Abstract
The oral microbiota plays an important role in the exogenous nitrate reduction pathway and is associated with heart and periodontal disease and cigarette smoking. We describe smoking-related changes in oral microbiota composition and resulting potential metabolic pathway changes that may explain smoking-related changes in disease risk. We analyzed health information and salivary microbiota composition among 1601 Cooperative Health Research in South Tyrol participants collected 2017–2018. Salivary microbiota taxa were assigned from amplicon sequences of the 16S-V4 rRNA and used to describe microbiota composition and predict metabolic pathways. Aerobic taxa relative abundance decreased with daily smoking intensity and increased with years since cessation, as did inferred nitrate reduction. Former smokers tended to be more similar to Never smokers than to Current smokers, especially those who had quit for longer than 5 years. Cigarette smoking has a consistent, generalizable association on oral microbiota composition and predicted metabolic pathways, some of which associate in a dose-dependent fashion. Smokers who quit for longer than 5 years tend to have salivary microbiota profiles comparable to never smokers.
Subject terms: Microbiology, Biomarkers, Health care, Risk factors
Introduction
Smoking is a risk factor for several complex, chronic diseases including but not limited to respiratory diseases1, periodontitis2–4, oropharyngeal cancers5,6 and cardiovascular diseases7. Recently, alterations to oral microbiota composition have been observed in cases of periodontitis8–11, squamous cells carcinoma12, cardiovascular diseases13,14 and in cigarette smokers15–19 (Supplementary File 1, Table 1). Therefore, it is possible that smoking related changes in the oral microbiota contribute to the etiology of one or more chronic health conditions. The oral microbiota performs several functions, including playing an important role in the exogenous nitrate reduction pathway and hence blood pressure regulation via nitric oxide (NO)20–23. Diets high in nitrate increase the presence of oral nitrate-reducing bacteria (NRB), the most prevalent of which are species in the Neisseria, Prevotella and Actinomyces genera24. when NRB are present, salivary nitrate reduction increases23,25. Whether tobacco consumption directly or indirectly alters the relative abundance of nitrate reducing bacteria remains to be explored; however, smoking was reported to inhibit uptake of blood-circulating nitrate into saliva26.
The salivary microbiota composition varies by smoking habits. A 2016 meta-analysis of 1204 USA citizens from two national cohorts found that compared to former or never smokers, smokers had a decreased relative abundance of Proteobacteria, an increase of Actinobacteria and a lower proportion of aerobic taxa after adjustment for age and sex15. A 2019 study set in New York city confirmed and extended those findings showing that, in contrast to former or never smokers (N = 86), the salivary microbiota of smokers (N = 86) showed higher abundance of genera Stomatobaculum, Megasphaera, Veillonella, Leptotrichia, Campylobacter and Treponema, and lower abundance of Neisseria, Lautropia, Haemophilus, Capnocytophaga16. Studies conducted in Saudi Arabia17, Asia27,28 and Europe19 reported comparable findings (Supplementary File 1, Figure 1). In a meta-analysis with 1204 Americans, Wu and colleagues uniquely found that the relative abundance of classes Betaproteobacteria, Gammaproteobacteria and Flavobacteriia was inversely correlated with the number of cigarettes smoked daily and directly correlated with the years since quitting smoking15. While associations between smoking status and salivary microbial composition have been previously characterized in Americans, no study has described associations of the salivary microbiota composition and metabolic potential with daily smoking intensity or years since quitting in a European population.
This study adds to our understanding of the associations of the salivary microbiota taxonomic and predicted metabolic functional composition with smoking status, intensity (grams/day) and history (years since cessation) in a large, novel, homogeneous Italian cohort aged 18–91: the Cooperative Health Research In South Tyrol (CHRIS)29 Microbiome study (CHRISMB). We hypothesized that we would observe results consistent with the literature and some novel insights attributable to the unique characteristics of CHRISMB and the large sample size. We additionally hypothesized that the nitrate reduction pathways could be less abundant in smokers, given the previous findings of decreases of taxa in the Neisseria and Haemophilus genera, which harbor several NRB species30.
Results
Characteristics of study population in relation to smoking
After exclusions (see “Methods” and Supplementary File 1, Tables 2 and 3 for details), CHRISMB consisted of 1601 individuals with an average age of 45 years (range 18–91) and had slightly more females (52.9%) than males. Most had 20 or more natural teeth (72.1%). Almost half (45%) were Current or Former smokers; cigarettes were the primary source of tobacco for all but 5 participants. Smokers were more frequently males and younger than Never or Former smokers (Table 1). Former smokers quit smoking 17.96 years, on average (Range 0–61; median 16). When stratified by age group, Current and Former smokers aged 41–60 years with higher lifetime exposure to smoke tended to have fewer teeth than smokers with a lower cumulative exposure (Supplementary File 1, Figure 2).
Table 1.
Distribution of selected demographic descriptors in relation to smoking status in the Cooperative Health Research in South Tyrol Microbiome (CHRISMB) study.
| Never (N = 880) |
Former (N = 395) |
Current (N = 326) |
CHRISMB (N = 1601) |
X2 p value | |
|---|---|---|---|---|---|
| Sex | 2.7e−07 | ||||
| Male | 356 (40.5%) | 222 (56.2%) | 173 (53.1%) | 751 (46.9%) | |
| Female | 524 (59.5%) | 173 (43.8%) | 153 (46.9%) | 850 (53.1%) | |
| Age category (years) | 3.6e−19 | ||||
| 18–30 | 238 (27.0%) | 41 (10.4%) | 130 (39.9%) | 409 (25.5%) | |
| 31–40 | 139 (15.8%) | 73 (18.5%) | 57 (17.5%) | 269 (16.8%) | |
| 41–50 | 196 (22.3%) | 75 (19.0%) | 64 (19.6%) | 335 (20.9%) | |
| 51–60 | 144 (16.4%) | 112 (28.4%) | 51 (15.6%) | 307 (19.2%) | |
| 61–70 | 93 (10.6%) | 57 (14.4%) | 23 (7.1%) | 173 (10.8%) | |
| 71+ | 70 (8.0%) | 37 (9.4%) | 1 (0.3%) | 108 (6.7%) | |
| N° teeth (self-reported) | 0.07 | ||||
| 0 | 50 (5.7%) | 23 (5.8%) | 16 (4.9%) | 89 (5.6%) | |
| 1–9 | 57 (6.5%) | 41 (10.4%) | 20 (6.1%) | 118 (7.4%) | |
| 10–19 | 117 (13.3%) | 74 (18.7%) | 48 (14.7%) | 239 (14.9%) | |
| 20+ | 656 (74.5%) | 257 (65.1%) | 242 (74.2%) | 1155 (72.1%) | |
| Gums health (self-reported) | 0.87 | ||||
| Excellent | 45 (5.1%) | 18 (4.6%) | 16 (4.9%) | 79 (4.9%) | |
| Very good | 188 (21.4%) | 79 (20.0%) | 64 (19.6%) | 331 (20.7%) | |
| Good | 291 (33.1%) | 124 (31.4%) | 87 (26.7%) | 502 (31.4%) | |
| Average | 229 (26.0%) | 84 (21.3%) | 99 (30.4%) | 412 (25.7%) | |
| Poor | 47 (5.3%) | 24 (6.1%) | 22 (6.7%) | 93 (5.8%) | |
| Very poor | 6 (0.7%) | 2 (0.5%) | 3 (0.9%) | 11 (0.7%) | |
| Missing | 74 (8.4%) | 64 (16.2%) | 35 (10.7%) | 173 (10.8%) | |
Per-column percentages were also reported in brackets. The whole cohort is included under the “CHRISMB” column. Significance was calculated as X2 test for categorical variables. Non-available measures were reported as “Missing”.
Salivary microbiota DNA sequencing of selected samples consisted of almost 36 million reads, with a median read count per sample of 22,308 (interquartile range: 11,884, full range 5283–65,837). After filtering by prevalence and minimum detection (see “Methods”), the dataset included 627 ASVs assigned to 82 genera (Supplementary File 1, Table 4).
Qualitative smoking habits are associated with compositional and functional profiles of salivary genera
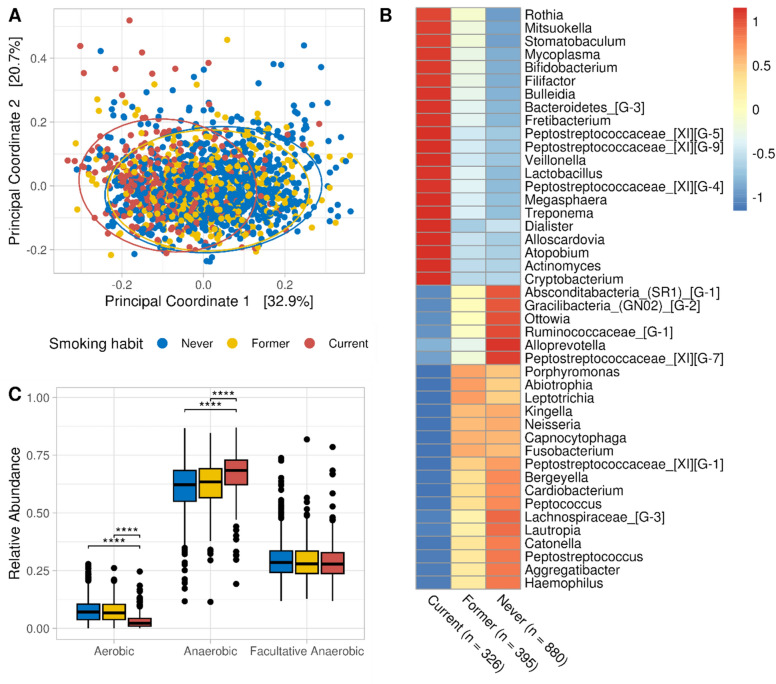
The microbiota composition of CHRISMB at phylum level was dominated by Firmicutes, followed by Bacteroidetes Proteobacteria, Fusobacteria and Actinomycetes; while at Genus level it was dominated by Prevotella, Streptococcus, Veillonella, Haemophilus, Neisseria (Supplementary File 1, Figure 3). The salivary microbiota was significantly associated with smoking (Fig. 1A, PERMANOVA R2 = 0.04, p = 0.001, 2000 permutations) as well as sex, age group and number of teeth, considering the marginal effect of all variables together (Supplementary File 1, Table 5). Alpha diversity was not significantly associated with smoking status (Supplementary File 1, Figure 4). Principal coordinate analysis and differential abundance analysis together suggested that the salivary microbiota of Former smokers was highly similar to Never smokers. Consensus-based differential abundance analysis identified 44 genera that were significantly different between Current smokers and Never smokers after adjusting for age, sex, and number of teeth (Fig. 1B). To investigate sex-dependent associations, we repeated the same consensus differential abundance analysis separately by sex, again adjusting for age and number of teeth. Despite finding sex-specific differentially abundant genera, all were in the set of 44 differentially abundant genera of the model adjusted for sex, age group, and number of teeth (Supplementary File 1, Figure 5). We annotated genera based on their oxygen requirements from a manually curated table by Calgaro et al.31, and observed that the relative abundance of aerobic taxa decreased consistently in smokers (from a median of 7% to 3%), in favor of anaerobes (Fig. 1C).
Figure 1.
Association between qualitative smoking habits (Never, Former and Current) and the salivary microbiota in the CHRISMB cohort. (A) Principal Coordinate Analysis on the Bray–Curtis dissimilarity at genus level. Confidence areas (95%) were drawn as ellipses. Group separations were mild but significant (PERMANOVA R2 = 0.04, p = 0.001, beta-dispersity p = 0.104). Axes x and y were chosen as the principal components which explained most of the overall microbiota variability, which is shown in square brackets. (B) Heatmap of the 44 genera differentially abundant between Current and Never smokers. Each genus was transformed to relative abundance and Z-score scaled. Red and blue colors indicate a higher and lower mean abundance, respectively, while yellow colors indicate no difference. Genera reported in the figure were differentially abundant (Benjamini–Hochberg q-value < 0.05, false discovery rate (FDR) = 5%, ALDEx2 Holm q-value < 0.05) in at least 4 out of 5 differential abundance methods (DESeq2, LinDA, MaAsLin2, ALDEx2, ANCOM-BC), adjusting for age (categorical), sex (binary) and number of teeth (categorical). (C) Relative abundance of aerobes, anaerobes, and facultative anaerobes in relation to smoking status. Statistical significance was calculated with pairwise Wilcoxon test adjusting p-values (q-values) for a 5% FDR with the Benjamini–Hochberg method (**q < 0.05; ***q < 0.001, ****q < 0.0001).
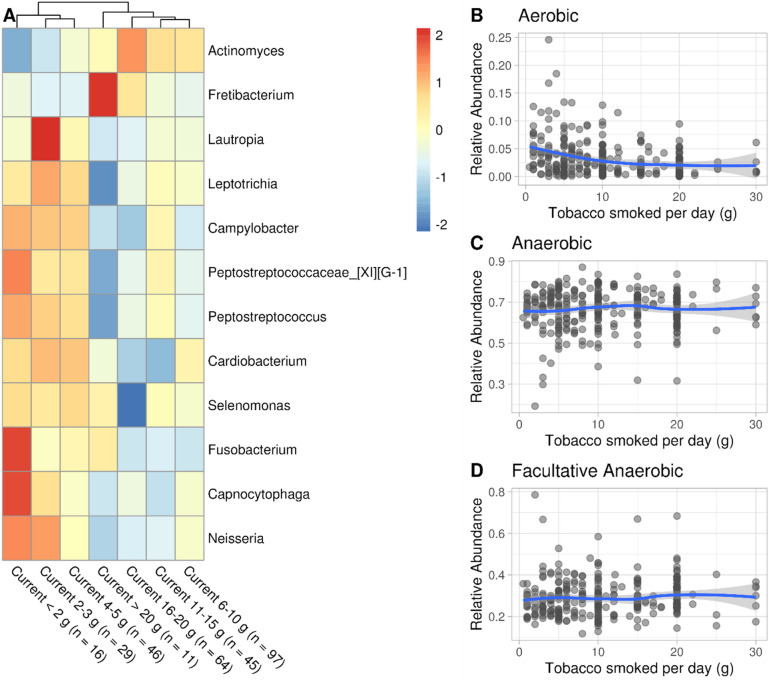
Several microbial genera associated with smoking habits are also associated with the grams of tobacco smoked daily
We regressed each genus against daily smoking intensity as multiples of 5 g per day (see “Methods”). Fretibacterium was positively associated with increases in daily smoking intensity and 10 with decreases (Fig. 2A). Except for Campylobacter and Selenomonas, the remaining 9 genera were also differentially abundant comparing Current against Never smokers (Fig. 1B). Additionally, the effect sizes estimated in the daily smoking intensity regression were highly correlated with the estimates obtained comparing Current against Never smokers (Pearson = 0.87, Supplementary File 1, Figure 6), suggesting that some genera associated with smoking against non-smoking were additionally associated with daily smoking intensity. The complete linkage hierarchical clustering in the pheatmap function tended to cluster heavier smokers together, further suggesting a dose effect (Fig. 2A). The mean relative abundance and variance of aerobes significantly decreased at the increasing daily smoking intensity (linear regression , p value = ; Supplementary File 1, Tables 6, 7), adjusted for age as continuous variable, sex and number of teeth; Figs. 2C, 3D), with a plateau at more than 10 g (Fig. 2B). Conversely, the relative abundance of anaerobes and facultative anaerobes slightly increased.
Figure 2.
Smokers’ (n = 308) daily smoking intensity is associated with relative abundance shifts of several genera and a decrease of aerobic taxa relative abundance. (A) Heatmap of genera significantly affected by daily smoking intensity. Genera were transformed to relative abundance and Z-score scaled to highlight relative differences in mean abundance in relation to the smoking intensity. Significant genera (Benjamini–Hochberg q-value < 0.05, FDR = 5%) were obtained modeling each genus in response to daily smoking intensity as multiples of 5 g per day as a semi-continuous variable, adjusting for age (continuous), sex and number of teeth in the DESeq2 negative binomial generalized linear model framework. (B, C, D) Relative abundance of aerobes, anaerobes and facultative anaerobes, respectively, in relation to the grams of tobacco smoked daily.
Figure 3.
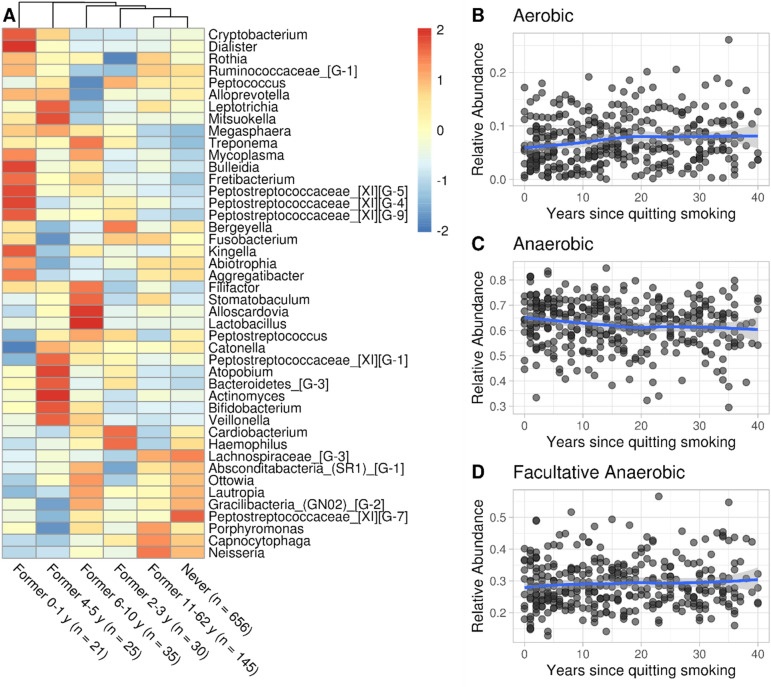
The salivary microbiota of individuals who quit smoking (n = 369) showed multiple-year perturbation and tends to resemble Never smokers’ profiles within 5 years. (A) Heatmap of the relationship between the years since quitting smoking and the mean relative abundance of genera previously found significantly associated with smoking (see Fig. 1). Taxa were transformed to relative abundance and scaled by row, to highlight differences in mean abundance in relation to bins of years since quitting to limit the low sample size of some categories. Complete linkage hierarchical clustering was used to cluster columns. Since Former smokers tend to be older and given the tendency of the elderly to lose teeth, we limited the visualization to people with 20 or more teeth. (B, C, D) Relative abundance of anaerobes, aerobes and facultative anaerobes in relation to years since quitting smoking.
Salivary microbiota of Former smokers who quit 5 years or longer tended to resemble Never smokers’ profiles
We studied the association between salivary genera of former smokers and the years since smoking cessation using the same model framework as the daily intensity regression (Fig. 2), with 1 year scale, finding no statistically significant association. We visualized the mean relative abundance of genera associated with smoking (Fig. 1B) in the Former smokers’ group with 20 or more natural teeth, grouping them by bins of years since quitting. We limited the visualization to individuals with 20 or more teeth to minimize the effect of tooth loss on the microbiota of Former smokers, who tended to be older than Current and Never smokers. Looking at the complete linkage hierarchical clustering, we noticed a gradual increase of similarity of Never smokers to Former smokers who quit for more years, except for the “Former 2–3 y” group. (Fig. 3A). The relative abundance of aerobes mildly increased in the first 20 years since quitting (, p value , adjusted for age, sex and number of teeth; Supplementary File 1, Tables 8, 9) (Fig. 3B).
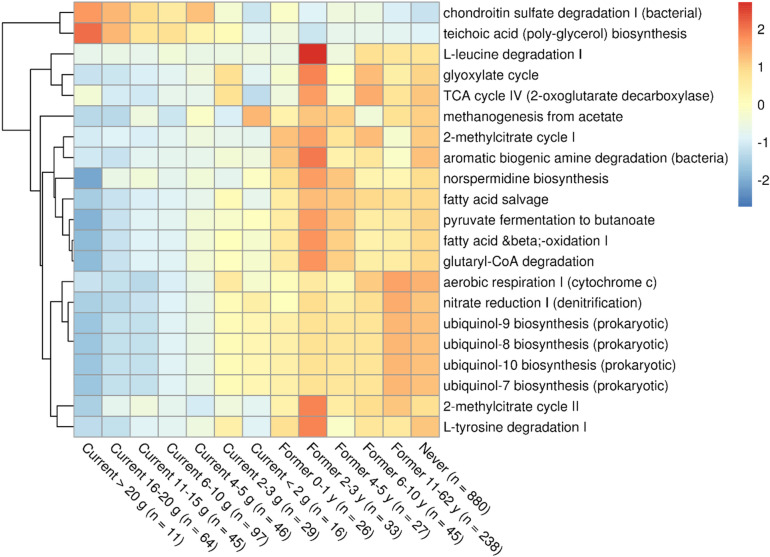
Predicted functional profiles associated with smoking highlighted a decrease of aerobic and nitrate reducing taxa
After predicting microbial pathway abundance with PICRUSt2, we identified pathways that were differentially abundant between Current and Never smokers using the same consensus method used for genus-level taxonomy. We identified 21 pathways, which we later visualized in relation to a gradient of smoking exposure, without clustering (Fig. 4). It should be noted that some of these were reconstructed from the same sets of predicted enzymes, therefore their correlation was 1 (e.g. Ubiquinol pathways). To avoid selection bias, we performed the analysis on all pathways regardless of their correlation and reported the correlation matrix of the significant ones in Supplementary File 1, Figure 7.
Figure 4.
Microbial metabolic pathways inferred with PICRUSt2 that were differentially abundant in relation to smoking exposure, adjusting for age, sex and number of teeth. Heatmap of the 21 differentially abundant pathways in Current against Never smokers contrasts. Each pathway was transformed to relative abundance and Z-score scaled. Groups were ordered based on decreasing exposure to smoking, from heavier smokers to Former smokers who quit for the most years. As a reference for absence of exposure to smoking, never smokers were included in the rightmost column. Red and blue colors indicate a higher and lower mean abundance, respectively, while yellow colors indicate no difference. Differential abundance analysis was performed with a consensus-based approach of 5 differential abundance methods (DESeq2, LinDA, MaAsLin2, ALDEx2, ANCOM-BC), modeling each pathway against smoking status and adjusting for age (categorical), sex (binary) and number of teeth (categorical). Pathways reported in the figure were differentially abundant (Benjamini–Hochberg q-value < 0.05, False Discovery Rate = 5%, ALDEx2 Holm q-value < 0.05) in at least 4 methods with an absolute effect size larger than 0.5.
Discussion
Summary of study and main results
We investigated the associations between salivary microbial genera and predicted metabolic pathways and smoking status, daily smoking intensity and years since cessation in CHRISMB, a convenience sample of 1601 adult participants in the CHRIS study in South Tyrol, Italy29. We confirmed previous findings regarding salivary microbiota compositional differences by smoking behavior. Additionally, we demonstrated that aerobic taxa varied with the frequency and intensity of smoking exposure, and that the salivary microbiota of Former smokers is generally more similar to the salivary microbiota of Never smokers, especially of those who quit longer than 5 years. Several aerobic or oxygen-requiring predicted microbial pathways decreased in smokers. The nitrate reduction pathway was significantly lower in smokers than in non-smokers. The decreases in nitrate reduction pathways among current smokers and increases in these pathways among former smokers is consistent with previous reports of decreases in cardiovascular events among former smokers32. This suggests that oral microbiota functional changes with smoking may be an additional explanation for changes in cardiovascular risk with changes in smoking habits.
Comparison with other studies
The relative abundance of salivary microbiota phyla of CHRISMB participants was comparable with the mean composition of a Japanese27 and Middle Eastern17: Firmicutes were the most abundant, followed by Bacteroidetes, Proteobacteria, Actinobacteria and Fusobacteria. Consistent with previous analyses in Americans of Caucasian, African and Hispanic ancestry15,16, and cohorts of middle17 and eastern Asian Ancestry18,27, Italian smokers had decreased abundance of Neisseria, Lautropia, Haemophilus, Capnocytophaga, and increased abundance of Atopobium, Megasphaera and Veillonella when compared to Never smokers (Fig. 1B). This suggests that cigarette smoking has a consistent and generalizable effect on the oral microbiota. We also identified 12 novel differentially abundant genera between Current and Never smokers: Alloscardovia, Bacteroidetes Genus 3, Bulleidia, Cryptobacterium, Fretibacterium, Mitsuokella, Parvimonas, Peptostreptococcaceae XI Genus 9 and Stomatobaculum were increased, while Absconditabacteria (SR1) Genus 1, Ottowia and Peptidiphaga were decreased (Supplementary File 1, Figure 1). Further work is required to determine whether these changes are specific to this work.
Salivary microbial genera composition and proportion of aerobes were strongly impacted by smoking
Of the 44 differentially abundant genera in smokers, compared to Never smokers, genera in the phylum Proteobacteria (N = 7) were decreased and Actinobacteria (N = 6) were increased among smokers. These two phyla harbor mostly aerobic and anaerobic taxa, respectively. Indeed, the proportion of aerobes was inversely proportional to the frequency and intensity of exposure to smoking (Figs. 1, 2, 3). We also predicted functional profiles based on our compositional data, observing an increase of Gram-positive associated pathways in smokers, in particular teichoic acid biosynthesis (Fig. 4), which we confirmed looking at the relative abundances of Gram staining of bacteria across smoking status (Supplementary File 1, Figure 8). Moreover, we observed a decrease in pathways associated with aerobes, such as nitrate reduction and ubiquinol synthesis, which is pivotal in the electron transport chain33, and a decrease of pathways that require oxygen and/or produce an excess reducing power, such as fatty acid oxidation. These findings support the hypothesis that smoking induces a hypoxic environment in the oral cavity.
A decreased abundance of the nitrate reduction pathway in smokers could be an effect of the decrease of genera Neisseria, Haemophilus, Kingella, which harbor several NRB. A decrease of NRB may have a detrimental effect on enterosalivary nitrate reduction34, which is a considerable source of blood nitrites for endogenous NO synthesis. Decreases in NO, which is a vasodilator35, might hinder gingival blood flow and increase stress over time, which could lead to higher chances of gingival recession and periodontal diseases36. Indeed, chondroitin sulfate degradation was increase in heavier smokers, which may be indicative of higher stress to the gingival connective tissue and increase the risk of periodontal diseases. NO deficiency has also been suggested as a risk factor for developing cardiovascular diseases37–39. Taken together, microbiota-derived NO depletion may increase the chance of developing periodontal and cardiovascular diseases in smokers, as recently reviewed40.
Some genera are statistically associated with daily smoking intensity but not with the years since smoking cessation
In addition to examining quantitative differences by Current smoking status, we tested for differences in bacterial composition by daily intensity of tobacco exposure (g/day) (Fig. 2). Extending observations by Wu et al.15 at lower taxonomic level and higher resolution of exposure variables, genera belonging to classes Betaproteobacteria (Lautropia, Neisseria), Gammaproteobacteria (Cardiobacterium) and Flavobacteriia (Capnocytophaga) were significantly decreased at increasing grams of tobacco smoked per day. Additionally, we found negative correlation with grams of tobacco smoked per day for genera in classes Clostridia (Peptostreptococcaceae Family XI—Genus 1, Peptostreptococcus), Epsilonproteobacteria (Campylobacter), Fusobacteriia (Fusobacterium, Leptotrichia) and Negativicutes (Selenomonas). Genera Actinomyces (class Actinobacteria) and Fretibacterium class Sinergistia) were significantly increased. The subsiding of smoking-related microbial taxa was in line with the observation of full recovery of cardiovascular health risk within 5 years since quitting41. It is possible that smoking induced oral microbiota alterations may last longer than 5 years (Fig. 3B,C,D), which would align with the subsiding of periodontal disease risks in smokers within 10 years42.
Study limitations
Major limitations of this study include the cross-sectional design and the lack of a professional assessment of the number of decayed, missing and filled teeth and gum health. While we controlled for age, sex, and number of teeth as potential confounders in our models, residual confounding is still possible due to, for instance, medications usage, diet and alcohol intake. Furthermore, some subgroup strata were small, and structural non-positivity could exist. Bacterial metabolic pathways inference was based solely on salivary microbiota composition. While it is encouraging that our results regarding changes in salivary microbiota composition with smoking habits are consistent with those of previous studies conducted among very different populations, prospective studies are required to more directly address whether oral microbiota play a mediating role in the onset of smoking-related chronic diseases.
Study strengths
Our analysis also has several strengths. The smoking questionnaire was detailed, allowing for high-resolution qualitative and quantitative characterization of smoking habits. We tested for a dose-dependent relationship between smoking and perturbation to the oral microbiota, supporting a causal relationship according to the Bradford Hill criteria43. This study cohort was particularly homogenous from the perspective of ethnicity, lifestyle and microbiota data generation, which should significantly limit confounding effects.
Our sample size was the largest to date to examine associations between smoking and the oral microbiota in a European population. While the salivary microbiota is a composite of multiple oral communities, saliva samples are easy to collect, making them ideal for large epidemiological cohorts and for future diagnostics and prognostics.
Conclusions
Smoking is associated with changed in the salivary microbiota composition often in a dose-dependent fashion. The salivary microbiota of people who quit smoking longer than 5 years resembled Never smokers’ profiles. Irrespective of the phylogeny, aerobic taxa are the most sensitive to smoking exposure. Decreased microbial nitrate reduction pathway abundance in smokers may provide an additional explanation for the effect of smoking on cardiovascular and periodontal diseases risk, a hypothesis which should be tested in future studies.
Materials and methods
Study ethical approval, design, and data collection
The CHRIS study was approved by the local Ethical Committee within the South Tyrol healthcare on April 19, 2011, and registered with code 21.2011. The legal base for personal data handling and protection was the informed consent explained to and signed by each participant. The personal data protection warrant of CHRIS constantly ensures that all data are handled and protected in full compliance with the European Regulation (EU 2016/679) and Italian law (D.L.vo 196/2003).
The CHRIS study includes adults of both sexes aged 18 and older. Participants were recruited starting in 2011 with extensive outreach including advertisements, electronic and paper mail to cover most people residing in the Vinschgau/Val Venosta district (South Tyrol, Italy). On the day of visit, participants answered lifestyle, dietary, general health, and socio-economic status questionnaires29. The CHRIS Salivary microbiota (CHRISMB) project is a convenience sample of CHRIS participants recruited between January 2017 and February 2018.
Epidemiological data generation
We defined age as the difference between the examination date and the birth date, rounded to the closest integer, and categorized age into six groups as shown in Table 1. CHRISMB participants filled in an adapted version of the World Health Organization oral health survey44, from which we extracted information about the number of natural teeth in 4 ranges: 0, 1–9, 10–19 and 20 or more. We derived smoking variables from smoking questionnaires harmonized from the European Community Respiratory Health Survey III questionnaire45. We defined qualitative smoking habits—“Never”, “Former”, “Current with reduction”—Current (R), and “Current without reduction”—Current (NR)—according to Murgia et al.46. Former smokers were smokers who quit for longer than 1 month prior to the visit. Current (R) were individuals who reported being smokers at the day of examination but that reduced the daily smoking intensity at least 1 month prior to the visit. Since we did not observe differences in the microbiota composition of Current (R) and Current (NR) (Supplementary File 1, Figure 9), we decided to aggregate the two smoking groups. For completeness, included in the supplement is a description of the study population showing the separate characteristics of the Current and Former smoker groups (Supplementary File 1, Table 10). Cigarettes were the primary source of tobacco, except for 5 participants. To include all sources of tobacco as one variable of smoking intensity, we converted the number of cigarettes, cigars, and cigarillos into grams of tobacco equivalents, respectively 1, 5 and 3 g (g), and converted g/week to g/day as previously proposed46,47. We defined “smoking history” as the difference between the age of the participant to CHRIS and the reported age at which the participant quit smoking, rounded to the closest integer.
Salivary microbiota data generation
Saliva sample collection and storage
CHRISMB participants were required to drink only water and fast from the night before. Additionally, they were required not to drink, eat or smoke within 1 h prior to the visit. During the visit, they provided 2–5 mL unstimulated saliva samples into Oragene OG-500 tubes. Within a few hours after the collection, samples were vortexed, split into 0.5 mL aliquots, and promptly stored at − 80 °C.
DNA extraction and sequencing
Salivary DNA extraction and sequencing were conducted by the University of Michigan microbiome core. DNA was extracted using the Eppendorf epMotion liquid handling system and Qiagen MagAttract PowerMicrobiome Kit protocol and quantified with the PicoGreen dsDNA Assay kit (Thermo Fisher Quant-iT, cat# P7589).We amplified the V4 hypervariable region of the 16S rRNA gene by polymerase chain reaction (PCR) using a dual indexing strategy48. PCR products were visualized using E-Gel 96 with 2% SYBR Safe DNA Gel Stain (Life Technologies cat# G7208-02). PCR products were then pooled and normalized using SequalPrep Normalization Plate Kit (Life Technologies, cat# A10510-01) following the manufacturer’s protocol for sequential elution.
The final pooled library consisted of equimolar amounts of each plate, normalized to the pooled plate at the lowest concentration. Sequencing libraries were prepared according to the Illumina MiSeq guidelines, adding phiX phage genome to ease diversity and quality control. Each of the 5 libraries contained 2 negative and 2 positive controls, respectively using water from the extraction step and commercially available DNA from communities of known composition from the PCR step (Zymo Research, cat# D6306). We sequenced reads on an Illumina MiSeq machine.
Sequencing data processing
Sequencing data processing
We assessed the sequencing quality of the 69,286,448 obtained reads using “MultiQC” (v. 1.7) to visually determine read trimming length. We performed FASTQ read trimming, filtering, and taxonomic assignment with the “DADA2” package (v. 1.14)49 in R (v. 3.6.0)50. This method generates a high-resolution sequence table of amplicon sequence variants (ASVs), each differing by at least one nucleotide. We removed the first 20 and last 8 nucleotides to eliminate primer and barcode sequences and to ensure homogeneity of ASV calling across batches. After these steps, we submitted 59,331,563 reads to the LearErrorRates step, separately for each run, using 1 × 108 bases as the learning rate parameter, which helps infer technical and real sequence differences. Then, we merged paired ends, resulting in 57,122,521 reads. Removal of chimeras using the consensus method resulted in an additional loss of 1.05% and 44,136,182 total reads used for taxonomic assignment. We assigned taxonomy from kingdom to genus level using the Bayesian classifier and the expanded Human Oral microbiome Database (eHOMD), while the species level was assigned using the 100% identity addSpecies strategy. To increase the likelihood of assignment at the species level, we enriched the eHOMD database with publicly available 16S rRNA FASTA sequences from known oral species in the genera Lactobacillus, Streptococcus, and Prevotella (Supplementary File 1, Table 11). We confirmed homogeneity across batches based on positive compositional profiles (Supplementary File 2).
Microbiota data preparation for analysis
We generated a phyloseq object starting from the counts table, taxonomic table and taxonomy tree using the Bioconductor package “phyloseq” (v. 1.42.0)51 and “ape” (v. 5.7). We retained only those taxa that were present with at least 10 reads in at least 1% of samples with the function core of the “microbiome” package (v. 1.20.0)52. We aggregated ASVs at the genus level with the tax_glom function in the GitHub package “speedyseq” (“mikemc/speedyseq”), a faster version of phyloseq for microbiome data manipulation.
Samples availability for statistical analysis
Participants with missing data on smoking habits (N = 4), number of teeth (N = 44) and antibiotic usage within 3 months prior to the visit (N = 83) or who reported taking antibiotics within 3 months prior to saliva collection (N = 191) were excluded, leaving 1601 analytic samples. Additionally, we excluded 17 smokers from the “Regression of microbial genera against smoking intensity” due to missing or inconsistent grams of tobacco smoked per day and 1 participant who declared smoking 60 cigarettes per day, which was far beyond the range of the rest of the data (0.5–30). We further excluded 4 participants from the analysis “Regression of microbial genera against smoking history” due to inconsistent or missing answers.
Statistical analysis
Unless reported otherwise, we performed all statistical analyses using R (v. 4.2.2) and RStudio Server (v. 2022.07.2).
Pairwise relationship between demographics
We tested the independence of smoking habits from age groups, sex, self-reported gum health and self-reported number of natural teeth using a χ2 test of independence with Yates’s correction for low-frequency groups. We considered traits with a p-value lower than 0.05 as statistically non-independent.
Beta diversity and dimensionality reduction visualization
We estimated between-sample microbiota dissimilarity transforming genera counts to relative abundance and calculating the Bray–Curtis dissimilarity with the distance function in “phyloseq”. We obtained eigenvectors with ordinate and visualized the two vectors explaining the most variance with plot_ordination, drawing 95% confidence interval ellipses with stat_ellipse in the “ggplot2” package (v. 3.4.0). We estimated the impact of smoking habits, number of teeth, sex and age group on the beta diversity using a permutational multivariate analysis of variance (PERMANOVA)53 with adonis2 in the “vegan” package (v. 2.6-4), with 2000 permutations considering the marginal effect of all variables. We ensured even intraclass dispersion of smoking status groups (Never, Former and Current) using betadisper followed by permutest, with 2000 permutations (Supplementary File 1, Table 5).
Differential abundance analysis in relation to smoking
To study the association between each oral genus abundance and smoking, we performed differential abundance analysis comparing Current with Never smokers adjusting for age group, sex and number of teeth. We performed a consensus based differential abundance analysis, as advised by Nearing et al.54, using 5 different methods having: DESeq2 (v. 1.38.2)55, LinDA (v. 1.1)56, MaAsLin2 (v. 1.12)57, ALDEx2 (v. 1.30)58 and ANCOMBC (v. 1.6.4)59.
We defined significant differentially abundant genera if Benjamini–Hochberg (BH) corrected q-values were below 0.05 in at least 4 out of 5 methods with a false discovery rate (FDR) = 5%60. We used Holm multiple testing correction in ALDEx2 as it was the only method implemented in its generalized linear model (GLM) framework.
Regression of microbial genera against smoking intensity
To study the compositional changes of microbial genera in response to the grams of tobacco smoked per day, we modeled each genus in against the grams of tobacco per day as a continuous variable in a Negative binomial GLM (DESeq2). We binned the daily tobacco smoked into multiples of 5 g as those were the most frequent answers (Supplementary File 1, Figure 10). We considered genera as significant when BH-corrected q-values were lower than 0.05 with FDR = 5%.
Regression of microbial genera against smoking history
To study the compositional changes of microbial genera in response to smoking history, we modeled each genus in response to years since smoking cessation as a continuous variable, at 1-year interval in a Negative binomial GLM (DESeq2). We considered genera as significant when BH-corrected q-values were lower than 0.05 with FDR = 5%.
Insights into the functional potential of the salivary microbiota
We inferred the functional potential of the oral microbiota at the ASV level using picrust2_pipeline.py with default parameters implemented in PICRUSt2 (v. 2.5)61. We investigated differential abundant pathways with the same strategy used for genera differential abundance. We considered pathways as significant if the absolute effect size was above 0.5 and the q-value below 0.05 in at least 4 methods. To further confirm the impact of smoking in relation to the proportion of aerobic taxa, we mapped each genus to a table of curated annotations of three oxygen metabolism classes: aerobic, anaerobic and facultative anaerobic31. We visualized the relative abundance of aerobes, anaerobes and facultative anaerobes in each sample with respect to smoking status with pairwise Wilcoxon tests, correcting p-values with BH (FDR = 5%).
Data and analysis scripts availability
CHRIS and CHRISMB data can be requested from https://chrisportal.eurac.edu/, upon approval of the researcher’s proposal by the CHRIS data access committee. Analysis scripts are freely accessible at https://github.com/g-antonello/CHRISMB-smoking-epidemiology.
Supplementary Information
Acknowledgements
We thank the CHRIS study coordinators and the personnel of the South Tyrol Healthcare Organization (SABES) for carrying out the data and sample collection in the CHRIS visit centre in Schlanders/Silandro. Special thanks to Deborah Mascalzoni and Massimiliano Pellegrini for providing the ethical and legal framework for CHRIS. The authors thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano for covering the Open Access publication costs.
Abbreviations
- ASV
Amplicon sequence variant
- BH
Benjamini–Hochberg
- CHRIS
Cooperative Health Research in South Tyrol
- CHRISMB
CHRIS microbiome project
- eHOMD
Expanded Human Oral Microbiome Database
- FDR
False discovery rate
- g
Gram/grams
- GLM
Generalized linear model
- NO
Nitric oxide
- NRB
Nitrate-reducing bacteria
- PCR
Polymerase chain reaction
Author contributions
G.A., designed and conducted the analyses, generated all figures and tables and wrote the manuscript. E.D. processed sequencing data into analysis-ready tables. F.B. C.F., B.F., N.S. provided regular feedback. R.M. and M.G. provided insights into metadata generation and statistical analysis strategies. C.F., C.P. and P.P. were crucial members during the population study and microbiota project design. All authors read, corrected and approved the final version of the manuscript.
Funding
The CHRIS study was funded by the Department of Innovation, Research and University of the Autonomous Province of Bolzano-South Tyrol and supported by the European Regional Development Fund (FESR1157). The CHRIS biobank was assigned the “Bioresource Research Impact Factor” code BRIF6107. The CHRISMB microbiota data generation was funded by the National Institute of Dental and Craniofacial Research grant number R01 DE014899.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Betsy Foxman and Christian Fuchsberger.
Contributor Information
Giacomo Antonello, Email: giacomo.antonello@eurac.edu.
Betsy Foxman, Email: bfoxman@umich.edu.
Christian Fuchsberger, Email: christian.fuchsberger@eurac.edu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-42474-7.
References
- 1.Vineis P. Smoking and impact on health. Eur. Respir. Rev. 2008;17:182–186. doi: 10.1183/09059180.00011002. [DOI] [Google Scholar]
- 2.Müller HP. Rauchen oder parodontale Gesundheit. Das Gesundh. 2000;62:400–408. doi: 10.1055/s-2000-12585. [DOI] [PubMed] [Google Scholar]
- 3.Nociti FH, Casati MZ, Duarte PM. Current perspective of the impact of smoking on the progression and treatment of periodontitis. Periodontolory. 2000;2015(67):187–210. doi: 10.1111/prd.12063. [DOI] [PubMed] [Google Scholar]
- 4.Ziukaite L, Slot DE, Loos BG, Coucke W, Van der Weijden GA. Family history of periodontal disease and prevalence of smoking status among adult periodontitis patients: A cross-sectional study. Int. J. Dent. Hyg. 2016;15:e28–e34. doi: 10.1111/idh.12224. [DOI] [PubMed] [Google Scholar]
- 5.Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, Preston-Martin S, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48:3282–3287. doi: 10.1007/s10552-007-9026-4. [DOI] [PubMed] [Google Scholar]
- 6.Gillison ML, Zhang Q, Jordan R, Xiao W, Westra WH, Trotti A, et al. Tobacco smoking and increased risk of death and progression for patients with p16-positive and p16-negative oropharyngeal cancer. J. Clin. Oncol. 2012;30:2102–2111. doi: 10.1200/JCO.2011.38.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Camp G. Cardiovascular disease prevention. Acta Clin. Belg.: Int. J. Clin. Lab. Med. 2014;69:407–411. doi: 10.1179/2295333714Y.0000000069. [DOI] [PubMed] [Google Scholar]
- 8.Lira R, Åkerman S, Klinge B, Boström EA, Gustafsson A. Salivary microbial profiles in relation to age, periodontal, and systemic diseases. PLoS ONE. 2018;13:1–14. doi: 10.1371/journal.pone.0189374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira LC, Nascimento JCR, Rêgo JMC, Canuto KM, Crespo-Lopez ME, Alvarez-Leite JI, et al. Apolipoprotein E, periodontal disease and the risk for atherosclerosis: A review. Arch. Oral Biol. 2019;98:204–212. doi: 10.1016/j.archoralbio.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Wade WG. The oral microbiome in health and disease. Pharmacol. Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Mason MR, Preshaw PM, Nagaraja HN, Dabdoub SM, Rahman A, Kumar PS. The subgingival microbiome of clinically healthy current and never smokers. ISME J. 2015;9:268–272. doi: 10.1038/ismej.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Börnigen D, Ren B, Pickard R, Li J, Ozer E, Hartmann EM, et al. Alterations in oral bacterial communities are associated with risk factors for oral and oropharyngeal cancer. Sci. Rep. 2017;7:17686. doi: 10.1038/s41598-017-17795-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Shi G. Smoking and microbiome in oral, airway, gut and some systemic diseases. J. Transl. Med. 2019;17:225. doi: 10.1186/s12967-019-1971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietiäinen M, Liljestrand JM, Kopra E, Pussinen PJ. Mediators between oral dysbiosis and cardiovascular diseases. Eur. J. Oral Sci. 2018;126(Suppl 1):26–36. doi: 10.1111/eos.12423. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016;10:2435–2446. doi: 10.1038/ismej.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beghini F, Renson A, Zolnik CP, Geistlinger L, Usyk M, Moody TU, et al. Tobacco exposure associated with oral microbiota oxygen utilization in the New York City Health and Nutrition Examination Study. Ann. Epidemiol. 2019;34:18–25.e3. doi: 10.1016/j.annepidem.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallès Y, Inman CK, Peters BA, Ali R, Wareth LA, Abdulle A, et al. Types of tobacco consumption and the oral microbiome in the United Arab Emirates Healthy Future (UAEHFS) Pilot Study. Sci. Rep. 2018;8:11327. doi: 10.1038/s41598-018-29730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia Y-J, Liao Y, He Y-Q, Zheng M-Q, Tong X-T, Xue W-Q, et al. Association between oral microbiota and cigarette smoking in the Chinese population. Front. Cell. Infect. Microbiol. 2021;11:658203. doi: 10.3389/fcimb.2021.658203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulsen CS, Nygaard N, Constancias F, Stankevic E, Kern T, Witte DR, et al. Association of general health and lifestyle factors with the salivary microbiota—Lessons learned from the ADDITION-PRO cohort. Front. Cell. Infect. Microbiol. 2022;12:1055117. doi: 10.3389/fcimb.2022.1055117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raizada MK, Joe B, Bryan NS, Chang EB, Dewhirst FE, Borisy GG, et al. Report of the National Heart, Lung, and Blood Institute Working Group on the role of microbiota in blood pressure regulation: Current status and future directions. Hypertension. 2017;70:479–485. doi: 10.1161/HYPERTENSIONAHA.117.09699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch CD, Gladwin MT, Freeman BA, Lundberg JO, Weitzberg E, Morris A. Enterosalivary nitrate metabolism and the microbiome: Intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. Free Radic. Biol. Med. 2017;105:48–67. doi: 10.1016/j.freeradbiomed.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryan NS, Tribble G, Angelov N. Oral microbiome and nitric oxide: The missing link in the management of blood pressure. Curr. Hypertens. Rep. 2017;19:33. doi: 10.1007/s11906-017-0725-2. [DOI] [PubMed] [Google Scholar]
- 23.Goh CE, Trinh P, Colombo PC, Genkinger JM, Mathema B, Uhlemann AC, et al. Association between nitrate-reducing oral bacteria and cardiometabolic outcomes: Results from ORIGINS. J. Am. Heart Assoc. 2019;8:e013324. doi: 10.1161/JAHA.119.013324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosier BT, Moya-Gonzalvez EM, Corell-Escuin P, Mira A. Isolation and characterization of nitrate-reducing bacteria as potential probiotics for oral and systemic health. Front. Microbiol. 2020;11:555465. doi: 10.3389/fmicb.2020.555465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burleigh MC, Liddle L, Monaghan C, Muggeridge DJ, Sculthorpe N, Butcher JP, et al. Salivary nitrite production is elevated in individuals with a higher abundance of oral nitrate-reducing bacteria. Free Radic. Biol. Med. 2018;120:80–88. doi: 10.1016/j.freeradbiomed.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 26.Knight TM, Forman D, Al-Dabbagh SA, Doll R. Estimation of dietary intake of nitrate and nitrite in Great Britain. Food Chem. Toxicol. 1987;25:277–285. doi: 10.1016/0278-6915(87)90123-2. [DOI] [PubMed] [Google Scholar]
- 27.Sato N, Kakuta M, Uchino E, Hasegawa T, Kojima R, Kobayashi W, et al. The relationship between cigarette smoking and the tongue microbiome in an East Asian population. J. Oral Microbiol. 2020;12:1742527. doi: 10.1080/20002297.2020.1742527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeshita T, Kageyama S, Furuta M, Tsuboi H, Takeuchi K, Shibata Y, et al. Bacterial diversity in saliva and oral health-related conditions: The Hisayama Study. Sci. Rep. 2016;6:22164. doi: 10.1038/srep22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattaro C, Goegele M, Mascalzoni D, Melotti R, Schwienbacher C, De Grandi A, et al. The Cooperative Health Research in South Tyrol (CHRIS) study: Rationale, objectives, and preliminary results. J. Transl. Med. 2015;13:348. doi: 10.1186/s12967-015-0704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyde ER, Andrade F, Vaksman Z, Parthasarathy K, Jiang H, Parthasarathy DK, et al. Metagenomic analysis of nitrate-reducing bacteria in the oral cavity: Implications for nitric oxide homeostasis. PLoS ONE. 2014;9:e88645. doi: 10.1371/journal.pone.0088645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calgaro M, Romualdi C, Waldron L, Risso D, Vitulo N. Assessment of statistical methods from single cell, bulk RNA-seq, and metagenomics applied to microbiome data. Genome Biol. 2020;21:191. doi: 10.1186/s13059-020-02104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mons U, Müezzinler A, Gellert C, Schöttker B, Abnet CC, Bobak M, et al. Impact of smoking and smoking cessation on cardiovascular events and mortality among older adults: Meta-analysis of individual participant data from prospective cohort studies of the CHANCES consortium. BMJ. 2015;350:h1551. doi: 10.1136/bmj.h1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernster L, Forsmark-Andrée P. Ubiquinol: An endogenous antioxidant in aerobic organisms. Clin. Investig. 1993;71:S60–S65. doi: 10.1007/BF00226842. [DOI] [PubMed] [Google Scholar]
- 34.Tribble GD, Angelov N, Weltman R, Wang B-Y, Eswaran SV, Gay IC, et al. Frequency of tongue cleaning impacts the human tongue microbiome composition and enterosalivary circulation of nitrate. Front. Cell. Infect. Microbiol. 2019;9:434270. doi: 10.3389/fcimb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pignatelli P, Fabietti G, Ricci A, Piattelli A, Curia MC. How periodontal disease and presence of nitric oxide reducing oral bacteria can affect blood pressure. Int. J. Mol. Sci. 2020;21:1–14. doi: 10.3390/ijms21207538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preshaw PM. Mouthwash use and risk of diabetes. Br. Dent. J. 2018;225:923–926. doi: 10.1038/sj.bdj.2018.1020. [DOI] [PubMed] [Google Scholar]
- 37.Naseem KM. The role of nitric oxide in cardiovascular diseases. Mol. Aspects Med. 2005;26:33–65. doi: 10.1016/j.mam.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Taddei S, Virdis A, Ghiadoni L, Salvetti G, Bernini G, Magagna A, et al. Age-related reduction of NO availability and oxidative stress in humans. Hypertension. 2001;38:274–279. doi: 10.1161/01.hyp.38.2.274. [DOI] [PubMed] [Google Scholar]
- 39.Ashley CT, Mayank A, Nathan SB. Nitric oxide and geriatrics: Implications in diagnostics and treatment of the elderly. J. Geriatr. Cardiol. 2012;8:230–242. doi: 10.3724/sp.j.1263.2011.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maki KA, Ganesan SM, Meeks B, Farmer N, Kazmi N, Barb JJ, et al. The role of the oral microbiome in smoking-related cardiovascular risk: A review of the literature exploring mechanisms and pathways. J. Transl. Med. 2022;20:584. doi: 10.1186/s12967-022-03785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallucci G, Tartarone A, Lerose R, Lalinga AV, Capobianco AM. Cardiovascular risk of smoking and benefits of smoking cessation. J. Thorac. Dis. 2020;12:3866–3876. doi: 10.21037/jtd.2020.02.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warnakulasuriya S, Dietrich T, Bornstein MM, Casals Peidró E, Preshaw PM, Walter C, et al. Oral health risks of tobacco use and effects of cessation. Int. Dent. J. 2010;60:7–30. doi: 10.1922/IDJ_2532Warnakulasuriya24. [DOI] [PubMed] [Google Scholar]
- 43.Hill AB. The environment and disease: association or causation? 1965. J. R. Soc. Med. 2015;108:32–37. doi: 10.1177/0141076814562718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.World Health Organization. Oral Health Surveys: Basic Methods. World Health Organization. https://play.google.com/store/books/details?id=8rEXDAAAQBAJ (2013).
- 45.Janson C, Anto J, Burney P, Chinn S, de Marco R, Heinrich J, et al. The European Community Respiratory Health Survey: What are the main results so far? European Community Respiratory Health Survey II. Eur. Respir. J. 2001;18:598–611. doi: 10.1183/09031936.01.00205801. [DOI] [PubMed] [Google Scholar]
- 46.Murgia F, Melotti R, Foco L, Gögele M, Meraviglia V, Motta B, et al. Effects of smoking status, history and intensity on heart rate variability in the general population: The CHRIS study. PLoS ONE. 2019;14:e0215053. doi: 10.1371/journal.pone.0215053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chinn S, Jarvis D, Melotti R, Luczynska C, Ackermann-Liebrich U, Antó JM, et al. Smoking cessation, lung function, and weight gain: A follow-up study. Lancet. 2005;365:1629–1635. doi: 10.1016/S0140-6736(05)66511-7. [DOI] [PubMed] [Google Scholar]
- 48.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.r-project.org/ (2022).
- 51.McMurdie PJ, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lahti L. SS. microbiome R package. https://bioconductor.org/packages/release/bioc/html/microbiome.html (2012).
- 53.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001;26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 54.Nearing JT, Douglas GM, Hayes MG, MacDonald J, Desai DK, Allward N, et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 2022;13:342. doi: 10.1038/s41467-022-28034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou H, He K, Chen J, Zhang X. LinDA: Linear models for differential abundance analysis of microbiome compositional data. Genome Biol. 2022;23:95. doi: 10.1186/s13059-022-02655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021;17:e1009442. doi: 10.1371/journal.pcbi.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandes AD, Reid JNS, Macklaim JM, McMurrough TA, Edgell DR, Gloor GB. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:1–13. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020;11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 61.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.