Abstract
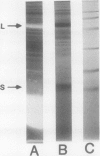
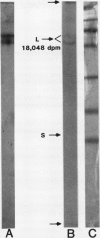
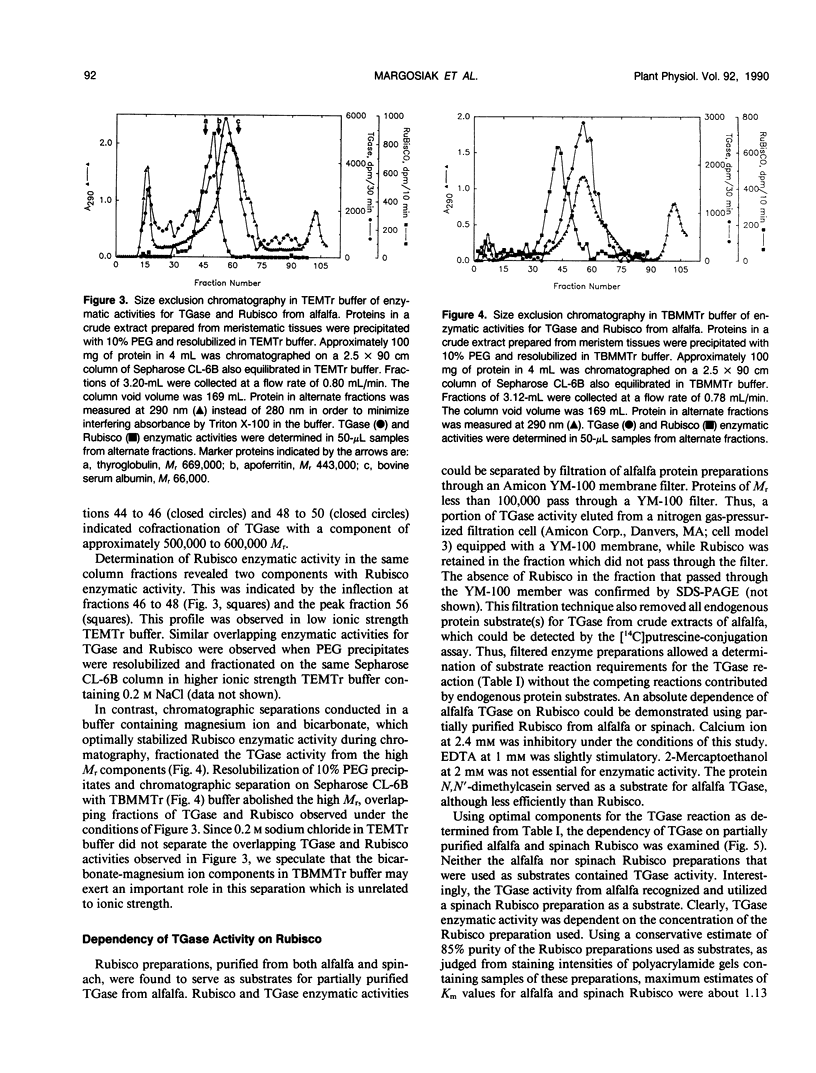
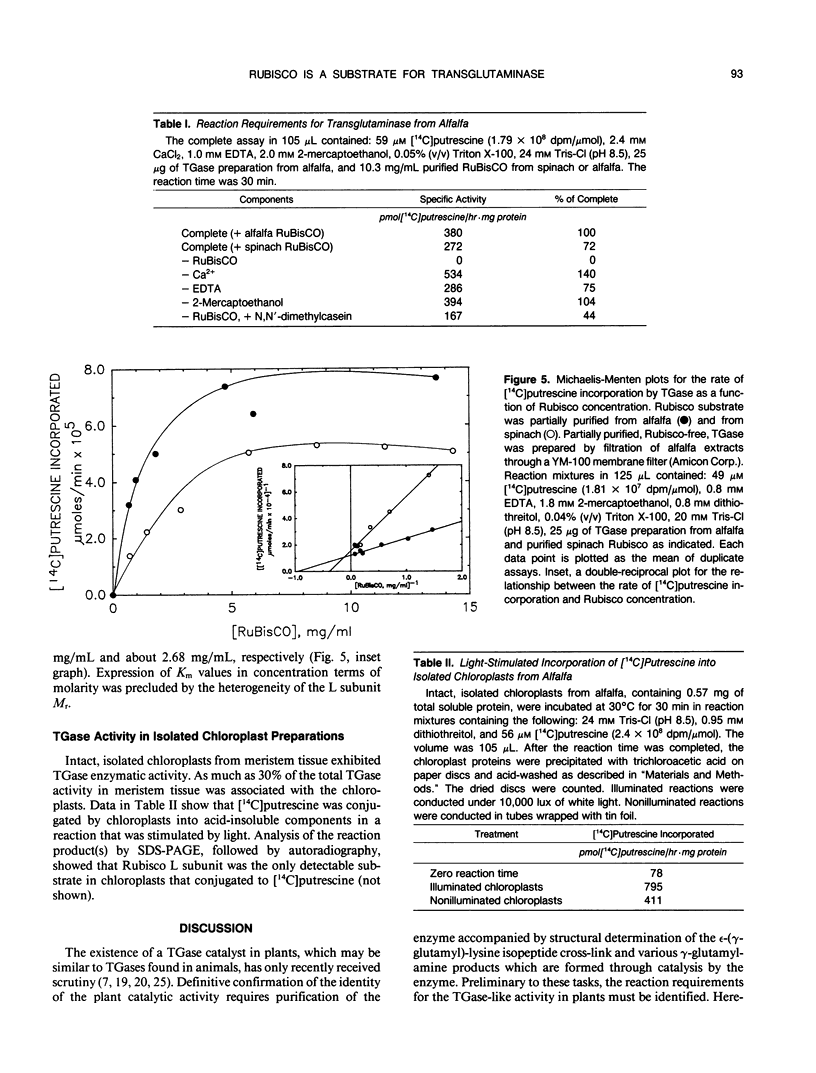
Extracts prepared from floral meristematic tissue of alfalfa (Medicago sativa L.) were investigated for expression of the enzyme transglutaminase in order to identify the major protein substrate for transglutaminase-directed modifications among plant proteins. The large polymorphic subunits of ribulose 1,5-bisphosphate carboxylase/oxygenase in alfalfa, with molecular weights of 52,700 and 57,600, are major substrates for transglutaminase in these extracts. This was established by: (a) covalent conjugation of monodansylcadaverine to the large subunit followed by fluorescent detection in SDS-polyacrylamide gels; (b) covalent conjugation of [14C]putrescine to the large subunit with detection by autoradiography; (c) covalent conjugation of monodansylcadaverine to the large subunit and demonstration of immunocross-reactivity on nitrocellulose transblot of the modified large subunit with antibody prepared in rabbits against dansylated-ovalbumin; (d) demonstration of a direct dependence of the rate of transglutaminase-mediated, [14C]putrescine incorporation upon the concentration of ribulose, 1,5-bisphosphate carboxylase/oxygenase from alfalfa or spinach; and (e) presumptive evidence from size exclusion chromatography that transglutaminase may cofractionate with native molecules of ribulose 1,5-bisphosphate carboxylase/oxygenase in crude extracts. Analysis of the primary structure of plant large subunit has revealed numerous potential glutaminyl and lysyl sites for transglutaminase-directed modifications of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chapman M. S., Suh S. W., Curmi P. M., Cascio D., Smith W. W., Eisenberg D. S. Tertiary structure of plant RuBisCO: domains and their contacts. Science. 1988 Jul 1;241(4861):71–74. doi: 10.1126/science.3133767. [DOI] [PubMed] [Google Scholar]
- Davies P. J., Davies D. R., Levitzki A., Maxfield F. R., Milhaud P., Willingham M. C., Pastan I. H. Transglutaminase is essential in receptor-mediated endocytosis of alpha 2-macroglobulin and polypeptide hormones. Nature. 1980 Jan 10;283(5743):162–167. doi: 10.1038/283162a0. [DOI] [PubMed] [Google Scholar]
- Folk J. E. Transglutaminases. Annu Rev Biochem. 1980;49:517–531. doi: 10.1146/annurev.bi.49.070180.002505. [DOI] [PubMed] [Google Scholar]
- Houtz R. L., Stults J. T., Mulligan R. M., Tolbert N. E. Post-translational modifications in the large subunit of ribulose bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1855–1859. doi: 10.1073/pnas.86.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icekson I., Apelbaum A. Evidence for transglutaminase activity in plant tissue. Plant Physiol. 1987 Aug;84(4):972–974. doi: 10.1104/pp.84.4.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juprelle-Soret M., Wattiaux-De Coninck S., Wattiaux R. Presence of a transglutaminase activity in rat liver lysosomes. Eur J Cell Biol. 1984 Jul;34(2):271–274. [PubMed] [Google Scholar]
- Kuehn G. D., McFadden B. A. Ribulose 1,5-diphosphate carboxylase from Hydrogenomonas eutropha and Hydrogenomonas facilis. I. Purification, metallic ion requirements, inhibition, and kinetic constants. Biochemistry. 1969 Jun;8(6):2394–2402. doi: 10.1021/bi00834a021. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lorand L., Campbell-Wilkes L. K., Cooperstein L. A filter paper assay for transamidating enzymes using radioactive amine substrates. Anal Biochem. 1972 Dec;50(2):623–631. doi: 10.1016/0003-2697(72)90074-7. [DOI] [PubMed] [Google Scholar]
- Lorand L., Conrad S. M. Transglutaminases. Mol Cell Biochem. 1984;58(1-2):9–35. doi: 10.1007/BF00240602. [DOI] [PubMed] [Google Scholar]
- Lorand L., Murthy S. N., Velasco P. T., Karush F. Identification of transglutaminase substrates in inside-out vesicles from human erythrocytes: immunoblotting with anti-dansyl antibody. Biochem Biophys Res Commun. 1986 Jan 29;134(2):685–689. doi: 10.1016/s0006-291x(86)80474-0. [DOI] [PubMed] [Google Scholar]
- Lorand L., Parameswaran K. N., Velasco P. T., Hsu L. K., Siefring G. E., Jr New colored and fluorescent amine substrates for activated fibrin stabilizing factor (Factor XIIIa) and for transglutaminase. Anal Biochem. 1983 Jun;131(2):419–425. doi: 10.1016/0003-2697(83)90193-8. [DOI] [PubMed] [Google Scholar]
- Lorand L., Rule N. G., Ong H. H., Furlanetto R., Jacobsen A., Downey J., Oner N., Bruner-Lorand J. Amine specificity in transpeptidation. Inhibition of fibrin cross-linking. Biochemistry. 1968 Mar;7(3):1214–1223. doi: 10.1021/bi00843a043. [DOI] [PubMed] [Google Scholar]
- Lorand L., Siefring G. E., Jr, Tong Y. S., Bruner-Lorand J., Gray A. J., Jr Dansylcadaverine specific staining for transamidating enzymes. Anal Biochem. 1979 Mar;93(2):453–458. doi: 10.1016/s0003-2697(79)80178-5. [DOI] [PubMed] [Google Scholar]
- Lorand L., Weissmann L. B., Epel D. L., Bruner-Lorand J. Role of the intrinsic transglutaminase in the Ca2+-mediated crosslinking of erythrocyte proteins. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4479–4481. doi: 10.1073/pnas.73.12.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer G. H. Ribulosebisphosphate carboxylase: amino acid sequence of a peptide bearing the activator carbon dioxide. Biochemistry. 1981 Mar 3;20(5):1236–1240. doi: 10.1021/bi00508a028. [DOI] [PubMed] [Google Scholar]
- McCurry S. D., Gee R., Tolbert N. E. Ribulose-1,5-bisphosphate carboxylase/oxygenase from spinach, tomato, or tobacco leaves. Methods Enzymol. 1982;90(Pt E):515–521. doi: 10.1016/s0076-6879(82)90178-1. [DOI] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Paonessa G., Metafora S., Tajana G., Abrescia P., De Santis A., Gentile V., Porta R. Transglutaminase-mediated modifications of the rat sperm surface in vitro. Science. 1984 Nov 16;226(4676):852–855. doi: 10.1126/science.6149619. [DOI] [PubMed] [Google Scholar]
- Peterson L. L., Wuepper K. D. Epidermal and hair follicle transglutaminases and crosslinking in skin. Mol Cell Biochem. 1984;58(1-2):99–111. doi: 10.1007/BF00240609. [DOI] [PubMed] [Google Scholar]
- Serafini-Fracassini D., Del Duca S., D'Orazi D. First evidence for polyamine conjugation mediated by an enzymic activity in plants. Plant Physiol. 1988 Jul;87(3):757–761. doi: 10.1104/pp.87.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Ashman H. G. Transglutaminases and the clotting of mammalian seminal fluids. Mol Cell Biochem. 1984;58(1-2):51–61. doi: 10.1007/BF00240604. [DOI] [PubMed] [Google Scholar]
- Yan S. B., Wold F. Neoglycoproteins: in vitro introduction of glycosyl units at glutamines in beta-casein using transglutaminase. Biochemistry. 1984 Jul 31;23(16):3759–3765. doi: 10.1021/bi00311a030. [DOI] [PubMed] [Google Scholar]