Abstract
It was recently shown that the white rot basidiomycete Pycnoporus cinnabarinus secretes an unusual set of phenoloxidases when it is grown under conditions that stimulate ligninolysis (C. Eggert, U. Temp, and K.-E. L. Eriksson, Appl. Environ. Microbiol. 62:1151–1158, 1996). In this report we describe the results of a cloning and structural analysis of the laccase-encoding gene (lcc3-1) expressed by P. cinnabarinus during growth under xylidine-induced conditions. The coding region of the genomic laccase sequence, which is preceded by the eukaryotic promoter elements TATA and CAATA, spans more than 2,390 bp. The corresponding laccase cDNA was identical to the genomic sequence except for 10 introns that were 50 to 60 bp long. A sequence analysis indicated that the P. cinnabarinus lcc3-1 product has a Phe residue at a position likely to influence the reduction-oxidation potential of the enzyme’s type 1 copper center. The P. cinnabarinus lcc3-1 sequence was most similar to the sequence encoding a laccase from Coriolus hirsutus (level of similarity, 84%).
By definition, laccases (p-diphenol:O2 oxidoreductase; EC 1.10.3.2) catalyze the oxidation of p-diphenols and the concurrent reduction of dioxygen to water, although the actual substrate specificities of laccases are often quite broad and vary with the enzyme source (11, 29). Laccases are members of the blue copper oxidase enzyme family characterized by having four cupric (Cu2+) ions coordinated such that each of the known magnetic species (type 1, type 2, and type 3) is associated with a single polypeptide chain. The Cu2+-binding domains are highly conserved in the blue copper oxidases, and the crystallographic structure of ascorbate oxidase, another member of this enzyme class, has provided a good model for the structure of the laccase active site (30, 31). This model has been supported by the results of numerous studies of the electron transfer reactions that occur between cupric ions during catalysis (35, 39, 40).
In contrast to our understanding of the electron transfer reactions that occur in laccases, relatively little is known about the physiological functions of these enzymes. Laccases have been implicated in pigmentation (1, 9), fruiting body formation (26), and pathogenicity (7, 45), as well as in lignin degradation (41) and biosynthesis (27). Very few of these functions have been experimentally proven, and only because of the availability of multiple gene sequences and crystallographic data has it been possible to speculate about how structure-function relationships may be important in the specific roles played by these enzymes (46). Some of this speculation has involved attempts to address the apparent contradictory functions of laccases in the synthesis and breakdown of lignin (3, 11).
To better understand the role of laccases in lignin degradation by white rot fungi, we studied the ligninolytic system of Pycnoporus cinnabarinus, a basidiomycete that produces an unusual set of ligninolytic enzymes. Just a single isoform of laccase, but no lignin peroxidase (LiP) or manganese peroxidase (MnP), was produced by this organism under conditions that stimulated lignin degradation (13). We wanted to determine more completely the pattern of phenoloxidase production in P. cinnabarinus, so the primary objective of this study was to analyze the structure of the P. cinnabarinus laccase gene and determine whether there are multiple laccase genes in the P. cinnabarinus genome.
MATERIALS AND METHODS
Organisms and reagents.
P. cinnabarinus PB (= ATCC 200478), an isolate recovered from decaying pine wood in the vicinity of Sydney, New South Wales, Australia, was maintained as described previously (13). Escherichia coli INVαF′ (One Shot competent cells) and the pCR2.1 vector used for direct cloning of PCR products were purchased from Invitrogen (San Diego, Calif.). Unless otherwise indicated, the enzymes used to manipulate DNA or RNA were obtained from Boehringer Mannheim (Indianapolis, Ind.), New England Biolabs (Beverly, Mass.), or Invitrogen (T4 DNA ligase) and were used according to the manufacturers’ instructions. All chemicals and reagents were at least analytical grade.
Oligonucleotides, probes, and primers.
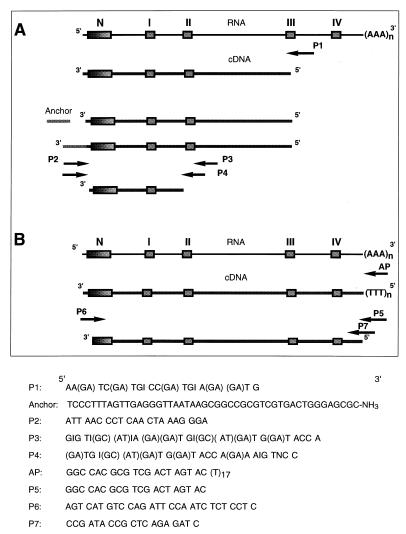
The sequences of most oligonucleotide primers used in this study are shown in Fig. 1; the exceptions are the sequences of the oligonucleotides used to isolate the P. cinnabarinus lcc3-1 promoter. A digoxigenin-labeled laccase probe was prepared by using primers P3 and P6 (Fig. 1). The AP oligonucleotide primer was purchased from Life Technologies (Bethesda, Md.). Other primers were synthesized at the Molecular Genetics Instrumentation Facility of the University of Georgia.
FIG. 1.
Strategy used for PCR cloning of the laccase-encoding cDNA from P. cinnabarinus and oligonucleotide primer sequences. The boxes indicate the regions encoding the N terminus of the mature protein and the Cu(II)-binding regions of the laccase that are highly conserved in blue copper oxidases. (A) Results of laccase-specific reverse transcription of P. cinnabarinus RNA and anchor-ligated PCR used to amplify the 450-bp fragment of the P. cinnabarinus laccase cDNA on which the gene-specific primer P6 was based. (B) After primer AP-primed reverse transcription of full-length P. cinnabarinus mRNAs, primer P5 was used in conjunction with primer P6 to amplify the laccase-encoding sequence. Primers P6 and P7 were used to clone the genomic laccase sequence.
RNA isolation.
P. cinnabarinus cultures grown for 3 days in modified Dodson medium (13) at 30°C on a rotary shaker (135 rpm) were induced with 2,5-xylidine (10 μM) as described previously (8). Longer cultivation times led to increased production of extracellular polysaccharides, which strongly interfered with RNA isolation. Fungal mycelia were collected by filtration, and then they were washed twice in sterile phosphate buffer (20 mM, pH 7.0) and frozen in liquid nitrogen before RNA was isolated by the method of Chomczynski and Sacchi (8). For Northern analyses, total RNA (10 μg) was separated on a 1.4% (wt/vol) agarose gel (32), transferred to Nytran-Plus membranes (Schleicher & Schuell, Keene, N.H.), and hybridized with labeled probes under high-stringency conditions as described below for the Southern blot analysis.
Genomic DNA isolation.
Mycelia from P. cinnabarinus grown in 250 ml of malt extract medium (15 g/liter, pH 5.0) at 30°C for 4 days were harvested, washed, and frozen in liquid N2 as described above. High-molecular-weight genomic DNA was isolated from frozen mycelia after grinding by using a plant DNA isolation kit (Boehringer) as recommended by the manufacturer.
cDNA synthesis, 5′ anchor ligation PCR, and PCR cloning.
Ligation-anchored PCR (42) was used to obtain full-length P. cinnabarinus laccase cDNA. Total RNA (1.0 μg) was primed by using a degenerate oligonucleotide primer (primer P1) designed to complement the third (from the amino terminus) copper-binding domain (domain III) that is conserved in laccases and other blue copper oxidases (Fig. 1A). Subsequent reverse transcription was performed with Superscript reverse transcriptase (Life Technologies), and an oligonucleotide anchor was ligated to the 5′ end of the resultant cDNA. Primer P2, which was complementary to the anchor sequence, was used in combination with two degenerate primers, primers P3 and P4, which were synthesized to match the second copper-binding domain (domain II) conserved in laccases, to amplify a 450-bp fragment of the P. cinnabarinus laccase gene. For PCR amplification of the anchor-ligated cDNA with primers P2 and P3, an aliquot (2 μl) of the ligation mixture was used as the template in a 25-μl reaction mixture containing Tfl polymerase (Epicentre Technologies, Madison, Wis.). For half-nested amplification of the first PCR product, a template (1 μl) was added to a reaction mixture containing primers P2 and P4 and Expand high-fidelity polymerase (Boehringer Mannheim). The resultant product was subcloned into the pCR2.1 vector (Invitrogen), and two clones were sequenced. From the resulting sequence, primer P6, an oligonucleotide primer whose sequence exactly matched the sequence of the 5′ untranslated region of the P. cinnabarinus laccase mRNA, was synthesized. After reverse transcription of total P. cinnabarinus RNA with primer AP, primer P5, which complemented a portion of the primer AP sequence, was used in combination with primer P6 and Expand polymerase to amplify the full-length P. cinnabarinus laccase cDNA.
To isolate genomic laccase sequences, exact-match primers P6 and P7 (the sequence of P7 corresponded to the sequence immediately downstream of the stop codon) were used in PCR mixtures (volume, 25 μl) containing genomic DNA (1 μg) and Expand polymerase. Amplified products that were approximately 2,100 bp long were cloned into the pCR2.1 vector, and two of the resultant clones were sequenced on both strands.
Isolation of the P. cinnabarinus lcc3-1 promoter region.
P. cinnabarinus genomic DNA was digested with KpnI, which cuts at positions 1050 and 1296 in the lcc3-1 gene. The cleavage products were circularized by ligation with T4 DNA ligase. The putative promoter region upstream of the laccase coding sequence was amplified by a two-step inverse PCR process by using primers PA (5′-CCACAGCGGCAAGAGAGACG-3′) and PB (5′-GAGGACAAAGGAGAGGAGAGATTGG-3′) (which were directed in the 5′ direction from nucleotides 343 and 319, respectively) and primer PC (5′-GATCACCCCCGCTCCTCTCA-3′) (which was directed in the 3′ direction from nucleotide 457). Sequential PCR amplifications were performed by starting with an aliquot (2 μl) of the ligation product in a 25-μl reaction mixture containing Expand polymerase. The resultant 1,400-bp fragment was subcloned into the pCR2.1 vector, and two of the resultant clones were sequenced on both strands.
DNA sequencing.
Nucleotide sequences were determined by using Taq polymerase cycle sequencing and an automated DNA sequencer (model ABI 377; Perkin-Elmer Corp., Foster City, Calif.) at the Molecular Genetics Instrumentation Facility on the University of Georgia campus. All cloned DNAs were sequenced on both strands, and the encoded amino acid sequences were predicted by using Gene Runner (Hastings Software, Hastings-on-the-Hudson, N.Y.). Sequences were aligned by using the CLUSTAL V algorithm (MegAlign; DNASTAR, Madison, Wis.).
Southern blot analysis.
Restriction endonuclease-digested DNA samples (10 μg) were separated on a 0.8% agarose gel and transferred to Nytran-Plus nylon membranes (Schleicher & Schuell) by using the procedure of Zhou et al. (49). When high-stringency conditions were used, hybridization was performed at 45°C with a DIG Easy Hyb solution (Boehringer Mannheim), the filters were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.0.15 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS) at 24°C and then in 0.1× SSC–0.5% SDS at 68°C. Unless indicated otherwise, when low-stringency conditions were used, hybridization was performed at 42°C with the same hybridization solution, and the filters were washed at 24°C in 2× SSC–0.5% SDS and then at 49°C in 0.1× SSC–0.5% SDS. Blots were developed by following the manufacturers’ instructions for chemiluminescent detection of digoxigenin-labeled probes with alkaline phosphatase-antibody conjugates (Boehringer Mannheim). Digoxigenin-labeled probes for the P. cinnabarinus laccase gene were prepared by using primers P2 and P3 to amplify a 450-bp fragment from the cloned cDNA.
Nucleotide sequence accession number.
The nucleotide sequence of P. cinnabarinus lcc3-1 reported in this paper has been deposited in the EMBL/GenBank database under accession no. AF025481.
RESULTS
Isolation of the laccase cDNA and characterization of the deduced protein.
The longest cDNA clone of the P. cinnabarinus laccase gene was 1,828 bp long without the poly(A) tail and contained a 1,554-bp open reading frame. Blots of the transcription products obtained from 3-day-old mycelium probed with a 450-bp digoxigenin-labeled fragment from the cDNA clone revealed that a single transcript about 1,800 bp long was produced (data not shown). Thus, the transcript size was consistent with the length predicted from the cDNA sequence.
G and C accounted for 58% of the nucleotides in the coding sequence, and the proportion of G and C in degenerate codon positions was high (66% GC, with C [46%] ≫ A [6%]). A similar or even more pronounced preference for pyrimidine bases has been found in other fungal genes (22, 36). Some examples of extreme codon bias in the P. cinnabarinus laccase are Leu (87% C/UUG/C versus 13% CUU), Val (80% GUG/C versus 20% GUA/U), and Phe (90% UUC versus 10% UUU).
The 21-amino-acid N-terminal sequence of the purified P. cinnabarinus laccase (13) was identical to residues 22 to 42 predicted from the open reading frame of the cDNA (Fig. 2). The putative 21-amino-acid signal sequence (Fig. 2) was followed by a sequence that could act as a peptidase recognition site as determined by the (−3, −1)-rule (43), which predicts that there is a small, uncharged amino acid residue (Ala) at position −1 relative to the cleavage site. Thus, the cleavage site sequence had the most common pattern, A-X-A, found in the C termini of signal peptides. The core region of the signal peptide is predominantly hydrophobic, which is typical for eukaryotic signal sequences (33).
FIG. 2.
Nucleotide sequence and deduced amino acid sequence of the P. cinnabarinus lcc3-1 gene. Putative CAAT and TATA boxes are indicated by open boxes. The N-terminal sequence of the purified laccase from P. cinnabarinus (13) is indicated by a shaded box. Possible N-glycosylation sites are underlined. Residues involved in binding Cu(II) ions are marked by single boxes. The numbers in italic type (1, 2, and 3) indicate with which of the three Cu(II) types the residues coordinate. The putative polyadenylation signal is underlined with a dotted line. Recognition sites for restriction endonucleases KpnI, HindIII, and BamHI, as well as the most prominent transcriptional start site (tsp), are indicated.
It was predicted that the mature laccase polypeptide secreted by P. cinnabarinus contained 497 amino acids and had a composite molecular mass of 53,871 Da. P. cinnabarinus laccase purified from culture supernatants was previously shown to have an apparent Mr of ca. 76,000 (as determined by SDS-polyacrylamide gel electrophoresis) or 81,000 (as determined by gel filtration) (13); thus, the observed and predicted Mr values for the deduced protein differed by about 30%. Glycosylation is one form of posttranslational processing that is probably responsible for at least some of the difference (38). The laccase contains six potential N-glycosylation sites (Asn-Xxx-Ser/Thr), at positions 72, 75, 229, 354, 362, and 455 of the deduced protein (Fig. 2), although for steric reasons, it seems unlikely that the sites at positions 72 and 75 are glycosylated simultaneously. On the other hand, the carbohydrate content reported previously for the secreted protein (13) (9%) is not sufficient to completely explain the differences between the predicted and observed molecular weights. The isoelectric point calculated for the cloned gene product (pI 4.5) also differs from the experimentally determined isoelectric point for the purified laccase (pI 3.7) (13).
The amino acid residues that act as Cu2+ ligands are highly conserved in all blue copper oxidases, including laccases (31). All of the expected Cu2+ ligands (10 His residues and one Cys residue) were present in the lcc3-1 coding sequence and are numbered in Fig. 2 on the basis of whether they coordinate with the type 1, type 2, or type 3 Cu2+ centers. Another residue (Phe), which is numbered in Fig. 2 to indicate interaction with the type 1 copper center and is located nearest the carboxyl terminus of the protein, varies in laccases from different sources and is considered a residue that is probably important in governing the reduction-oxidation potential of type 1 copper centers (46).
Isolation and structural analysis of the laccase genomic sequence.
A portion of the laccase gene was amplified from P. cinnabarinus DNA by using oligonucleotide primers whose sequences correspond to sequences identified in the 5′ and 3′ untranslated regions of the mRNA (Fig. 2). The 2,390-bp coding region was 526 bp longer than the corresponding cDNA sequence, and a comparison of the sequences revealed 10 short introns (length, 50 to 60 bp) in the genomic sequence.
Inverse PCR was used to isolate the laccase gene promoter. Circularized KpnI digests of P. cinnabarinus genomic DNA were amplified with primers based on the genomic laccase sequence (primers PA [nucleotide 343], PB [nucleotide 319], and PC [nucleotide 457]). This approach yielded an additional 1,400 bp of upstream sequence, and 240 bp of this sequence, including putative CAAT and TATA promoter elements, is shown in Fig. 2. An analysis of 5′ RACE (random amplification of cDNA ends)-amplified cDNAs strongly suggested that transcription of the laccase gene starts 68 bp upstream of the translational start site (nucleotide 240). Thus, the location of the putative TATA element in this promoter is similar to the locations found in several other fungal genes, in which the TATA box is generally located 30 to 60 nucleotides upstream from the transcriptional start site (20). Paired TATA and CAAT elements have been identified in other fungal laccase promoters (Tvi [21, 22], Po [19], Ch [24], and Pa [17]); however, although the order of these motifs is conserved, their absolute positions vary. In genes of filamentous fungi, pyrimidine-rich sequences often directly precede the transcriptional start site, particularly in highly expressed genes (2, 20); however, such sequences were not found in the P. cinnabarinus laccase promoter. A putative polyadenylation signal, AATAA, which is a slight variation of the consensus polyadenylation signal sequence AATAAA (37), was found 167 bp downstream of the laccase stop codon.
The 10 introns in the laccase gene were in good agreement with respect to the consensus sequence predicted for the 5′ splice sites of eukaryotic genes, GT(a/g)NG(c/t) (2). Only introns 6 (T at position 3) and 3 (T at position 5) exhibited slight variations. The consensus sequence for 3′ splice sites, (c/t)N(c/t)AG, also matched, except for position 1 in introns 5 and 7 (A) and introns 6 and 9 (G). The overall exon-intron structure of the lcc3-1 gene was very similar to the structure determined for the Coriolus hirsutus laccase gene (24).
Laccase gene family.
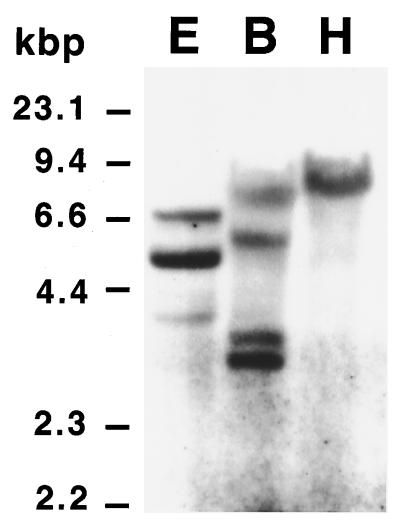
Southern blot analysis was used to estimate the number of laccase genes in the P. cinnabarinus genome. Genomic DNA that had been digested with EcoRI, BamHI, and HindIIII was hybridized to a 450-bp digoxigenin-labeled cDNA probe spanning the region from the start of the open reading frame to the second Cu-binding domain. None of these restriction enzymes cut in the probe sequence. In HindIII digests, the laccase probe hybridized to a single band of approximately 8.6 kb (Fig. 3), but three EcoRI fragments (4.1, 5.5, and 6.7 kb) and four BamHI fragments (3.7, 3.9, 6.1, and 8.1 kb) were detected by the probe. Each of the fragments in the latter digests was large enough to contain a complete copy of the laccase gene.
FIG. 3.
Southern blot of P. cinnabarinus genomic DNA. Total DNA from P. cinnabarinus was digested with EcoRI (lane E), BamHI (lane B), or HindIII (lane H). The resultant DNA fragments (10 μg per lane) were resolved by agarose gel electrophoresis and blotted onto a nylon membrane. The filter was probed with a 450-bp digoxigenin-labeled fragment of P. cinnabarinus lcc3-1. The relative mobilities of HindIII-restricted lambda DNA fragments are indicated on the left.
Similarity to other laccase sequences.
The results of a comparison of the amino acid sequence of the P. cinnabarinus laccase (encoded by the P. cinnabarinus lcc3-1 gene) with all laccase sequences available in databases are shown in Table 1. P. cinnabarinus lcc3-1 is most closely related to C. hirsutus phe1 (84.0% similarity), followed by Trametes villosa lcc1 (83.0% similarity) and lac2 of the unidentified basidiomycete strain CECT 20197 (82.6% similarity). The laccases isolated so far from the ligninolytic basidiomycetes are highly conserved (>58% similarity). In general, sequence similarity follows phylogenetic position: basidiomycetous laccases (36 to 84% similarity) > ascomycetous laccases (23 to 25% similarity) > plant laccases (18 to 20% similarity), with the notable exception of the Aspergillus nidulans laccase (17% similarity).
TABLE 1.
Levels of similarity between P. cinnabarinus lcc3-1 and other fungal and plant laccase genesa
| Organism and geneb | % Similarity to gene:
|
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | |
| 1. P.c. lcc3-1 | 84.0 | 83.0 | 75.3 | 60.9 | 67.6 | 64.5 | 68.0 | 73.7 | 82.6 | 78.4 | 76.0 | 62.5 | 58.5 | 58.3 | 43.6 | 42.5 | 37.1 | 36.1 | 35.7 | 25.5 | 25.1 | 23.4 | 20.1 | 17.8 | 21.2 | 18.7 | 19.9 | 18.3 | 17.4 | |
| 2. C.h. po1 | 91.0 | 79.4 | 60.9 | 68.1 | 64.0 | 68.7 | 77.6 | 86.3 | 80.6 | 79.5 | 63.5 | 61.3 | 61.2 | 44.4 | 42.9 | 36.7 | 34.6 | 35.8 | 23.8 | 21.7 | 24.6 | 20.2 | 20.8 | 22.9 | 22.5 | 21.3 | 19.0 | 17.1 | ||
| 3. T.vi. lcc1 | 77.1 | 60.3 | 68.3 | 64.0 | 68.8 | 77.1 | 84.6 | 80.0 | 78.3 | 62.5 | 58.1 | 59.8 | 43.8 | 42.7 | 37.5 | 35.4 | 36.9 | 24.8 | 23.1 | 24.2 | 20.8 | 20.6 | 22.1 | 22.5 | 21.9 | 19.8 | 17.1 | |||
| 4. T.vi. lcc2 | 62.4 | 65.1 | 62.8 | 65.7 | 84.2 | 76.3 | 75.1 | 72.9 | 60.5 | 59.9 | 59.7 | 43.4 | 42.2 | 36.6 | 35.1 | 39.1 | 23.9 | 23.1 | 23.7 | 20.0 | 21.2 | 23.1 | 22.5 | 20.4 | 20.0 | 12.1 | ||||
| 5. T.vi. lcc3 | 59.2 | 52.0 | 59.4 | 63.6 | 60.9 | 61.3 | 59.0 | 59.4 | 51.2 | 52.6 | 43.8 | 43.3 | 40.8 | 38.1 | 37.4 | 22.8 | 22.0 | 23.3 | 19.0 | 19.7 | 20.3 | 20.1 | 21.6 | 19.5 | 17.3 | |||||
| 6. T.vi. lcc4 | 69.4 | 99.4 | 66.3 | 66.3 | 67.3 | 67.3 | 61.9 | 61.2 | 60.0 | 44.4 | 43.3 | 39.8 | 36.9 | 37.3 | 26.0 | 21.7 | 22.5 | 17.9 | 19.0 | 20.8 | 19.8 | 21.7 | 19.6 | 16.2 | ||||||
| 7. T.vi. lcc5 | 69.8 | 63.8 | 63.1 | 63.9 | 64.0 | 57.5 | 52.9 | 48.0 | 40.6 | 39.8 | 33.8 | 33.8 | 33.0 | 22.8 | 22.6 | 21.1 | 15.6 | 19.9 | 19.4 | 20.5 | 20.1 | 15.4 | 14.6 | |||||||
| 8. T.v. lcc1 | 66.9 | 66.9 | 67.9 | 67.3 | 62.3 | 61.5 | 60.4 | 44.2 | 43.1 | 40.0 | 36.7 | 37.5 | 26.0 | 23.8 | 22.3 | 17.9 | 19.0 | 20.6 | 20.0 | 21.7 | 19.6 | 16.2 | ||||||||
| 9. B.20197 lac1 | 76.1 | 73.0 | 73.5 | 58.6 | 58.6 | 58.8 | 44.5 | 43.2 | 38.9 | 36.8 | 39.3 | 24.9 | 25.6 | 23.7 | 21.6 | 22.7 | 23.1 | 23.1 | 22.7 | 17.9 | 16.6 | |||||||||
| 10. B.20197 lac2 | 77.1 | 76.6 | 61.0 | 60.0 | 59.8 | 44.2 | 42.1 | 36.5 | 35.0 | 36.0 | 22.3 | 24.2 | 23.1 | 21.2 | 21.3 | 22.7 | 22.3 | 21.0 | 18.8 | 15.4 | ||||||||||
| 11. B.20197 lac3 | 72.3 | 60.7 | 61.6 | 60.7 | 44.2 | 43.1 | 37.4 | 36.8 | 38.2 | 26.5 | 22.3 | 26.0 | 22.1 | 22.5 | 24.6 | 23.5 | 22.5 | 20.6 | 11.5 | |||||||||||
| 12. B.PM1 lac1 | 64.4 | 60.0 | 59.8 | 43.9 | 43.5 | 40.2 | 36.6 | 37.5 | 23.8 | 24.4 | 23.2 | 19.0 | 22.8 | 23.6 | 23.6 | 20.3 | 18.4 | 14.5 | ||||||||||||
| 13. P.r. lac | 55.8 | 56.8 | 41.5 | 41.7 | 36.1 | 31.9 | 38.4 | 23.4 | 23.0 | 23.5 | 18.4 | 19.7 | 19.0 | 20.6 | 20.8 | 17.3 | 16.4 | |||||||||||||
| 14. P.o. pox1 | 89.8 | 43.5 | 42.9 | 35.5 | 35.9 | 37.6 | 22.3 | 21.9 | 23.1 | 18.7 | 18.9 | 17.8 | 17.6 | 18.9 | 19.1 | 14.4 | ||||||||||||||
| 15. P.o. pox2 | 43.8 | 43.1 | 36.0 | 36.6 | 38.2 | 22.7 | 23.3 | 22.7 | 17.4 | 17.6 | 18.4 | 17.6 | 19.7 | 18.9 | 13.3 | |||||||||||||||
| 16. A.b. lcc1 | 93.1 | 39.4 | 39.0 | 41.0 | 24.4 | 22.1 | 25.0 | 19.6 | 18.8 | 20.0 | 19.4 | 18.3 | 19.6 | 14.8 | ||||||||||||||||
| 17. A.b. lcc2 | 40.2 | 39.2 | 41.7 | 24.0 | 22.7 | 24.6 | 19.6 | 19.6 | 20.2 | 19.8 | 18.7 | 19.2 | 18.3 | |||||||||||||||||
| 18. R.s. lcc2 | 62.4 | 46.1 | 18.9 | 21.7 | 20.5 | 21.2 | 20.3 | 19.8 | 21.5 | 22.3 | 20.7 | 16.5 | ||||||||||||||||||
| 19. R.s. lcc3 | 42.2 | 23.4 | 23.4 | 21.2 | 19.5 | 19.1 | 18.2 | 18.5 | 21.5 | 19.5 | 15.6 | |||||||||||||||||||
| 20. R.s. lcc4 | 22.6 | 23.4 | 23.0 | 20.3 | 21.1 | 19.6 | 20.0 | 21.8 | 22.2 | 13.4 | ||||||||||||||||||||
| 21. P.a. lac2 | 52.6 | 59.8 | 16.8 | 19.0 | 18.9 | 19.7 | 18.9 | 16.1 | 11.5 | |||||||||||||||||||||
| 22. C.pa. lac-1 | 53.8 | 19.5 | 18.3 | 17.7 | 19.0 | 21.5 | 18.6 | 14.4 | ||||||||||||||||||||||
| 23. N.c. lac | 16.8 | 16.9 | 20.1 | 20.0 | 16.3 | 15.6 | 16.1 | |||||||||||||||||||||||
| 24. L.t. Lac2-1 | 77.4 | 75.3 | 77.4 | 54.0 | 39.8 | 14.9 | ||||||||||||||||||||||||
| 25. L.t. Lac2-2 | 85.0 | 91.1 | 56.7 | 42.1 | 14.2 | |||||||||||||||||||||||||
| 26. L.t. Lac2-3 | 88.9 | 56.4 | 42.5 | 14.3 | ||||||||||||||||||||||||||
| 27. L.t. Lac2-4 | 57.5 | 42.8 | 15.0 | |||||||||||||||||||||||||||
| 28. N.t. pTL3 | 46.9 | 15.6 | ||||||||||||||||||||||||||||
| 29. A.p. | 14.7 | |||||||||||||||||||||||||||||
| 30. A.n. yA | ||||||||||||||||||||||||||||||
Calculated by using the CLUSTAL method with data for the inferred proteins.
Abbreviations (and references from which data were obtained or accession no.): P.c., Pycnoporus cinnabarinus; C.h., Coriolus hirsutus (24); T.vi., Trametes villosa (47); T.v., Trametes versicolor (22); B.20197, basidiomycete strain CECT 20197 (28); B.PM1, basidiomycete strain PM1 (10); P.r., Phlebia radiata (38); P.o., Pleurotus ostreatus (19); A.b., Agaricus bisporus (34); R.s., Rhizoctonia solani (44); P.a., Podospora anserina (17); C.pa., Cryphonectria parasitica (7); N.c., Neurospora crassa (18); L.t., Liriodendron tulipifera (GenBank accession no. U73103-106); N.t., Nicotiana tabacum (23); A.p., Acer pseudoplatanus (25); A.n., Aspergillus nidulans (1).
DISCUSSION
Structural similarity of laccases.
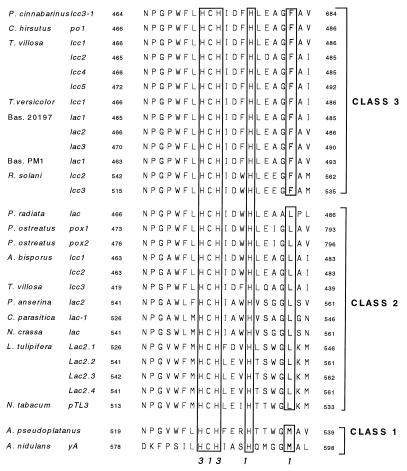
A laccase-encoding gene and its corresponding cDNA were cloned from P. cinnabarinus, and the gene product was shown to correspond to a laccase previously isolated from ligninolytic cultures of this organism (13). The single cysteine residue and 10 histidine residues that bind the four catalytic cupric ions in all blue copper oxidases, including laccases, were conserved in the P. cinnabarinus gene. Studies of type 1 copper centers have shown that an additional residue 10 amino acids downstream of the conserved cysteine can have a major effect on the redox potential of the cupric ion (6). This residue was found to be a phenylalanine residue in the P. cinnabarinus laccase, but leucine and methionine residues have been found in the laccase sequences of other fungi and plants (Fig. 4). The results of site-directed mutagenesis studies performed with azurin (6, 31), as well as work done by Xu et al. (46), support the hypothesis that laccases harboring phenylalanine residues at this position should have type 1 copper centers with high redox potentials, whereas the copper centers of laccases with methionines at this position should have low redox potentials. On the basis of the three known possible residues at the critical position, we categorized the known laccase sequences into classes 1, 2, and 3 in order of postulated increasing redox potential. This classification is also coordinated with the proposed Lac1 (Met)-Lac2 (Leu)-Lac3 (Phe) nomenclature recently submitted by one of us (J.F.D.D.) to the Commission for Plant Gene Nomenclature (CPGN) for plant laccase genes.
FIG. 4.
Alignment of the amino acid sequences constituting the copper-binding domain closest to the carboxyl terminus in laccases from a variety of sources. The residues in boxes are associated with the type 1 and type 3 copper centers, as indicated by the numbers at the bottom. It has been predicted that the residues in the box farthest to the right (M, L, and F) influence the redox characteristics of the enzyme, and so these residues provide the basis for assigning laccases to class 1, 2, or 3. The numbers on either side of the sequences indicate the positions of the amino acids within the laccase polypeptide, starting from the translational start site. Annotated laccase gene sequences that lead to a putative protein sequence with an anomalous Cu-4 center, including the Cryptococcus neoformans (45) (GenBank accession no. L22866) and Rhizoctonia solani lcc4 (44) (GenBank accession no. Z54277) sequences, are not shown. T. versicolor, Trametes versicolor; Bas., basidiomycete; P. anserina, Podospora anserina; L. tulipifera, Liriodendron tulipifera; N. tabacum, Nicotiana tabacum; A. pseudoplatanus, Acer pseudoplatanus.
The A. nidulans (class 1) laccase exhibits very low levels of similarity to all other known laccases, including those isolated from plants. This probably reflects the very specialized function of this laccase (i.e., spore morphogenesis) (1). In contrast, the Botrytis cinerea laccase, whose nucleotide sequence is not known, has been implicated in the detoxification of phytoalexins (4). These examples highlight the variety of specialized physiological functions for which laccases have evolved in fungi. It remains to be seen, however, whether multiple laccases that have different physiological functions can be isolated from the same organism.
Multiple laccase genes in P. cinnabarinus.
When grown in liquid culture, P. cinnabarinus secreted a single laccase isozyme (13). Stimulation of laccase secretion by adding 2,5-xylidine, a strategy previously shown to induce laccase production in a wide variety of basidiomycetes (5), did not result in production of additional bands in isoelectric focusing gels. Likewise, reverse transcriptase PCR analyses did not reveal any additional laccase transcripts when the fungus was exposed to 2,5-xylidine. The N-terminal sequences deduced for all of the cDNA-encoded laccases isolated were identical to the sequence previously determined for the laccase purified from cultures.
On the other hand, Southern analysis revealed the presence of a small gene family encoding laccases in P. cinnabarinus. Whereas the laccase probe hybridized to four BamHI fragments, it hybridized to only a single 8.6-kb HindIII fragment, suggesting that there is a laccase gene cluster in P. cinnabarinus. As P. cinnabarinus PB is a dikaryon, allelic variants of the laccase gene are expected, and the presence of four nonallelic laccase genes is highly unlikely since all of them would have to be located on the same 8.6-kbp HindIII fragment.
The copy numbers of laccase genes vary among fungi. A laccase gene family in which the genes encoding two of five laccases were located on the same chromosome was found in T. villosa (47, 48), and three laccase genes were found to be clustered within approximately 11 kb of each another in Rhizoctonia solani (44). Pleurotus ostreatus and Agaricus bisporus each contain at least two different laccase genes (19, 34), while allelic forms of a single laccase gene have been found in C. hirsutus, as well as Neurospora crassa (18, 24). Single copies of laccase genes were also found in Phlebia radiata and Cryphonectria parasitica (7, 38).
In P. cinnabarinus, the laccase purified from liquid cultures was found to be important not only for lignin degradation (14, 16) but also for synthesis of the phenoxazinone pigments which give the fruiting bodies a red color (15). In fact, a more precise name for the laccase from P. cinnabarinus is 3-hydroxyanthranilate:O2 oxidoreductase. The phenoxazinone pigments and, consequently, the laccase activity can also be linked to the antimicrobial activity of this organism (12). Thus, the product of one laccase gene in P. cinnabarinus appears to serve two separate, but interwoven, functions in this fungus. It remains to be determined under what conditions the other laccase genes are expressed and what physiological function(s) they perform under those conditions.
ACKNOWLEDGMENTS
Alexandria Tristram provided valuable technical advice and assistance.
This research was supported by a fellowship from the Alexander von Humboldt-Stiftung (to C.E.) supplemented with funds from the University of Georgia Research Foundation, as well as by grant RR 50778F from the National Science Foundation.
REFERENCES
- 1.Aramayo R, Timberlake W E. Sequence and molecular structure of the Aspergillus nidulans yA (laccase) gene. Nucleic Acids Res. 1990;18:3415. doi: 10.1093/nar/18.11.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballance D J. Sequences important for gene expression in filamentous fungi. Yeast. 1986;2:229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- 3.Bao W, O’Malley D M, Whetten R, Sederoff R R. A laccase associated with lignification in loblolly pine xylem. Science. 1993;260:672–674. doi: 10.1126/science.260.5108.672. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Nun N, Mayer A M. Cucurbitacins—repressors of induction of laccase formation. Phytochemistry. 1989;28:1369–1371. [Google Scholar]
- 5.Bollag J-M, Leonowicz A. Comparative studies of extracellular fungal laccases. Appl Environ Microbiol. 1984;48:849–854. doi: 10.1128/aem.48.4.849-854.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canters G W, Gilardi G. Engineering type-1 copper sites in proteins. FEBS Lett. 1993;325:39–48. doi: 10.1016/0014-5793(93)81410-2. [DOI] [PubMed] [Google Scholar]
- 7.Choi G H, Larson T G, Nuss D L. Molecular analysis of the laccase gene from the chestnut blight fungus and selective suppression of its expression in an isogenic hypovirulent strain. Mol Plant-Microbe Interact. 1992;5:119–128. doi: 10.1094/mpmi-5-119. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extractions. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Clutterbuck A J. Absence of laccase from yellow-spored mutants of Aspergillus nidulans. J Gen Microbiol. 1972;70:423–425. doi: 10.1099/00221287-70-3-423. [DOI] [PubMed] [Google Scholar]
- 10.Coll P M, Tabernero C, Santamaría R, Pérez P. Characterization and structural analysis of the laccase I gene from the newly isolated ligninolytic basidiomycete PM1 (CECT 2971) Appl Environ Microbiol. 1993;59:4129–4135. doi: 10.1128/aem.59.12.4129-4135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean J F D, Eriksson K-E L. Laccase and the deposition of lignin in vascular plants. Holzforschung. 1994;48:21–38. [Google Scholar]
- 12.Eggert C. Laccase is responsible for antimicrobial activity of Pycnoporus cinnabarinus. Microbiol Res. 1997;152:315–318. doi: 10.1016/S0944-5013(97)80046-8. [DOI] [PubMed] [Google Scholar]
- 13.Eggert C, Temp U, Eriksson K-E L. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl Environ Microbiol. 1996;62:1151–1158. doi: 10.1128/aem.62.4.1151-1158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggert C, Temp U, Eriksson K-E L. Laccase is essential for lignin degradation by the white-rot fungus Pycnoporus cinnabarinus. FEBS Lett. 1997;407:89–92. doi: 10.1016/s0014-5793(97)00301-3. [DOI] [PubMed] [Google Scholar]
- 15.Eggert C, Temp U, Dean J F D, Eriksson K-E L. Laccase-mediated formation of the phenoxazinone derivative, 3-hydroxyanthranilic acid. FEBS Lett. 1995;376:202–206. doi: 10.1016/0014-5793(95)01274-9. [DOI] [PubMed] [Google Scholar]
- 16.Eggert C, Temp U, Dean J F D, Eriksson K-E L. A fungal metabolite mediates oxidation of non-phenolic lignin structures and synthetic lignin by laccase. FEBS Lett. 1996;391:144–148. doi: 10.1016/0014-5793(96)00719-3. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Larrea J, Stahl U. Isolation and characterization of a laccase gene from Podospora anserina. Mol Gen Genet. 1996;252:539–551. doi: 10.1007/BF02172400. [DOI] [PubMed] [Google Scholar]
- 18.Germann U A, Müller G, Hunziker P E, Lerch K. Characterization of two allelic forms of Neurospora crassa laccase. J Biol Chem. 1988;263:885–896. [PubMed] [Google Scholar]
- 19.Giardina P, Cannio R, Martirani L, Marzullo L, Palmieri G, Sannia G. Cloning and sequencing of a laccase gene from the lignin-degrading basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 1995;61:2408–2413. doi: 10.1128/aem.61.6.2408-2413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 21.Iiumura Y, Takenouchi K, Nakamura M, Kawai S, Katayama Y, Morohoshi N. Cloning and sequence analysis of laccase genes and its use for an expression vector in Coriolus versicolor. In: Kuwahara M, Shimada M, editors. Biotechnology in the pulp and paper industry. Kyoto, Japan: Uni Publisher; 1992. pp. 27–30. [Google Scholar]
- 22.Jönsson L, Sjöström K, Häggström I, Nyman P O. Characterization of a laccase gene from the white-rot fungus Trametes versicolor and structural features of basidiomycete laccases. Biochim Biophys Acta. 1995;1251:210–215. doi: 10.1016/0167-4838(95)00104-3. [DOI] [PubMed] [Google Scholar]
- 23.Kiefer-Meyer M-K, Gomord V, O’Connell A, Halpin C, Faye L. Cloning and sequence analysis of laccase-encoding cDNA clones from tobacco. Gene. 1996;178:205–207. doi: 10.1016/0378-1119(96)00381-2. [DOI] [PubMed] [Google Scholar]
- 24.Kojima Y, Tsukuda Y, Kawai Y, Tsukamoto A, Sugiura J, Sakaino M, Kita Y. Cloning, sequence analysis, and expression of ligninolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J Biol Chem. 1990;265:15224–15230. [PubMed] [Google Scholar]
- 25.LaFayette P R, Eriksson K-E L, Dean J F D. Nucleotide sequence of a cDNA clone encoding an acidic laccase from sycamore maple (Acer pseudoplatanus L.) Plant Physiol. 1995;107:667–668. doi: 10.1104/pp.107.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leatham G F, Stahmann M A. Studies on the laccase of Lentinus edodes: specificity, localization and association with the development of fruiting bodies. J Gen Microbiol. 1981;125:147–157. [Google Scholar]
- 27.Liu L, Dean J F D, Friedman W E, Eriksson K-E L. A laccase-like phenoloxidase is correlated with lignin biosynthesis in Zinnia elegans stem tissues. Plant J. 1994;6:213–224. [Google Scholar]
- 28.Mansur M, Suárez T, Fernández-Larrea J B, Brizuela M A, González A E. Identification of a laccase gene family in the new lignin-degrading basidiomycete CECT 20197. Appl Environ Microbiol. 1997;63:2637–2646. doi: 10.1128/aem.63.7.2637-2646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer A M, Harel E. Polyphenol oxidases in plants. Phytochemistry. 1979;33:765–767. [Google Scholar]
- 30.Messerschmidt A, Rossi A, Ladenstein R, Huber R, Bolognesi M, Gatti G, Marchesini A, Petruzelli R, Finazzi-Agro A. X-ray crystal structure of the blue oxidase ascorbate oxidase from zucchini. Analysis of the polypeptide fold and a model of the copper sites and ligands. J Mol Biol. 1989;206:513–529. doi: 10.1016/0022-2836(89)90498-1. [DOI] [PubMed] [Google Scholar]
- 31.Messerschmidt A, Huber R. The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships. Eur J Biochem. 1990;187:341–352. doi: 10.1111/j.1432-1033.1990.tb15311.x. [DOI] [PubMed] [Google Scholar]
- 32.Palle R, Murphy N B. Northern hybridization: rapid and simple electrophoretic conditions. Nucleic Acids Res. 1993;21:2783–2784. doi: 10.1093/nar/21.11.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlman D, Halvorson H O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983;167:391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- 34.Perry C R, Smith M, Britnell C H, Wood D A, Thurston C F. Identification of two laccase genes in the cultivated mushroom Agaricus bisporus. J Gen Microbiol. 1993;139:1209–1218. doi: 10.1099/00221287-139-6-1209. [DOI] [PubMed] [Google Scholar]
- 35.Peyratout C S, Severns J C, Holm S R, McMillin D R. EPR studies of ligand binding to the type2/type3 cluster in tree laccase. Arch Biochem Biophys. 1994;314:405–411. doi: 10.1006/abbi.1994.1460. [DOI] [PubMed] [Google Scholar]
- 36.Pribnow D G, Mayfield M B, Nipper N J, Gold M H. Characterization of a cDNA encoding manganese peroxidase from the lignin-degrading basidiomycete Phanerochaete chrysosporium. J Biol Chem. 1989;264:5036–5040. [PubMed] [Google Scholar]
- 37.Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 38.Saloheimo M, Niku-Paavola M-L, Knowles J K C. Isolation and structural analysis of the laccase gene from the lignin-degrading fungus Phlebia radiata. J Gen Microbiol. 1991;137:1537–1544. doi: 10.1099/00221287-137-7-1537. [DOI] [PubMed] [Google Scholar]
- 39.Shin W, Sundaram U M, Cole J L, Zhang H H, Hedman B, Hodgson K O, Solomon E I. Chemical and spectroscopic definition of the peroxide-level intermediate in the multicopper oxidase: relevance to the catalytic mechanism of dioxygen to water. J Am Chem Soc. 1996;118:3202–3215. [Google Scholar]
- 40.Solomon E, Lowery M D. Electronic structure contributions to function in bioorganic chemistry. Science. 1993;259:1575–1581. doi: 10.1126/science.8384374. [DOI] [PubMed] [Google Scholar]
- 41.Thurston C F. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- 42.Troutt A B, McHeyzer-Williams M G, Pulendran B, Nossal G J V. Ligation anchored PCR: a simple amplification technique with single-sided specificity. Proc Natl Acad Sci USA. 1992;89:9823–9825. doi: 10.1073/pnas.89.20.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahleitner J A, Xu F, Brown K M, Brown S H, Golightly E J, Halkier T, Kauppinen S, Pederson A, Schneider P. The identification and characterization of four laccases from the plant pathogenic fungus Rhizoctonia solani. Curr Genet. 1995;29:395–403. doi: 10.1007/BF02208621. [DOI] [PubMed] [Google Scholar]
- 45.Williamson P R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu F, Shin W, Brown S H, Wahleitner J A, Sundaram U M, Salomon E I. A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim Biophys Acta. 1996;1292:303–311. doi: 10.1016/0167-4838(95)00210-3. [DOI] [PubMed] [Google Scholar]
- 47.Yaver D S, Xu F, Golightly E J, Brown K M, Brown S H, Rey M W, Schneider P, Halkier T, Mondorf K, Dalbøge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Environ Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaver D S, Golightly E J. Cloning and characterization of three laccase genes from the white-rot basidiomycete Trametes villosa: genomic organization of the laccase gene family. Gene. 1996;181:95–102. doi: 10.1016/s0378-1119(96)00480-5. [DOI] [PubMed] [Google Scholar]
- 49.Zhou M Y, Xue D, Gomez-Sanchez P, Gomez-Sanchez C S. Improved downward capillary transfer for blotting of DNA and RNA. BioTechniques. 1994;16:58–59. [PubMed] [Google Scholar]