and detection with RT-PCR overcame these limitations, as sample clarification was not required. In addition, the detection of viable parasites was defined by the presence or absence of the correct RT-PCR product, thus overcoming the difficulties often encountered with microscopy caused by sample debris masking fluorescence, interference from autofluorescing algae, or misinterpretation of vital dye stain patterns caused by sample exposure to chlorine (30, 31, 33, 34). However, one obvious limitation of RT-PCR is that it is not strictly quantitative, although a most probable number format may be used if enumeration is required (43).
This RT-PCR method was able to detect low numbers of Giardia cysts and C. parvum oocysts artificially spiked into river and creek water concentrates with packed pellets of 100 μl. Oligo(dT)25 magnetic beads overcame potential RT-PCR inhibition and proved to be a convenient and efficient mRNA purification system.
An RNA IPC was developed to guard against false-negative results. It was added to all samples analyzed at a very low concentration (10 fg), and the effect each sample had on the efficiency of mRNA capture and RT-PCR could be evaluated by visually comparing the intensities of the IPC RT-PCR products of samples and controls. Equivalent intensities gave confidence that negative results were valid and not due to magnetic bead capture failure, RNA degradation, or inhibition of the RT or PCR.
The test detected only viable organisms since mRNA was selectively recovered from samples and previously RT-PCR-positive samples were rendered negative by heat or formalin treatment, thus demonstrating that amplifiable mRNA was not maintained in cysts treated in either of these ways. Our observation that heat-inactivated cysts did not produce or maintain mRNA is in contrast with that by another investigator, who reported detection of giardin mRNA after heat treatment (23). This discrepancy may be related to experimental differences such as the time between inactivation of cysts and mRNA extraction.
The high sensitivity of the assay was further demonstrated by a comparison of RT-PCR with IF-DAPI-PI microscopy in 29 nonspiked water samples. The RT-PCR was found to be the more sensitive detection method, detecting the presence of viable Giardia in four times as many river and creek samples as did IF-DAPI-PI microscopy. Interestingly, the intensity of the PCR products from these analyses (Fig. 3) suggested viable cyst concentrations greater than 20 cysts per sample concentrate, and yet only one viable cyst was detected in the same concentrate by IF-DAPI-PI microscopy. This suggests possible losses of cysts throughout the density gradient clarification process and nondetection by IF microscopy. Due to the inability of the Cryptosporidium fluorescent antibody to specifically stain C. parvum oocysts, it was not possible to compare the detection of viable C. parvum in 3% of samples by RT-PCR with the detection of viable Cryptosporidium in 14% of samples by microscopy. However, a lack of C. parvum RT-PCR detection sensitivity can be discounted, given the results of spiking experiments. The difference in detection rates suggests that viable Cryptosporidium species other than C. parvum may have been present in those water samples.
To address this possibility, the Cryptosporidium HSP genus primer set recently described (29) was used to analyze 16 water samples. All samples were positive with the correct-size PCR product. However, these primer sequences appear to have cross-reactivity with hsp genes from a range of other organisms which may be expected to occur naturally in water. This genus PCR is therefore likely to be of diagnostic value only in conjunction with Cryptosporidium species-specific probes, such as the CPHSP2475 C. parvum probe (29).
The results presented in this paper demonstrate the effectiveness of using Diamond filter concentration in conjunction with RT-PCR for detection of viable waterborne Giardia and Cryptosporidium. The use of magnetic bead mRNA capture with RT-PCR to directly screen sample concentrates for parasites significantly enhances the quality of pathogen incidence data compared to that with microscopy-based detection. First, low theoretical detection limits, <0.01 cysts or oocysts/liter or better, are conveniently achieved, as large volumes (>100 liters) are easily sampled and analyzed. Secondly, actual detection limits are lower in that RT-PCR is more sensitive than IF microscopy. Thirdly, detection of viable parasites is possible. Other benefits include batch sample processing, objective assessments, and short analysis times. For example, analysis of 15 water sample concentrates takes one operator 6 h by RT-PCR compared with 3 to 4 days by IF microscopy.
More work is now required to standardize PCR-based methods between laboratories and to demonstrate the portability and reliability of this technology for environmental pathogen monitoring.
ACKNOWLEDGMENTS
This work was substantially supported by funding from the Government of Victoria through the Water Bureau, Department of Natural Resources and Environment, and in part by the Urban Water Research Association, Australia.
We also thank Peter Cox and Malcolm Warnecke for critical review of the manuscript and Joanne O’Toole and Kate Hines for advice and expert technical assistance.
REFERENCES
- 1.Abbaszadegan M, Huber M S, Gerba C P, Pepper I L. Detection of viable Giardia cysts by amplification of heat shock-induced mRNA. Appl Environ Microbiol. 1997;63:324–328. doi: 10.1128/aem.63.1.324-328.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.American Public Health Association. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C: American Public Health Association; 1989. [Google Scholar]
- 4.American Society for Testing and Materials. Proposed test method for Giardia cysts and Cryptosporidium oocysts in low-turbidity water by a fluorescence procedure. Annu Book ASTM Stand. 1991;11.01:925–934. [Google Scholar]
- 5.Anguish L J, Ghiorse W C. Computer-assisted laser scanning and video microscopy for analysis of Cryptosporidium parvum oocysts in soil, sediment, and feces. Appl Environ Microbiol. 1997;63:724–733. doi: 10.1128/aem.63.2.724-733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anonymous. November 1997 draft. Method 1622: Cryptosporidium in water by filtration/IMS/FA. EPA 821-R-97-021. U.S. Environmental Protection Agency, Washington, D.C.
- 7.Campbell A, Smith H. Immunomagnetic separation of Cryptosporidium oocysts from water samples: round robin comparison of techniques. Water Sci Technol. 1997;35:397–401. [Google Scholar]
- 8.Campbell A T, Robertson L J, Smith H V. Viability of Cryptosporidium parvum oocysts: correlation of in vitro excystation with inclusion or exclusion of fluorogenic vital dyes. Appl Environ Microbiol. 1992;58:3488–3493. doi: 10.1128/aem.58.11.3488-3493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell A T, Robertson L J, Smith H V, Girdwood R W A. Viability of Cryptosporidium parvum oocysts concentrated by calcium carbonate flocculation. J Appl Bacteriol. 1994;76:638–639. doi: 10.1111/j.1365-2672.1994.tb01663.x. [DOI] [PubMed] [Google Scholar]
- 10.Clancy J L, Gollnitz W D, Tabib Z. Commercial labs: how accurate are they? J Am Water Works Assoc. 1994;86:89–97. [Google Scholar]
- 11.Deng M Q, Cliver D O, Mariam T W. Immunomagnetic capture PCR to detect viable Cryptosporidium parvum oocysts from environmental samples. Appl Environ Microbiol. 1997;63:3134–3138. doi: 10.1128/aem.63.8.3134-3138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dykes A C, Juranek D D, Lorenz R A, Sinclair S P, Jakubowski W, Davies R B. Municipal waterborne giardiasis: an epidemiologic investigation. Beavers implicated as a possible reservoir. Ann Intern Med. 1980;92:165–170. doi: 10.7326/0003-4819-92-2-165. [DOI] [PubMed] [Google Scholar]
- 13.Gibson U E M, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 14.Gilgen M, Wegmüller B, Burkhalter P, Bühler H-P, Müller U, Lüthy J, Candrian U. Reverse transcription PCR to detect enteroviruses in surface water. Appl Environ Microbiol. 1995;61:1226–1231. doi: 10.1128/aem.61.4.1226-1231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas C N, Crockett C S, Rose J B, Gerba C P, Fazil A M. Assessing the risk posed by oocysts in drinking water. J Am Water Works Assoc. 1996;88:131–136. [Google Scholar]
- 16.Jakubowski W, Boutros S, Faber W, Fayer R, Ghiorse W, LeChevallier M, Rose J, Schaub S, Singh A, Stewart M. Environmental methods for Cryptosporidium. J Am Water Works Assoc. 1996;88:107–121. [Google Scholar]
- 17.Jenkins M B, Anguish L J, Bowman D D, Walker M J, Ghiorse W C. Assessment of a dye permeability assay for determination of inactivation rates of Cryptosporidium parvum. Appl Environ Microbiol. 1997;63:3844–3850. doi: 10.1128/aem.63.10.3844-3850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent G P, Greenspan J R, Herndon J L, Mofenson L M, Harris J S, Eng T R, Waskin H A. Epidemic giardiasis caused by a contaminated public water supply. Am J Public Health. 1988;78:139–143. doi: 10.2105/ajph.78.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laxer M A, Timblin B K, Patel R J. DNA sequences for the specific detection of Cryptosporidium parvum by the polymerase chain reaction. Am J Trop Med Hyg. 1991;45:688–694. doi: 10.4269/ajtmh.1991.45.688. [DOI] [PubMed] [Google Scholar]
- 21.LeChevallier M W, Norton W D, Siegel J E, Abbaszadegan M. Evaluation of the immunofluorescence procedure for detection of Giardia cysts and Cryptosporidium oocysts in water. Appl Environ Microbiol. 1995;61:690–697. doi: 10.1128/aem.61.2.690-697.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisle J T, Rose J B. Cryptosporidium contamination of water in the USA and UK: a mini-review. J Water SRT—Aqua. 1995;44:103–117. [Google Scholar]
- 23.Mahbubani M H, Bej A K, Perlin M, Schaefer III F W, Jakubowski W, Atlas R M. Detection of Giardia cysts by using the polymerase chain reaction and distinguishing live from dead cysts. Appl Environ Microbiol. 1991;57:3456–3461. doi: 10.1128/aem.57.12.3456-3461.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahbubani M H, Bej A K, Perlin M H, Schaefer III F W, Jakubowski W, Atlas R M. Differentiation of Giardia duodenalis from other Giardia spp. by using polymerase chain reaction and gene probes. J Clin Microbiol. 1992;30:74–78. doi: 10.1128/jcm.30.1.74-78.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangiapan G, Vokurka M, Schouls L, Cadranel J, Lecossier D, Van Embden J, Hance A. Sequence capture-PCR improves detection of mycobacterial DNA in clinical specimens. J Clin Microbiol. 1996;34:1209–1215. doi: 10.1128/jcm.34.5.1209-1215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan U M, O’Brien P A, Thompson R C A. The development of diagnostic PCR primers for Cryptosporidium using RAPD-PCR. Mol Biochem Parasitol. 1996;77:103–108. doi: 10.1016/0166-6851(96)02577-7. [DOI] [PubMed] [Google Scholar]
- 27.Nahrstedt A, Gimbel R. A statistical method for determining the reliability of the analytical results in the detection of Cryptosporidium and Giardia in water. J Water SRT—Aqua. 1996;45:101–111. [Google Scholar]
- 28.Payment P, Bérubé A, Perreault D, Armon R, Trudel M. Concentration of Giardia lamblia cysts, Legionella pneumophila, Clostridium perfringens, human enteric viruses, and coliphages from large volumes of drinking water, using a single filtration. Can J Microbiol. 1989;35:932–935. doi: 10.1139/m89-154. [DOI] [PubMed] [Google Scholar]
- 29.Rochelle P A, Ferguson D M, Handojo T J, De Leon R, Stewart M H, Wolfe R L. An assay combining cell culture with reverse transcriptase PCR to detect and determine the infectivity of waterborne Cryptosporidium parvum. Appl Environ Microbiol. 1997;63:2029–2037. doi: 10.1128/aem.63.5.2029-2037.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodgers M R, Flanigan D J, Jakubowski W. Identification of algae which interfere with the detection of Giardia cysts and Cryptosporidium oocysts and a method for alleviating this interference. Appl Environ Microbiol. 1995;61:3759–3763. doi: 10.1128/aem.61.10.3759-3763.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saby S, Sibille I, Mathieu L, Paquin J L, Block J C. Influence of water chlorination on the counting of bacteria with DAPI (4′,6-diamidino-2-phenylindole) Appl Environ Microbiol. 1997;63:1564–1569. doi: 10.1128/aem.63.4.1564-1569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sauch J F, Flanigan D, Galvin M L, Berman D, Jakubowski W. Propidium iodide as an indicator of Giardia cyst viability. Appl Environ Microbiol. 1991;57:3243–3247. doi: 10.1128/aem.57.11.3243-3247.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schupp D G, Erlandsen S L. A new method to determine Giardia cyst viability: correlation of fluorescein diacetate and propidium iodide staining with animal infectivity. Appl Environ Microbiol. 1987;53:704–707. doi: 10.1128/aem.53.4.704-707.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepherd K M, Wyn-Jones A P. An evaluation of methods for the simultaneous detection of Cryptosporidium oocysts and Giardia cysts from water. Appl Environ Microbiol. 1996;62:1317–1322. doi: 10.1128/aem.62.4.1317-1322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuber A P, Grondin V J, Klinger K W. A simplified procedure for developing multiplex PCRs. Genome Res. 1995;5:488–493. doi: 10.1101/gr.5.5.488. [DOI] [PubMed] [Google Scholar]
- 37.Smith H V, Hayes C R. The status of UK methods for the detection of Cryptosporidium spp. oocysts and Giardia spp. cysts in water concentrates. Water Sci Technol. 1997;35:369–376. [Google Scholar]
- 38.Solo-Gabriele H, Neumeister S. US outbreaks of cryptosporidiosis. J Am Water Works Assoc. 1996;88:76–86. [Google Scholar]
- 39.Stinear T, Matusan A, Hines K, Sandery M. Detection of a single viable Cryptosporidium parvum oocyst in environmental water concentrates by reverse transcription-PCR. Appl Environ Microbiol. 1996;62:3385–3390. doi: 10.1128/aem.62.9.3385-3390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoflet E S, Koeberl D D, Sarkar G, Sommer S S. Genomic amplification with transcript sequencing. Science. 1988;239:491–494. doi: 10.1126/science.3340835. [DOI] [PubMed] [Google Scholar]
- 41.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teunis P F M, Medema G J, Kruidenier L, Havelaar A H. Assessment of the risk of infection by Cryptosporidium or Giardia in drinking water from a surface water source. Water Res. 1997;31:1333–1346. [Google Scholar]
- 43.Toranzos G A, Alvarez A J, Dvorsky E A. Application of the polymerase chain reaction technique to the detection of pathogens in water. Water Sci Technol. 1993;27:207–210. [Google Scholar]
- 44.Tsai Y-L, Olson B H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992;58:754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vesey G, Slade J S, Byrne M, Shepherd K, Dennis P J, Fricker C R. Routine monitoring of Cryptosporidium oocysts in water using flow cytometry. J Appl Bacteriol. 1993;75:87–90. doi: 10.1111/j.1365-2672.1993.tb03413.x. [DOI] [PubMed] [Google Scholar]
- 46.Vesey G, Slade J S, Byrne M, Shepherd K, Fricker C R. A new method for the concentration of Cryptosporidium oocysts from water. J Appl Bacteriol. 1993;75:82–86. doi: 10.1111/j.1365-2672.1993.tb03412.x. [DOI] [PubMed] [Google Scholar]
- 47.Wagner-Wiening C, Kimmig P. Detection of viable Cryptosporidium parvum oocysts by PCR. Appl Environ Microbiol. 1995;61:4514–4516. doi: 10.1128/aem.61.12.4514-4516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeates C, Gillings M R, Davison A D, Altavilla N, Veal D A. PCR amplification of crude microbial DNA extracted from soil. Lett Appl Microbiol. 1997;25:303–307. doi: 10.1046/j.1472-765x.1997.00232.x. [DOI] [PubMed] [Google Scholar]