Abstract
A two-step PCR protocol was used to identify and sequence a family 11 xylanase gene from Dictyoglomus thermophilum Rt46B.1. Family 11 xylanase consensus fragments (GXCFs) were amplified from Rt46B.1 genomic DNA by using different sets of consensus PCR primers that exhibited broad specificity for conserved motifs within fungal and/or bacterial family 11 xylanase genes. On the basis of the sequences of a representative sample of the GXCFs a single family 11 xylanase gene (xynB) was identified. The entire gene sequence was obtained in the second step by using genomic walking PCR to amplify Rt46B.1 genomic DNA fragments upstream and downstream of the xynB GXCF region. The putative XynB peptide (Mr, 39,800) encoded by the Rt46B.1 xynB open reading frame was a multidomain enzyme comprising an N-terminal catalytic domain (Mr, 22,000) and a possible C-terminal substrate-binding domain (Mr, 13,000) that were separated by a short serine-glycine-rich 23-amino-acid linker peptide. Seven xylanases which differed at their N and C termini were produced from different xynB expression plasmids. All seven xylanases exhibited optimum activity at pH 6.5. However, the temperature optima of the XynB xylanases varied from 70 to 85°C. Pretreatment of Pinus radiata and eucalypt kraft-oxygen pulps with XynB resulted in moderate xylan solubilization and a substantial improvement in the bleachability of these pulps.
The xylans are the major hemicelluloses in differentiated hardwood and are also abundant in the secondary cell walls of gymnosperms and the primary cell walls of grasses. Hence, the xylans represent a considerable reservoir of fixed carbon in nature (6). Structurally, the xylans are a complex and highly variable family of polysaccharides which are based on a β-1,4-linked backbone of xylopyranosyl residues substituted with 4-O-methyl-glucuronosyl, 4-O-arabinosyl, and acetic acid side groups. The type and degree of substitution vary from source to source, and as a consequence, many cellulolytic and hemicellulolytic microorganisms produce an assortment of xylanases that are used to cleave and debranch the xylan backbone in order to make efficient use of lignocellulosic substrates. Endo-1,4-β-d-xylanases (EC 3.2.1.8) are responsible for random cleavage of the xylan backbone and hence have industrial significance because it may be possible to use them in the pulp and paper industry to manufacture bleached kraft pulp with reduced consumption of bleaching chemicals (8, 33).
The xylanases have been grouped into two unrelated families (family F or 10 and family G or 11) on the basis of primary sequence homologies (17). However, based on the functional clustering of xylanase catalytic properties and the existence of high-pI and low-pI family 11 xylanases, it is clear that the xylanases can be further segregated within these two families on the basis of additional functional and physicochemical criteria (2, 21, 36). β-1,4-Xylanases are highly modular in structure, like most other β-glycanases responsible for the metabolism of plant carbohydrate polymers, and the catalytic domains of family 10 and 11 xylanases are associated with a wide range of noncatalytic domains (14). Typically, the noncatalytic domains of the microbial multidomain xylanases that have been characterized apparently have roles in either substrate binding (25), cell wall association (20), or protein-protein interactions (16). However, other functions, including thermal stabilization and initiation of plant nodulation responses, have also been described (3, 5, 10, 23). A striking feature of many microbial xylanolytic systems is the presence of multiple family 10 and/or 11 xylanases which are produced as discrete gene products (34). Detailed comparisons of the catalytic properties of xylanases isolated from multixylanase systems have revealed distinctions in the hydrolytic activities of the individual xylanases present, including differences in the yields, rates of hydrolysis, and hydrolytic products obtained from different xylan substrates that vary in complexity. These observations suggest that xylanase multiplicity is a mechanism employed by both bacteria and fungi to enhance their xylanolytic capabilities on complex xylan substrates.
The Dictyoglomus isolate Rt46B.1 is an extremely thermophilic, strictly anaerobic bacterium which has been identified as a strain of Dictyoglomus thermophilum on the basis of morphological, biochemical, and genetic characteristics (27). Previously, a family 10 xylanase gene (xynA) was found in Rt46B.1 by using traditional gene library construction and screening procedures (13). The recombinant xylanase (XynA) produced from the cloned Rt46B.1 xynA gene exhibited optimal endoxylanase activity at 85°C and pH 6.5 and had a half-life of more than 24 h in the absence of substrate under these conditions. Native xylanases have also been purified from the culture supernatants of various thermophilic and strictly anaerobic Dictyoglomus strains and have been shown to have good activity over a broad pH range (pH 5.5 to near pH 9.0) at 80°C. Pretreatment of pine and birch kraft pulps with a Dictyoglomus sp. strain B1 xylanase preparation enhanced the efficacy of a one-stage peroxide delignification procedure and increased the final pulp brightness by 2 ISO units (22, 28).
We describe here the identification, cloning, and expression of a second xylanase gene (xynB) from D. thermophilum Rt46B.1, obtained by using a two-step PCR approach. We also describe the use of the peptide encoded by xynB (XynB) in elemental chlorine-free (ECF) and total chlorine-free (TCF) bleaching of eucalpyt kraft-oxygen pulp and ECF bleaching of Pinus radiata kraft-oxygen pulp.
MATERIALS AND METHODS
Bacterial strain.
Escherichia coli JM101 [Δ(lac-proAB) thi-1 supE44 F′ (traD36 proAB+ lacZΔM15)] was used as the bacterial host in all DNA cloning and expression studies.
PCRs.
PCRs were performed in 50-μl reaction mixtures containing 100 ng of forward primer, 100 ng of reverse primer, each deoxynucleoside triphosphate at a concentration of 0.25 mM, 1 U of Taq DNA polymerase, 2.5 mM MgCl2, and 0.1 volume of Taq DNA polymerase PCR buffer (Perkin-Elmer). Approximately 10 ng of DNA was used as the template. The PCRs were performed with a Perkin-Elmer model 2400 GeneAmp apparatus by using 30-s denaturation, annealing (55 to 60°C), and primer extension steps.
Consensus PCRs.
Family 11 (formerly family G [17]) xylanase consensus fragments (GXCFs) were amplified from Rt46B.1 genomic DNA by using either the xynGF-xynGR or the newGF-newGR family 11 xylanase consensus primer pair, as shown in Table 1. The PCRs were performed for 35 cycles consisting of 94°C for 1 min, 37°C for 1 min, and 72°C for 1 min with primers xynGF and xynGR and for 35 cycles consisting of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min with primers newGF and newGR. The xynGF-xynGR and newGF-newGR primer pairs gave amplification products that were approximately 330 and 160 bp long, respectively.
TABLE 1.
Family 11 xylanase consensus primers
| Primer | Sequence (5′ to 3′) |
|---|---|
| xynGF | TAT NTG RST NTM TAT GGW TGG |
| Y L – – Y G Wa | |
| xynGR | GAA GGN TAC CAA AGN AGC GG |
| E G Y Q S S | |
| newGF | GAR TAY TAY RTY GTY GAM MGY TGG GG |
| E Y Y I I – – W | |
| newGR | ACY TTY NMS CAG TAC TGG AGY GTY CG |
| T F – Q Y W S V |
The sequences below the primer sequences are the amino acid sequences encoded. –, no single amino acid encoded due to the presence of degenerate nucleotides in the codon.
GWPCRs.
The rationale behind the design and implementation of the genomic walking PCR (GWPCR) technique used has been described previously (25).
(i) Linker assembly.
The general-purpose synthetic DNA linker with NcoI and blunt termini (NcoI blunt linker) was assembled by annealing 75 pmol of the NcoI blunt upper-strand oligonucleotide (berg41; 5′ CAT GGC GCA GGA AAC AGC TAT GAC CGG T 3′) with 75 pmol of the universal lower-strand oligonucleotide (DS43; 5′ CGC GTC CTT TGT CGA TAC TGG CCA 3′) in 50 μl of TE buffer (10 mM Tris-HCl, 0.1 mM EDTA; pH 7.5) at 50°C for 30 min following a 4-min denaturation step at 94°C.
(ii) Linker library construction.
Rt46B.1 restriction fragments were generated by incubating 1 μg of RNaseA-treated Rt46B.1 genomic DNA overnight at 37°C with 20 U of restriction endonuclease NcoI, DraI, EcoRV, HincII, HpaI, PvuII, or SspI. Rt46B.1 restriction fragment linker libraries were then prepared by ligating 5 μl of digested Rt46B.1 DNA with 1 μl of the NcoI blunt DNA linker in a 10-μl overnight reaction mixture by using T4 DNA ligase in 0.1 volume of the buffer supplied by the manufacturer (Boehringer Mannheim). The Rt46B.1 NcoI restriction fragments were extracted with phenol due to the regeneration of the NcoI restriction site upon linker ligation. Following ligation, the library volume was brought to 50 μl with TE buffer.
(iii) GWPCRs.
GWPCRs were performed in standard 50-μl PCR mixtures by using 12 pmol of linker primer (DS43 for the NcoI library, berg41 for the blunt-end libraries), 12 pmol of the forward walking primer (dictGF; 5′ GGT ACT ATT GAT CAA ATT ACT CTT TGT GTT G 3′) or 12 pmol of the reverse walking primer (dictGR; 5′ GTA ATT GGC GTC CAC CAG GTG CAA CC 3′), and 1 μl of the appropriate Rt46B.1 restriction fragment linker library. GWPCRs were performed with a Perkin-Elmer Cetus DNA thermal cycler as described previously (25). The xynB genomic walking primers were positioned so that they provided maximum novel sequence information from the xynB GWPCR products and minimum overlap between the xynB GXCF and GWPCR products so that an accurate sequence alignment could be obtained. In addition, to help ensure the high fidelity of the PCRs, the lengths of the walking primers were optimized with respect to their GC compositions for a target theoretical melting temperature of 72°C, the theoretical melting temperature of the linker primer (berg41). This was done so that the walking and linker primers would exhibit similar annealing profiles; consequently, the likelihood of background amplification from mismatched primers was reduced.
DNA sequencing.
DNA sequencing was carried out with an Applied Biosystems model 373A(stretch) automated DNA sequencer by using dye primer and dye terminator chemistries. M13mp18 was used as the vector for sequencing all of the Rt46B.1-derived PCR fragments. GXCFs and GWPCR fragments amplified from Rt46B.1 genomic DNA were purified from the reaction mixtures by using a High Pure PCR product purification system (Boehringer Mannheim, Auckland, New Zealand) and then were treated for 30 min at 37°C with the Klenow fragment of E. coli DNA polymerase I (1 U; BRL Life Technologies, Auckland, New Zealand), T4 polynucleotide kinase (1 U; Boehringer Mannheim), and T4 DNA polymerase (0.1 U; Boehringer Mannheim) in the presence of deoxyribonucleoside triphosphates (each at a concentration of 0.25 mM) and 0.1 volume of T4 DNA ligase buffer (Boehringer Mannheim) to make the termini of the fragments blunt. The end-repaired PCR fragments were then separated on 1% low-melting-temperature agarose (BRL Life Technologies) gels in TAE buffer and subsequently were gel purified by using the BresaClean DNA purification system (Bresatec, Adelaide, South Australia). The final Rt46B.1 DNA fragments were cloned nondirectionally into the phosphatase-treated SmaI site of M13mp18 replicative-form DNA, and single-stranded bacteriophage DNAs were prepared by the methods described by Sambrook et al. (29). The sequence analysis software of the Genetics Computer Group (7) installed in a Silicon Graphics Indigo 2 computer was used to analyze the Rt46B.1 GXCF and GWPCR sequence data.
Construction of recombinant pJLA602 plasmids.
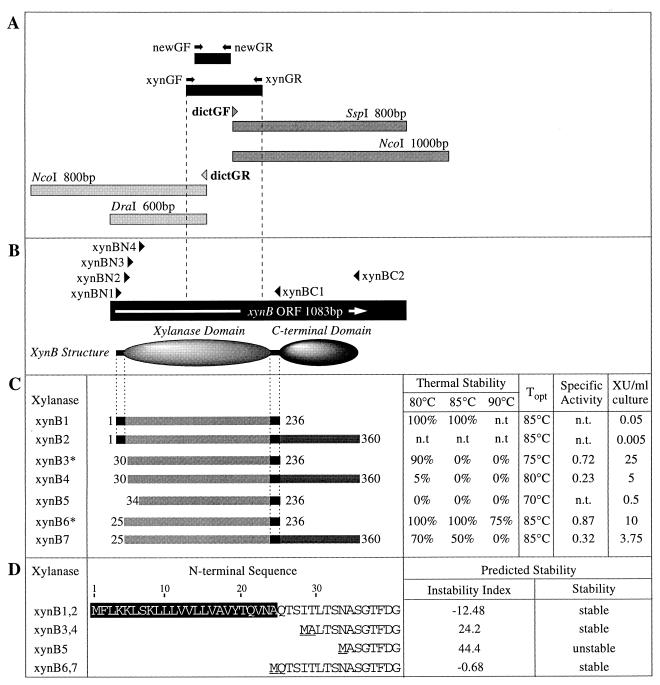
Plasmid pJLA602 is a heat-inducible, Apr expression vector containing the bacteriophage lambda left and right promoters and the cI875 temperature-sensitive lambda repressor gene (30). At permissive temperatures (temperatures less than 37°C), transcription from PLPR is blocked by the cI875 protein. The plasmid is induced by increasing the growth temperature to 42°C. Seven xynB pJLA602 recombinant plasmids were prepared by using different combinations of the four forward and two reverse xynB PCR primers (Table 2), as shown in Fig. 1C. The xynB PCR primers were designed to allow directional in-frame ligation of the xynB PCR fragments into pJLA602 plasmid DNA linearized with either NcoI-BamHI or SphI-BamHI. The four 5′ xynB PCR primers contained translational initiation codons at different 5′ positions and yielded XynB enzymes having N termini either (i) at the beginning of the XynB leader peptide (xynBN1), (ii) immediately upstream of the XynB leader peptide cleavage site (xynBN2), (iii) three residues downstream of the XynB leader peptide cleavage site (xynBN3), or (iv) five residues downstream of the XynB leader peptide cleavage site (xynBN4).
TABLE 2.
Rt46B.1 xynB PCR primers
| Primer | Sequencea |
|---|---|
| xynBN1 | 5′ GGTGTGTACCATGGTTCTTAAAAAACTTAG 3′ |
| xynBN2 | 5′ GCTCAAACGTCCATGGCACTAACAAGTAATGC 3′ |
| xynBN3 | 5′ CTAACAACCATGGCAAGCGGTACTTTTGATGGC 3′ |
| xynBN4 | 5′ GCAAGTTAGCATGCAAACGTCTATAACACTAAC 3′ |
| xynBC1 | 5′ CCACTACTGGATCCACTACTTCCACTACTGC 3′ |
| xynBC2 | 5′ CCTCCCGGATCCTAAACTTCCCCCTCCTTAC 3′ |
The underlined portions of the sequences are restriction sites.
FIG. 1.
Overview of cloning and sequencing of the Rt46B.1 xynB gene and characterization of the XynB xylanase. (A) Upstream and downstream GWPCR products obtained with the dictGR and dictGF walking primers, respectively. The xynB GXCFs amplified by the xynGF-xynGR and newGF-newGR consensus PCR primer pairs are indicated by black boxes, and their respective positions are indicated by grey shading in panels A and B. (B) Diagrammatic representation of the XynB domain structure compared to the 1,083-bp xynB open reading frame (ORF) (indicated by the arrow). The positions of the four forward (xynBN1, xynBN2, xynBN3, and xynBN4) and two reverse (xynBC1 and xynBC2) xynB PCR primers used for amplification of xynB fragments during construction of recombinant pJLA602 expression plasmids are also shown (positions not to scale). (C) Domain structures of the seven XynB xylanases, indicating the N- and C-terminal coordinates of each enzyme, and their thermal stabilities, temperature optima (Topt), specific activities (in XU per microgram of enzyme), and yields (in XU per milliliter of culture). The values for thermal stability at 80, 85, and 90°C are the percentages of the initial xylanase activity remaining following 8 h of incubation in 12.5 mM BTP buffer in the absence of substrate. The leader and interdomain linker peptides present in the domain structures of the various XynB enzymes are indicated by black boxes. n.t., not tested. (D) Putative N-terminal peptide sequences of the XynB1-XynB2, XynB3-XynB4, XynB5, and XynB6-XynB7 xylanases and the predicted stabilities (instability indexes) of the eight N-terminal residues of each sequence, as predicted by the ProtParam amino acid analysis software (see Materials and Methods).
The xynB PCR fragments were purified from the reaction mixtures by using the High Pure PCR purification system (Boehringer Mannheim), and 1 μg of purified DNA was then digested with either NcoI plus BamHI or SphI plus BamHI, as appropriate for the forward PCR primer used. The digested DNAs were subsequently gel purified from 1% low-melting-temperature agarose gels as described above and ligated into either NcoI-BamHI- or SphI-BamHI-linearized pJLA602 plasmid DNA.
The recombinant xynB:pJLA602 plasmids encoding the XynB1, XynB3, XynB5, and XynB6 xylanases were sequenced on both strands to confirm that there were not any PCR-derived base changes in the DNAs.
WWW-based sequence analysis software.
The following two peptide sequence analysis tools with hyperlinks to the ExPASy molecular biology server (URL: http://expasy.hcuge.ch/www/expasy-top.html) were used during the analysis of the XynB peptide sequence: (i) SignalP for predicting the XynB leader peptide cleavage site (URL: http://www.cbs.dtu.dk/services/signalP) and (ii) ProtParam for predicting the stability of the XynB N-terminal peptide sequences (URL: http://expasy.hcuge.ch/sprot/protparam.html).
Production of XynB xylanase samples.
For large-scale production of XynB, 1,200 ml of Luria-Bertani medium was supplemented with ampicillin at a final concentration of 100 μg/ml and was inoculated with 12 ml of an overnight culture (grown at 32°C) of an E. coli strain harboring the desired recombinant xynB:pJLA602 plasmid. The culture was incubated in a 5-liter flask with rapid shaking for approximately 3 h at 32°C until the absorbance at 600 nm (A600) was 1.0, and then the flask was transferred to a 42°C water bath and incubated for 3 h. Following induction, the cells were harvested by mild centrifugation, resuspended in MilliQ-purified water, and lysed by passage through a French pressure cell. The resulting whole-cell extracts were then heat treated at 75 to 85°C for 30 min and centrifuged at high speed to remove the denatured host proteins. The final extracts were stored at 4°C for immediate use or at −20°C for extended storage or were freeze-dried for indefinite storage and delivery.
Purification of XynB xylanases.
The XynB1, XynB3, XynB4, and XynB6 xylanases were purified to electrophoretic homogeneity by cation-exchange chromatography. A 2.5-ml sample of a heat-treated E. coli cell extract prepared in 10 mM sodium acetate (pH 5.5) was passed through a type PD-10 Sephadex G-25 column (Pharmacia, Auckland, New Zealand) to remove the low-molecular-weight compounds. The PD-10 column fraction (3.5 ml) was then applied to a type S15 SartoBind cation-exchange disk (Sartorius Australia, Victoria, Australia) attached to a Pharmacia fast protein liquid chromatography (FPLC) workstation. The S15 disk was washed with 15 ml of equilibration buffer (10 mM sodium acetate, pH 5.5), and then the bound proteins were eluted with a 0 to 0.3 M NaCl gradient. Rt46B.1 XynB proteins eluted at approximately 0.05 M NaCl. Protein concentrations were estimated with a type BCA protein quantification kit (Pierce, Rockford, Ill.).
Assays for xylanase activity.
Qualitative endo-1,4-β-d-xylanase assays were performed with transformant E. coli colonies harboring recombinant xynB:pJLA602 plasmids by using the Congo red procedure of Teather and Wood (32).
The reducing sugars generated from 0.25% oat spelt xylan solutions by XynB were measured by using the p-hydroxybenzoic acid hydrazide (PHBAH) colorimetric assay (19). Samples (10 μl) of cell extract containing approximately 0.005 international xylanase unit (XU) (1 XU was defined as the amount of xylanase activity that resulted in the release of 1.0 μmol of xylose reducing sugar per min [1]) were added to 190-μl portions of a buffered 0.25% oat spelt xylan solution and incubated in 1.5-ml microcentrifuge tubes at the desired temperature for 10 min. Following incubation, the volume of each sample was brought to 600 μl with PHBAH, the samples were boiled 5 min, and the A405 of a 200-μl portion of each sample was determined in a flat-bottom microtiter tray. A net A405 of 1.000 was equivalent to 0.067 μmol of xylose. Sodium acetate was used to buffer solutions at pH values between 4.0 and 6.0, 1,3-bis(tris[hydroxymethyl]-methylamino)propane (BTP) was used to buffer solutions at pH values between 6.0 and 9.0, and 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) was used to buffer solutions at pH values between 9.0 and 11.0. For XynB pH and temperature activity profiles, assays were performed in triplicate.
Temperature stability assays.
All XynB temperature stability assays were performed in 12.5 mM BTP buffer (pH 6.5) in the absence of substrate. XynB samples were incubated in tightly capped 1.5-ml microcentrifuge tubes. At the appropriate time points, the residual xylanase activity was measured immediately by using triplicate 10-μl samples and the PHBAH assay described above. The percentage of remaining activity was expressed relative to the residual xylanase activity of a control xylanase sample kept either on ice or at room temperature.
Hydrolysis of P. radiata kraft pulp.
Oven-dried P. radiata kraft-oxygen pulp was used as the substrate for the XynB kraft pulp hydrolysis studies. Prior to hydrolysis, the pulp was washed at 70°C to rinse it and to liberate the pulp fibers. The washed pulp (0.4 g, dry weight) was incubated with 20 XU of XynA or XynB in 100 ml of 12.5 mM BTP buffer (pH 6.5) in a 500-ml flask at 70 to 80°C for 6 h with occasional shaking. The total reducing sugar contents were measured by using triplicate 500-μl samples of filtered hydrolysate and the PHBAH colorimetric assay described above. A net A405 of 1.0 was equivalent to 0.075 μmol of reducing sugar (measured as xylose).
Pretreatment of eucalypt kraft-oxygen pulp with XynB.
A freeze-dried sample of XynB3 was dissolved in water to obtain a final enzyme concentration of 500 XU/ml. Pulp was treated with the xylanase in plastic bags by using a pulp concentration of 6%. The pulp pH was adjusted to 7.0 with sulfuric acid, xylanase was added, and the mixture was incubated at 75°C for 2 h. The reference pulp (control) was treated under identical conditions but without xylanase.
ECF and TCF bleaching of XynB-treated eucalypt pulp.
ECF bleaching involved a D-EO-D-D procedure, where D is chlorine dioxide treatment and EO is oxygen-reinforced alkaline extraction. All steps were carried out with a pulp concentration of 10%. The initial D step was performed in sealed plastic bags at 70°C for 70 min with an applied active chlorine multiple of 0.05, 0.1, 0.15, or 0.22 and with no adjustment of the pH. The EO step was performed in stainless steel vessels at 90°C for 30 min under 780 kPa of oxygen with a sodium hydroxide charge of 1.5% (pulp basis). The final two D steps were performed in sealed plastic bags at 70°C for 4 h by using 1.32% active chlorine in each step. The pH was adjusted by adding sulfuric acid or sodium hydroxide at the beginning so that the final pH was 3.5 to 4.0.
TCF bleaching involved a Q-PO procedure, where Q is EDTA chelation and PO is pressurized hydrogen peroxide treatment. The conditions used for the Q step were pH 6.0, 2 h, 53°C, and a pulp concentration of 10%. The PO step was performed in Teflon-lined vessels pressurized to 500 kPa with oxygen. The pulp (consistency, 10%) was mixed with a solution containing 3% hydrogen peroxide, 1.5% sodium hydroxide, 2% sodium silicate, 0.2% diethylenetriaminepentaacetic acid (DTPA), and 1% magnesium sulfate and incubated at 115°C for 2 h.
The active chlorine multiple used in these tests was defined as the percentage of chlorine used in the first chlorination step divided by the kappa number of the unbleached pulp.
Pretreatment of P. radiata kraft-oxygen pulp with XynB.
The pH of kraft-oxygen pulp (15.4 kappa) was adjusted to 7.0 by adding sulfuric acid and equilibrating the preparation overnight. Enzyme treatment with XynB was performed at a consistency of 5% in plastic bags at 75°C for 120 min by using an enzyme charge of 10 XU/g of pulp. The reference pulp was prepared by using no enzyme. Following treatment, the pulp filtrates were analyzed for individual and total carbohydrates and UV absorbance (4).
ECF bleaching of XynB-treated P. radiata pulp.
P. radiata pulp was ECF bleached at a pulp consistency of 10% in plastic bags by using a D-E-D procedure, where D is chlorine dioxide treatment and E is alkaline extraction. In the first D step, chlorine dioxide was applied to the pulp at a chlorine multiple of 0.15, 0.20, or 0.25. In the E step 1.0, 1.5, and 2.0% sodium hydroxide were used. The reaction temperature was 70°C, and the reaction was performed for 60 min. The final D step was performed for 180 min at 70°C in the presence of 0.4% sodium hydroxide and 0.8% chlorine dioxide. The final pulp preparations were washed and adjusted to pH values of ∼5.0 with sulfuric acid before ISO brightness (ISO 3688-1977) was measured.
Nucleotide sequence accession number.
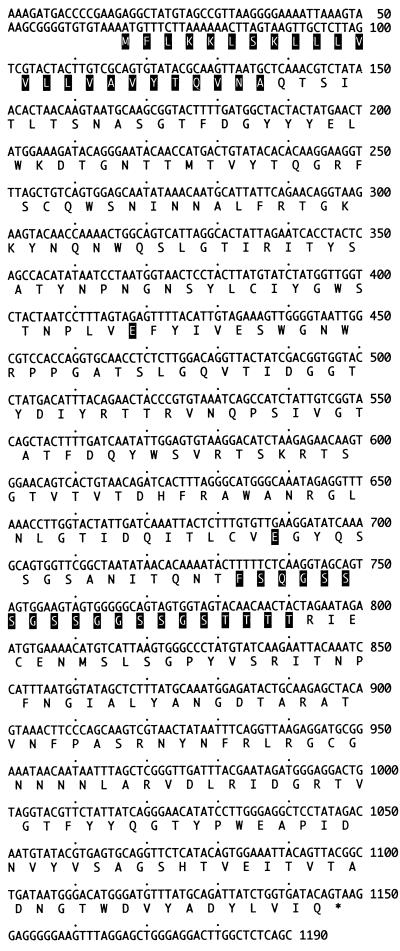
The complete contiguous 1,190-bp sequence generated from the xynB GWPCR fragments which encompasses the 1,083-bp D. thermophilum Rt46B.1 xynB open reading frame (coding sequence from bases 67 to 1149) has been deposited in the GenBank database under accession no. U76545.
RESULTS AND DISCUSSION
Two-step PCR cloning of the Rt46B.1 xynB gene.
The two-step PCR approach used for identification and cloning of the Rt46B.1 xynB gene involved a consensus PCR step (step one) to identify any family 11 xylanase genes in the Rt46B.1 genome, followed by a GWPCR step (step two) to sequence family 11 xylanases.
As the xylanase gene identification step used in this approach is PCR based, it is very sensitive and permits direct analysis of the xylanase gene repertoire in an organism without the necessity of performing time-consuming (and often insensitive) gene library construction and screening procedures. Furthermore, the sequence information obtained from the resulting xylanase consensus fragments is invaluable in planning subsequent gene-cloning steps, particularly when novel genes are sought. The sequence information can be used to decide whether a new gene is sufficiently different to warrant proceeding with the subsequent cloning steps.
Step one: consensus PCR.
GXCFs which migrated at a single apparent molecular size, 330 bp, were amplified from a sample of Rt46B.1 genomic DNA with the xynGF-xynGR consensus PCR primer pair. Nucleotide sequencing of four Rt46B.1 GXCFs revealed a single sequence which exhibited homology to the sequences of other bacterial family 11 xylanases in the GenBank and EMBL databases. Subsequent sequencing of six Rt46B.1 GXCFs amplified with the newGF-newGR consensus primer pair failed to reveal any additional family 11 xylanase genes. In accordance with the previously assigned name of the family 10 xylanase gene from Rt46B.1, xynA, the Rt46B.1 family 11 xylanase gene was named xynB.
Step two: GWPCR.
To allow amplification of DNA fragments upstream and downstream of the Rt46B.1 xynB GXCF region, the dictGR reverse and dictGF forward genomic walking primers were designed based on the sequence of the xynB GXCF.
The following two upstream xynB GWPCR products were amplified from Rt46B.1 restriction fragment linker libraries by PCR with the dictGR walking primer and the berg41 linker primer: (i) an 800-bp fragment from the NcoI library and (ii) a 600-bp fragment from the DraI library. Likewise, the following two downstream xynB GWPCR products were amplified with the dictGF-berg41 primer pair: (i) an 800-bp fragment from the SspI library and (ii) a 1,000-bp fragment from the NcoI library. The nucleotide sequence data generated from these upstream and downstream xynB GWPCR fragments were combined with the xynB GXCF sequence data to generate a 1,190-bp consensus sequence which encompassed a complete 1,086-bp xynB open reading frame (Fig. 2).
FIG. 2.
Sequence of the Rt46B.1 xynB gene and the encoded peptide (XynB). The leader peptide, interdomain linker peptide, and putative catalytic residues are highlighted in the XynB peptide sequence.
The GWPCR xynB cloning step was simplified because a single apparent family 11 xylanase gene was present in the Rt46B.1 genome. If additional family 11 xylanase genes had been present, greater care would have been required in the placement of the genomic walking primers in order to ensure that competition for the primers did not occur between the family 11 xylanase genes.
Analysis of the xynB nucleotide sequence.
The 1,086-bp Rt46B.1 xynB open reading frame (Fig. 2) encoded a 39,800-Mr XynB peptide comprising an N-terminal family 11 xylanase domain (Mr, 22,000), a central 21-amino-acid serine-glycine-rich linker peptide, and a 13,000-Mr C-terminal domain (Fig. 1B). The mature N terminus of XynB was located downstream of a 23-amino-acid leader peptide, as predicted by the SignalP signal peptide analysis software and supported by the results of a multiple sequence analysis performed with other bacterial family 11 xylanases.
The full-length XynB peptide was similar in sequence and architecture to XynD (81% identity) (unpublished data) from the thermophile Rt69B.1 and to XynY (58% identity) from a Bacillus species (35). The XynB C-terminal domain exhibited additional homology (37% identity) to the C-terminal region of the XynA xylanase-arabinofuranosidase from Bacillus polymyxa (15).
Overexpression of XynB in E. coli.
Seven XynB xylanases which differed at their N and C termini were prepared from different recombinant xynB:pJLA602 expression plasmids in order to assess the contributions of the XynB C-terminal domain and N-terminal regions to the characteristics of the enzyme (Fig. 1C). The final yields of the seven XynB xylanases varied considerably, as judged by both xylanase activity assays and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the xylanase preparations (data not shown). The XynB leader peptide made the enzyme toxic for E. coli, as bacterial cells producing XynB1 and XynB2 started to lyse within 1 h of induction, which resulted in low final yields of these enzymes. Endoxylanases are typically extracellular enzymes and, accordingly, carry an N-terminal signal peptide for posttranslational export across the cytoplasmic membrane. Consequently, heterologous expression of genes encoding xylanase preproteins in E. coli can result in periplasmic accumulation of the mature enzymes (18, 20, 26). Hence, it is likely that the apparent toxicity of XynB1 and XynB2 for E. coli occurred either during or after export of these peptides to the periplasm. Alternatively, cytoplasmic accumulation of uncleaved XynB peptides in E. coli may have been toxic.
The final yields of the remaining XynB xylanases (XynB3, XynB4, XynB5, XynB6, and XynB7) (Fig. 1C) varied over a 50-fold range from the lowest yield (XynB5) to the highest yield (XynB3). As expected, the levels of the full-length XynB enzymes (XynB2, XynB4, and XynB7) were lower than the levels of their single-domain counterparts due to the increased time and energy demands associated with processing of the longer xynB transcripts. The markedly different levels of the single-domain XynBs (XynB3, XynB5, and XynB6) were not expected given that these xylanases were produced by the same bacterial strain with the same expression system. The pJLA602 expression plasmid possessed all of the necessary features for high-level expression in E. coli; hence, the factors affecting the synthesis, stability, and translation of the more or less identical xynB3, xynB5 and xynB6 transcripts should be largely independent of the actual xynB coding regions. Therefore, it appears that the differences in the levels of XynB3, XynB5, and XynB6 were due to differences in the N-terminal peptide sequences of these xylanases, which affected either the translational or posttranslational stabilities of the peptides. The reduced structural stability of XynB5 may also have contributed to a reduction in the translational or posttranslational stability of this xylanase compared to the XynB3 and XynB6 single-domain enzymes.
XynB characterization.
All seven xylanases exhibited optimal xylanolytic activity at pH 6.5; however, the temperature optima of these enzymes varied over a 15°C range, from 70 to 85°C (Fig. 1C). The thermal stabilities of the different XynBs also differed substantially. The incremental deletion of a mature N-terminal peptide sequence had an additive effect on the thermolability of XynB, which suggested that several key residues which were essential for maintaining the structural integrity of the enzyme were located in the XynB N-terminal region. However, it is interesting that the predicted stabilities of the N-terminal peptides of XynB3-XynB4, XynB5, and XynB6-XynB7 were related to the observed temperature optima of these enzymes insofar as a higher predicted instability index corresponded to a lower temperature optimum. Therefore, it was possible that the thermal stabilities of the different XynB xylanases were affected, at least in part, by the intrinsic stabilities of their N-terminal peptides.
Curiously, under the conditions employed for the temperature stability assays, the XynB C-terminal domain decreased the thermal stability of the enzyme. However, while addition of oat spelt xylan resulted in only a slight improvement in the temperature stability of XynB3 and XynB6 (data not shown), it appears that these enzymes are stable in the presence of lignocellulosic substrates given that XynB3 and XynB4 released equal amounts of reducing sugars from P. radiatus kraft pulp following extended incubation at 75°C (see below).
The most stable engineered XynB enzyme tested, XynB6, exhibited 50% activity at pH values between 5.0 and 8.0 and at temperatures between 65 and >100°C (at pH 6.5) (data not shown). Specific activity data were determined for the XynB3, XynB4, XynB6, and XynB7 xylanases (Fig. 1C). The XynB6 xylanase had a slightly higher specific activity than XynB3, which may have been a consequence of the increased structural stability of XynB6 compared to XynB3. In addition, we observed that the XynB4 and XynB7 full-length xylanases had lower specific activities than their single-domain counterparts (XynB3 and XynB6, respectively). The approximately twofold differences in the molecular masses of XynB4 and XynB7 compared to XynB3 and XynB6 could not entirely explain the observed threefold differences in the specific activities of these enzymes. Hence, we concluded that the presence of the C-terminal domain slightly inhibited the action of the XynB xylanase domain, perhaps due to steric reasons.
Characterization of the XynB C-terminal domain.
The domain structure of XynB was similar in overall architecture to the domain structure of the two-domain family 11 xylanases from Streptomyces lividans (XynB) (31) and Thermomonospora fusca (TfxA) (12), each of which possesses a C-terminal family 11 cellulose-binding domain consisting of approximately 80 residues. By analogy, one suggested function of the 118-residue XynB C-terminal domain was substrate binding. A kinetic analysis of the single-domain XynB3 and full-length XynB4 xylanases indicated that XynB4 had considerably greater affinity for oat spelt xylan than XynB3 did (the Km values on oat spelt xylan were 0.05% for XynB4 and 0.2% for XynB3). However, following incubation of XynB4 with the insoluble fraction of oat spelt xylan at both high (70°C) and low (4°C) temperatures, the enzymatic activity could not be sequestered from the solution. A similar absence of binding was observed when XynB4 was incubated with crystalline cellulose (Avicel). Therefore, while the XynB C-terminal domain did appear to enhance the affinity of XynB4 for oat spelt xylan, no physical binding was observed between this enzyme and insoluble xylan or cellulose.
Kraft pulp hydrolysis by XynB.
The use of XynB as a nonchemical bleaching agent in the production of bleached kraft pulp requires activity on the modified residual xylans present in kraft pulp. One method of assessing the activity of a xylanase on kraft pulp is to measure the reducing sugars released from the pulp following enzymatic hydrolysis. However, it should be noted that there has not yet been any conclusive evidence showing a direct correlation between the release of reducing sugars during enzyme treatment and enhancement of pulp bleaching.
Both XynB3 and XynB4 (50 XU/g of pulp) released 0.0625 μmol of net reducing sugar per g (dry weight) of pulp following 6 h of incubation at 75°C with 0.4 g of P. radiata kraft-oxygen pulp. By comparison, an equivalent amount of Rt46B.1 XynA xylanase released 0.0875 μmol of reducing sugar per g (dry weight) of pulp. However, the composition of the reducing sugars released by XynA was quite different from the composition of the reducing sugars released by XynB. Paper chromatography (11) of XynA and XynB kraft pulp hydrolysates showed that the family 10 xylanase (XynA) released predominantly xylose, xylobiose, and xylotriose, whereas the family 11 xylanase (XynB3) released a number of high-molecular-weight xylooligosaccharides in addition to xylobiose and xylotriose (data not shown). Chromatographs of the XynA and XynB oat spelt xylan hydrolysates were identical to the respective chromatographs of the P. radiata kraft-oxygen pulp hydrolysates. Hence, XynA and XynB exhibited similar catalytic properties with the fiber-bound xylans of kraft pulp, as they did with the soluble xylans present in oat spelt xylan solutions.
The range of high-molecular-weight xylooligosaccharides released by XynB3 from kraft pulp and oat spelt xylan may indicate that the action of XynB3 is inhibited by side chains in the xylan backbone. In addition, the general absence of xylose in the XynB3 xylan hydrolysates suggested that the XynB3 active site can hydrolyze only xylooligosaccharides that are larger than xylotriose. Indeed, only XynA was able to cleave xylotriose to generate xylobiose and xylose. These differences in the hydrolytic properties of XynA and XynB are consistent with the differences observed with other family 10 and 11 xylanases belonging to multixylanase systems, in which the family 10 enzymes in general appear to possess a more elaborate repertoire of hydrolytic capabilities with respect to their action on branched xylan substrates (2, 9, 21, 34).
Use of XynB3 in ECF and TCF bleaching of eucalypt kraft pulp.
The high thermal stability of XynB3 at its optimum temperature, 75°C, and the very high yields attained with the xynB3 expression plasmid made this enzyme an ideal candidate for laboratory-scale bleaching trials. Previous studies (unpublished data) showed that the Rt46B.1 XynA family 10 xylanase had a negligible effect on the bleachability of eucalypt kraft pulp when ECF and TCF bleaching technologies were used. Consequently, in view of the different catalytic and physicochemical properties of XynA and XynB, a preliminary study was performed to assess the benefits of using XynB in ECF and TCF bleaching of eucalypt kraft pulp.
Treatment of eucalypt kraft-oxygen pulp with XynB3 at enzyme doses of up to 10 XU/g of pulp resulted in reductions in the pulp kappa number of up to 0.8 U following a D-EO ECF bleaching procedure. At XynB3 doses greater than 10 XU/g of pulp, there was a levelling off in the kappa number reduction to a maximum of 1.0 U as the enzyme dose approached 30 XU/g of pulp. The final pulp brightness of XynB3-treated pulp at an enzyme dose of 10 XU/g of pulp increased 1.5% ISO units (from 88.5 to 90% ISO with respect to the reference pulp) following a D-EO-D-D ECF bleaching procedure. This increase in brightness is significant, since in the absence of xylanase pretreatment, a kraft-oxygen pulp brightness of 90% ISO could be attained only by modifying the actual ECF bleaching procedure. XynB3-treated pulp (10 XU/g of pulp) could be bleached to a target brightness of 88% ISO by using 3.4% ClO2 (as active chlorine) in a D-EO-D-D procedure, compared with 4.0% ClO2 for untreated pulp. The savings in chlorine dioxide was 15% of the chemical charge used for the untreated pulp, or 6 kg of active chlorine per tonne of pulp.
The XynB3 xylanase was also able to enhance the bleachability of eucalypt kraft-oxygen pulp in a TCF bleaching procedure. Treatment of the pulp at a xylanase dose of 10 XU/g increased the final pulp brightness from 82.2 to 86.6% ISO following a Q-PO TCF bleaching procedure.
Bleaching studies were also performed to compare the effect of XynB pretreatment on pulp bleachability to the effects of the commercial xylanase Irgazyme-40 (Ciba-Geigy Australia Ltd.) and a noncommercial xylanase, DCPX. At an enzyme dose of 10 XU/g of pulp, all three xylanases decreased the bleached pulp yield by 3% (based on the yield of unbleached pulp); however, the final pulp brightness values obtained with the xylanases following a D-EO-D-D ECF bleaching procedure were 89.2% ISO for DCPX, 89.5% ISO for Irgazyme-40, and 89.8% ISO for XynB3. Hence, at the same enzyme dose XynB3 was the most effective of the three xylanases tested.
Use of XynB3 in ECF bleaching of P. radiata kraft pulp.
Following pretreatment of P. radiata kraft-oxygen pulp with XynB3 (10 XU/g of pulp), 7 mg of carbohydrate (xylose-arabinose, 12:1) per g of pulp was solubilized, which was comparable to the net xylan attack observed with commercial xylanase preparations. UV-absorbing materials (net A280, 2.08) were also solubilized from the kraft pulp following XynB3 treatment.
The brightness of XynB3-treated pulp was consistently greater than the brightness of the reference pulp following D-E-D bleaching performed with different active chlorine multiples (Table 3). At the lowest chemical charge, pulp treated with XynB3 had a brightness 6 to 8% ISO greater than brightness of the reference pulp. However, as determined previously with eucalypt pulp, the increase in brightness that resulted from XynB3 treatment decreased as the chlorine multiple increased. In general, the ability of XynB to improve D-E-D bleaching was similar to the ability observed with a range of commercial xylanase products which act in the temperature range from 40 to 55°C.
TABLE 3.
D-E-D bleaching of XynB3-treated P. radiata kraft-oxygen pulp
| Step 1 active chlorine multiple | Step 3 reference pulp brightness (%ISO) | Step 3 XynB3-treated pulp brightness (%ISO) |
|---|---|---|
| 0.15 | 66.6 | 74.2 (7.6)a |
| 0.20 | 74.7 | 80.7 (6.0) |
| 0.25 | 81.7 | 82.2 (0.5) |
The values in parentheses are the net increases in brightness of the XynB3-treated pulp preparations compared with the reference pulp preparations.
In summary, the XynB xylanase was able to enhance the bleachability of eucalypt kraft-oxygen pulp in ECF and TCF bleaching procedures and the bleachability of P. radiata kraft-oxygen pulp in ECF bleaching procedures. This xylanase appears to be well-suited to the high-temperature applications encountered during kraft pulping and bleaching procedures as a result of its high temperature optimum and temperature stability.
ACKNOWLEDGMENTS
This work was supported in part by grants from the University of Auckland Research Committee and the Foundation for Research, Science and Technology, Wellington, New Zealand.
REFERENCES
- 1.Bailey M J, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;23:257–270. [Google Scholar]
- 2.Biely P, Kluepfel D, Morosoli R, Shareck F. Mode of action of three endo-β-1,4-xylanases of Streptomyces lividans. Biochim Biophys Acta. 1993;1162:246–254. doi: 10.1016/0167-4838(93)90288-3. [DOI] [PubMed] [Google Scholar]
- 3.Black G W, Hazlewood G P, Millward-Sadler S J, Laurie I L, Gilbert H J. A modular xylanase containing a novel xylan-specific binding domain. Biochem J. 1995;307:191–195. doi: 10.1042/bj3070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark T A, Allison R W, Kibblewhite R P. Effects of enzymatic modification on radiata pine kraft fibre wall chemistry and physical properties. Appita J. 1997;50:329–335. [Google Scholar]
- 5.Clarke J, Davidson K, Gilbert H J, Fontes C M G A, Hazlewood G P. A modular xylanase from mesophilic Cellulomonas fimi contains the same cellulose-binding domain and thermostabilising domain as xylanases from thermophilic bacteria. FEMS Microbiol Lett. 1996;139:27–35. doi: 10.1111/j.1574-6968.1996.tb08175.x. [DOI] [PubMed] [Google Scholar]
- 6.Dekker R F H. Biodegradation of hemicelluloses. In: Higuchi T, editor. Biosynthesis and biodegradation of wood components. Orlando, Fla: Academic Press; 1985. pp. 505–533. [Google Scholar]
- 7.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlop-Jones N, Grönberg V. Recent developments in the application of xylanase enzymes in elemental chlorine-free (ECF) and total chlorine-free (TCF) bleaching. Pulp Pap Can. 1995;96:20–24. [Google Scholar]
- 9.Elegir G, Sykes M, Jeffries T W. Differential and synergistic action of Streptomyces endoxylanases in prebleaching of kraft pulps. Enzyme Microb Technol. 1995;17:954–959. [Google Scholar]
- 10.Fontes C M, Hazlewood G P, Morag E, Hall J, Hirst B H, Gilbert H J. Evidence for a general role for non-catalytic thermostabilising domains in a xylanase from thermophilic bacteria. Biochem J. 1995;307:151–158. doi: 10.1042/bj3070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry S C. The growing plant cell wall. Essex, United Kingdom: Longman Scientific and Technical; 1988. [Google Scholar]
- 12.Ghangas G S, Hu Y, Wilson D B. Cloning of a Thermomonospora fusca xylanase gene and its expression in Escherichia coli and Streptomyces lividans. J Bacteriol. 1989;171:2963–2969. doi: 10.1128/jb.171.6.2963-2969.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbs M D, Reeves R A, Bergquist P L. Cloning, sequencing, and expression of a xylanase gene from the extreme thermophile Dictyoglomus thermophilum Rt46B.1 and activity of the enzyme on fiber-bound substrate. Appl Environ Microbiol. 1995;61:4403–4408. doi: 10.1128/aem.61.12.4403-4408.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilkes N R, Henrissat B, Kilburn D G, Miller R C J, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosables M J, Perez-Gonzalez J A, Gonzales R, Navarros A. Two beta-glycanase genes are clustered in Bacillus polymyxa: molecular cloning, expression and sequence analysis of genes encoding a xylanase and an endo-beta-(1,3)-(1,4)-glucanase. J Bacteriol. 1991;173:7705–7710. doi: 10.1128/jb.173.23.7705-7710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grépinet O, Chebrou M-C, Béguin P. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J Bacteriol. 1988;170:4582–4588. doi: 10.1128/jb.170.10.4582-4588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Lowe S E, Zeikus G J. Gene cloning, sequencing, and biochemical characterization of endoxylanase from Thermoanaerobacterium saccharolyticum B6A-RI. Appl Environ Microbiol. 1993;59:3134–3137. doi: 10.1128/aem.59.9.3134-3137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lever M. Colorimetric and fluorometric carbohydrate determination with p-hydroxybenzoic acid hydrazide. Biochem Med. 1973;7:274–281. doi: 10.1016/0006-2944(73)90083-5. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Gherardini F C, Matuschek M, Bahl H, Weigel J. Cloning, sequencing, and expression of the gene encoding a large S-layer-associated endoxylanase from Thermoanaerobacterium sp. strain JW/SL-YS 485 in Escherichia coli. J Bacteriol. 1996;178:1539–1547. doi: 10.1128/jb.178.6.1539-1547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maringer U, Wong K K Y, Saddler J N, Kubicek C P. A functional comparison of two pairs of β-1,4-xylanases from Trichoderma harzianum E58 and Trichoderma reesei RUT C-30. Biotechnol Appl Biochem. 1995;21:49–65. [Google Scholar]
- 22.Mathrani I M, Ahring B K. Thermophilic and alkalophilic xylanases from several Dictyoglomus isolates. Appl Microbiol Biotechnol. 1992;38:23–27. [Google Scholar]
- 23.Millward-Sadler S J, Davidson K, Hazlewood G P, Black G W, Gilbert H J, Clarke J H. Novel cellulose-binding domains, NodB homologues and conserved modular architecture in xylanases from the aerobic soil bacteria Pseudomonas fluorescens subsp. cellulosa and Cellvibrio mixtus. Biochem J. 1995;312:39–48. doi: 10.1042/bj3120039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millward-Sadler S J, Poole D M, Henrissat B, Hazlewood G P, Clarke J H, Gilbert H J. Evidence for a general role for high-affinity non-catalytic cellulose binding domains in microbial plant cell wall hydrolases. Mol Microbiol. 1994;11:375–382. doi: 10.1111/j.1365-2958.1994.tb00317.x. [DOI] [PubMed] [Google Scholar]
- 25.Morris D D, Reeves R A, Gibbs M D, Saul D J, Bergquist P L. Correction of the β-mannanase domain of the celC pseudogene from Caldicellulosiruptor saccharolyticus and activity of the gene product on kraft pulp. Appl Environ Microbiol. 1995;61:2262–2269. doi: 10.1128/aem.61.6.2262-2269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paradis F W, Zhu H, Krell P J, Phillips J P, Forsberg C W. The xynC gene from Fibrobacter succinogenes S85 codes for a xylanase with two similar catalytic domains. J Bacteriol. 1993;175:7666–7672. doi: 10.1128/jb.175.23.7666-7672.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel B K, Morgan H W, Daniel R M. Isolation of an extremely thermophilic chemoorganotrophic anaerobe similar to Dictyoglomus thermophilum from a New Zealand hot spring. Arch Microbiol. 1987;147:21–24. [Google Scholar]
- 28.Rättö M, Mathrani I, Ahring B, Viikari L. Application of thermostable xylanases of Dictyoglomus sp. in enzymatic treatment of kraft pulps. Appl Microbiol Biotechnol. 1994;41:130–133. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schauder B, Blöcker H, Frank R, McCarthy J E G. Inducible expression vectors incorporating the Escherichia coli atpE transcriptional initiation region. Gene. 1987;52:279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- 31.Shareck F, Roy C, Yaguchi M, Morosoli R, Kluepfel D. Sequences of three genes specifying xylanases in Streptomyces lividans. Gene. 1991;107:75–82. doi: 10.1016/0378-1119(91)90299-q. [DOI] [PubMed] [Google Scholar]
- 32.Teather R M, Wood P J. Use of Congo red polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viikari L, Kantelinen A, Sundquist J, Linko M. Xylanases in bleaching: from an idea to the industry. FEMS Microbiol Rev. 1994;13:335–350. [Google Scholar]
- 34.Wong K K Y, Tan L U L, Saddler J N. Multiplicity of β-1,4-xylanases in microorganisms: functions and applications. Microbiol Rev. 1988;52:305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J H, Park Y S, Yum D Y, Kim J M, Kong I S, Bai D H. Nucleotide sequence and analysis of a xylanase gene (xynS) from alkali-tolerant Bacillus sp. YA-14 and comparison with other xylanases. J Microbiol Biotechnol. 1993;3:139–145. [Google Scholar]
- 36.Zhu H, Paradis F W, Krell P J, Phillips J P, Forsberg C W. Enzymatic specificities and modes of action of the two catalytic domains of the xynC xylanase from Fibrobacter succinogenes S85. J Bacteriol. 1994;176:3885–3894. doi: 10.1128/jb.176.13.3885-3894.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]