Abstract
Essential oils contain a variety of volatile metabolites, and are expected to be utilized in wide fields such as antimicrobials, insect repellents and herbicides. However, it is difficult to foresee the effect of oil combinations because hundreds of compounds can be involved in synergistic and antagonistic interactions. In this research, it was developed and evaluated a machine learning method to classify types of (synergistic/antagonistic/no) antibacterial interaction between essential oils. Graph embedding was employed to capture structural features of the interaction network from literature data, and was found to improve in silico predicting performances to classify synergistic interactions. Furthermore, in vitro antibacterial assay against a standard strain of Staphylococcus aureus revealed that four essential oil pairs (Origanum compactum—Trachyspermum ammi, Cymbopogon citratus—Thujopsis dolabrata, Cinnamomum verum—Cymbopogon citratus and Trachyspermum ammi—Zingiber officinale) exhibited synergistic interaction as predicted. These results indicate that graph embedding approach can efficiently find synergistic interactions between antibacterial essential oils.
Subject terms: Cheminformatics, Drug discovery and development, Oils, Natural variation in plants, Secondary metabolism, Antibiotics, Virtual drug screening
Introduction
Plants produce and emit diverse volatile organic compounds (VOCs). Humans have found value in the VOCs, and extracted them as essential oils (EOs) by distillation or expression. EOs have been extracted from approximately 3000 plants, and widely used for pharmaceutical, agronomic, food, sanitary, cosmetic and perfume industries1. In the last decades, VOCs were elucidated to be involved in protection against pathogens, defense against herbivores, attraction of pollinators and plant–plant signaling2. However, it is still uncertain how diverse VOCs cooperatively fulfill their functions under each physiological condition.
Although a large number of EOs and VOCs have been reported to show pharmacological activities3,4, development of bioactive products from them is still a challenging task. Many studies have shown that combined EOs exhibit stronger/weaker effects (hereinafter referred to as “EO–EO interaction”) than expected5,6. Unfortunately, the causal relationship of the EO–EO interaction is not clear because tens to hundreds of VOCs can be involved in the interaction. Thus, EO products occasionally fail to show the expected activity even though they are generally used in combination.
Advances in machine learning have made significant progress in predicting biologically important pairs such as protein–protein interaction7, drug–target interaction8 and drug–drug interaction9 in the last decades. Traditional approaches represent interaction pair as a numerical vector by operating corresponding (molecular or protein) descriptors, and consider the prediction task as a binary classification problem of the presence/absence of interaction. These classification-based approaches have shown good results for many applications including our previous study on drug–target interaction10. However, these approaches are unable to capture complex interactions if the descriptors do not depict characteristics of the interactions. Recently, graph embedding approaches have gained attraction in biomedical fields in order to capture structural features of the interaction network11. A systematic comparison on drug–drug interaction showed the graph embedding methods achieved competitive performance without using biological features12.
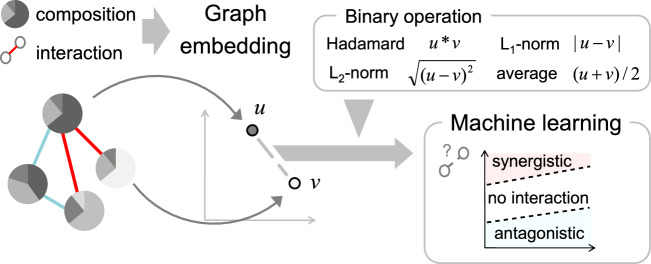
In the present paper, it was developed a machine learning method to predict EO–EO interactions using the graph embedding. The interactions were represented as a network structure with EOs and VOCs as nodes, and their synergistic/antagonistic interactions as edges (Fig. 1). The network structure and oil composition data was integrated to the graph embedding algorithm to encode the nodes as numerical vectors. The edge features were constructed from pairs of the learned node representations with either of binary operators, and were inputted to a machine learning algorithm to classify synergistic/antagonistic/no-interaction pairs. The in silico classification performance was evaluated by cross-validation, a statistical method of evaluating learning algorithms. Furthermore, in vitro antibacterial assay was performed for EO pairs predicted as synergistic by the machine learning model.
Figure 1.
Overview of the graph embedding method to predict interaction between essential oils.
Results
Graph embedding and machine learning of EO–EO interaction
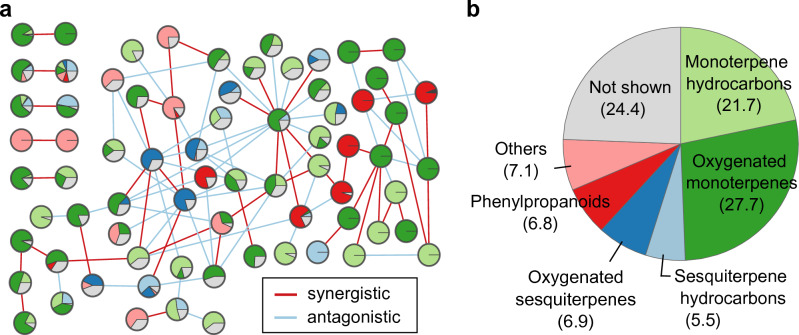
The three-class classifier was successfully constructed using graph embedding from antibacterial interaction data composed of 46 synergistic, 53 antagonistic and 172 no interactions between EOs (Supplementary Tables S1 and S2 online). The network structure and chemical composition of EOs were visualized in Fig. 2 for better understanding on the interaction data.
Figure 2.
(a) Network structure of antibacterial interaction data on Staphylococcus aureus. Each edge is colored by synergistic (red) or antagonistic (light blue) interaction. Each node has a pie chart with the chemical composition divided into chemical categories shown in (b) for better visualization. (b) Mean composition of essential oils in the interaction data. Values in parentheses indicate the mean percentage composition.
Output probability for synergistic-versus-rest and antagonistic-versus-rest classifications were evaluated by ten-fold cross-validation with receiver operating characteristic (ROC) curve to visualize the relative trade-offs between the true positive rate and false positive rate. Among four (Hadamard, L1-norm, L2-norm and average) binary operators, average operator showed the best area under the ROC curve (AUC) for both classifications (Supplementary Table S3 online). Furthermore, for the synergistic-versus-rest classification, the operator also showed the best partial AUCs (AUC0.5 = 0.211 and AUC0.2 = 0.048). Therefore, the average operator was selected for further validation to find unknown synergistic EO–EO interactions. The graph embedding method performed significantly better in AUC (0.615 vs 0.556, p = 1.1 × 10−3), AUC0.5 (0.211 vs 0.164, p = 1.7 × 10−4) and AUC0.2 (0.048 vs 0.033, p = 3.8 × 10−5) for the synergistic classification than those performed without graph embedding (Table 1). However, no significant differences (p > 0.01) were observed for the antagonistic-versus-rest classification.
Table 1.
AUC and partial AUCs obtained by ten-fold cross-validation.
| Classification | Metric | Method | |
|---|---|---|---|
| Graph embedding | Classification-based | ||
| Synergistic-versus-rest | AUC | 0.615 ± 0.020 | 0.556 ± 0.040 |
| AUC0.5 | 0.211 ± 0.016 | 0.164 ± 0.023 | |
| AUC0.2 | 0.048 ± 0.004 | 0.033 ± 0.006 | |
| Antagonistic-versus-rest | AUC | 0.576 ± 0.014 | 0.550 ± 0.033 |
| AUC0.5 | 0.150 ± 0.010 | 0.159 ± 0.017 | |
| AUC0.2 | 0.020 ± 0.004 | 0.024 ± 0.003 | |
Values are means ± SD of 10 iterations, and the significantly better results are highlighted in bold (paired t-test, p < 0.01).
AUC areas under the receiver operating characteristic (ROC) curve.
Prediction of synergistic interaction between available EOs
The probability of synergistic/antagonistic interaction between all possible pairs of the commercially available 84 EOs (Supplementary Table S4 online) were calculated using the classifier constructed above. The classifier predicted 2,088 EO pairs as synergistic when Youden index (= 0.351) was used as the threshold probability. Sixteen EO pairs (Table 2) were randomly selected from them for following gas chromatography/mass spectrometry (GC/MS) analysis and in vitro antibacterial assay.
Table 2.
Observed antibacterial interaction between essential oil pairs predicted as synergistic.
| Essential oilA | Essential oilB | Probability | MICA | MICB | MICmix | FICI | |
|---|---|---|---|---|---|---|---|
| Synergistic | Antagonistic | (mg/mL) | (mg/mL) | (mg/mL) | |||
| C. citratus | O. compactum | 0.669 | 0.075 | 0.83 | 0.5 | 0.5 | 0.80 (N) |
| O. compactum | M. balsamum | 0.629 | 0.161 | 0.5 | > 4 | 1 | 1.0–1.1 (N) |
| C. citratus | M. balsamum | 0.607 | 0.123 | 0.83 | > 4 | 2 | 1.2–1.5 (N) |
| O. compactum | T. ammi | 0.585 | 0.190 | 0.5 | 0.5 | 0.25 | 0.50 (S) |
| T. vulgare | O. compactum | 0.584 | 0.182 | 0.5 | 0.5 | 0.5 | 1.0 (N) |
| C. citratus | T. ammi | 0.565 | 0.146 | 0.83 | 0.5 | 0.4 | 0.64 (N) |
| T. vulgare | C. citratus | 0.562 | 0.140 | 0.5 | 0.83 | 0.5 | 0.80 (N) |
| T. ammi | L. petersonii | 0.541 | 0.159 | 0.5 | 1 | 0.5 | 0.75 (N) |
| T. vulgare | L. petersonii | 0.538 | 0.152 | 0.5 | 1 | 1 | 1.5 (N) |
| C. citratus | T. dolabrata | 0.537 | 0.111 | 0.83 | 0.5 | 0.125 | 0.20 (S) |
| C. grevei | M. balsamum | 0.535 | 0.190 | 1 | > 4 | > 4 | > 2.5 (A/N) |
| C. verum | O. compactum | 0.504 | 0.280 | 0.5 | 0.5 | 0.5 | 1.0 (N) |
| C. verum | C. citratus | 0.497 | 0.218 | 0.5 | 0.83 | 0.25 | 0.40 (S) |
| L. petersonii | Z. officinale | 0.487 | 0.132 | 1 | > 4 | 2 | 1.0–1.3 (N) |
| T. ammi | Z. officinale | 0.415 | 0.285 | 0.5 | > 4 | 0.25 | 0.25–0.28 (S) |
| T. vulgare | Z. officinale | 0.415 | 0.274 | 0.5 | > 4 | 0.5 | 0.50–0.56 (N) |
The FICI was interpreted as S: synergistic (FICI ≤ 0.5); N: no interaction (0.5 < FICI < 4); A: antagonistic (FICI ≥ 4). Observed synergistic interactions are highlighted in bold.
MICmix denotes a summation of two oil concentratations in a mixture. C. citratus: Cymbopogon citratus, O. compactum: Origanum compactum, M. balsamum: Myroxylon balsamum var. pereirae, T. ammi: Trachyspermum ammi, T. vulgaris: Thymus vulgaris ct. thymol, L. petersonii: Leptospermum petersonii, T. dolabrata: Thujopsis dolabrata, C. grevei: Cedrelopsis grevei, C. verum: Cinnamomum verum, Z. officinale: Zingiber officinale.
Gas chromatography/mass spectrometry analysis of selected EOs
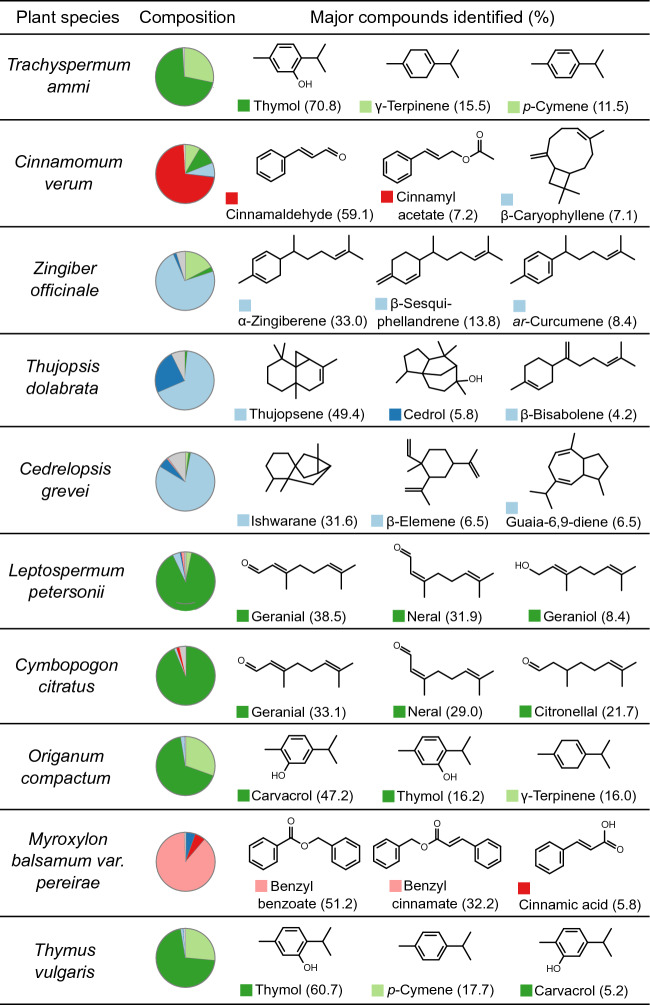
In order to obtain more comprehensive composition data for the selected EO pairs, EOs from Trachyspermum ammi, Cinnamomum verum, Zingiber officinale, Thujopsis dolabrata, Cedrelopsis grevei, Leptospermum petersonii, Cymbopogon citratus, Origanum compactum, Myroxylon balsamum var. pereirae and Thymus vulgaris ct. thymol were analyzed by GC/MS. The most dominant constituents of the EOs were thymol (70.8%), cinnamaldehyde (59.1%), α-zingiberene (33.0%), thujopsene (49.4%), ishwarane (31.6%), geranial (38.5%), geranial (33.1%), carvacrol (47.2%), benzyl benzoate (51.2%) and thymol (60.7%), respectively (Fig. 3). Furthermore, 7, 19, 38, 43, 41, 33, 20, 25, 10 and 21 VOCs from the EOs were respectively characterized by the GC/MS analysis (Supplementary Table S5 online). The classification results of the 16 EO pairs were reproduced by inputting the chemical composition obtained by GC/MS analysis instead of those from EO suppliers.
Figure 3.
Chemical composition of the selected essential oils. Values in parentheses are the percentage of the total peak area obtained from the total ion current (TIC) chromatogram. Pie charts represent the chemical composition divided into chemical categories shown in Fig. 2b.
In vitro antibacterial assay
Broth microdilution was performed to determine the types of antibacterial interaction between the predicted EO pairs. The EOs and 1:1 mixtures of the EO pairs showed minimum inhibitory concentration (MIC) range of 0.5 to > 4 mg/mL and 0.125 to > 4 mg/mL, respectively (Table 2). MIC for thymol (positive control) was 0.25 mg/mL, which was equivalent to literature data (0.03 v/v %13). No inhibition of bacterial growth was observed in the negative control.
Four EO pairs (O. compactum—T. ammi, C. citratus—T. dolabrata, C. verum—C. citratus and T. ammi—Z. officinale) exhibited fractional inhibitory concentration indices (FICIs) less than or equal to 0.5, namely, synergistic interaction. In particular, the C. citratus—T. dolabrata combination showed the lowest MIC (0.125 mg/mL) which was lower than that of thymol, and its FICI reached 0.20. Meanwhile, the other 12 pairs showed antagonistic or no interactions.
Discussion
Artificial intelligence has been applied to classify bioactive EOs using chemical composition data, and has shown good predicting performance14,15. However, as far as we know, its application to EO–EO interaction is not yet reported. The difficulty of EO–EO interaction prediction lies in the huge number of chemical constituent pairs to be analyzed compared with the sample number of known EO–EO interactions. In this study, we confronted this problem with graph embedding to compensate the shortage by adding network structure data of the interaction. This strategy worked well for synergistic-versus-rest classification in the cross-validation. The possible reason is that there exists antibacterial contribution of trace constituents absent in the composition data. In fact, several blends of major constituents were known to show much weaker antibacterial activity than original EOs16. On the other hand, the graph embedding approach did not show better performance for antagonistic-versus-rest classification in this research. This result suggests that the major components may play key roles in antagonistic actions although the mode of actions is not well known16.
The precision obtained by antibacterial assay (4 / 16 = 25%) was apparently low, but the frequency of synergistic interaction should be taken into consideration. It is generally difficult to infer the frequency of EO–EO interactions from the literature data because EO pairs with no interactions tend to be considered as negative results, and to be not reported. An indicative study was performed by Orchard et al. testing 247 EO combinations against three reference strains of Staphylococcus aureus (ATCC 25923) and methicillin-resistant Staphylococcus aureus (ATCC 43300 and ATCC 33592), which resulted in observation of 6, 9 and 14 synergistic interactions, respectively17. Assuming that synergism is observed at the same level, our method is expected to detect more synergistic pairs (4 / 16) than random sampling (6 to 14/247).
Predicting interaction against out-of-sample (not learned) EOs is a critical issue because our training data covers just 54 EOs, namely, most of the available EOs lack the interaction data. Furthermore, for each plant species, chemical composition varies under environmental conditions such as temperature, carbon dioxide, lighting and soil fertility18. In this study, the graph embedding method successfully detected synergistic interactions for the out-of-sample EOs (T. ammi, T. dolabrata and Z. officinale) and for EOs from different sources (C. verum, O. compactum and C. citratus). This result indicates that the proposed approach is applicable to a wide variety of EOs.
The molecular mechanism of action provides insights to understand the synergistic and antagonistic interactions. Previous studies on EOs pointed out the involvement of hydrophobicity which is responsible for the disruption of bacterial cell membrane16,19. For example, carvacrol and p-cymene are considered to act synergistically by expanding cell membrane, which results in the destabilization of the membrane20. This mechanism may contribute to the synergistic interaction we have found between O. compactum (composed of 47.2% carvacrol) and T. ammi (composed of 11.5% p-cymene). However, the other three interactions (C. citratus—T. dolabrata, C. verum—C. citratus and T. ammi—Z. officinale) are not explained by known interactions between the major constituents. Enrichment of the mechanism information of VOCs will not only provide interpretation of the assay results but also improve the predicting performance of graph embedding approach by incorporating the network structure of VOC–target interactions into the embedding.
Finally, the graph embedding approaches have potential limitations. The first is that the embedding is generally performed in a black-box fashion, which makes difficult to understand which VOCs contribute to the interaction. Feature extraction with wrapper method (e.g. recursive feature elimination) may resolve the issue. The second limitation concerns triple or more combination. The method described in this research is based on binary combination for model simplification. Further assay data and statistical theories focused on multiple combination are needed.
Our study suggests that graph embedding approach can efficiently find synergistic interactions between antibacterial EOs. Application of machine learning for other bioactive EO–EO interaction will be evaluated in future research.
Methods
Data
Literature search on antibacterial interaction among EOs and VOCs was performed using PubMed21 and Google scholar (https://scholar.google.com) in April 2021. The keywords “synergy”, “synergistic”, “antagonistic”, “antimicrobial” and “antibacterial” were used for the search. The tested organisms were restricted to Staphylococcus aureus, the most targeted bacteria for exploring antibacterial activity of plant extracts22. Cytoscape23 (ver. 3.9.1) was used to visualize the EO–EO interaction data.
Chemical composition data of commercially available 84 EOs were retrieved from homepages of product suppliers in Japan. We excluded EOs rich in monoterpene hydrocarbons because their antibacterial effects seemed to be much weaker than other constituents24.
Reagents
Essential oils from Trachyspermum ammi, Cinnamomum verum, Cedrelopsis grevei, Cymbopogon citratus, Origanum compactum, Myroxylon balsamum var. pereirae and Thymus vulgaris ct. thymol were purchased from Kenso Igakusha Co., Ltd. Essential oils from Zingiber officinale, Thujopsis dolabrata and Leptospermum petersonii were purchased from TREE OF LIFE Co., Ltd. Acetone for gas chromatography was purchased from KISHIDA CHEMICAL Co., Ltd, Japan. Dimethyl sulfoxide (DMSO) and thymol (special grade, purity 100.0%) were purchased from FUJIFILM Wako Pure Chemical Corporation, Japan. A series of n-alkane standards (C9 to C40) was purchased from GL Sciences Inc., Tokyo, Japan. Mueller–Hinton II broth was purchased from Becton, Dickinson and Company, USA. Staphylococcus aureus (NBRC 12,732) for antibacterial activity tests were from the National Institute of Technology and Evaluation, Biological Resource Center (NBRC), Japan.
Graph embedding
The network structure and oil composition data were inputted to attri2vec25, a graph embedding algorithm to encode the nodes as numerical vectors. The number and the size of hidden layer were set to 1 and 16, respectively. Walk length was set to 3, number of walk was set to 3, batch size was set to 32, epochs was set to 50 and learning rate of Adam optimizer was set to 0.01. Binary cross-entropy was chosen as loss function. StellarGraph library (https://github.com/stellargraph/stellargraph) was used for the attri2vec implementation. The edge features were constructed from pairs of the learned node representations with four binary operators (Hadamard, L1-norm, L2-norm and average)26. For comparison with a classification-based method, the oil composition data without graph embedding was used to construct the edge features.
Machine learning of EO–EO interaction
The edge features constructed above were inputted to multinomial logistic regression with L-BFGS method27 to classify the three types (synergistic/antagonistic/no-interaction) of interactions. Output probability for synergistic and antagonistic classes were evaluated by receiver operating characteristic (ROC) curve28, respectively. We repeated ten-fold cross-validation 10 times, and used a paired two-tailed t-test to determine whether there is any difference in area under the ROC curve (AUC) between the two methods. The partial AUCs were calculated using ‘pROC’ (ver. 1.18.0) R package.
Prediction of synergistic interaction between available EOs
The probability of synergistic/antagonistic interaction between all possible pairs of the commercially available 84 EOs were calculated using chemical composition data provided by suppliers and the classifier constructed above. Youden index29 obtained by the cross-validation was used to set cut-off probability. Sixteen EO pairs were selected for following evaluation. The EOs corresponding to the selected pairs were purchased from the suppliers.
Gas chromatography/mass spectrometry (GC/MS) analysis
Chemical characterization was performed in the same manner as reported by the authors30 using gas chromatograph coupled with mass spectrometer model QP2010 (Shimadzu, Kyoto, Japan). Essential oils were dissolved in acetone (2 μL/mL). This solution (1 μL) was injected in split mode (1:50 ratio) onto a DB-5MS column (30 m × 0.25 mm i.d. × 0.25 μm film thickness, Agilent, USA). The injection temperature was set at 270 °C. The oven temperature was started at 60 °C for 1 min after injection and then increased at 10 °C/min to 180 °C for 1 min, increased at 20 °C/min to 280 °C for 3 min followed by an increase at 20 °C/min to 325 °C, where the column was held for 20 min. Mass spectra were obtained in the range of 20 to 550 m/z. Essential oil components were identified based on a search (National Institute of Standards and Technology, NIST 14), the calculation of retention indices relative to homologous series of n-alkane, and a comparison of their mass spectra libraries with data from the mass spectra in the literature31,32.
In vitro antibacterial assay
The essential oil alone and the 1:1 combinations were tested using the broth microdilution assay in the same manner reported by the authors30. A stock solution of each essential oil (dissolved to a concentration of 40 mg/mL in DMSO) was diluted to 4 mg/mL by Mueller–Hinton II broth medium, followed by serial dilution by the medium to lower concentrations (2, 1, 0.5, 0.25, 0.125, 0.0625, 0.0313, 0.0156 and 0.0078 mg/mL). Thymol, a known antibacterial agent, was dissolved and diluted in the same way to ensure microbial susceptibility (positive control). The oils were all tested in triplicate. Staphylococcus aureus NBRC 12,732 was inoculated onto normal agar plates, and cultured for 24 h at 35 ± 1 °C. The bacterial suspensions were diluted by saline to obtain 0.5 McFarland turbidity equivalent (ca. 108 colony forming units per mL (CFU/mL)), and were further diluted 10 times (ca. 107 CFU/mL). 0.1 mL of essential oil-containing medium and 5 μL inoculum were added to sterile micro-titre plates. 10% (v/v) DMSO in the medium was used to determine if the solvent exhibited any antibacterial effect (negative control). The micro-titre plates were incubated for 18 to 24 h at 35 ± 1 °C. Based on the opacity and color change in each well, the lowest concentration capable of inhibiting the growth was determined as minimum inhibitory concentration (MIC).
The type of interaction was determined using fractional inhibitory concentration (FIC), a widely accepted means of measuring the interactions33, followed by calculating FIC index (FICI) through the equations below:
where
and
The FICI values were interpreted as follows:
Supplementary Information
Acknowledgements
This research was supported financially by the Kayamori Foundation of Informational Science Advancement (K32 ken XXV 577).
Author contributions
H.Y. conceived the idea of the study. H.Y., A.S., M.N., Y.N. and S.T. developed the machine learning method, and conducted statistical analyses. K.H., M.F., T.O. and M.M. validated the proposed method, and contributed to the interpretation of the results. H.Y. drafted the original manuscript. K.M. supervised the conduct of this study. All authors reviewed the manuscript draft, revised it critically on intellectual content, and approved the final version of the manuscript to be published.
Data availability
Python scripts are available at https://github.com/yabuuchi-hiroaki/graph-embedding-eo-eo-interaction. All other relevant data are within the paper and its Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-46377-5.
References
- 1.Bakkali F, Averbeck S, Averbeck D, Idaomar M. Biological effects of essential oils–a review. Food Chem. Toxicol. 2008;46:446–475. doi: 10.1016/j.fct.2007.09.106. [DOI] [PubMed] [Google Scholar]
- 2.Dudareva N, Klempien A, Muhlemann JK, Kaplan I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013;198:16–32. doi: 10.1111/nph.12145. [DOI] [PubMed] [Google Scholar]
- 3.Bunse M, et al. Essential oils as multicomponent mixtures and their potential for human health and well-being. Front. Pharmacol. 2022;13:956541. doi: 10.3389/fphar.2022.956541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalemba D, Kunicka A. Antibacterial and antifungal properties of essential oils. Curr. Med. Chem. 2003;10:813–829. doi: 10.2174/0929867033457719. [DOI] [PubMed] [Google Scholar]
- 5.Lesgards JF, Baldovini N, Vidal N, Pietri S. Anticancer activities of essential oils constituents and synergy with conventional therapies: A review. Phytother. Res. 2014;28:1423–1446. doi: 10.1002/ptr.5165. [DOI] [PubMed] [Google Scholar]
- 6.Bassolé IHN, Juliani HR. Essential oils in combination and their antimicrobial properties. Molecules. 2012;17:3989–4006. doi: 10.3390/molecules17043989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu X, Feng C, Ling T, Chen M. Deep learning frameworks for protein–protein interaction prediction. Comput. Struct. Biotechnol. J. 2022;20:3223–3233. doi: 10.1016/j.csbj.2022.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Ru X, Song R. Application of machine learning for drug–target interaction prediction. Front. Genet. 2021;12:680117. doi: 10.3389/fgene.2021.680117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han K, et al. A review of approaches for predicting drug–drug interactions based on machine learning. Front. Pharmacol. 2022;12:814858. doi: 10.3389/fphar.2021.814858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yabuuchi H, et al. Analysis of multiple compound–protein interactions reveals novel bioactive molecules. Mol. Syst. Biol. 2011;7:472. doi: 10.1038/msb.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson W, et al. To embed or not: Network embedding as a paradigm in computational biology. Front. Genet. 2019;10:381. doi: 10.3389/fgene.2019.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue X, et al. Graph embedding on biomedical networks: Methods, applications and evaluations. Bioinformatics. 2020;36:1241–1251. doi: 10.1093/bioinformatics/btz718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reichling J, Suschke U, Schneele J, Geiss HK. Antibacterial activity and irritation potential of selected essential oil components—Structure–activity relationship. Nat. Prod. Commun. 2006;1:1003–1012. [Google Scholar]
- 14.Daynac M, Cortes-Cabrera A, Prieto JM. Application of artificial intelligence to the prediction of the antimicrobial activity of essential oils. Evid. Based Complement. Alternat. Med. 2015;2015:561024. doi: 10.1155/2015/561024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Attar NE, Hassan MK, Alghamdi OA, Awad WA. Deep learning model for classification and bioactivity prediction of essential oil-producing plants from Egypt. Sci. Rep. 2017;10:65–78. doi: 10.1038/s41598-020-78449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyldgaard M, Mygind T, Meyer RL. Essential oils in food preservation: Mode of action, synergies, and interactions with food matrix components. Front. Microbiol. 2012;3:12. doi: 10.3389/fmicb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orchard A, Viljoen A, van Vuuren S. Wound pathogens: Investigating antimicrobial activity of commercial essential oil combinations against reference strains. Chem. Biodivers. 2018;15:e1800405. doi: 10.1002/cbdv.201800405. [DOI] [PubMed] [Google Scholar]
- 18.Pant P, Pandey S, Dall’Acqua S. The influence of environmental conditions on secondary metabolites in medicinal plants: A literature review. Chem. Biodivers. 2021;18:e2100345. doi: 10.1002/cbdv.202100345. [DOI] [PubMed] [Google Scholar]
- 19.Langeveld WT, Veldhuizen EJA, Burt SA. Synergy between essential oil components and antibiotics: A review. Crit. Rev. Microbiol. 2014;40:76–94. doi: 10.3109/1040841X.2013.763219. [DOI] [PubMed] [Google Scholar]
- 20.Ultee A, Bennik MHJ, Moezelaar R. The phenolic hydroxyl group of carvacrol is essential for action against the food-borne pathogen Bacillus cereus. Appl. Environ. Microbiol. 2002;68:1561–1568. doi: 10.1128/AEM.68.4.1561-1568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayers EW, et al. Database resources of the national center for biotechnology information in 2023. Nucleic Acids Res. 2023;51:D29–D38. doi: 10.1093/nar/gkac1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chassagne F, et al. A systematic review of plants with antibacterial activities: A taxonomic and phylogenetic perspective. Front. Pharmacol. 2021;11:586548. doi: 10.3389/fphar.2020.586548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.İşcan G. Antibacterial and anticandidal activities of common essential oil constituents. Rec. Nat. Prod. 2017;11:374–388. [Google Scholar]
- 25.Zhang D, Jie Y, Zhu X, Zhang C. Attributed network embedding via subspace discovery. Data Min. Knowl. Disc. 2019;33:1953–1980. doi: 10.1007/s10618-019-00650-2. [DOI] [Google Scholar]
- 26.Grover, A. & Leskovec, J. Node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD '16), 855–864 (2016). [DOI] [PMC free article] [PubMed]
- 27.Liu DC, Nocedal J. On the limited memory BFGS method for large scale optimization. Math. Program. 1989;45:503–528. doi: 10.1007/BF01589116. [DOI] [Google Scholar]
- 28.Fawcett T. An introduction to ROC analysis. Pattern Recog. Lett. 2006;27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 29.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Yabuuchi H, et al. Virtual screening of antimicrobial plant extracts by machine-learning classification of chemical compounds in semantic space. PLOS ONE. 2023;18:e0285716. doi: 10.1371/journal.pone.0285716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babushok VI, Linstrom PJ, Zenkevich IG. Retention indices for frequently reported compounds of plant essential oils. J. Phys. Chem. Ref. Data. 2011;40:043101. doi: 10.1063/1.3653552. [DOI] [Google Scholar]
- 32.Adams RP. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry. 3. Allured Publishing Corp.; 1995. [Google Scholar]
- 33.van Vuuren S, Viljoen A. Plant-based antimicrobial studies—Methods and approaches to study the interaction between natural products. Planta Med. 2011;77:1168–1182. doi: 10.1055/s-0030-1250736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Python scripts are available at https://github.com/yabuuchi-hiroaki/graph-embedding-eo-eo-interaction. All other relevant data are within the paper and its Supplementary Information files.